Abstract
Hematoma expansion occasionally occurs in patients with intracerebral hemorrhage (ICH), associating with poor outcome. Multimodal neural networks incorporating convolutional neural network (CNN) analysis of images and neural network analysis of tabular data are known to show promising results in prediction and classification tasks. We aimed to develop a reliable multimodal neural network model that comprehensively analyzes CT images and clinical variables to predict hematoma expansion. We retrospectively enrolled ICH patients at four hospitals between 2017 and 2021, assigning patients from three hospitals to the training and validation dataset and patients from one hospital to the test dataset. Admission CT images and clinical variables were collected. CT findings were evaluated by experts. Three types of models were developed and trained: (1) a CNN model analyzing CT images, (2) a multimodal CNN model analyzing CT images and clinical variables, and (3) a non-CNN model analyzing CT findings and clinical variables with machine learning. The models were evaluated on the test dataset, focusing first on sensitivity and second on area under the receiver operating curve (AUC). Two hundred seventy-three patients (median age, 71 years [59–79]; 159 men) in the training and validation dataset and 106 patients (median age, 70 years [62–82]; 63 men) in the test dataset were included. Sensitivity and AUC of a CNN model were 1.000 (95% confidence interval [CI] 0.768–1.000) and 0.755 (95% CI 0.704–0.807); those of a multimodal CNN model were 1.000 (95% CI 0.768–1.000) and 0.799 (95% CI 0.749–0.849); and those of a non-CNN model were 0.857 (95% CI 0.572–0.982) and 0.733 (95% CI 0.625–0.840). We developed a multimodal neural network model incorporating CNN analysis of CT images and neural network analysis of clinical variables to predict hematoma expansion in ICH. The model was externally validated and showed the best performance of all the models.
Keywords: Intracerebral hemorrhage, Hematoma expansion, Prediction, Multimodal neural network
Subject terms: Cerebrovascular disorders, Stroke
Introduction
Spontaneous intracerebral hemorrhage (ICH) is a severe form of stroke with a high mortality rate1. In its early clinical stage, 20–30% of patients experience hematoma expansion, leading to neurological deterioration or the need for surgical treatment2,3. Accurate stratification of expansion risk on admission therefore guides patient management, including admission to the intensive care unit and transfer from remote areas to specialist care.
CT image analysis plays an important role in predicting hematoma expansion. Several CT findings, such as blend sign or intrahematoma hypodensities, have been identified as potential indicators of hematoma expansion4–10. Scoring systems incorporating both CT findings and clinical variables have also been proposed11–13. However, their sensitivity and C-statistics have been found to be low to moderate in external validation and remain unsatisfactory for clinical application14–16. Machine learning models based on CT findings and clinical variables have been proposed, demonstrating superior predictive ability compared to the traditional scoring systems14,17. However, a major drawback of both the scoring systems and machine learning models is that prediction requires CT findings to be evaluated by human experts.
Recently, a convolutional neural network (CNN) has been applied to image analysis in ICH18–23, with superior predictive ability of hematoma expansion compared to previously proposed CT findings and scoring systems18. CNNs can directly interpret the image data without the evaluation of human experts, overcoming the drawbacks of the scoring systems and machine learning models. Furthermore, in other fields, a multimodal neural network that combines CNN analysis of images and neural network analysis of clinical information such as age, sex, medical history, etc. has been introduced for disease type or risk classification tasks, which has demonstrated better performance than CNN analysis of images alone24,25.
Here, we hypothesized that it would be feasible to create a multimodal neural network model incorporating CNN analysis of CT images on admission and neural network analysis of clinical variables to predict hematoma expansion in acute ICH, and that the model would outperform CNN analysis of CT images alone and be superior to machine learning analysis of CT findings and clinical variables.
Methods
Patients
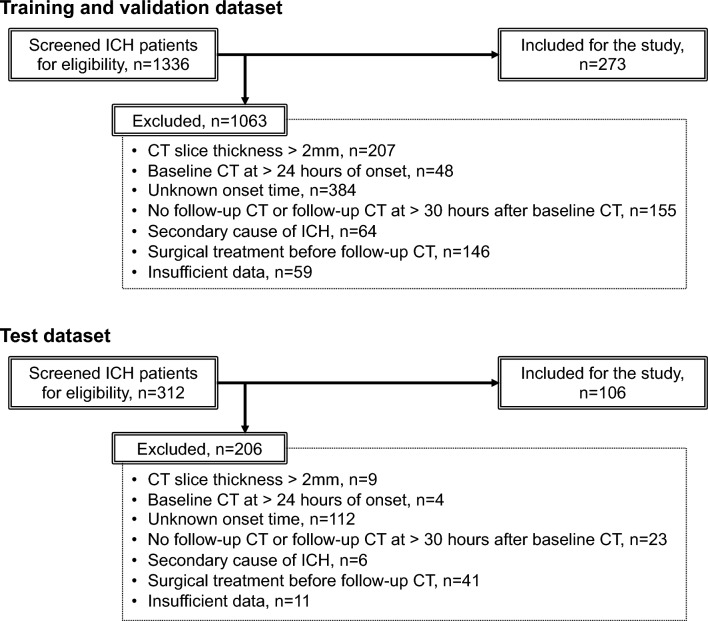
We retrospectively reviewed consecutive patients with spontaneous ICH, aged ≥ 18 years, admitted to a university hospital and three community hospitals in Japan between January 2017 and December 2021. Patients were included if they underwent a baseline CT scan with a thickness of 2.0 mm or less within 24 h of onset and a follow-up CT scan within 30 h of the baseline CT scan. Patients were excluded if they had a secondary cause of ICH (e.g., tumor, aneurysm, arteriovenous malformation, arteriovenous fistula, and hemorrhagic transformation of ischemic infarction), head trauma, or surgical hematoma evacuation before the follow-up CT scan, or if they had missing data. Patients from a university hospital and two community hospitals were assigned to training and validation dataset, while patients from the other community hospital were assigned to a test dataset (Fig. 1).
