Abstract
Sepsis progression is significantly associated with the disruption of gut eubiosis. However, the modulatory mechanisms of gut microbiota operating during sepsis are still unclear. Herein, we investigated how gut commensals impact sepsis development in a pre-clinical model. Cecal ligation and puncture (CLP) surgery was used to establish polymicrobial sepsis in mice. Mice depleted of gut microbiota by an antibiotic cocktail (ABX) exhibited a significantly higher level of mortality than controls. As determined by metabolomics analysis, ABX treatment has depleted many metabolites, and subsequent supplementation with l-rhamnose (rhamnose, Rha), a bacterial carbohydrate metabolite, exerted profound immunomodulatory properties with a significant enhancement in macrophage phagocytosis, which in turn improved organ damage and mortality. Mechanistically, rhamnose binds directly to and activates the solute carrier family 12 (potassium-chloride symporter), member 4 (SLC12A4) in macrophages and promotes phagocytosis by activating the small G-proteins, Ras-related C3 botulinum toxin substrate1 (Rac1) and cell division control protein 42 homolog (Cdc42). Interestingly, rhamnose has enhanced the phagocytosis capacity of macrophages from sepsis patients. In conclusion, by identifying SLC12A4 as the host interacting protein, we disclosed that the gut commensal metabolite rhamnose is a functional molecular that could promote the phagocytosis capacity of macrophages and protect the host against sepsis.
Key words: Gut microbiota, Sepsis, Rhamnose, SLC12A4, GTP-Rac1, GTP-Cdc42, Macrophage, Phagocytosis
Graphical abstract
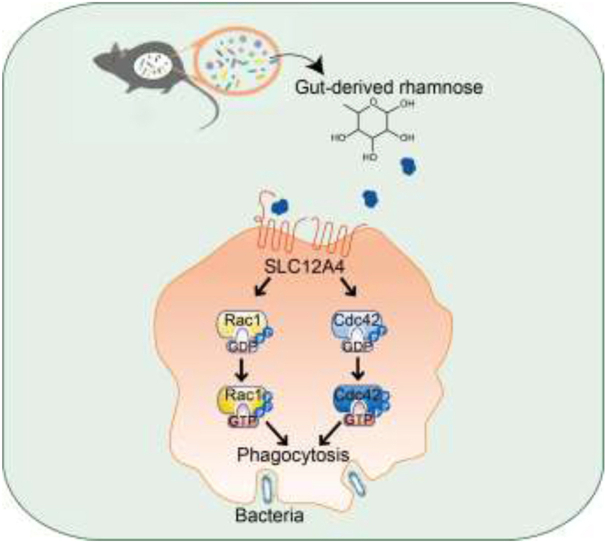
The depletion of gut microbiota leads to increased mortality in sepsis, which is associated with decreased gut-derived rhamnose. The rhamnose enhances the macrophage's phagocytic activity and protects against sepsis by modulating the SLC12A4/Cdc42 and Rac1 axis.
1. Introduction
Sepsis-induced multiple organ failure and death are the leading medical problems worldwide1, 2, 3. The pathogenesis of sepsis is quite complex. Upon infection, pathogen-associated molecular patterns trigger the activation of the innate immune system. Macrophages play a central role in orchestrating innate immune responses including phagocytosis and bacterial killing during sepsis4, 5, 6, 7. These responses are critical not only for eliminating the infection but also in preventing tissue injury and disruptions to the host organs during sepsis.
The complex process of phagocytosis is mediated by a large variety of receptors, such as Fcγ receptors (FcγR) or the complement receptors8. The phagocytic process can broadly be divided into several steps, which begin with an initial cup formation that depends on the rapid polymerization of F-actin. The cup formation step is followed by pseudopod extension, which accompanies a continuous production of F-actin polymerization. The last step of phagocytosis is the internalization with the closure of the phagocytic cup9. Many studies demonstrated that the Rho family GTPases, cell division control protein 42 homolog (Cdc42) and Ras-related C3 botulinum toxin substrate1 (Rac1) have significant regulatory functions in the formation of phagocytic cups10. In addition, activation of Cdc42 and Rac1 leads to actin cytoskeletal remodeling during phagocytosis. Enhancing bacterial clearance during the onset of sepsis remains one of the most efficient strategies in clinics.
