Abstract
The understanding of dengue virus pathogenesis has been hampered by the lack of in vitro and in vivo models of disease. The study of viral factors involved in the production of severe dengue, dengue hemorrhagic fever (DHF), versus the more common dengue fever (DF), have been limited to indirect clinical and epidemiologic associations. In an effort to identify viral determinants of DHF, we have developed a method for comparing dengue type 2 genomes (reverse transcriptase PCR in six fragments) directly from patient plasma. Samples for comparison were selected from two previously described dengue type 2 genotypes which had been shown to be the cause of DF or DHF. When full genome sequences of 11 dengue viruses were analyzed, several structural differences were seen consistently between those associated with DF only and those with the potential to cause DHF: a total of six encoded amino acid charge differences were seen in the prM, E, NS4b, and NS5 genes, while sequence differences observed within the 5′ nontranslated region (NTR) and 3′ NTR were predicted to change RNA secondary structures. We hypothesize that the primary determinants of DHF reside in (i) amino acid 390 of the E protein, which purportedly alters virion binding to host cells; (ii) in the downstream loop (nucleotides 68 to 80) of the 5′ NTR, which may be involved in translation initiation; and (iii) in the upstream 300 nucleotides of the 3′ NTR, which may regulate viral replication via the formation of replicative intermediates. The significance of four amino acid differences in the nonstructural proteins NS4b and NS5, a presumed transport protein and the viral RNA polymerase, respectively, remains unknown. This new approach to the study of dengue virus genome differences should better reflect the true composition of viral RNA populations in the natural host and permit their association with pathogenesis.
Due to the global growth of populations, urbanization, and the spread of the main mosquito vector, Aedes aegypti, dengue diseases are a major, emerging problem with the cocirculation of different virus serotypes, increased frequency of epidemics, and the introduction of dengue hemorrhagic fever (DHF) in areas where it was not previously known. At present, it is estimated that more than 50 million human dengue infections occur annually and that, since 1958, more than 60,000 children have died due to DHF (24).
Dengue viruses, which belong to the genus Flavivirus (family Flaviviridae), have four distinct antigenic types (serotypes 1 to 4). Dengue virus infections can be subclinical or cause illnesses ranging from a mild, flu-like syndrome with rash (dengue fever [DF]) to a severe and sometimes fatal disease, characterized by capillary leakage, thrombocytopenia, and sometimes hypovolemic shock (DHF). However, due to the lack of in vivo and in vitro correlates of virulence, the precise mechanism(s) by which dengue viruses cause severe disease is still not understood. It has been shown by epidemiologic and clinical associations that both immunologic and viral factors determine the severity of disease (26, 62). Infection with one dengue virus serotype does not provide protective immunity against the others, and sequential (heterotypic) infection has been shown to increase virus replication and thus the probability of developing DHF by a process known as antibody-dependent enhancement (25, 39). Host immune factors, such as cytokines, interferon, and activated complement have been suggested to increase the capillary permeability, the hallmark of DHF (41). Recent events in this hemisphere suggest the importance of viral factors. Despite cocirculation of several dengue serotypes in the Americas, it was not until the Cuban epidemic of 1981 that the first DHF cases occurred, and this coincided with the introduction of a new genetic type of dengue virus serotype 2 (DEN-2) (58). Phylogenetic studies demonstrate different genetic types of DEN-2; the “native” American genotype has been associated thus far with mild disease (DF), while the introduced Southeast Asian genotype coincided with the appearance of DHF in four different countries (60). Other support comes from epidemiologic studies in Peru, where over a period of 4 years (1993 to 1997), active surveillance for DF cases revealed that, in spite of secondary infection rates of up to 75%, no DHF cases have been detected; this country has not yet imported the Southeast Asian genotype of DEN-2 (70). Our search for viral factors of severe disease has therefore focused on DEN-2 viruses.
As with other flaviviruses, the dengue virus RNA genome is a single-stranded positive-sense genome of approximately 10,700 bases in length, surrounded by a nucleocapsid and covered by a lipid envelope containing the envelope and the membrane proteins. The genome contains a single open reading frame, which encodes a precursor polyprotein and is flanked by two nontranslated regions (5′ and 3′ NTR). Co- and posttranslational proteolytic cleavage of the precursor results in the formation of three structural proteins, capsid (C), membrane (M), and envelope (E), and seven nonstructural proteins, NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5 (8). For other flaviviruses, including yellow fever (33), Japanese encephalitis (7, 27), Murray Valley (43), Louping ill (34), and tick-borne encephalitis (TBE) (29) viruses, most molecular markers for pathogenicity have been localized in the E gene. For dengue, several studies of cloned or cell culture-passaged wild and attenuated viruses have been performed, but each report described different sites of nucleotide and amino acid changes possibly associated with virulence (3, 38, 48–50, 53). Most of these studies suggested that sites in the prM, E, NS1, and NS3 genes or the NTRs are of importance. Since studies relating markers such as mouse neurovirulence, cell culture growth characteristics, and immunogenicity in mice are of limited value in assessing human disease potential, the viral determinants of severe dengue are still unclear. We have determined the full nucleotide sequences of 11 viruses directly from clinical samples from DF and DHF patients in Thailand and the Americas in an attempt to define genome structural differences that might be correlated with pathogenesis.
MATERIALS AND METHODS
Viruses.
DEN-2 viruses were obtained from acute-phase plasma collected from patients enrolled in ongoing prospective clinical studies (Thailand) (35, 59) or epidemiologic surveillance programs (Venezuela, Mexico, and Peru) during 1981 to 1997. Sample aliquots were used to infect C6/36 mosquito cells and were identified as DEN-2 by indirect fluorescent antibody tests with type-specific monoclonal antibodies. Virus titers were determined for the Thai plasma samples by mosquito probit analysis (16), based on 10-fold dilutions: 20 Toxorhynchites splendens mosquitoes were injected per plasma dilution, and indirect fluorescent antibody testing was performed with 15 of the survivors (69). Mosquito-infectious doses per milliliter ranged between 6.9 and 8.55 log10. Virus isolation and passage histories of samples used in this study and previously published strains (16681, NGC, 1409, PR159S1, ThNHp7, and ThNH11) used for comparison are given in Table 1.
TABLE 1.
Comparison of DEN-2 viruses by sequence analysis
| Strain | Passage historya | Location | Yr | Clinical statusb | GenBank accession no. |
|---|---|---|---|---|---|
| NGC | SMB 38, C6/36 1, CRFK 1 | New Guinea | 1944 | DF | M32941 |
| 16681 | BS-C-1 ?, LLC-MK2 8, Monk.1, Mosq. 2, PGMK 1, C6/36 4 | Bangkok, Thailand | 1964 | DHF | U87411 |
| PR159S1 | PGMK 19, C6/36 1 | Puerto Rico | 1969 | DF | M32953 |
| 1318 | Mosq. 2, C6/36 1 | Puerto Rico | 1981 | DF | AF100149 |
| 044 | C6/36 2 | Tapachula, Chiapas, Mexico | 1983 | DF | AF100151 |
| 1409 | C6/36 3, LLC-MK2 1 | Jamaica | 1983 | DHF | M20558 |
| 348600 | C6/36 3 | Tumaco, Nariño, Colombia | 1986 | DF | AF100458 |
| Ven2 | AP61 2 | Maracay, Aragua, Venezuela | 1987 | DF | AF100465 |
| Mara4 | None | Maracay, Aragua, Venezuela | 1990 | DHF | AF100466 |
| Mara10 | None | Maracay, Aragua, Venezuela | 1990 | DHF | AF100145 |
| 102692 | None | Aragua, Venezuela | 1991 | DHF | AF100150 |
| 0131 | None | Navojoa, Sonora, Mexico | 1992 | DF | AF100469 |
| 0147 | C6/36 1 | Chinipas, Chihuahua, Mexico | 1992 | DF | AF100148 |
| ThNHp7/93 | C6/36 3 | Nakhon Phanom, Thailand | 1993 | DSS | AF022434 |
| ThNH11/93 | C6/36 3 | Nakhon Phanom, Thailand | 1993 | DF | AF022437 |
| K0008 | None | Kamphaeng Phet, Thailand | 1994 | DHF | AF100459 |
| K0010 | None | Kamphaeng Phet, Thailand | 1994 | DF | AF100460 |
| CO371 | None | Bangkok, Thailand | 1995 | DF | AF100461 |
| CO390 | None | Bangkok, Thailand | 1995 | DHF | AF100462 |
| 383 | None | Tamazunchale, S. L. Potosi, Mexico | 1995 | DHF | AF100147 |
| IQT1797 | None | Iquitos, Loreto, Peru | 1995 | DF | AF100467 |
| IQT2913 | None | Iquitos, Loreto, Peru | 1996 | DF | AF100468 |
| CO166 | None | Bangkok, Thailand | 1996 | DF | AF100463 |
| CO167 | None | Bangkok, Thailand | 1996 | DHF | AF100464 |
| 926 | None | Acapulco, Guerrero, Mexico | 1997 | DHF | AF100146 |
SMB, suckling mouse brain; C6/36, Aedes albopictus cell line; CRFK, Crandell’s feline kidney cell line; BS-C-1, grivet monkey kidney cell line; ?, number of passages unknown; LLC-MK2, rhesus monkey kidney cell line; Monk., rhesus macaque monkey; Mosq., mosquito inoculation; PGMK, primary green monkey kidney cell line; AP61, Aedes pseudoscutellaris cell line; None, acute-phase plasma of patient.