Figure 1.
Flow diagram of patient selection. ICH = intracerebral hemorrhage.
This study was approved by the following institutional review boards: Mie Chuo Medical Center institutional review board [permit number: MCERB-202321], Matsusaka Chuo General Hospital institutional review board [permit number: 325], Suzuka Kaisei Hospital institutional review board [permit number: 2020-05], and Mie University Hospital institutional review board [permit number: T2023-7]. Because this was a retrospective study, separate informed patient consent was waived by the following institutional review boards: Mie Chuo Medical Center institutional review board [permit number: MCERB-202321], Matsusaka Chuo General Hospital institutional review board [permit number: 325], Suzuka Kaisei Hospital institutional review board [permit number: 2020-05], and Mie University Hospital institutional review board [permit number: T2023-7]. All study protocols and procedures were conducted in accordance with the Declaration of Helsinki. This manuscript was prepared according to the standards for reporting of diagnostic accuracy (STARD) statement.
Clinical variables
Demographic and clinical data on admission were collected. The following variables were recorded: age, sex, medical history (hypertension, diabetes mellitus, dyslipidemia, ICH, cerebral infarction, and ischemic heart disease), anticoagulant use, antiplatelet use, systolic and diastolic blood pressure, Glasgow Coma Scale, white blood cell count, hemoglobin, platelet count, prothrombin time-international normalized ratio (PT-INR), serum creatinine, serum total bilirubin, and time from onset to baseline CT scan.
Image acquisition and segmentation
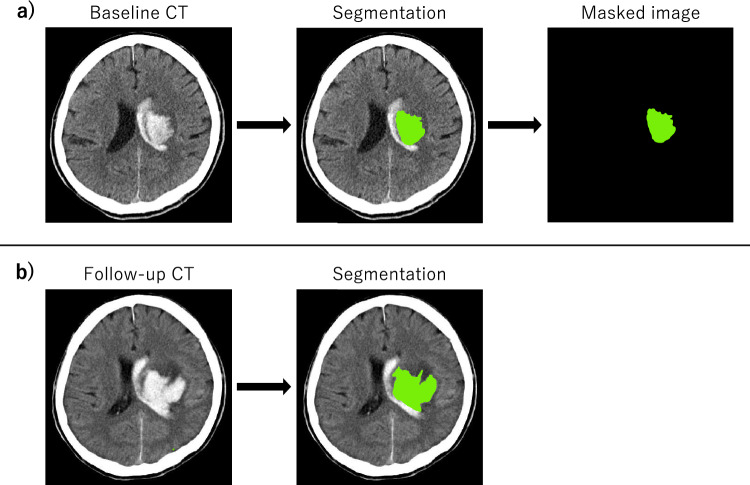
CT scans were performed in the supine position at 120 kVp with a thickness of 0.5–2.0 mm and an image shape of 512 × 512 or greater; the images were exported as original images in the Digital Imaging and Communications in Medicine (DICOM) format. For baseline and follow-up CT scans, intraparenchymal hematomas were manually segmented by two raters, board-certified stroke specialists with more than 15 years of experience, using 3D Slicer, with hematoma volumes calculated by planimetry (Fig. 2). Intraventricular hematomas were neither segmented nor included in the hematoma volume. Hematoma expansion was defined as a volume increase between baseline and follow-up CT scans greater than 6 cm3 or 33% of baseline volume11–13,15,16. According to this definition, all the patients were labeled as having hematoma expansion or no hematoma expansion. On the baseline CT, segmented areas were marked with a value of 1 and other areas were marked with 0; the images were exported as masked images in DICOM format for the CNN analysis (Fig. 2).
Figure 2.
Baseline (a) and follow-up (b) CT images of a case with hematoma expansion. Intraparenchymal hematomas were manually segmented (green areas), with hematoma volumes computed by planimetry. (a) For baseline images, segmented areas were marked as 1 and other areas were marked as 0, which were exported as masked images.
Evaluation of CT findings at baseline
As CT findings in predicting hematoma expansion, blend sign, intrahematoma hypodensities, and irregular shape were evaluated. Blend sign was defined as the blending of a relatively hypoattenuating area with an adjacent hypoattenuating area within the hematoma6,7,9. Intrahematoma hypodensities were defined as the presence of any hypodense region encapsulated within the hematoma and separated from the surrounding parenchyma7,10. Irregular hematoma shape was defined as having 2 or more edge irregularities4,5,7. These were assessed independently by two raters; prior to assessment, the raters were trained using at least 10 patients with ICH that were not included in this study. In case of disagreement, the findings were reassessed by both raters together until a consensus was reached. Hematoma location and intraventricular hematoma extension were also evaluated.
Statistical analysis
Categorical variables were summarized as counts with percentages and compared using Fisher's exact test. Continuous variables were summarized as mean with standard deviation or median with interquartile range and compared using Student's t-test or Mann–Whitney U-test, depending on the distribution of the variable assessed by the Shapiro–Wilk test. Sensitivity, specificity, accuracy, and area under the receiver operating curve (AUC) were calculated along with the 95% confidence intervals derived from the actual and predicted labels. P values less than 0.05 were considered significant. Statistical analyses were performed using EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan)26.