Solute carrier family 12 member 4 (SLC12A4), also known as KCC1, is generally considered to maintain intracellular Cl– concentration for the normal functioning of cellular physiological activities11, 12, 13. Although SLC12A4 has been well characterized in sickle cell disease, its other biological functions remain to be discovered. Recently, loss of SLC12A4 was shown to inhibit the uptake of cellular debris by phagocytes14. However, the pathophysiologic role of SLC12A4 in immune cells and related diseases remains largely unclear.
The gut microbiota is recognized as one of the key upstream regulators of sepsis development15. Traditionally, sepsis is known to disrupt the gut barrier functions, and in turn intestinal bacteria may enter the circulatory system to boost the systemic inflammatory responses16. We previously reported the presence of significant disturbances to enteric eubiosis in sepsis patients and such dysbiosis may serve as the driver of organ damage17. In contrast, intestinal microbes also exert beneficial effects. The gut microbiota plays a critical role in maintaining homeostasis of the host immune system18. Gut-derived products, including sugars and short-chain fatty acids (SCFA), can modulate the immune system both within and outside the gut19,20. As the main cell type of innate immunity, macrophage functions could be regulated by multiple metabolites from gut commensals, which in turn could affect the susceptibility to sepsis21. These observations increased our understanding of the modulatory effects of gut microbiota on sepsis. However, there are still several fundamental questions that are not fully answered, for example, how gut commensal microbiota participates in sepsis development is still unknown. Thus, identifying the specific microbial products that directly impact sepsis could be crucial to developing novel treatments for sepsis patients. To address these questions, in this study, we evaluated the contribution of gut microbiota to cecal ligation and puncture (CLP)-induced polymicrobial sepsis and explored the regulatory mechanisms related to a gut-derived protective compound, l-rhamnose (rhamnose, Rha). Apart from promoting bacterial phagocytosis, rhamnose prevented sepsis-induced organ damage and death. In addition, rhamnose enhanced the phagocytic capacity of macrophages by directly binding to and activating SLC12A4. Thus, our current work broadened our understanding regarding host-gut microbiota interaction in the context of sepsis and provided a novel potential therapeutic strategy for treating sepsis patients.
2. Materials and methods
2.1. Animal model
Male C57BL/6 wild-type mice (6–8 weeks old) were purchased from GemPharmatech Co., Ltd. (Nanjing, China). All the mice were housed in polystyrene cages under a 12 h light/12 h dark cycle and given food and water ad libitum. All experiments were approved by the Animal Care and Use Committee of Southern Medical University (Approval number: SMUL2021094).
Experimental for antibiotic cocktail (ABX) treatment, mice were pretreated with ABX, including ampicillin sodium salt (400 mg/kg, Cat# A800429, Macklin, China), metronidazole (400 mg/kg, Cat# M813526, Macklin, China), neomycin sulfate (400 mg/kg, Cat# N814740, Macklin, China), and vancomycin (200 mg/kg, Cat# V105495, Aladdin, China) or PBS (Phosphate buffer solution, Cat# C10010500BT, Gibco, USA) for three days followed by further experiments.
Experimental for the sepsis model, after anesthetization of mice, two punctures were performed in the cecum with an 18- or 21-gauge needle to induce lethal or moderate CLP-induced sepsis22, respectively. Specifically, for the moderate CLP-induced sepsis model which was used for ABX pretreatment experiments, a single through-and-through cecal puncture was performed with a 21-gauge needle, and tissue was collected at 16 h after CLP. For the lethal CLP-induced sepsis model, an 18-gauge needle was used and tissue samples were collected at 12 h after CLP. Experimental for rhamnose intervention, in moderate CLP model, mice were pretreated with ABX for three days and then PBS or rhamnose (74 mg/kg, Cat# R108982, Aladdin, China) dissolved in PBS was used 2 h before moderate CLP surgery. In the lethal CLP model, mice were also orally administrated with PBS or rhamnose 2 h prior to lethal CLP surgery. All the survival rate of septic mice was observed for up to 72 h.
Experimental for the cecal content injection model, the cecal content obtained from C57BL/6 mice was dissolved at 0.25 g/mL in sterile PBS and centrifuged at 4 °C for 1 min (1000 rpm, Allegra X-30; Beckman, CA, USA) to collect the supernatant. Mice were injected intraperitoneally (i.p.) with a single dose of cecal supernatant (100 μL) and 16 h later, mice were sacrificed and the tissue samples were collected for further analysis. The survival rate of infected mice was observed for up to 72 h.