DSS, dengue shock syndrome.
RNA extraction and RT-PCR.
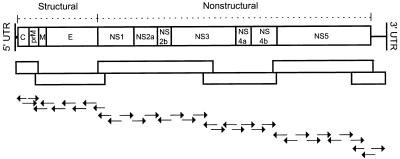
Total RNA was extracted from 200 to 500 μl of plasma or cell culture supernatant by using Trizol LS (GIBCO BRL, Gaithersburg, Md.) according to the manufacturer’s recommendations. Ethanol-precipitated RNA was recovered by centrifugation and air-dried. The RNA pellet was resuspended in 35 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) and used as a template in reverse transcriptase PCR (RT-PCR). Synthetic oligonucleotide primer pairs were designed based on published sequence data for strains 16681, New Guinea C, Jamaica 1409, and PR159S1 and were optimized over the course of the study to generate six overlapping double-stranded DNA (dsDNA) products spanning genome nucleotide (nt) regions 11 to 681, 618 to 2602, 2406 to 6342 (or 2409 to 5751), 5442 to 7522, 7476 to 10322 (or 7243 to 10327), and 9948 to 10695. Diagrams of the genome amplification and sequencing strategies are shown in Fig. 1.
FIG. 1.
Schematic of the DEN-2 RNA genome showing (from top to bottom) the genome organization, the RT-PCR strategy (overlapping PCR fragments are shown as blocks), and the sequencing strategy. Arrows pointing to the right indicate regions sequenced 3′→5′ on the minus-sense strand; arrows pointing to the left indicate regions sequenced 3′→5′ on the plus-sense strand. UTR, untranslated region.
To amplify each fragment, a mixture of 5 μl of RNA, 50 pM of corresponding sense and antisense PCR primers, and diethyl pyrocarbonate-treated water (to a total volume of 50 μl) was incubated at 85°C for 5 min and chilled on ice. A one-tube reaction mixture containing 3.3× PCR buffer II (Perkin-Elmer, Branchburg, N.J.), 200 μM each deoxynucleotide, 1.1 mM Mg(OAc)2, 10 mM dithiothreitol, 40 U of RNasin, 10 U of avian myeloblastosis virus RT (Promega, Madison, Wis.), and 3 U of rTth DNA polymerase (Perkin-Elmer) was added. The reverse transcription reaction was performed at 42°C for 60 min. Thermocycling began with a “hot start” at 94°C for 2 min, and the PCR conditions (30 cycles) differed according to primer pair characteristics and the expected fragment size. Oligonucleotide primers used for amplification (RT-PCR) and the respective thermocycling conditions are given in Table 2. Extension times were increased for the last 20 cycles by 10 s/cycle, and a final extension was performed at 72°C for 15 min. Reaction mixtures were stored at 4°C until further processing.
TABLE 2.
Oligonucleotide primers used for amplification (RT-PCR) and thermocycling conditions
| Primer namea | Sequence (5′→3′) | Genome positionb | Length (nt) | Thermocycling conditon
|
||
|---|---|---|---|---|---|---|
| Melting | Annealing | Extensionc | ||||
| D2/11V | CTACGTGGACCGACAAAGACAG | 11–32 | 670 | 94°C for 30 s | 55°C for 30 s | 70°C for 2 min |
| D2/662 | GGTACAAGTCCCATAGGTTA | 662–681 | ||||
| D2/618V | ACCAGAAGACATAGATTGTTGGTGC | 618–642 | 1,984 | 94°C for 35 s | 61°C for 35 s | 70°C for 2 min |
| D2/2578 | TTACTGAGCGGATTCCACAGATGCC | 2578–2602 | ||||
| D2/2406V | AGTCATGGTGCAGGCCGATAGTG | 2406–2428 | 3,345 | 94°C for 40 s | 62°C for 40 s | 70°C for 3 min |
| D2/5728 | AATGTCAGTTGTAACCACGAAATC | 5728–5751 | ||||
| D2/2406V-IId | AGTTATGGTGCAAGCCGATAGTG | 2406–2428 | 3,345 | 94°C for 40 s | 62°C for 40 s | 70°C for 3 min |
| D2/5728d | AATGTCAGTTGTAACCACGAAATC | 5728–5751 | ||||
| D2/2409Ve | GGGAGCTATGGTGCAGGCTGATA | 2409–2431 | 3,342 | 94°C for 40 s | 62°C for 40 s | 70°C for 3 min |
| D2/5728-IIe | AATGTCAGTTGTAACCACGAAGTC | 5728–5751 | ||||
| D2/5442V | TGAGGCAGCTGGGATTTTTATGAC | 5442–5465 | 2,080 | 94°C for 35 s | 60°C for 35 s | 70°C for 2 min |
| D2/7500 | CTCCGGCCAAGTAACTCCCTCTA | 7500–7522 | ||||
| D2/7243V | GGCATCATGAAAAACCCAACTGT | 7243–7265 | 3,084 | 94°C for 40 s | 62°C for 40 s | 70°C for 3 min |
| D2/10304 | TACTATGGCTTAACTCGACCTGAC | 10304–10327 | ||||
| D2/9948V | AACCTGGTCCATACATGCTAAACA | 9948–9971 | 747 | 94°C for 30 s | 60°C for 30 s | 70°C for 2 min |
| D2/10676 | CATTTTCTGGCGTTCTGTGC | 10676–10695 | ||||
Primer names with an V indicate a viral-sense orientation; names without indicate a complementary sense orientation.
Genome positions are given according to the published sequence of strain 16881 (17).
Thermocycling occurred 30 times, with extension times increased for the last 20 cycles for 10 s/cycle.
Primer modification for Ven2, 0131, IQT1797, and IQT2913.
Primer modification for Mara4 and 102692.
Sequencing of PCR fragments.
For automated sequencing, spin column-purified (Qiagen, Chatsworth, Calif.) dsDNA fragments were analyzed by the cycle-sequencing dye terminator method. The Big Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer, Applied Biosystems, Foster City, Calif.) was chosen because it contains high-sensitivity dye and a variant of Thermus aquaticus DNA polymerase that has no 5′→3′ exonuclease activity. For each sequencing reaction, approximately 50 to 100 ng purified DNA was combined with 3.2 pmol of primer and a reaction cocktail containing the four dye-labeled dideoxynucleotide terminators. Cycle sequencing parameters used were as described in the manufacturer’s protocol (25 cycles of 96°C for 30 s, 50°C for 60 s, and 60°C for 4 min). The reaction mixture was column purified (Centri-Sep, Princeton Separations, Adelfia, N.J.), and the DNA was dried in a vacuum centrifuge for 20 min. The pellet was resuspended in 16-μl of template suppression reagent, heated for 2 min at 95°C, and kept on ice until loaded on the sequencer, an Applied Biosystems Prism 310, by using a short capillary (47 cm by 50 μm [inside diameter]) and Performance Optimized Polymer 6 (Perkin-Elmer, Applied Biosystems).
Nucleotide and amino acid sequence analysis.
Overlapping nucleic acid sequences were combined for analysis and edited with the aid of the Lasergene software package (DNASTAR, Inc., Madison, Wis.). Prior to phylogenetic analysis, DEN-2 virus nucleic acid sequences were aligned with the DEN-2 strain 16681 virus by using the multiple sequence alignment methods CLUSTAL and JOTUN-HEIN, within the MEGALIGN program (DNASTAR, Inc.). With our protocol, we were able to determine up to 10,666 nt of the entire DEN-2 genome, encoding a polyprotein of 3,391 amino acids (aa). Due to the small quantities of viral RNA contained in the human plasma samples, we were not able to determine the 11 nt at the 5′ end and 47 nt at the 3′ end; here, we have assumed that these regions are conserved and identical to those in virus strain 16681 (38). The predicted amino acid sequences were analyzed by using algorithms within the PROTEAN software package (DNASTAR, Inc.): Chou-Fasman (10) and Garnier-Robson (19) for secondary structure predictions, Kyte-Doolittle (42) for hydropathy prediction, and Jameson-Wolf (32) and Hopp-Woods (30) for prediction of potential antigenic determinants.