Designing models for prediction
Three types of models were designed to predict hematoma expansion: (1) a CNN model, (2) a multimodal CNN model, and (3) a non-CNN model. A CNN model used CT images as input. A multimodal CNN model used CT images and clinical variables as input, combining CNN analysis of CT images and neural network analysis of clinical variables. A non-CNN model used human-assessed CT findings and clinical variables as input, which were analyzed with machine learning. For each type, several models were devised with different algorithm or input.
CNN Model 1 used unmodified original CT images as input. CNN Model 2 used only intraparenchymal hematoma images, which were generated from original and masked images (Fig. 3).
Figure 3.
Intraparenchymal hematoma images generated from original and masked images.
Multimodal CNN Model 1 used all available clinical variables as one input. For Multimodal CNN Model 2, we first analyzed all clinical variables with univariate analyses between expansion and no expansion cases in the training and validation dataset, and used the clinical variables associated with hematoma expansion with statistical significance. For both models, the original images or the intraparenchymal hematoma images, whichever performed better in the comparison between CNN Model 1 and 2, were used as the other input.
Non-CNN Model 1 and 2 employed k-nearest neighbors as algorithm, while Non-CNN Model 3 and 4 employed fully connected neural network. Non-CNN Model 1 and 3 used all available CT findings and clinical variables as input, whereas non-CNN Model 2 and 4 used CT findings and clinical variables significantly associated with hematoma expansion in univariate analyses between expansion and no expansion cases in the training and validation dataset.
Preprocessing
All processing was done with Keras (version 2.12.0), a deep learning application programming interface in Python, running with 40 GB of GPU memory. All of the code in this study is available on GitHub (https://github.com/AI-neurosurg/Multimodal-network-for-predicting-hematoma-expansion-in-ICH).
For clinical variables, standardization was first performed for continuous variables in the training and validation dataset. Standardization of the test dataset was then performed based on the mean and standard deviation in the training and validation dataset.
Prior to image processing, all DICOM files were converted to Neuroimaging Informatics Technology Initiative (NIfTI) files. Preprocessing for CNN was performed separately for original and masked images. For the original images, the following steps were executed: (1) density scaling, (2) reslicing, (3) pixel size unification, and (4) resizing. First, after extracting the brain and hematoma by thresholding the Hounsfield units between 0 and 100, the pixel values were scaled between 0 and 1 by dividing the values by 100. Second, the images were resliced with a new slice thickness of 2 mm, and the new number of slices was set to 80, with all slices at or beyond the 81st position from the most cranial slice being deleted. Third, the axial pixel sizes were unified to 0.5 × 0.5 mm, because the image magnification varied between CT scans. Since all image shapes became slightly smaller than 512 × 512, padding was performed to keep the image shape at 512 × 512. Fourth, resizing was done by changing the image shape from 512 × 512 to 256 × 256 to fit the GPU memory. The preprocessed original images were used as input to CNN Model 1. For the masked images, the above steps were executed from the second to the fourth, since the masked images were already binary, either 0 or 1. Intraparenchymal hematoma images were generated from preprocessed original and masked images for CNN Model 2 (Fig. 3).
From the training and validation dataset, 70% were randomly assigned to the training set and the rest to the validation set. To balance the ratio of expansion cases to no expansion cases, data augmentation and random oversampling were applied only to expansion cases in the training set. Data augmentation was conducted in CNN and multimodal CNN models, where images were flipped and rotated 30 degrees. Random oversampling was performed in non-CNN models.
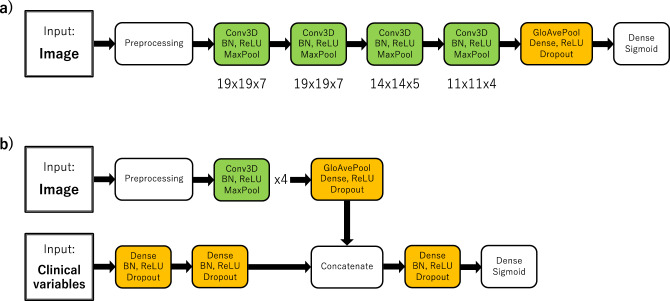
Model architecture
CNN Model 1 and 2 were composed of four 3-dimensional convolutional layer blocks with batch normalization, ReLU activation function, and max pooling, followed by a dense layer block with global average pooling, ReLU activation function, and dropout (Fig. 4a). At the end, a final dense layer with sigmoid activation function was placed. The kernel sizes in the convolutional layers were 19 × 19 × 7, 19 × 19 × 7, 14 × 14 × 5, and 11 × 11 × 4 consecutively. Multimodal CNN Model 1 and 2 consisted of two parts: an image part and a clinical-variables part (Fig. 4b). The image part had the same architecture as the CNN Models except for the final block. The clinical-variables part was composed of two dense layer blocks with batch normalization, ReLU activation function, and dropout. The image and clinical-variables parts were concatenated in the middle, followed by a dense layer block with batch normalization, ReLU activation function and dropout and a final dense layer with sigmoid activation function.
Figure 4.
(a) The CNN models were composed of four 3-dimensional convolutional layer blocks and one dense layer block, followed by a final dense layer block with sigmoid activation function. The kernel sizes of the convolutional layers were 19 × 19 × 7, 19 × 19 × 7, 14 × 14 × 5, and 11 × 11 × 4, respectively. (b) The multimodal CNN models consisted of an image part and a clinical-variables part. The architecture of the image part was the same as in the CNN models (a), except for the last block. The clinical-variables part was consisted of two dense layer blocks. These two parts were concatenated, followed by a dense layer block and a final dense layer block.