For the bacteria injection model, Escherichia coli NCTC12900 (Shiga toxin negative O157:H7) was grown in Luria Broth (LB, Cat# L3022, Sigma, USA) overnight at 37 °C until it reached an exponential phase, centrifuged, washed, and resuspended in sterile PBS. A single injection was administered at an infectious dose of 8.4 × 107 CFU to induce peritonitis. 16 h later mice were sacrificed for further study.
2.2. Bacterial load assay
For measuring the bacterial load from septic mice, blood and peritoneal lavage fluid were collected and enumerated by plating serial dilutions in the blood agar plates, followed by incubation at 37 °C for 24 h under aerobic or anaerobic conditions.
2.3. Patients
All subjects gave written informed consent prior to enrolment in the study. The inclusion criteria were patients (1) aged between 18 and 80 years, and (2) diagnosed with sepsis according to the current third international consensus protocol (sepsis 3.0)23. The exclusion criteria were patients who were (1) pregnant or lactating during the study period, (2) aged <18 or >80 years, (3) having an interrupted treatment regimen or (4) lacking complete information. The study involving human participants was approved by the Medical Ethics Committee of Nanfang Hospital, Southern Medical University (Approval number: NFEC-2021-139). All subjects in the studies participated voluntarily and informed consent was obtained before participation.
2.4. RNA extraction and bulk transcriptome analysis
The PLF cells collected from mice were centrifuged at 1000 rpm for 10 min at 4 °C (Allegra X-30; Beckman, CA, USA), resuspended in TRIzol™ Reagent (Cat# 15596018, Invitrogen, USA), and total RNA was extracted. The RNA Nano 600 Assay kit for the Bioanalyzer 2100 system (Agilent Technologies, CA, USA) was used for checking the quantity and quality of RNA. Library preparation, clustering, and sequencing were completed by Novogene (Beijing, China) using the Illumina Novaseq 6000 platform. Raw reads in fastq format were processed before mapping to the reference genome using Hiset2 v4.8.2. Read numbers were quantified for each gene using htseq v0.13.5, and FPKM was used to quantify the gene expression level. The difference in the gene expression levels between the groups was determined using DESeq1.30.124. Genes with a log2 (Fold change) absolute value greater than 1 were considered to be significantly different. The pathways were analyzed by KEGG pathway annotations included in the KEGG pathway database (http://www.genome.jp/kegg/pathway.html).
2.5. 16S rDNA sequencing analysis
Stool samples were collected, and DNA was extracted by phenol–chloroform method. Bacterial primers targeting the V4 region (forward: 5′-GTGTGYCAGCMGCCGCGGTAA-3′; reverse: 5′-GCGGACTACNVGGGTWTCTAAT-3′) of the 16S rDNA were used for qPCR. The V4 region was sequenced on an Illumina NovaSeq 6000 System (Illumina). Paired end reads and the sequences of the tags were compared with FLASH assembly (V1.2.7, http://ccb.jhu.edu/software/FLASH/) by using the Greengenes database (https://greengenes.secondgenome.com/) and UCHIME algorithm. QIIME (V1.9.1)25 was used for quality filtering of the raw tags. OTU clustering and species annotation were performed to obtain biodiversity and taxonomic composition data.
2.6. Metabolomics analysis and high-performance liquid chromatography (HPLC) analysis
Metabolomics analysis was performed by Metabo-Profile (Shanghai, China). Lyophilized stools were homogenized with zirconium oxide beads in sterile water for 3 min. Then, the metabolite extraction was performed with 120 μL of methanol containing an internal standard. Finally, metabolite analysis was performed by using an ultra-performance liquid chromatography coupled to a tandem mass spectrometry (UPLC–MS/MS) system (ACQUITY UPLC-Xevo TQ-S, Waters Corp., Milford, MA, USA). The raw UPLC–MS/MS data were processed using the TMBQ software (v1.0, Metabo-Profile, Shanghai, China) to do peak integration, calibration, and quantification for each metabolite. Thereafter, principal component analysis (PCA) was performed by using the self-developed platform iMAP (v1.0, Metabo-Profile, Shanghai, China).