Nucleotide and amino acid mutation rates.
To obtain nucleotide and amino acid mutation rates for the DEN-2 genome, we used data from Thai samples. The total number of nucleotide and amino acid changes of each of the two samples from year 1994 were compared to those of each of the two samples from 1995. The differences were averaged and divided by the total lengths of the genome and polyprotein, respectively. This procedure was repeated with data from samples collected in 1995 and 1996. The nucleotide and amino acid mutation rate for the observed 2-year period (1994 to 1996) resulted from the average of the rates obtained for each year.
Phylogenetic analyses.
Phylogenetic analyses were done with the PAUP* program, with uniform character weights, TBR branch swapping, and a heuristic search for the most parsimonious trees (68). Sequences from representatives of the other three serotypes (1, 2, and 4) (18, 52, 74) were used to root the trees. The reliability of the inferred phylogenetic tree was estimated by the bootstrap method, with 100 replications (15).
Secondary structure analysis of the NTRs.
RNA secondary structure predictions of the 5′ and 3′ NTRs were performed with the mfold (75) and the RNASTAR (22) programs. The latter simulates the RNA folding process, using a genetic algorithm, and can predict tertiary structures such as pseudoknots, while the former does not. The resulting predicted folding patterns were graphically represented with the loopDloop program (20). The RNASTAR-generated secondary structures were compared by the RNAdistance program (64) of the Vienna RNA package (28), and the resulting distance matrix was analyzed with PAUP* (68) by using the distance optimality criterion and heuristic search settings. Phylogenetic trees reflecting the relationship between the predicted RNA secondary structures were then compared to those generated by comparison of full genome nucleotide sequences.
Secondary structure analysis of the 3′ NTR was performed with a total of 19 DEN-2 virus nucleotide sequences. To determine the consistency of the predicted RNA structures and to investigate the possible influence of cell culture passage on these structures, we included eight additional viruses from the Americas: four unpassaged samples (Mara10, 102692, 383, and 926) and four low-passage strains (1318, 044, 348600, and 0147), obtained from both DF and DHF patients (Table 1).
Nucleotide sequence accession number.
The nucleotide sequences reported in this study have been deposited in the GenBank database under the accession no. AF100145 to AF100151 and AF100458 to AF100469.
RESULTS
Nucleotide and amino acid sequence similarities between all samples and previously published strains 16681, ThNH7, ThNHp11, NGC, PR159S1, and 1409 are shown in a matrix (Table 3).
TABLE 3.
Comparison of amino acid and nucleotide sequences of the full genome of DEN-2 virusesa
| % Nucleotide similarity of genome | % Amino acid similarity of genome
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NGC | 16681 | ThNH7 | ThNHp11 | K0008 | K0010 | CO371 | CO390 | CO166 | CO167 | PR159S1 | 1409 | Ven2 | Mara4 | 0131 | IQT1797 | |
| NGC | 98.7 | 98.3 | 98.4 | 98.3 | 98.2 | 98.3 | 98.4 | 98.2 | 98.1 | 97.6 | 98.2 | 97.4 | 97.9 | 97.3 | 97.3 | |
| 16681 | 97.0 | 98.8 | 98.9 | 98.9 | 98.8 | 98.9 | 98.9 | 98.7 | 98.7 | 97.3 | 98.0 | 97.2 | 97.8 | 97.1 | 97.1 | |
| ThNH7 | 95.4 | 96.8 | 99.4 | 99.6 | 99.6 | 99.7 | 99.5 | 99.6 | 99.6 | 97.1 | 97.6 | 96.9 | 97.5 | 96.9 | 96.9 | |
| ThNHp11 | 95.3 | 96.7 | 98.2 | 99.4 | 99.4 | 99.5 | 99.7 | 99.3 | 99.3 | 97.2 | 97.8 | 97.1 | 97.5 | 97.0 | 97.1 | |
| K0008 | 95.4 | 96.8 | 98.8 | 98.1 | 99.8 | 99.8 | 99.5 | 99.9 | 99.9 | 97.1 | 97.7 | 97.0 | 97.6 | 96.9 | 97.0 | |
| K0010 | 95.4 | 96.8 | 98.7 | 98.1 | 99.2 | 99.7 | 99.4 | 99.7 | 99.7 | 97.1 | 97.6 | 96.9 | 97.5 | 96.8 | 96.9 | |
| CO371 | 95.3 | 96.8 | 98.5 | 98.1 | 98.8 | 98.7 | 99.6 | 99.7 | 99.6 | 97.2 | 97.8 | 97.1 | 97.7 | 97.0 | 97.1 | |
| CO390 | 95.4 | 96.8 | 98.4 | 98.8 | 98.4 | 98.4 | 98.5 | 99.4 | 99.4 | 97.3 | 97.8 | 97.1 | 97.6 | 97.1 | 97.1 | |
| CO166 | 95.2 | 96.6 | 98.7 | 97.9 | 99.6 | 99.0 | 98.7 | 98.4 | 99.9 | 97.0 | 97.6 | 96.8 | 97.5 | 96.8 | 96.8 | |
| CO167 | 95.1 | 96.6 | 98.5 | 98.1 | 99.3 | 98.6 | 98.3 | 98.1 | 99.6 | 97.0 | 97.5 | 96.8 | 97.4 | 96.8 | 96.8 | |
| PR159S1 | 92.0 | 91.3 | 90.4 | 90.6 | 90.5 | 90.5 | 90.4 | 90.6 | 90.4 | 90.4 | 96.9 | 99.3 | 96.6 | 99.1 | 99.2 | |
| 1409 | 95.3 | 94.5 | 93.2 | 93.3 | 93.3 | 93.3 | 93.3 | 93.3 | 93.1 | 93.1 | 90.7 | 96.9 | 99.1 | 96.8 | 96.7 | |
| Ven2 | 91.5 | 90.9 | 90.0 | 90.2 | 90.2 | 90.1 | 90.1 | 90.2 | 90.1 | 90.1 | 98.5 | 90.3 | 96.6 | 99.4 | 99.4 | |
| Mara4 | 95.0 | 94.3 | 93.1 | 93.0 | 93.3 | 93.3 | 93.3 | 93.1 | 93.1 | 93.1 | 90.5 | 98.4 | 90.1 | 96.4 | 96.4 | |
| 0131 | 91.4 | 90.8 | 90.0 | 90.1 | 90.1 | 90.0 | 90.0 | 90.1 | 90.1 | 90.0 | 98.2 | 90.2 | 98.5 | 90.0 | 99.2 | |
| IQT1797 | 91.4 | 90.8 | 90.1 | 90.2 | 90.2 | 90.1 | 90.1 | 90.2 | 90.1 | 90.1 | 97.9 | 90.3 | 98.2 | 90.1 | 97.8 | |
Viruses listed in Table 1 were compared for sequence similarity by using the MEGALIGN program (DNASTAR, Inc.).
Southeast Asian genotype sequence analysis.
Analysis of six Thai viruses revealed nucleotide and amino acid sequence similarities of 98.1 to 99.6% and 99.4 to 99.9%, respectively; nucleotide similarities to a 1964 Thai isolate, strain 16681, ranged from 96.6 to 96.8%. This suggests that, during a 30-year period in Thailand, the prevalent DEN-2 virus population has fixed up to only 3.4% mutations. However, based on samples taken during the period we observed, 1994 to 1996, the mutation rate per year was 1.5%, which is much higher than was observed for other dengue virus serotypes (e.g., 0.12 to 0.14% per year for DEN-3) (11). It could be that because we are not passaging virus after patient sampling, we are seeing the actual distribution of virus variants, with a concomitant rise in observed mutations (i.e., no population selection due to cell culture or other propagation). Amino acid variation was 0.5% per year, or one-third of that of nucleotides, suggesting that nucleotide variation was selective, with a preference for the third codon position. Nucleotide sequence variation was present as single-base substitutions scattered throughout the entire length of the genome, except for the 5′ NTR, which was identical to the 5′ NTR of reference strain 16681. No nucleotide deletions or insertions were detected within the coding region, although within the 3′ NTR, all Thai samples except CO390 show an insertion at position 10392, bringing their total genome length to 10,724 bases (versus 10,723 bases for strain 16681). The majority of nucleotide substitutions occurred at third codon positions, and, overall, only 6.9 to 16.3% of the nucleic acid changes resulted in amino acid changes. Analysis of the predicted polyprotein revealed that amino acid substitutions occurred throughout the coding region, with the majority (23 of 28) in the nonstructural proteins. None of these were consistent among DF or DHF samples and could not be correlated with disease outcome. Within the structural region, the C and prM genes were conserved for all samples except CO390 (2 aa differences in C-305 and C-309, and 1 aa difference in prM-16). When comparing the six Thai samples as a group, the numbers of amino acid differences among them per gene were as follows: C, 2 of 114 (1.75%); prM, 1 of 91 (1.10%); M, 0 of 75; E, 2 of 495 (0.40%); NS1, 6 of 352 (1.70%); NS2a, 7 of 281 (3.21%); NS2b, 0 of 130; NS3, 4 of 618 (0.65%); NS4a, 2 of 150 (1.33%); NS4b, 2 of 248 (0.81%); and NS5, 2 of 900 (0.22%).