In non-CNN Model 1 and 2, k-nearest neighbors from scikit-learn machine learning library (version 1.2.2) was adopted. A hyperparameter of the number of neighbors were chosen from 3, 5, 7, 9, and 11. Non-CNN Model 3 and 4 were composed of three dense layer blocks with batch normalization, ReLU activation function, and dropout, followed by a final dense layer block with sigmoid activation function.
The labels to predict in the models were hematoma expansion or no hematoma expansion. No missing data were treated in the models because all the required data, including images and clinical variables, were complete for all patients.
Training, validation and test
For CNN Models, Multimodal CNN Models, and Non-CNN Model 3 and 4, 70 epochs of training were performed with a batch size of 2, where binary cross-entropy and Adam were used for the loss function and optimizer, respectively27. For Adam, the following settings were used: learning rate = 0.001, beta 1 = 0.9, beta 2 = 0.999, and epsilon = 1e-07. The cut-off value of 0.5 was used to binarize the cases. At each epoch, sensitivity and area under the receiver operating curve (AUC) were calculated with the validation dataset to monitor the training process. Five trained model weights from all epochs that had better sensitivity and AUC in validation were selected and used for testing, where the final test result was derived from the weights with the highest sensitivity.
For Non-CNN Model 1 and 2, the training and validation dataset were fitted to the k-nearest neighbors algorithm while changing the hyperparameter of the number of neighbors, to which the test dataset was applied and the final result was derived from the number of neighbors with the highest sensitivity.
Ethical approval
This study was approved by the following institutional review boards: Mie Chuo Medical Center institutional review board [permit number: MCERB-202321], Matsusaka Chuo General Hospital institutional review board [permit number: 325], Suzuka Kaisei Hospital institutional review board [permit number: 2020-05], and Mie University Hospital institutional review board [permit number: T2023-7]. Because this was a retrospective study, separate informed patient consent was waived by the following institutional review boards: Mie Chuo Medical Center institutional review board [permit number: MCERB-202321], Matsusaka Chuo General Hospital institutional review board [permit number: 325], Suzuka Kaisei Hospital institutional review board [permit number: 2020-05], and Mie University Hospital institutional review board [permit number: T2023-7]. All study protocols and procedures were conducted in accordance with the Declaration of Helsinki. This manuscript was prepared according to the standards for reporting of diagnostic accuracy (STARD) statement.
Results
After applying the inclusion and exclusion criteria, 273 patients were assigned to the training and validation dataset, while 106 patients were assigned to the test dataset. Patient characteristics of the study population are shown in Table 1. Their row data are stored in OSF in comma-separated values format (https://osf.io/jmnzs).
Table 1.
Characteristics of the study population.
| Training and validation (n = 273) | Test (n = 106) | P value | |
|---|---|---|---|
| Age (years) | 71 (59–79) | 72 (62–82) | 0.099 |
| Sex (male) | 159 (57.9) | 63 (59.4) | 0.817 |
| Medical history | |||
| Hypertension | 162 (59.3) | 74 (69.8) | 0.060 |
| Diabetes mellitus | 58 (21.3) | 30 (28.3) | 0.175 |
| Dyslipidemia | 93 (34.1) | 37 (34.9) | 0.904 |
| Intracerebral hemorrhage | 18 (6.6) | 7 (6.6) | 1.000 |
| Cerebral infarction | 30 (11.0) | 14 (13.2) | 0.593 |
| Ischemic heart disease | 15 (5.5) | 7 (6.6) | 0.633 |
| Anticoagulant use | 30 (11.0) | 10 (9.4) | 0.714 |
| Antiplatelet use | 34 (12.5) | 25 (23.6) | 0.011 |
| Systolic blood pressure (mmHg) | 187.6 ± 33.3 | 182.6 ± 33.6 | 0.193 |
| Diastolic blood pressure (mmHg) | 105.7 ± 23.3 | 104.2 ± 23.1 | 0.580 |
| Glasgow Coma Scale | 14 (11–15) | 15 (12–15) | 0.046 |
| White blood cell count (106/mL) | 7.60 (5.80–9.60) | 7.85 (6.15–9.78) | 0.163 |
| Hemoglobin (mg/dL) | 13.9 ± 2.1 | 13.3 ± 1.8 | 0.005 |
| Platelet count (106/mL) | 216.2 ± 59.3 | 225.1 ± 68.0 | 0.212 |
| PT-INR | 0.96 (0.92–1.02) | 0.99 (0.95–1.05) | 0.003 |
| Serum creatinine (mg/dL) | 0.74 (0.59–0.91) | 0.72 (0.58–0.90) | 0.589 |
| Serum total bilirubin (mg/dL) | 0.7 (0.5–0.9) | 0.7 (0.5–1.0) | 0.770 |
| Time from onset to baseline CT scan (h) | 1 (1–3) | 2 (1–4) | < 0.001 |
| CT findings at baseline | |||
| Blend sign | 25 (9.2) | 5 (4.7) | 0.203 |
| Intrahematoma hypodensities | 118 (43.2) | 33 (31.3) | 0.035 |
| Irregular hematoma shape | 160 (58.6) | 56 (52.8) | 0.355 |
| Hemorrhage location | |||
| Brain stem | 17 (6.2) | 3 (2.8) | 0.304 |
| Cerebellum | 11 (4.0) | 12 (11.3) | 0.014 |
| Thalamus | 81 (29.7) | 35 (33.0) | 0.537 |
| Basal ganglia | 117 (42.9) | 36 (34.0) | 0.130 |
| Lobe | 47 (17.2) | 20 (18.9) | 0.764 |
| Intraventricular hematoma extension | 103 (37.7) | 46 (43.4) | 0.349 |
| Baseline hematoma volume, mL | 12.2 (5.6–30.2) | 10.2 (3.6–19.9) | 0.031 |
| Hematoma expansion | 54 (19.8) | 14 (13.2) | 0.179 |
Data are presented as n (%), mean ± standard deviation, or median (interquartile range).