For HPLC analysis of rhamnose, 100 μL plasma was extracted with acetonitrile and then centrifuged at 12,000 rpm (Allegra X-30; Beckman, CA, USA) for 15 min at 4 °C to collect supernatant. 10 μL supernatant was injected into high-performance liquid chromatography (Agilent, USA). The mobile phase was acetonitrile and water at a volume ratio of 4:1.
2.7. Single cell transcriptomic analysis
Mouse peripheral blood mononuclear cells (PBMCs) from three groups of moderate CLP-induced septic mice (CLP; CLP + ABX; CLP + ABX + Rha, n = 3/group) were collected. Pre-treatment for single cell transcriptomic sequencing was based on the standard procedure used in the 10 × Genomics Chromium Single Cell platform 5.0.1 with default parameters. Raw data obtained by Cell Ranger analysis was aligned with the mouse reference genome mm39. Cell quantitative matrix was imported to R 4.0.3. The data were filtered using SoupX 1.5.226 and DoubletFinder 2.0.327 package to remove all the cell-free RNA and doublet cells. Low-quality cells were filtered according to the following criteria: 1) 200 < nFeature_RNA <5000; 2) 0 < nCount_RNA <30,000; 3) percent of mitochondrial genes <15%; 4) percentage of hemoglobin genes <0.01% and 5) percentage of ribosome genes <50%. After filtering, a total of 72,368 cells with high quality from 9 mice were obtained. Gene expression matrices were normalized and integrated by using Seurat package 4.0.628. Using harmony package version 0.1.029 to remove batch effect. The differentially expressed genes from each cluster were extracted, and KEGG enrichment analysis was performed using the clusterProfiler package 3.18.130. The gene set enrichment analysis (GSEA) pathway enrichment analysis was explained by normalized enrichment score (NES) using GSEA version 1.20.031. Extract the non-zero expressed gene matrix of each group. The gene set variation analysis (CSVA) was performed by the GSVA package1.42.032. Annotated gene set in the Molecular Signatures Database (MSigDB, http://software.broadinstitute.org/gsea/msigdb/index.jsp) was selected as the reference gene sets. To evaluate the enrichment of each group, this study calculated the mean enrichment score of each group to represent this group enrichment.
2.8. Plasma biochemical analysis
The alanine aminotransferase (ALT), aspartate aminotransferase (AST) and creatinine (Cr) concentrations in the plasma were determined by using alanine aminotransferase (Cat# C009-3-1), aspartate aminotransferase (Cat# C010-3-1), and creatinine (sarcosine oxidase) (Cat# C011-2-1) assay kits from Nanjing Jiancheng Bioengineering Institute (Nanjing, China).
2.9. Cytokines analysis
TNF-α (Cat# EMC102a.96), IL-1β (Cat# EMC001b.96), and IL-6 (Cat# EMC004.96) concentrations in the plasma were measured by using ELISA kits from NeoBioscience (Shenzhen, China).
2.10. Quantitative real-time PCR
Total RNA from tissues or cells was extracted using a total RNA isolation kit according to the manufacturer's instructions (Vazyme Biotech Co., Ltd., Cat# RC112-01, China), and used for cDNA synthesis with a reverse transcription kit (Cat# FSQ-101, Toyobo, Japan). All the qPCRs were performed using SYBR Green Real-time PCR Master Mix (Cat# QPK-201, Toyobo, Japan) with the ABI 7500 Real-Time PCR System (Applied Biosystems, USA). Threshold cycle numbers were normalized against the 18S values, and the relative fold change was analyzed according to the comparative CT (ΔΔCt) method. All the primers used in the study are listed in the Supporting Information Table S1.
2.11. Transfection with plasmids
BMDMs were transfected with control siRNA or Slc12a4 siRNA (50 μmol/L, mouse Slc12a4-siRNA: 5′-CCACTTCGCTGGTGTACTT-3′) for 72 h. THP-1 derived macrophages were transfected with control siRNA or human Slc12a4-siRNA: 5′-GCCCAAAGGTATCGTCTCT-3′) for 48 h. Control siRNA was purchased from RiboBio Co., Ltd. (Guangzhou, China) by using a riboFECT CP Transfection Kit (166T) (Cat# C10511-05, RiboBio, Guangzhou, China) and used for subsequent experiments. SLC12A4-overexpressing (Cat# EX-Y3536-M98, Gene ID: 6560) and control (Cat# EX-NEG-M98) plasmids were constructed by iGene Biotechnology Co., Ltd. (Guangzhou, China). Plasmids overexpressing SLC12A4 or control plasmids (1.5 μg/mL) were transfected into BMDMs and THP-1 derived macrophages using Lipofectamine 3000 for 48 h (Cat# L3000015, Invitrogen, USA).