American genotype sequence analysis.
The samples representing the American genotype had nucleotide and amino acid sequence similarities ranging from 97.8 to 98.5% and 99.1 to 99.4%, respectively. Within the entire coding area, no nucleotide deletions or insertions were observed. However, within the 5′ NTR, both Peruvian samples (IQT1797 and IQT2913) had a deletion at position 76, and within the 3′ NTR, all samples had deletions at positions 10276 to 10283 (8 nt), 10299, and 10388, bringing their total genome length to 10,712 bases. A nucleotide insertion was seen at position 10612 in both Peruvian samples. As in the Thai samples, the majority of nucleotide differences were single-base substitutions, occurring in the third position of the codons; only 10.36 to 12.42% of the nucleic acid changes resulted in amino acid changes. When comparing the five samples as a group, the numbers of amino acid differences among them per gene were as follows: C, 0 of 114; prM, 1 of 91 (1.10%); M, 1 of 75 (1.33%); E, 2 of 495 (0.40%); NS1, 6 of 352 (1.70%); NS2a, 4 of 281 (1.83%); NS2b, 0 of 130; NS3, 8 of 618 (1.29%); NS4a, 0 of 150; NS4b, 1 of 248 (0.40%); and NS5, 14 of 900 (1.55%).
Comparison of two genotypes.
Because the sequence analysis of the Southeast Asian genotype viruses did not reveal any specific changes consistently correlated with disease outcome and their hydrophilicity profiles, calculated by the method of Kyte and Doolittle (42), were virtually superimposable, we compared the Southeast Asian genotype viruses to American genotype viruses, since the former have the potential to cause DHF while the latter are associated only with DF (60, 70). All amino acid residues with predicted biologic or enzymatic activities in the encoded polypeptide were conserved among all the viruses: cysteine residues in prM, E, and NS1; N-linked glycosylation sites at prM-67, E-69, and NS1-207; the NS1/NS2a cleavage motif in NS1 (14); the substrate binding pocket of viral protease in NS3 (54); and an N-terminal sequence motif in NS5, found in other S-adenosylmethionine-utilizing methyltransferases (40). A total of 55 aa changes were consistently detected between the two genotypes, while only 6 of these resulted in a charge difference and 5 resulted in a side chain difference (polar versus nonpolar). A summary of amino acid substitutions that confer a change in charge or side chain polarity is given in Table 4. Computer analysis of the amino acid changes at prM-28 (Glu→Lys) and prM-31 (Val→Thr) revealed a change of hydrophilicity and predicted antigenicity for the samples belonging to the American genotype. The amino acid charge difference at E-390 (Asn→Asp) did not change the hydrophilicity or antigenicity profiles. Within the nonstructural genes, the amino acid substitutions at positions NS1-128 (Ser→Leu) and NS3-567 (Ile→Thr) did result in a change of hydrophilicity and predicted antigenicity, while a charge change at NS4b-17 from neutral to positive (Ser→His) did not change the secondary structure. Five of the 8 aa changes detected within the nonstructural genes were localized in NS5. Three amino acid substitutions at NS5-645 (Asn→Asp), NS5-676 (Ser→Arg/Lys), and NS5-800 (Lys→Ser) with a charge change did not change the structure, while the changes at NS5-271 (Ile→Thr) and NS5-819 (Gln→Leu) resulted in significant changes in the predicted hydrophilicity and antigenicity profiles.
TABLE 4.
Summary of consistent amino acid changes between Southeast Asian and American genotype viruses
| Gene | Position no. | Change of amino acida
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ThNHp11 (DF) | K00010 (DF) | CO371 (DF) | CO166 (DF) | ThNH7 (DSS)b | K0008 (DHF) | CO390 (DHF) | CO167 (DHF) | Mara4 (DHF) | 102692 (DHF) | Ven2 (DF) | 0131 (DF) | IQT1797 (DF) | IQT2913 (DF) | ||
| prM | 28 | E | E | E | E | E | E | E | E | E | E | K | K | K | K |
| 31 | V | V | V | V | V | V | V | V | V | V | T | T | T | T | |
| E | 390 | N | N | N | N | N | N | N | N | N | N | D | D | D | D |
| NS1 | 128 | S | S | S | S | S | S | S | S | S | S | L | L | L | L |
| NS3 | 567 | I | I | I | I | I | I | I | I | I | I | T | T | T | T |
| NS4b | 17 | S | S | S | S | S | S | S | S | S | S | H | H | H | H |
| NS5 | 271 | I | I | I | I | I | I | I | I | I | — | T | T | T | T |
| 645 | N | N | N | N | N | N | N | N | N | — | D | D | D | D | |
| 676 | S | S | S | S | S | S | S | S | S | — | R | R | K | K | |
| 800 | K | K | K | K | K | K | K | K | K | — | S | S | S | S | |
| 819 | Q | Q | Q | Q | Q | Q | Q | Q | Q | — | L | L | L | L | |
Charge changes are italicized, and side chain changes are in boldface. Negative charge amino acids, E (glutamic acid) and D (aspartic acid); positive charge amino acids, K (lysine), H (histidine), and R (arginine); neutral amino acids with polar side chains, V (valine), L (leucine), and I (isoleucine); neutral amino acids with nonpolar side chains, T (threonine), N (asparagine), S, (serine), and Q (glutamine). —, not done.
DSS, dengue shock syndrome.
Phylogenetic analysis of full genome sequences.
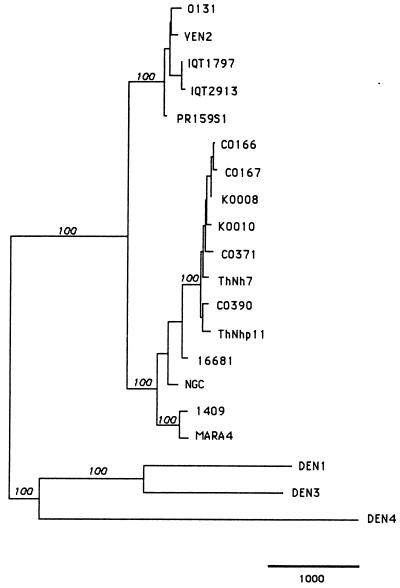
Maximum parsimony analysis of the entire genomes resulted in an evolutionary tree with branching patterns proportional to the genetic relatedness among strains (Fig. 2). These results did not differ substantially from previously published limited nucleotide sequence comparisons (full genes, or parts of genes, e.g., 240 nt at the E/NS1 gene junction). All DEN-2 viruses segregated, as expected, into two different groups, each representing a distinct genotype, as defined previously (58). The main difference seen in this analysis from previous phylogenetic studies is that statistical measures of significance of monophyletic groupings (bootstrap values) reached 100%, as would be expected from inclusion of the total available sequence data. Within the Thai samples, there was neither an obvious segregation according to geographic region (the two study locations are located about 360 km apart) nor segregation according to clinical outcome (DF versus DHF). However, all samples obtained in the Americas that were associated with DF only segregated to one phylogenetic group that was clearly distinct from the Southeast Asian genotype.
FIG. 2.
Phylogenetic tree generated by maximum parsimony analysis of nucleotide sequences from the entire genome of 11 DEN-2 viruses, sequences of previously published strains and representatives of DEN-1, -3, and -4. Viruses are listed by strain number (Table 1). Branch lengths (proportional to the bar, which equals 1,000) represent the number of nucleotide substitutions between the viruses over the 10,724 nt of the genome used for comparison. Bootstrap values are shown in italics above the branches.
Predicted RNA secondary structure of the 5′ NTR.
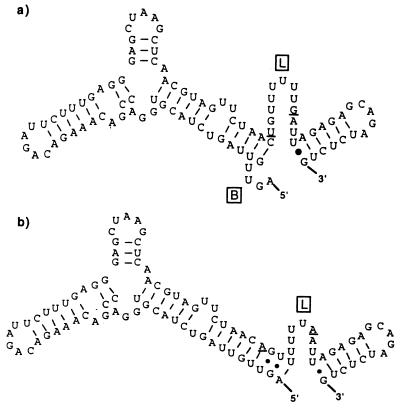
Within the Thai samples, all nucleotide sequences were conserved in this region and were identical to those of reference strain 16681, so the predicted RNA structures did not change. Within the American genotype viruses, two consistent nucleotide changes at positions 69 and 77 (A→T and A→G, respectively) were observed compared to the Southeast Asian genotype viruses. The 97 nt of the 5′ NTR were predicted to form a long stem structure with a top loop (between nt 29 and 35), a short loop (between nt 44 and 55), and a 3′-terminal loop preceeding the translation initiation codon (Fig. 3). Despite the fact that sequences obtained from the first 68 nt were identical among all viruses, those belonging to the American genotype had a distinct sequence at nt 69 and 77 which changed the predicted secondary structure, with a 4-nt bulge at the 5′ terminus, resulting in a reduced stem length, and a longer 3′-terminal loop of 12 nt (nt 68 to 80 versus 72 to 80 for the Southeast Asian genotype).