PT-INR = prothrombin time-international normalized ratio.
On CT findings, intrahematoma hypodensities, hematoma location, and hematoma volume were statistically significant in univariate analyses between expansion and no expansion cases in the training and validation dataset; these were used as input in Non-CNN Model 2 and 4. On clinical variables, anticoagulant use, systolic and diastolic blood pressure, PT-INR, and time from onset to baseline CT were significant and used as input in Multimodal CNN Model 2 and Non-CNN Model 2 and 4.
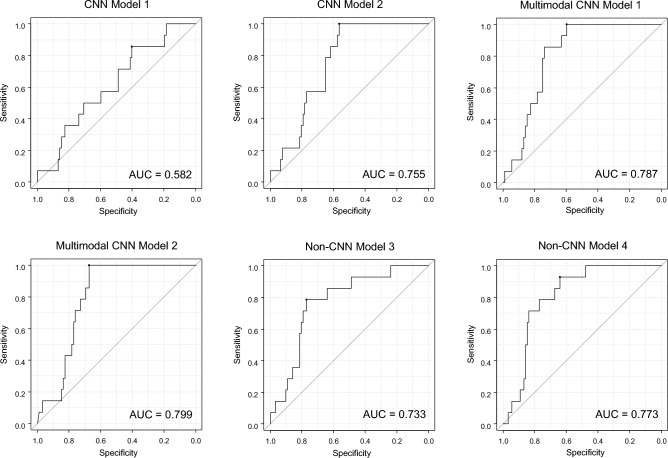
The performance of each model is shown in Table 2 and Fig. 5. In CNN Model 1 and 2, sensitivity was the same, but specificity and AUC were higher in CNN Model 2. Therefore, intraparenchymal hematoma images, rather than original images, were used as input of Multimodal CNN Model 1 and 2 (Fig. 3). The number of neighbors of 7 achieved the highest sensitivity for Non-CNN Model 1 and 2.
Table 2.
Test results for predicting hematoma expansion in each model.
| Sensitivity | Specificity | Accuracy | AUC | |
|---|---|---|---|---|
| CNN Model 1 | 1.000 (0.768–1.000) | 0.163 (0.094–0.255) | 0.274 (0.191–369) | 0.582 (0.544–0.614) |
| CNN Model 2 | 1.000 (0.768–1.000) | 0.511 (0.404–0.617) | 0.575 (0.476–0.671) | 0.755 (0.704–0.807) |
| Multimodal CNN Model 1 | 0.857 (0.572–0.982) | 0.717 (0.614–0.806) | 0.736 (0.641–0.817) | 0.787 (0.682–0.893) |
| Multimodal CNN Model 2 | 1.000 (0.768–1.000) | 0.598 (0.490–0.699) | 0.615 (0.552–0.741) | 0.799 (0.749–0.849) |
| Non-CNN Model 1 | 0.571 (0.289–0.823) | 0.761 (0.661–0.844) | 0.736 (0.641–0.817) | 0.666 (0.525–0.808) |
| Non-CNN Model 2 | 0.643 (0.351–0.872) | 0.696 (0.591–0.787) | 0.689 (0.591–0.775) | 0.669 (0.531–0.808) |
| Non-CNN Model 3 | 0.857 (0.572–0.982) | 0.609 (0.501–0.709) | 0.642 (0.543–0.732) | 0.733 (0.625–0.840) |
| Non-CNN Model 4 | 0.786 (0.493–0.953) | 0.761 (0.661–0.844) | 0.764 (0.672–0.841) | 0.773 (0.653–0.893) |
Data are presented as value (95% confidence interval).
AUC = area under the receiver operating characteristic curve.
Figure 5.
Receiver operating curves for each model in Table 2, except for Non-CNN Model 1 and 2. These models were excluded because they do not return continuous values as a prediction.
Sensitivity was higher for CNN Models and Multimodal CNN Models than for Non-CNN Models. In particular, CNN Model 1 and 2 and Multimodal CNN Model 2 achieved a sensitivity of 1.000 (95% confidence interval 0.768–1.000). AUC was above 0.75 for CNN Model 2, Multimodal CNN Model 1 and 2, and Non-CNN Model 2, with the Multimodal CNN Model 2 having the highest AUC. Specificity was highest for Non-CNN Model 1 and 4. Accuracy was highest for Non-CNN Model 4.
Multimodal CNN Model 2 showed the highest sensitivity and AUC of all models. Its model weights are stored in OSF in HDF5 format (419 MB, https://osf.io/wm768).
Discussion
A multimodal neural network model incorporating CNN analysis of CT images and neural network analysis of clinical variables showed a sensitivity of 1.000 for predicting hematoma expansion in spontaneous ICH. The model outperformed CNN analysis of CT images alone and machine learning analysis of CT findings and clinical variables. It is a highly complete model that utilizes all available patient information. The multimodal model would be beneficial in clinical practice, as it effectively identifies patients who require thorough and intensive care after admission.