2.12. Confocal microscopy
For confocal analysis, GFP-E. coli was prepared as previously described33. BMDMs or THP-1 derived macrophages were grown on glass-bottom cell culture dishes (Cat# 801002, NEST Biotechnolog, China) and cells were pretreated with rhamnose (100 μmol/L) for 18 h and then incubated with GFP-E. coli at 37 °C in 5% CO2 for 45 min with a multiplicity of infection (MOI) of 60. After washing with PBS for three times, the cell membrane was labeled with Wheat germ agglutinin, (WGA, Cat# W11262, Thermo Fisher Scientific, USA) for 30 min, and cell nucleus was labeled with Hoechst 33342 (Cat# GC63515, GlpBio, USA) for 10 min. The cells were fixed with 4% paraformaldehyde and phagocytosis was observed using a Nikon C1 spectral confocal microscope (Nikon, Tokyo, Japan).
Experimental for rhamnose binding, biotinylated rhamnose (Bio-Rha) was synthesized and served as a molecular probe in combination with streptavidin bonded commercialized quantum dots (QD605s, Cat# QS605, Wuhan jiayuan quantum dots Co., Ltd., Wuhan, China), After a 30 min pre-incubation, probes were added to BMDMs for 30 min in situ to visualize the expression of rhamnose interacted proteins on the cell membrane surface by using confocal microscopy.
2.13. Phagocytosis and bacteria killing assay
For phagocytosis assay, BMDMs or THP-1 derived macrophages were pretreated with rhamnose (100 μmol/L) or PBS for 18 h and then incubated with E. coli at 37 °C in 5% CO2 for 20 min or 45 min with a MOI of 60. For neutrophils phagocytosis, cells were pretreated with rhamnose or PBS for 2 h and then incubated with E. coli at 37 °C in 5% CO2 for 45 min with a MOI of 60. After infection, cells were washed three times with gentamycin (25 μg/mL, Cat# E003632, Sigma, USA)-containing PBS to clear the extracellular bacteria. Cells were lysed with 0.1% TritonX-100 (Cat# X100, Sigma, USA) and plated on LB agar overnight. The bacterial load as CFUs were assessed by manual counting. For phagocytosis assay by flow cytometry, after GFP-E. coli was infected, cells were washed three times with PBS containing 25 μg/mL gentamycin. Cells were fixed with 4% paraformaldehyde (Cat# P1110, Solarbio, China), and then tested by flow cytometry for mean fluorescence intensity (MFI) of GFP.
For bacterial killing assay34, BMDMs or THP-1 cells were divided into two aliquots. One aliquot was infected with E. coli for 20 min or 45 min. Then cells were washed in PBS containing 25 μg/mL gentamycin to remove extracellular bacteria, and then cells were lysed with 0.1% TritonX-100, then cell lysis was collected and labeled as t = 0. The other aliquot of cells was treated with gentamycin (25 μg/mL)-containing culture medium for 4 h. Subsequently, the infected cells were washed with PBS to remove the bacteria remained in the supernatants. Next, the cell lysates were prepared using 0.1% Triton-X-100. Serial dilutions of the cell lysates were diluted in LB and plated on LB agar plates overnight at 37 °C and the bacterial colonies were counted manually. The bactericidal activity was calculated as the percentage of killed bacteria compared to t = 0 according to Eq. (1):
| (1) |
2.14. Construction of biotinylated-rhamnose and human proteome microarray assays
Biotinylated-rhamnose was produced by Shanghai Nafu Technology Co., Ltd. (Shanghai, China). The human proteome microarray (CDI Laboratories, Baltimore, MD, USA) Rhamnose-interacting protein analysis was done by Wayen Biotechnologies Inc. (Shanghai, China). Briefly, human proteome microarrays (HuProt™ 20K) were blocked with a blocking buffer [Tween 20 (PBS-T) containing 5% BSA], and 10 μmol/L biotinylated-rhamnose was incubated with the proteome microarrays at room temperature (RT) for 1 h. After washing, the microarrays were incubated with streptavidin-Cy5 at a dilution of 1:1000 (Cat# SA1011, Thermo Fisher Scientific, USA) for 20 min at RT. GenePix 4000B (Axon Instruments, Sunnyvale, CA, USA) was used for scanning and recording the results. GenePix Pro 6.0 was used for data analysis.