FIG. 3.
Predicted RNA secondary structures formed by the complete 5′ NTR of two genotypes of DEN-2: sample 0131, representing the American genotype (a), and sample K0010, representing the Southeast Asian genotype (b). Nucleotide differences between the genotypes (positions 69 and 77) are underlined. B, 5′-terminal bulge; L, 3′-terminal loop.
Predicted RNA secondary structure of the 3′ NTR.
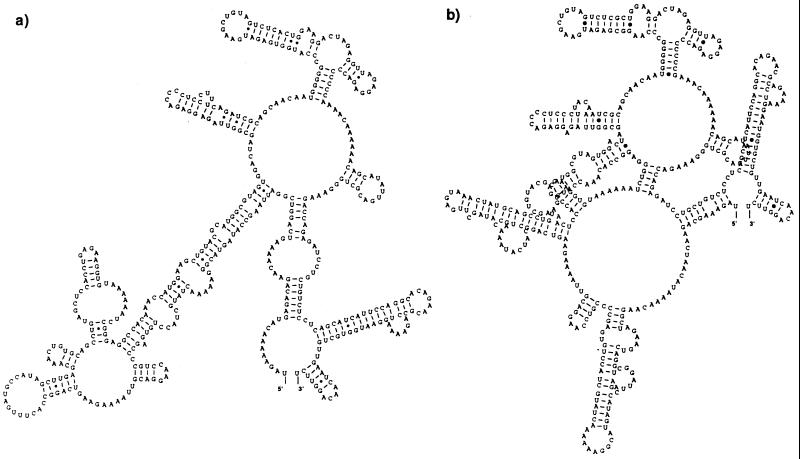
Within the Thai samples, strain CO390 had 5 nt differences compared to the rest of the group (nt 10386 [C→T], nt 10407 [T→A], nt 10425 [A→G], nt 10438 [A→G], and nt 10448 [A→G]), and strain CO167 had one difference at nt 10554 (G→A). These nucleotide differences did not segregate with disease association nor did they alter the predicted secondary stem-loop structure. Within viruses of the American genotype, strain Ven2 had 5 nt differences compared to the rest of the group (nt 10315 [T→C], nt 10418 [G→A], nt 10427 [T→C], nt 10609 [A→G], and nt 10638 [T→C]), strain 1318 had four differences (nt 10290 [A→T], nt 10291 [G→A], nt 10292 [A→G], and nt 10569 [T→C]), both IQT1797 and IQT2913 had four differences (nt 10305 [T→C], nt 10386 [C→T], nt 10611 [C→A], and nt 10612 [A→C]), strain 044 had three differences (nt 10315 [T→C], nt 10609 [A→G], and nt 10648 [T→C]), strain 0131 had two differences (nt 10523 [A→T] and nt 10550 [A→T]), and strain 0147 had two differences (nt 10487 [C→T] and nt 10523 [A→T]). This comparison revealed two characteristics that distinguished viruses across genotypes: all American samples have an 8-nt deletion at the beginning of the 3′ NTR (nt 10276 to 10283) immediately downstream from the stop codon along with two more interspersed deletions at positions 10299 and 10388, and they have consistent nucleotide differences at positions 10297 (G→A), 10331 (A→G), 10388 (A→T), 10390 (A→G), and 10527 (A→G) compared to the Thai samples as a group. Secondary structure predictions using all 443 to 455 nt of the 3′ NTR revealed characteristic conformations that segregated the genotypes (Fig. 4); only the RNASTAR predictions are shown here. The 100 nt at the 3′ terminus were conserved for all viruses studied, and their predicted folding pattern formed a characteristic stem-loop structure described previously (56). However, for the American genotype, a drastically different structure was predicted for the area upstream of nt 10498, with a very long stem and loop. The Southeast Asian genotype virus RNA folded in a much more compact pattern, with the possibility for more tertiary structure interactions. To obtain a quantitative estimate of RNA structure differences, an algorithm that compares stems and loops (RNAdistance) was used to assign distance values and to generate a distance matrix of secondary structure relationships. The distance matrix and its graphic representation, a phylogenetic tree (data not shown), displayed the same relationships between viruses of the Southeast Asian and American genotype as when comparing full genome sequences (Fig. 2).
FIG. 4.
Predicted RNA secondary structures formed by the complete 3′ NTR of two genotypes of DEN-2: sample 0131, representing the American genotype (a), and sample K0010, representing the Southeast Asian genotype (b).
DISCUSSION
Prior studies of wild and attenuated dengue viruses have suggested that genetic differences among strains of the four serotypes can be associated with attenuation, virulence, and/or epidemic potential. Despite the fact that various numbers of nucleotide and amino acid differences were found in coding or noncoding regions of the genome, no specific site(s) could be correlated with attenuation or severe disease in humans. In vitro (e.g., plaque size) and in vivo (e.g., mouse neurovirulence and monkey viremia) markers used to pinpoint probable virulence determinants have been shown to be imperfect models of human disease. This is the first report of genetic differences between two DEN-2 genotypes that have been associated with distinct clinical presentations in humans: the Southeast Asian genotype with DF and DHF and the American genotype with DF only. The new approach of full genome sequencing directly from patient plasma avoids any selection of virus variants by cultivation or cloning methods and is thus significantly different from previous studies. Therefore, the resulting nucleotide and/or amino acid differences we have reported here should better reflect the virus population in the host and permit association with pathogenesis.
It was not surprising that, within the Thai samples, no specific genome structures could be consistently correlated with disease outcome (DF versus DHF); previous studies comparing DEN-2 viruses from Thailand revealed similar results (3, 49). This observation supports the conclusion that all viruses belonging to the Southeast Asian genotype have the potential to cause severe disease; only epidemiologic associations provided the first indication that transmission of this genotype is directly associated with the occurrence of severe dengue (60). Ongoing studies in Peru reveal that no cases of DHF have occurred in a population with high secondary infection rates, and this is probably explained by the fact that only the American genotype of DEN-2 is being transmitted in this region (70). We have shown that a comparison of viruses from genetic groups with distinct clinical and epidemiologic associations can better identify structural differences that correlate with pathogenesis potential.
Significance of one E glycoprotein difference.
The observed amino acid difference between the Southeast Asian and American genotype viruses at position E-390 is of major importance. The E glycoprotein, which is exposed on the surface of the dengue virion, represents the dominant virus antigen, conferring protective immune responses by eliciting neutralizing, hemagglutination-inhibiting, antifusion, and virus-enhancing antibodies and is responsible for virus attachment, virus-specific membrane fusion in acid pH endosomes, and virus assembly (9, 61). An analogous situation has been described for other flaviviruses: single-amino-acid substitutions in the envelope protein at positions 52 (27), 138 (66), 271, and 336 in Japanese encephalitis virus (7); at position 390 in Murray Valley virus (44); at positions 104 and 107 in a tick-borne encephalitis–DEN-4 chimera (53); and at positions 171 and 384 for TBE virus (47) were shown to have a dramatic effect on virulence in vitro or in vivo. Sanchez et al. reported that clones of a DEN-2 virus (Mexican strain) with a single-amino-acid change at position E-390 from Asp to His were virulent when inoculated intracerebrally into suckling mice (63). Mutations from Asp to Asn conferred attenuation; however, this correlation is not seen in the Southeast Asian genotype viruses we analyzed (Asn) compared to those belonging to the American genotype (Asp). An attenuated DEN-2 vaccine candidate did not differ from the parental virus strain in the E gene sequence (38). These observations on attenuation differ from our results in that the phenomenon we are attempting to measure deals with the severity of disease (from DF to DHF) versus the presence or absence of disease.
It is generally accepted that most antigenic determinants reactive with antibodies are exposed on the surface of the protein (2, 67) and hence are hydrophilic. Based on this notion, algorithms which calculate hydrophilicity have been used to predict antigenicity (30, 42). Most of the neutralization-resistant variants of flaviviruses obtained so far have amino acid substitutions which cause a change in charge (27, 34, 46), and it is known that charged residues are important in the interaction of antigenic sites with antibodies. An analysis of the envelope glycoprotein shows that residue E-390 is located in a highly hydrophilic region; Asn and Asp have the same hydrophilicity value, but the first is neutral in charge while the latter is acidic.