For clinicians treating ICH, the greatest concern in predicting hematoma expansion is missing expansion cases because they may experience neurological deterioration and require careful observation in the intensive care units2,3. Not missing a single case at risk is critical in stroke care. Therefore, the goal of the prediction in this study was set to achieve higher sensitivity while balancing with AUC. In binary classification, the binary cross-entropy is usually used as the loss function, and training is aimed at minimizing the loss function. However, a low value of binary cross-entropy loss does not always equate to high sensitivity. Thus, for testing, we did not simply select the model weights with the lowest loss value in validation, but selected those with better sensitivity and AUC. However, this selection might have enhanced the model performance.
The Multimodal CNN Model 2, which used CT images and selected clinical variables as input, showed the best performance. The superior performance of Multimodal CNN Model 2 compared to CNN Model 2 underscores the importance of clinical information in predicting hematoma expansion. Here, not all clinical variables were necessary, only 5 were used. The fact that the model works with fewer inputs is critical to its practical use, as it can sometimes be difficult to collect sufficient information in clinical settings. Furthermore, the superiority of multimodal neural network models over non-CNN models underscores that CNN analysis outperforms human-based CT findings evaluation, even when combined with clinical information, in predicting hematoma expansion.
One of the most challenging aspects in the development of CNN and multimodal CNN models was the size of the kernels in the convolutional layers. Typically, a kernel size of 3 or at most 5 is used because a larger kernel size consumes more computational power28. However, in preliminary experiments, we observed a divergence in the training process with a kernel size of 3 or 5, even when adding layers or increasing the number of kernels. The voxel size of the CT images was 0.5 × 0.5 × 2.0 mm, where the kernel size of 3 or 5 may have been too small to extract features from the hematoma. The larger kernel sizes up to 19 × 19 × 7 worked effectively in this study; we could not confirm the kernel size in other studies that used CNN to predict hematoma expansion because the programming codes were not disclosed18–21,29.
Several considerations have been suggested for the soundness of research using artificial intelligence (AI) techniques, such as the use of an external test set for the final report, transparency of algorithms, etc.30,31. However, many clinical studies have not actually followed these basic considerations. In this study, the model was trained on data from several hospitals and tested on external data. Multiple models were created for comparison. Clinical information, algorithm programming code, and model weights were disclosed to make our results verifiable and the model reproducible.
To date, there are 2 studies that predicted hematoma expansion in ICH by analyzing both CT images and clinical information with CNN29,32. In one study, hematoma features extracted from CNN and radiomics and clinical variables were integrally analyzed with support vector machine29. It achieved sensitivity of 0.83 and AUC of 0.95; however, the testing method was not described in detail and patient data from a single hospital were used for both training and testing29. The other analyzed CNN-derived hematoma features and clinical variables using multivariate logistic regression, achieving sensitivity of 0.76 and AUC of 0.8332. This is also a single-center study, and the sensitivity is low to use the model in clinical practice. In our study, we achieved a sensitivity of 1.00, which is critical for clinical use in stroke management.
CNN Model 2 using intraparenchymal hematoma images outperformed CNN Model 1 using unmodified original CT images. Therefore, intraparenchymal hematoma images were used as input for multimodal models. However, their segmentations were performed manually by humans in this study because automated segmentation remains unsatisfactory in some cases with an inaccurate differentiation between intraparenchymal and intraventricular hematoma33–35. When more accurate segmentation of intraparenchymal hematoma becomes possible, and clinical variables can be automatically collected from the medical record, this prediction task could be fully automated.
Several limitations should be noted. First, although a perfect sensitivity of 1.000 was achieved in a multimodal neural network model while balancing AUC, the lower limit of the confidence interval was 0.768, indicating that more cases are needed for more reliable model validation. Second, although the external dataset from another hospital was used for testing, validation with various patient demographics is required to further ensure the reliability of the developed models. Third, the comparisons of the model performance were not supported by statistical significance testing; instead, simple comparisons were conducted among the models. More cases are also required to statistically demonstrate significant differences based on 95% confidence intervals. Fourth, the images were resized to 256 × 256 to fit the GPU memory. Analysis at the original 512 × 512 size may yield better results. Fifth, only k-nearest neighbors and fully connected neural networks were employed for non-CNN models. Other machine learning models may have performed better, but logistic regression, support vector machines, random forests, and gradient boosting were inferior to k-nearest neighbors in the previous report predicting hematoma expansion14. Sixth, this is a retrospective study. Validation with prospective data is required as a future step. Seventh, to apply the models to clinical practice, systems that comprehensively capture patient information, including images and clinical variables, are required. Last, although the clinical variables that are included in the study were generally collected from the patients in the clinical setting for stroke care, they are sometimes unavailable. A model capable of handling missing data may be beneficial.
Conclusion
We developed a multimodal neural network model incorporating CNN analysis of CT images and neural network analysis of clinical variables to predict hematoma expansion in acute spontaneous ICH. The model was externally validated and outperformed CNN analysis of CT images alone and machine learning analysis of CT findings and clinical variables. The multimodal model achieved sufficient performance with a sensitivity of 1.000 to potentially enable decision support in clinical settings; it effectively identifies patients who require thorough and intensive care after admission. The algorithm programming code and model weights are available for verification and public use. To ensure the reliability of the models, validation with prospective datasets for various patient demographics is necessary as a future step. Furthermore, a model capable of handling missing data or systems that comprehensively capture patient information would be required to enable widespread use of predictive models in clinical practice.