2.15. SLC12A4 binding experiments
An anti-SLC12A4 antibody pre-coated 96-well microplates purchased from ELK Biotechnology Co., Ltd. was used (Cat# ELK9749, Wuhan, China). A recombinant human SLC12A4 (rhSLC12A4) protein was synthesized by Crystalobio Biopharma (Shenzhen, China). rhSLC12A4 was added to the pre-coated 96-well microplate at 1 μg/mL for 80 min. After washing the plates, bio-rhamnose was added at 10, 50, 100, or 500 μmol/L and incubated for 50 min at 37 °C. Free rhamnose was added at 10 mmol/L concentration to determine the reduction in the interactions with the bio-rhamnose molecule.
2.16. Docking studies
The crystal structure of the SLC12A4 complex was obtained from the protein data bank (PDB ID: 6kkr) and the structure of rhamnose was obtained from PubChem (PubChem CID: 25310). Induced-fit docking of Rha to each potential binding site on SLC12A4 was performed using Schrodinger-Maestro software (version 11.1). Processing of the protein structures was performed with the ‘Protein Preparation Wizard’. Converting of ligands from 2D to 3D structures was performed using ‘LigPrep’. ‘Binding site detection’ was used to find the protein binding sites. Receptor grid generation was used to choose a grid box, which enclosed the whole binding site and dock ligand. Then rhamnose was docked into the presumptive binding sites, while other parameters were set to default during the docking analysis. The docking images and docking scores were exported for further evaluation.
2.17. Surface plasmon resonance (SPR) analysis
The binding affinity of rhamnose to rhSLC12A4 was determined using the PlexArray® HT System (PlexArray, USA) with a 3D Dextran sensor chip (Cat# PX201, Plexera Bioscience, USA). rhSLC12A4 protein was dissolved in PBS, immobilized on the chip and activated with 10 mmol/L NiSO4. Different concentrations of rhamnose, including 5000, 2500, 1000, 750 and 500 μmol/L were prepared with the running buffer (PBS). Sensor and sample plates were placed in the instrument and the reaction signal was collected for analysis with PlexArray HT specialized software (Plexera Bioscience, USA).
2.18. Intracellular chloride detection
BMDMs were incubated with rhamnose for 5 min, cells were then labelled with 10 mmol/L MQAE (N-(ethoxycarbonylmethyl)-6-methoxyquinolinium bromide, Cat# HY-D0090, MCE, China) for 1 h in DMEM medium. Cells were then washed three times with Cl− containing buffer (HBSS) and MQAE signals were monitored by flow cytometry at excitation wavelength of 355 nm and an emission wavelength of 460 nm. FlowJo software (v.10, Tree Star, USA) was used to analyze the data, which was expressed as mean fluorescence intensity (MFI).
2.19. Statistical analysis
Prism 8.3 software (GraphPad, USA) was used for data analysis. Data are presented as the mean ± standard error of mean (SEM). The sample numbers (n) are showed in the figure legends. The survival curves were analyzed by the log-rank test. Two-tailed unpaired student t-test were used for two groups analysis. One-way analysis of variance (ANOVA) was used for three or more groups analysis. P-values less than 0.05 were considered as statistically significant, ns, denotes not significant.