In a manner analogous to the recently solved molecular structure of the amino-terminal fragment of the TBE virus E glycoprotein homodimer (57), the dengue E glycoprotein is expected to fold into three distinct functional domains, named I, II, and III, which correlate well with recently described dengue virus E antigenic domains C, A, and B (61). The observed difference in residue E-390 is localized in the C-terminal domain III (aa 303 to 395) on the G-sheet (aa 388 to 394) which forms, together with the C and the F sheet, the outer lateral surface of the dimer. This is important because the lateral surface of domain III is suggested to contain residues implicated as determinants for host range, tropism, and virulence in different flaviviruses (57). The observed amino acid change at position E-390 is also localized within one of the putative glycosaminoglycan binding motifs at aa 284 to 310 and 386 to 411 (9). These motifs have been shown to specifically bind DEN-2 virus (Tonga, 1974) to the host cell surface, and blockage of this receptor with an antagonist prevented infection. Since glycosaminoglycan binding motifs in proteins are predicted by defining multiple regions enriched for basic amino acids (17), it must be determined whether a change from acidic Asp (American genotype) to neutral Asn (Southeast Asian genotype) enhances DEN-2 virus cell attachment.
Significance of prM differences.
While monoclonal antibodies specific for the prM protein of DEN-3 and DEN-4 viruses have been shown to passively protect mice against challenge with both homologous and heterologous dengue viruses (36), prM monoclonal antibodies did not display neutralizing activity in vitro (4). The comparison of DEN-2 strain 16681 to its vaccine derivative, strain PDK-53, revealed an amino acid change from Asp to Val at position prM-29 (38). We identified 2 aa, at prM-28 and prM-31, that distinguished the Southeast Asian from the American genotype; however, since the antigenic structure of prM is not known and the locations of protective epitopes have not been identified, the role of these amino acid changes is difficult to assess.
Significance of NS4b and NS5 differences.
Although some structural properties of dengue virus NS4b and NS5 have been elucidated, little is known about their antigenic structure. For NS4a and NS4b proteins it was shown that, despite marked amino acid sequence heterogeneity, their hydrophilicity profiles are remarkably conserved among the dengue viruses and flaviviruses in general. In terms of function, both NS4 proteins may be involved in membrane localization of NS3 and NS5 replication complexes via protein-protein interaction (8). The observed residue change at NS4b-17 did not affect the hydrophobicity profiles, and its importance in protein function is unknown. Because the NS5 gene presumably encodes a protein with important enzymatic functions, the RNA polymerase (40), there was conservation of its structure, especially in the presumed active sites. Of 5 aa substitutions observed here, 3, at NS5-645 (Asn→Asp), NS5-676 (Ser→Arg/Lys), and NS5-800 (Lys→Ser), conferred a charge change, but were not predicted to change the structure, while the side chain changes at NS5-271 (Ile→Thr) and NS5-819 (Gln→Leu) resulted in significant differences in hydrophilicity and antigenicity. However, since the three-dimensional structure or antigenic function of the NS5 protein has not been determined, the relative contribution of these changes to infectivity and immunogenicity is unknown.
Significance of NTR differences.
The NTRs of several positive-strand RNA viruses are predicted to fold into stem-loop structures that interact with viral or cellular proteins (1, 5, 12, 23, 51). At the 5′ NTR, ribosomes bind to the 5′ terminus of the positive strand to initiate translation, and replicase binds to the 3′ terminus of the minus strand to initiate transcription of positive strands, thus regulating replication. Mutations that modify these structures, and thereby alter the RNA-protein interactions, have been shown to affect virulence or cause attenuation (31, 37). A single-nucleotide mutation in the 5′ NTR of poliovirus is associated with neurovirulence (13), and attenuation was correlated with disruption of its secondary structure (45). Engineered mutations and deletions in the 5′ NTR of a full-length DEN-4 cDNA clone were shown to restrict DEN-4 virus growth in cell culture and in inoculated mosquitoes (6); most mutations within the long stem structure were lethal, but RNA transcripts containing deletions in the loop or short stem regions were usually infectious. However, a mutant bearing deletions in the 3′-terminal loop was least efficient in translation. Our results show that an A-to-U mutation that distinguishes Southeast Asian from American genotype viruses at position 69 was predicted to change the secondary structure of the viral RNA: a 4-nt-long bulge was formed at the 5′ terminus of the American genotype, which reduced the length of the stem, but increased the length of the 3′-terminal loop. Whether the presence of this small bulge could reduce translation efficiency remains unclear; bulges as small as 1 nt have been shown to reduce RNA-protein interactions in other viral systems (71, 73).
The 3′ NTR contains sequences essential for virus replication and growth, serving as signals for the initiation of minus-strand synthesis, and possibly packaging in yellow fever and DEN-4 (23, 50). A stable secondary structure motif, formed by the 3′-terminal 100 nt, was described for all mosquito-borne flaviviruses studied to date (21, 23, 56, 65, 72). Within this region and farther upstream, three highly conserved sequences, termed CS1, CS2, and RCS2, are thought to be functionally important elements of the 3′ NTR (23). While conservation of the 3′-terminal region is assumed to be essential for flaviviruses, there is evidence that regions farther upstream determine virus replication efficiency. Deletions introduced into full-length DEN-4 cDNA clones that did not extend beyond the 3′-terminal 113 nt were viable when transfected into cells in culture, but they exhibited a range of growth restrictions (50). In the 19 viruses we compared, a conserved region of 110 nt was found at the 3′ terminus, which folded in a form seen in other flaviviruses (56). However, a striking size and sequence heterogeneity was observed in the 300-nt upstream region that allowed us to distinguish the genotypes, based on nucleotide alignments. All American genotype samples revealed deletions in the region immediately downstream of the stop codon as well as 5 interspersed nt differences when compared to the Southeast Asian genotype; secondary structure predictions yielded drastically different conformations that were characteristic for each genotype. The importance of these structures in defining dengue virus pathogenicity is unknown, but 3′ NTR structure has also been shown to correlate with virulence in yellow fever (55) and TBE (56) viruses.
With the approach used here, focusing on consistent differences between two genotypes, we have reduced the background “noise” (e.g., single nucleotides) that may not be directly involved in determining pathogenicity. Virus virulence characteristics have been shown to be due to the synergistic effect of various genomic loci; it is possible that dengue pathogenesis is the result of cohesive activity of the sites we have pinpointed here, in addition to host immunologic factors. The amino acid substitutions within the coding region, which may affect antigenicity or cell attachment, and nucleotide changes within the NTRs, affecting secondary structure and thereby replication, may be the viral determinants of severe dengue in humans. Genome differences in nonstructural genes or in the NTRs potentially altering replication efficiency could possibly be measured most effectively in terms of human viremias. In addition, we are currently deriving modified infectious clones to test these hypotheses, by using reverse genetics, although we are limited to imperfect models of human disease for testing phenotype. Further sequence analyses may also confirm whether the structures we have found are maintained consistently in other dengue transmission cycles. From our data it could be concluded that, for vaccine design, it would be advantageous to manipulate the NTRs of the genome to reduce viral replication, whereas high immunogenicity would be ensured by maintaining the native structure of the viral proteins.
ACKNOWLEDGMENTS
This research was supported by NIH grants AI01124 and AI34533 and the Army and Navy Medical Research and Material Command.
We thank Siripen Kalayanarooj, Suchitra Nimmannitya, Saroj Suntayakorn, Ananda Nisalak, Duilia Tovar, Dalia Hernandez, Alan Rothman, Sharone Green, Mark Sharp, and Terry Hawkins for virus typing, clinical grading, shipment, and computer support.