Author contributions
S.T. designed and conceptualized study. S.T., K.T., T.A., Y.N., T.Y., T.K., and M.I. collected and analyzed the data. S.T. drafted the manuscript. O.U.A., A.H., F.I., H.S., and D.F. interpreted the data and revised the manuscript. All authors reviewed the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
Row data of patient characteristics are stored in OSF in comma-separated values format (https://osf.io/jmnzs). All code in this study is available on GitHub (https://github.com/AI-neurosurg/Multimodal-network-for-predicting-hematoma-expansion-in-ICH). Model weights of the best-performing multimodal convolutional neural network model are stored in OSF in HDF5 format (419 MB, https://osf.io/wm768).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Feigin VL, Lawes CM, Bennett DA, Barker-Collo SL, Parag V. Worldwide stroke incidence and early case fatality reported in 56 population-based studies: A systematic review. Lancet Neurol. 2009;8:355–369. doi: 10.1016/S1474-4422(09)70025-0. [DOI] [PubMed] [Google Scholar]
- 2.Leira R, Davalos A, Silva Y, Gil-Peralta A, Tejada J, Garcia M, et al. Early neurologic deterioration in intracerebral hemorrhage: Predictors and associated factors. Neurology. 2004;63:461–467. doi: 10.1212/01.WNL.0000133204.81153.AC. [DOI] [PubMed] [Google Scholar]
- 3.Delcourt C, Huang Y, Arima H, Chalmers J, Davis SM, Heeley EL, et al. Hematoma growth and outcomes in intracerebral hemorrhage: The INTERACT1 study. Neurology. 2012;79:314–319. doi: 10.1212/WNL.0b013e318260cbba. [DOI] [PubMed] [Google Scholar]
- 4.Blacquiere D, Demchuk AM, Al-Hazzaa M, Deshpande A, Petrcich W, Aviv RI, et al. Intracerebral hematoma morphologic appearance on noncontrast computed tomography predicts significant hematoma expansion. Stroke. 2015;46:3111–3116. doi: 10.1161/STROKEAHA.115.010566. [DOI] [PubMed] [Google Scholar]
- 5.Barras CD, Tress BM, Christensen S, MacGregor L, Collins M, Desmond PM, et al. Density and shape as CT predictors of intracerebral hemorrhage growth. Stroke. 2009;40:1325–1331. doi: 10.1161/STROKEAHA.108.536888. [DOI] [PubMed] [Google Scholar]
- 6.Sporns PB, Schwake M, Schmidt R, Kemmling A, Minnerup J, Schwindt W, et al. Computed tomographic blend sign is associated with computed tomographic angiography spot sign and predicts secondary neurological deterioration after intracerebral hemorrhage. Stroke. 2017;48:131–135. doi: 10.1161/STROKEAHA.116.014068. [DOI] [PubMed] [Google Scholar]
- 7.Morotti A, Boulouis G, Romero JM, Brouwers HB, Jessel MJ, Vashkevich A, et al. Blood pressure reduction and noncontrast CT markers of intracerebral hemorrhage expansion. Neurology. 2017;89:548–554. doi: 10.1212/WNL.0000000000004210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q, Zhang G, Xiong X, Wang XC, Yang WS, Li KW, et al. Black hole sign: Novel imaging marker that predicts hematoma growth in patients with intracerebral hemorrhage. Stroke. 2016;47:1777–1781. doi: 10.1161/STROKEAHA.116.013186. [DOI] [PubMed] [Google Scholar]
- 9.Li Q, Zhang G, Huang YJ, Dong MX, Lv FJ, Wei X, et al. Blend sign on computed tomography: Novel and reliable predictor for early hematoma growth in patients with intracerebral hemorrhage. Stroke. 2015;46:2119–2123. doi: 10.1161/STROKEAHA.115.009185. [DOI] [PubMed] [Google Scholar]
- 10.Boulouis G, Morotti A, Brouwers HB, Charidimou A, Jessel MJ, Auriel E, et al. Association between hypodensities detected by computed tomography and hematoma expansion in patients with intracerebral hemorrhage. JAMA Neurol. 2016;73:961–968. doi: 10.1001/jamaneurol.2016.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morotti A, Dowlatshahi D, Boulouis G, Al-Ajlan F, Demchuk AM, Aviv RI, et al. Predicting intracerebral hemorrhage expansion with noncontrast computed tomography: The BAT score. Stroke. 2018;49:1163–1169. doi: 10.1161/STROKEAHA.117.020138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Arima H, Al-Shahi Salman R, Woodward M, Heeley E, Stapf C, et al. Clinical prediction algorithm (BRAIN) to determine risk of hematoma growth in acute intracerebral hemorrhage. Stroke. 2015;46:376–381. doi: 10.1161/STROKEAHA.114.006910. [DOI] [PubMed] [Google Scholar]
- 13.Brouwers HB, Chang Y, Falcone GJ, Cai X, Ayres AM, Battey TW, et al. Predicting hematoma expansion after primary intracerebral hemorrhage. JAMA Neurol. 2014;71:158–164. doi: 10.1001/jamaneurol.2013.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanioka S, Yago T, Tanaka K, Ishida F, Kishimoto T, Tsuda K, et al. Machine learning prediction of hematoma expansion in acute intracerebral hemorrhage. Sci. Rep. 2022;12:12452. doi: 10.1038/s41598-022-15400-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poli L, Leuci E, Costa P, De Giuli V, Caria F, Candeloro E, et al. Validation and comparison of noncontrast CT scores to predict intracerebral hemorrhage expansion. Neurocrit. Care. 2020;32:804–811. doi: 10.1007/s12028-019-00797-2. [DOI] [PubMed] [Google Scholar]