3. Results
3.1. Gut microbes defend the hosts against polymicrobial sepsis
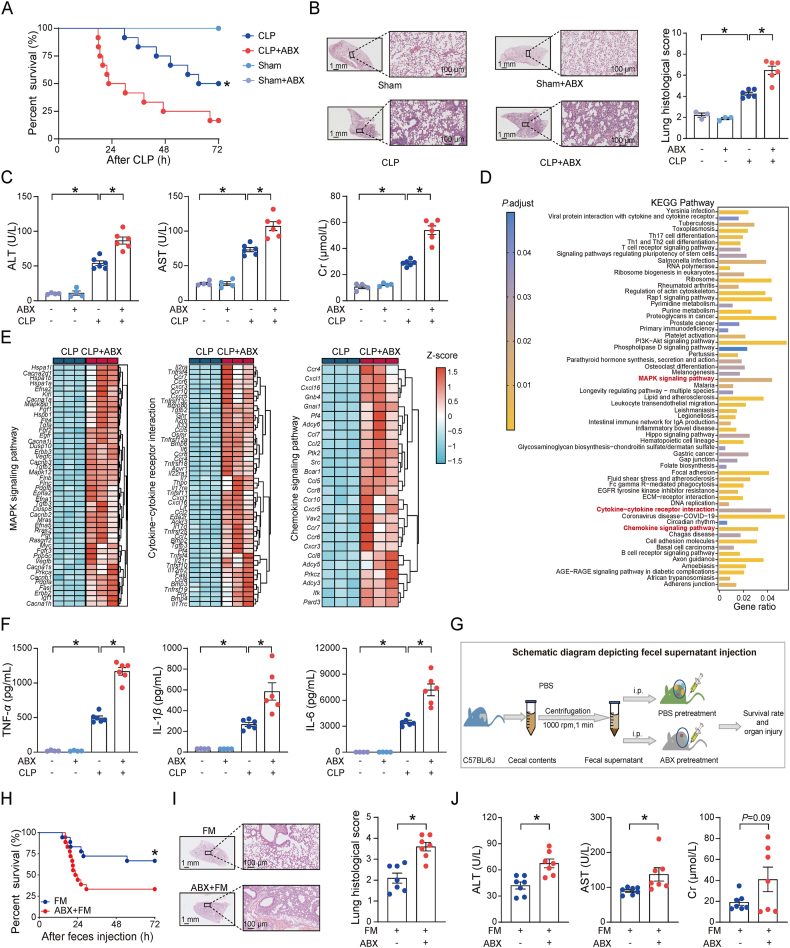
We initially studied the contribution of gut microbes to polymicrobial sepsis by evaluating how antibiotics (ABX) cocktail-pretreated mice responded to the CLP challenge. As presented in Fig. 1A, ABX pretreatment significantly decreased the survival rate of mice after CLP surgery. The lung, a most frequently injured organ in sepsis, had significantly more severe pathological changes in the ABX pretreated mice than no-ABX pretreated mice (Fig. 1B). In addition, plasma biochemical analysis of alanine aminotransferase (ALT), aspartate aminotransferase (AST) and creatinine (Cr) levels, confirmed the severity of organ damage in ABX pretreated mice compared to no-ABX pretreated mice (Fig. 1C). These data demonstrate the role of gut microbiota depletion in augmenting polymicrobial sepsis, which is partially in agreement with previous studies showing the protective property of commensal microbes against infections with specific pathogens35.
Figure 1.
Gut microbes protect against CLP-induced mortality and organ damage. (A) Survival curve after moderate cecal ligation and puncture (CLP) or sham surgery. Mice received a daily oral gavage PBS or ABX cocktail (ABX) for three days followed by moderate CLP or sham surgery (Sham, n = 4/group; CLP, n = 12/group). (B) Representative lung H&E images and histological scores of lung tissues. Mice were pretreated with PBS or ABX for 3 days and then performed moderate CLP or sham surgery. Tissue samples were collected at 16 h after surgery (Sham, n = 3/group; CLP, n = 6/group). Insets are low magnification images, Scale bar = 1 mm; high magnification view (200 × ), Scale bar = 100 μm. (C) Plasma ALT, AST, and Cr levels at 16 h after moderate CLP or sham surgery (sham, n = 4/group; CLP, n = 6). Experimental design as in Fig. 1B. (D) KEGG enrichment analysis of differentially expressed genes between CLP + ABX and CLP groups in the peritoneal lavage fluid (PLF) cells. Red color shows characteristic inflammatory pathways. Mice were pretreated with ABX for three days followed by moderate CLP surgery and 16 h later PLF cells were collected for Bulk RNA sequencing (n = 3/group). (E) Significant up-regulation of genes involved in MAPK signaling pathway, cytokine–cytokine receptor interactions and chemokine signaling pathway in ABX pretreated septic mice (n = 3/group). Experimental design as in Fig. 1D. (F) Plasma TNF-α, IL-1β and IL-6 protein levels in mice at 16 h after moderate CLP or sham surgery (Sham, n = 4/group; CLP, n = 6/group). Experimental design as in Fig. 1B. (G) Schematic diagram depicting fecal material (cecal content) injection protocol. Mice were injected intraperitoneally (i.p.) with a single dose of fecal material (FM, cecal content) supernatant (100 μL) and tissue samples were collected for subsequent experiments. (H) Survival curve of control (FM) and ABX-pretreated mice (ABX + FM) after fecal material (cecal content) supernatant injection (n = 18/group). Experimental design as in Fig. 1G. (I) Representative H&E images and histological scores of lung tissues at 16 h after fecal material (cecal content) supernatant injection. Insets are low magnification images, Scale bar = 1 mm; high magnification view (200 × ), Scale bar = 100 μm. n = 7/group. Experimental design as in Fig. 1G. (J) Plasma ALT, AST, and Cr level in fecal materials injected mice (n = 7/group). Experimental design as in Fig. 1G. Data are expressed as mean ± SEM. The survival curves were analyzed by the log-rank test. P values were determined by a two-tailed unpaired Student's t-test for two groups, one-way analysis of variance (ANOVA) analysis was used for three or more groups. ∗P < 0.05 was considered statistically significant. n: indicates number of samples.