REFERENCES
- 1.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′ end of the viral RNA. Cell. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berzofsky J A. Intrinsic and extrinsic factors in protein antigenic structure. Science. 1985;229:932–940. doi: 10.1126/science.2410982. [DOI] [PubMed] [Google Scholar]
- 3.Blok J, Gibbs A J, McWilliam S M, Vitarana U T. NS1 gene sequences from eight dengue-2 viruses and their evolutionary relationships with other dengue-2 viruses. Arch Virol. 1991;118:209–223. doi: 10.1007/BF01314031. [DOI] [PubMed] [Google Scholar]
- 4.Bray M, Lai C-J. Dengue virus premembrane and membrane proteins elicit a protective immune response. Virology. 1991;185:505–508. doi: 10.1016/0042-6822(91)90809-p. [DOI] [PubMed] [Google Scholar]
- 5.Brinton M A, Dispoto J H. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology. 1988;162:290–299. doi: 10.1016/0042-6822(88)90468-0. [DOI] [PubMed] [Google Scholar]
- 6.Cahour A, Pletnev A, Vazeille-Falcoz M, Rosen L, Lai C-J. Growth-restricted dengue virus mutants containing deletions in the 5′ noncoding region of the RNA genome. Virology. 1995;207:68–76. doi: 10.1006/viro.1995.1052. [DOI] [PubMed] [Google Scholar]
- 7.Cecilia D, Gould E A. Nucleotide changes responsible for loss of neuroinvasiveness in Japanese encephalitis virus neutralization-resistant mutants. Virology. 1991;181:70–77. doi: 10.1016/0042-6822(91)90471-m. [DOI] [PubMed] [Google Scholar]
- 8.Chambers T J, Chang S H, Galler R, Rice C M. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Maguire T, Marks R M. Demonstration of binding of dengue virus envelope protein to target cells. J Virol. 1996;70:8765–8772. doi: 10.1128/jvi.70.12.8765-8772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou P Y, Fasman G D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol. 1978;45:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 11.Chungue E. Molecular epidemiology of dengue viruses. In: Saluzzo J F, Dodet B, editors. Factors in the emergence of arbovirus diseases. Paris, France: Elsevier; 1997. pp. 93–101. [Google Scholar]
- 12.Day S P, Murphy P, Brown E A, Lemon S M. Mutations within the 5′ nontranslated region of hepatitis A virus RNA which enhance replication in BS-C-1 cells. J Virol. 1992;66:6533–6540. doi: 10.1128/jvi.66.11.6533-6540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans D M A, Dunn G, Minor P D, Schild G C, Cann A J, Stanway G, Almond J W, Currey K, Maizel J V J. Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature. 1985;314:448–550. doi: 10.1038/314548a0. [DOI] [PubMed] [Google Scholar]
- 14.Falgout B, Chanock R, Lai C-J. Proper processing of dengue virus nonstructural glycoprotein NS1 requires the N-terminal hydrophobic signal sequence and the downstream nonstructural protein NS2a. J Virol. 1989;63:1852–1860. doi: 10.1128/jvi.63.5.1852-1860.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 16.Finney D J. Probit analysis. Cambridge, United Kingdom: Cambridge University Press; 1971. [Google Scholar]
- 17.Fromm J R, Hileman R E, Caldwell E E, Weiler J M, Linhardt R J. Differences in the interaction of heparin with arginine and lysine and the importance of these basic amino acids in the binding of heparin to acidic fibroblast growth factor. Arch Biochem Biophys. 1995;323:279–287. doi: 10.1006/abbi.1995.9963. [DOI] [PubMed] [Google Scholar]
- 18.Fu J, Tan B H, Yap E H, Chan Y C, Tan Y H. Full-length cDNA sequence of dengue type 1 virus (Singapore strain S275/90) Virology. 1992;188:935–958. doi: 10.1016/0042-6822(92)90560-c. [DOI] [PubMed] [Google Scholar]
- 19.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple method for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert, D. G. 9 April 1997, posting date. [Online.] loopDloop in Java, a Java application for visualizing RNA secondary structure, v.2.07b. ftp://iubio.bio.indiana.edu/molbio/loopdloop/java/. [2 April 1999, last date accessed.]
- 21.Grange T, Bouloy M, Girard M. Stable secondary structures at the 3′-end of the genome of yellow fever virus (17D vaccine strain) FEBS Lett. 1985;188:159–163. doi: 10.1016/0014-5793(85)80895-4. [DOI] [PubMed] [Google Scholar]
- 22.Gultyaev A P, van Batenburg F H D, Pleij C W A. The computer simulation of RNA folding pathways using a genetic algorithm. J Mol Biol. 1995;250:37–51. doi: 10.1006/jmbi.1995.0356. [DOI] [PubMed] [Google Scholar]
- 23.Hahn C S, Hahn Y S, Rice C M, Lee E, Dalgarno L, Strauss E G, Strauss J H. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J Mol Biol. 1987;198:33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- 24.Halstead S B. Epidemiology of dengue and dengue hemorrhagic fever. In: Gubler D J, Kuno G, editors. Dengue and dengue hemorrhagic fever. Wallingford, United Kingdom: CAB International; 1997. pp. 23–44. [Google Scholar]
- 25.Halstead S B. Observations related to pathogenesis of dengue hemorrhagic fever. VI. Hypotheses and discussion. Yale J Biol Med. 1970;42:350–362. [PMC free article] [PubMed] [Google Scholar]
- 26.Halstead S B. Pathogenesis of dengue: challenges to molecular biology. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 27.Hasegawa H, Yoshida M, Shiosaka T, Fujita S, Kobayashi Y. Mutations in the envelope protein of Japanese encephalitis virus affect entry into cultured cells and virulence in mice. Virology. 1992;191:158–165. doi: 10.1016/0042-6822(92)90177-q. [DOI] [PubMed] [Google Scholar]
- 28.Hofacker I L, Fontana W, Stadler P F, Bonhoeffer S, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh Chem. 1994;125:167–188. [Google Scholar]
- 29.Holzmann H, Heinz F X, Mandl C W, Guirakhoo F, Kunz C. A single amino acid substitution in envelope protein E of tick-borne encephalitis virus leads to attenuation in the mouse model. J Virol. 1990;64:5156–5159. doi: 10.1128/jvi.64.10.5156-5159.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopp T P, Woods K R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iizuka N, Kohara M, Hagino-Yamagishi K, Abe S, Komatsu T, Tago K, Arita M, Nomoto A. Construction of less neurovirulent polioviruses by introducing deletions into the 5′ noncoding sequence of the genome. J Virol. 1989;63:5354–5363. doi: 10.1128/jvi.63.12.5354-5363.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jameson B A, Wolf H. The antigenic index: a novel algorithm for predicting antigenic determinants. CABIOS. 1988;4:181–186. doi: 10.1093/bioinformatics/4.1.181. [DOI] [PubMed] [Google Scholar]
- 33.Jennings A D, Whitby J E, Minor P D, Barrett A D T. Comparison of the nucleotide and deduced amino acid sequences of the wild-type French viscerotropic strain of yellow fever virus and the live vaccine strain, French neurotropic vaccine derived from it. Virology. 1993;192:692–695. doi: 10.1006/viro.1993.1090. [DOI] [PubMed] [Google Scholar]
- 34.Jiang W R, Lowe A, Higgs S, Reid S, Gould E A. Single amino acid codon changes detected in louping ill virus antibody-resistant mutants with reduced neurovirulence. J Gen Virol. 1993;74:931–935. doi: 10.1099/0022-1317-74-5-931. [DOI] [PubMed] [Google Scholar]
- 35.Kalayanarooj S, Vaughn D W, Nimmannitya S, Green S, Suntayakorn S, Kunentrasai N, Viramitrachai W, Ratanachu-eke S, Kiatpolpoj S, Innis B L, Rothmann A L, Nisalak A, Ennis F A. Early clinical and laboratory indicators of acute dengue illness. J Infect Dis. 1997;176:313–321. doi: 10.1086/514047. [DOI] [PubMed] [Google Scholar]
- 36.Kaufman B M, Summers P L, Dubois D R, Houston Cohen W, Gentry M K, Timchak R L, Burke D S, Eckels K H. Monoclonal antibodies for dengue virus prM glycoprotein protect mice against lethal dengue infection. Am J Trop Med Hyg. 1989;41:576–580. doi: 10.4269/ajtmh.1989.41.576. [DOI] [PubMed] [Google Scholar]
- 37.Kinney R M, Chang G-J, Tsuchiya K R, Sneider J M, Roehrig J T, Woodward T M, Trent D W. Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5′-noncoding region and the E2 envelope glycoprotein. J Virol. 1993;67:1269–1277. doi: 10.1128/jvi.67.3.1269-1277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinney R M, Butrapet S, Chang G-J J, Tsuchiya K R, Roehrig J T, Bhamarapravati N, Gubler D. Construction of infectious cDNA clones for Dengue 2 virus: strain 16681 and its attenuated vaccine derivate, strain PDK-53. Virology. 1997;230:300–308. doi: 10.1006/viro.1997.8500. [DOI] [PubMed] [Google Scholar]