- 16.Huynh TJ, Aviv RI, Dowlatshahi D, Gladstone DJ, Laupacis A, Kiss A, et al. Validation of the 9-point and 24-point hematoma expansion prediction scores and derivation of the PREDICT A/B scores. Stroke. 2015;46:3105–3110. doi: 10.1161/STROKEAHA.115.009893. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Xu H, Chen Q, Zhang T, Sheng W, Huang Q, et al. Prediction of hematoma expansion in spontaneous intracerebral hemorrhage using support vector machine. EBioMedicine. 2019;43:454–459. doi: 10.1016/j.ebiom.2019.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong JW, Jin YJ, Song ZJ, Lin B, Lu XH, Chen F, et al. Deep learning for automatically predicting early haematoma expansion in Chinese patients. Stroke Vasc. Neurol. 2021;6:610–614. doi: 10.1136/svn-2020-000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Z, Zhu Y, Lu X, Wu D, Fan X, Shen J, et al. Deep learning-based prediction of hematoma expansion using a single brain computed tomographic slice in patients with spontaneous intracerebral hemorrhages. World Neurosurg. 2022. [DOI] [PubMed]
- 20.Teng L, Ren Q, Zhang P, Wu Z, Guo W, Ren T. Artificial intelligence can effectively predict early hematoma expansion of intracerebral hemorrhage analyzing noncontrast computed tomography image. Front. Aging Neurosci. 2021;13:632138. doi: 10.3389/fnagi.2021.632138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo DC, Gu J, He J, Chu HR, Dong N, Zheng YF. External validation study on the value of deep learning algorithm for the prediction of hematoma expansion from noncontrast CT scans. BMC Med. Imaging. 2022;22:45. doi: 10.1186/s12880-022-00772-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu M, Wang Y, Tian J, Feng H. Application of deep learning and radiomics in the prediction of hematoma expansion in intracerebral hemorrhage: A fully automated hybrid approach. Diagn. Interv. Radiol. 2024. [DOI] [PMC free article] [PubMed]
- 23.Feng C, Ding Z, Lao Q, Zhen T, Ruan M, Han J, et al. Prediction of early hematoma expansion of spontaneous intracerebral hemorrhage based on deep learning radiomics features of noncontrast computed tomography. Eur. Radiol. 2024;34:2908–2920. doi: 10.1007/s00330-023-10410-y. [DOI] [PubMed] [Google Scholar]
- 24.Gessert N, Nielsen M, Shaikh M, Werner R, Schlaefer A. Skin lesion classification using ensembles of multi-resolution EfficientNets with meta data. MethodsX. 2020;7:100864. doi: 10.1016/j.mex.2020.100864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J, Foroughi Pour A, Deirawan H, Daaboul F, Aung TN, Beydoun R, et al. Integrative deep learning analysis improves colon adenocarcinoma patient stratification at risk for mortality. EBioMedicine. 2023;94:104726. doi: 10.1016/j.ebiom.2023.104726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanda Y. Investigation of the freely available easy-to-use software 'EZR' for medical statistics. Bone Marrow Transpl. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980. 2014.
- 28.Géron A. Hands-on machine learning with Scikit-Learn, Keras and TensorFlow : concepts, tools, and techniques to build intelligent systems. 3rd ed: O'Reilly; 2022.
- 29.Bo R, Xiong Z, Huang T, Liu L, Chen Z. Using radiomics and convolutional neural networks for the prediction of hematoma expansion after intracerebral hemorrhage. Int. J. Gen. Med. 2023;16:3393–3402. doi: 10.2147/IJGM.S408725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akay EMZ, Hilbert A, Carlisle BG, Madai VI, Mutke MA, Frey D. Artificial intelligence for clinical decision support in acute ischemic stroke: A systematic review. Stroke. 2023;54:1505–1516. doi: 10.1161/STROKEAHA.122.041442. [DOI] [PubMed] [Google Scholar]
- 31.Bluemke DA, Moy L, Bredella MA, Ertl-Wagner BB, Fowler KJ, Goh VJ, et al. Assessing radiology research on artificial intelligence: A brief guide for authors, reviewers, and readers-from the radiology editorial board. Radiology. 2020;294:487–489. doi: 10.1148/radiol.2019192515. [DOI] [PubMed] [Google Scholar]
- 32.Lee H, Lee J, Jang J, Hwang I, Choi KS, Park JH, et al. Predicting hematoma expansion in acute spontaneous intracerebral hemorrhage: Integrating clinical factors with a multitask deep learning model for non-contrast head CT. Neuroradiology. 2024;66:577–587. doi: 10.1007/s00234-024-03298-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharrock MF, Mould WA, Hildreth M, Ryu EP, Walborn N, Awad IA, et al. Bayesian deep learning outperforms clinical trial estimators of intracerebral and intraventricular hemorrhage volume. J. Neuroimaging. 2022;32:968–976. doi: 10.1111/jon.12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kok YE, Pszczolkowski S, Law ZK, Ali A, Krishnan K, Bath PM, et al. Semantic segmentation of spontaneous intracerebral hemorrhage, intraventricular hemorrhage, and associated Edema on CT images using deep learning. Radiol. Artif. Intell. 2022;4:e220096. doi: 10.1148/ryai.220096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao X, Chen K, Wu G, Zhang G, Zhou X, Lv C, et al. Deep learning shows good reliability for automatic segmentation and volume measurement of brain hemorrhage, intraventricular extension, and peripheral edema. Eur. Radiol. 2021;31:5012–5020. doi: 10.1007/s00330-020-07558-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Row data of patient characteristics are stored in OSF in comma-separated values format (https://osf.io/jmnzs). All code in this study is available on GitHub (https://github.com/AI-neurosurg/Multimodal-network-for-predicting-hematoma-expansion-in-ICH). Model weights of the best-performing multimodal convolutional neural network model are stored in OSF in HDF5 format (419 MB, https://osf.io/wm768).