- 39.Kliks S C, Nisalak A, Brandt W E, Wahl L, Burke D S. Antibody-dependent enhancement of dengue virus growth in human monocytes as a risk factor for dengue hemorrhagic fever. Am J Trop Med Hyg. 1989;40:444–451. doi: 10.4269/ajtmh.1989.40.444. [DOI] [PubMed] [Google Scholar]
- 40.Koonin E V. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and gamma 2 protein of reovirus. J Gen Virol. 1993;74:733–740. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- 41.Kurane I, Rothman A, Livingston P, Green S, Gagnon S, Janus J, Innis B, Nimmannitya S, Nisalak A, Ennis F. Immunopathologic mechanisms of dengue hemorrhagic fever and dengue shock syndrome. Arch Virol Suppl. 1994;9:59–64. doi: 10.1007/978-3-7091-9326-6_7. [DOI] [PubMed] [Google Scholar]
- 42.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 43.Lobigs M, Arthur C E, Mullbacher A, Blanden R V. The flavivirus nonstructural protein NS3 is a dominant source of cytotoxic T cell peptide determinants. Virology. 1994;202:195–201. doi: 10.1006/viro.1994.1335. [DOI] [PubMed] [Google Scholar]
- 44.Lobigs M, Usha R, Nestorowicz A, Marshall I D, Weir R C, Dalgarno L. Host cell selection of Murray Valley encephalitis virus variants altered at an RGD sequence in the envelope protein and in mouse virulence. Virology. 1990;176:587–595. doi: 10.1016/0042-6822(90)90029-q. [DOI] [PubMed] [Google Scholar]
- 45.MacAdam A J, Ferguson G, Stone J B D, Skuce R, Almond J W. Correlation of RNA secondary structure and attenuation of Sabin vaccine of poliovirus in tissue culture. Virology. 1992;189:415–422. doi: 10.1016/0042-6822(92)90565-7. [DOI] [PubMed] [Google Scholar]
- 46.Mandl C W, Guirakhoo F, Holzmann H, Heinz F X, Kunz C. Antigenic structure of the flavivirus envelope protein E at the molecular level, using tick-borne encephalitis virus as a model. J Virol. 1989;63:564–571. doi: 10.1128/jvi.63.2.564-571.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mandl C W, Heinz F X, Kunz C. Sequence of the structural proteins of tick-borne encephalitis virus (western subtype) and comparative analysis with other flaviviruses. Virology. 1988;166:197–205. doi: 10.1016/0042-6822(88)90161-4. [DOI] [PubMed] [Google Scholar]
- 48.Mangada M N, Igarashi A. Sequences of terminal non-coding regions from four dengue-2 viruses isolated from patients exhibiting different disease severities. Virus Genes. 1997;14:5–12. doi: 10.1023/a:1007914520454. [DOI] [PubMed] [Google Scholar]
- 49.Mangada M N M, Igarashi A. Molecular and in vitro analysis of eight dengue type 2 viruses isolated from patients exhibiting different disease severities. Virology. 1998;244:458–466. doi: 10.1006/viro.1998.9093. [DOI] [PubMed] [Google Scholar]
- 50.Men R, Bray M, Clark D, Chanock R M, Lai C-J. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J Virol. 1996;70:3930–3937. doi: 10.1128/jvi.70.6.3930-3937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niesters H G M, Strauss J H. Defined mutations in the 5′ nontranslated sequence of Sindbis virus RNA. J Virol. 1990;64:4162–4168. doi: 10.1128/jvi.64.9.4162-4168.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osatomi K, Sumiyoshi H. Complete nucleotide sequence of dengue type 3 virus genome RNA. Virology. 1990;176:643–647. doi: 10.1016/0042-6822(90)90037-r. [DOI] [PubMed] [Google Scholar]
- 53.Pletnev A G, Bray M, Lai C-J. Chimeric tick-borne encephalitis and dengue type 4 viruses: effects of mutations on neurovirulence in mice. J Virol. 1993;67:4956–4963. doi: 10.1128/jvi.67.8.4956-4963.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Preugschat F, Lenches E M, Strauss J H. Flavivirus enzyme-substrate interactions studied with chimeric proteinases: identification of an intragenic locus important for substrate recognition. J Virol. 1991;65:4749–4758. doi: 10.1128/jvi.65.9.4749-4758.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Proutski V, Gaunt M W, Gould E A, Holmes E C. Secondary structure of the 3′-untranslated region of yellow fever virus: implications for virulence, attenuation and vaccine development. J Gen Virol. 1997;78:1543–1549. doi: 10.1099/0022-1317-78-7-1543. [DOI] [PubMed] [Google Scholar]
- 56.Rauscher S, Flamm C, Mandl C W, Heinz F X, Stadler P F. Secondary structure of the 3′-noncoding region of flavivirus genomes: comparative analysis of base pairing probabilities. RNA. 1997;3:779–791. [PMC free article] [PubMed] [Google Scholar]
- 57.Rey F A, Heinz F X, Mandl C, Kunz C, Harrison S C. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 58.Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174:479–493. doi: 10.1016/0042-6822(90)90102-w. [DOI] [PubMed] [Google Scholar]
- 59.Rico-Hesse R, Harrison L, Nisalak A, Vaughn D, Kalayanarooj S, Green S, Rothman A, Ennis F. Molecular evolution of dengue type 2 virus in Thailand. Am J Trop Med Hyg. 1998;58:96–101. doi: 10.4269/ajtmh.1998.58.96. [DOI] [PubMed] [Google Scholar]
- 60.Rico-Hesse R, Harrison L, Salas R, Tovar D, Nisalak A, Ramos C, Boshell J, de Mesa M R, Nogueira R, Travassos da Rosa A. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 1997;230:244–251. doi: 10.1006/viro.1997.8504. [DOI] [PubMed] [Google Scholar]
- 61.Roehrig J T, Bolin R A, Kelly R G. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology. 1998;246:317–328. doi: 10.1006/viro.1998.9200. [DOI] [PubMed] [Google Scholar]
- 62.Rosen L. “The Emperor’s New Clothes” revisited, or reflections on the pathogenesis of dengue hemorrhagic fever. Am J Trop Med Hyg. 1977;26:337–343. doi: 10.4269/ajtmh.1977.26.337. [DOI] [PubMed] [Google Scholar]
- 63.Sanchez I J, Ruiz B H. A single nucleotide change in the E protein gene of dengue virus 2 Mexican strain affects neurovirulence in mice. J Gen Virol. 1996;77:2541–2545. doi: 10.1099/0022-1317-77-10-2541. [DOI] [PubMed] [Google Scholar]
- 64.Shapiro B A, Zhang K. Comparing multiple RNA secondary structures using tree comparison. CABIOS. 1990;6:309–318. doi: 10.1093/bioinformatics/6.4.309. [DOI] [PubMed] [Google Scholar]
- 65.Shi P-Y, Brinton M A, Veal J M, Zhong Y Y, Wilson W D. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry. 1996;35:4222–4230. doi: 10.1021/bi952398v. [DOI] [PubMed] [Google Scholar]
- 66.Sumiyoshi H, Tignor G H, Shope R E. Characterization of a highly attenuated Japanese encephalitis virus generated from molecularly cloned cDNA. J Infect Dis. 1995;171:1144–1151. doi: 10.1093/infdis/171.5.1144. [DOI] [PubMed] [Google Scholar]
- 67.Sutcliffe J G, Shinnick T M, Green N, Lerner R A. Antibodies that react with predetermined sites on proteins. Science. 1983;219:660–666. doi: 10.1126/science.6186024. [DOI] [PubMed] [Google Scholar]
- 68.Swofford D. PAUP*, Phylogenetic Analysis Using Parsimony (*and Other Methods), 4 ed. Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- 69.Vaughn D W, Green S, Kalayanarooj S, Innis B L, Nimmannitya S, Suntayakorn S, Rothman A L, Ennis F A, Nisalak A. Dengue in the early febrile phase: viremia and antibody responses. J Infect Dis. 1997;176:322–330. doi: 10.1086/514048. [DOI] [PubMed] [Google Scholar]
- 70.Watts, D., K. Porter, R. Putvatana, R. Vasquez, C. Calampa, C. Hayes, and S. Halstead. Failure of secondary dengue 2 genotype I infection to cause dengue hemorrhagic fever. Submitted for publication. [DOI] [PubMed]
- 71.Weber K, Konisberg W. Proteins of the RNA phages. In: Zinder N D, editor. RNA phages. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1975. pp. 51–84. [Google Scholar]
- 72.Wengler G, Castle E. Analysis of structural properties which possibly are characteristic for the 3′-terminal sequence of genomic RNA of flaviviruses. J Gen Virol. 1986;67:1183–1188. doi: 10.1099/0022-1317-67-6-1183. [DOI] [PubMed] [Google Scholar]
- 73.Wu H-N, Uhlenbeck O C. Role of a bulged A residue in a specific RNA-protein interaction. Biochemistry. 1987;26:8221–8227. doi: 10.1021/bi00399a030. [DOI] [PubMed] [Google Scholar]
- 74.Zhao B, Mackow E, Buckler-White A, Markoff L, Chanock R M, Lai C J, Makino Y. Cloning full-length dengue type 4 viral DNA sequences: analysis of genes coding for structural proteins. Virology. 1986;155:77–88. doi: 10.1016/0042-6822(86)90169-8. [DOI] [PubMed] [Google Scholar]
- 75.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]