Next, we assessed the systemic inflammation after CLP challenge to explore the impact of gut microbiota on sepsis progression. Bulk RNA-seq analysis of peritoneal lavage fluid (PLF) cells revealed the significant differences in the overall expression profiles of the CLP and CLP + ABX groups (Supporting Information Fig. S1A), with an enrichment of differentially expressed genes (DEGs) in many inflammation-related pathways, such as chemokine signaling pathway, mitogen-activated protein kinase (MAPK) signaling pathway and cytokine–cytokine receptor interactions (Fig. 1D). In addition, the expression level of the major genes participating in these three pathways was significantly upregulated in the CLP + ABX group (Fig. 1E). Notably, the enzyme-linked immunosorbent assay (ELISA) results indicated that plasma cytokines were markedly enhanced in the CLP + ABX group compared to the CLP group (Fig. 1F). The elevated levels of key cytokine and chemokine genes in PLF cells was detected in the CLP + ABX mice (Fig. S1B). These results demonstrate a significant increase in sepsis-induced immunological abnormalities after gut microbiota depletion in the mice.
ABX could directly kill and break down the bacteria, thereby causing a large accumulation of bacterial fragments including bacterial DNA, bacterial flagella et al. Theoretically, more bacterial fragments could be rapidly accumulated in ABX pretreated group than no-ABX group from the “punched hole” of the intestine to the abdominal cavity and cause higher mortality in mice after CLP. Thus, the increased bacterial debris in ABX group could be a confounder that contributed to the increased mortality observed in CLP model. To best exclude the possibility of higher mortality observed in ABX pretreated mice not due to increased bacterial debris, we employed the cecal content injection experiment to further validate our findings. In brief, the mouse cecal content from several specific pathogen-free (SPF) mice was collected and resuspended in sterile phosphate-buffered saline (PBS), followed by centrifugation to remove the larger particles. Then, the fecal supernatant was directly injected into mice pretreated with or without ABX to mimic the polymicrobial sepsis model (Fig. 1G). In turn, ABX pretreated mice and no-ABX pretreated mice were intraperitoneally injected with equal amounts of fecal supernatant to induce polymicrobial sepsis. This model did not directly “hurt” the intestine and would not allow the direct leakage of intestinal contents from the gut. Interestingly, results were similar to the CLP model, ABX pretreated mice also had a higher mortality rate (Fig. 1H) and more severe lung pathological alterations (Fig. 1I) than no-ABX pretreated mice. As expected, plasma concentrations of ALT, AST, Cr, and the levels of systemic inflammation in ABX-pretreated mice were higher than no-ABX pretreated mice (Fig. 1J, Fig. S1C and S1D). These results further confirmed the host protective function of gut microbes against polymicrobial sepsis in the animal models.
3.2. Microbial metabolite rhamnose protects against sepsis
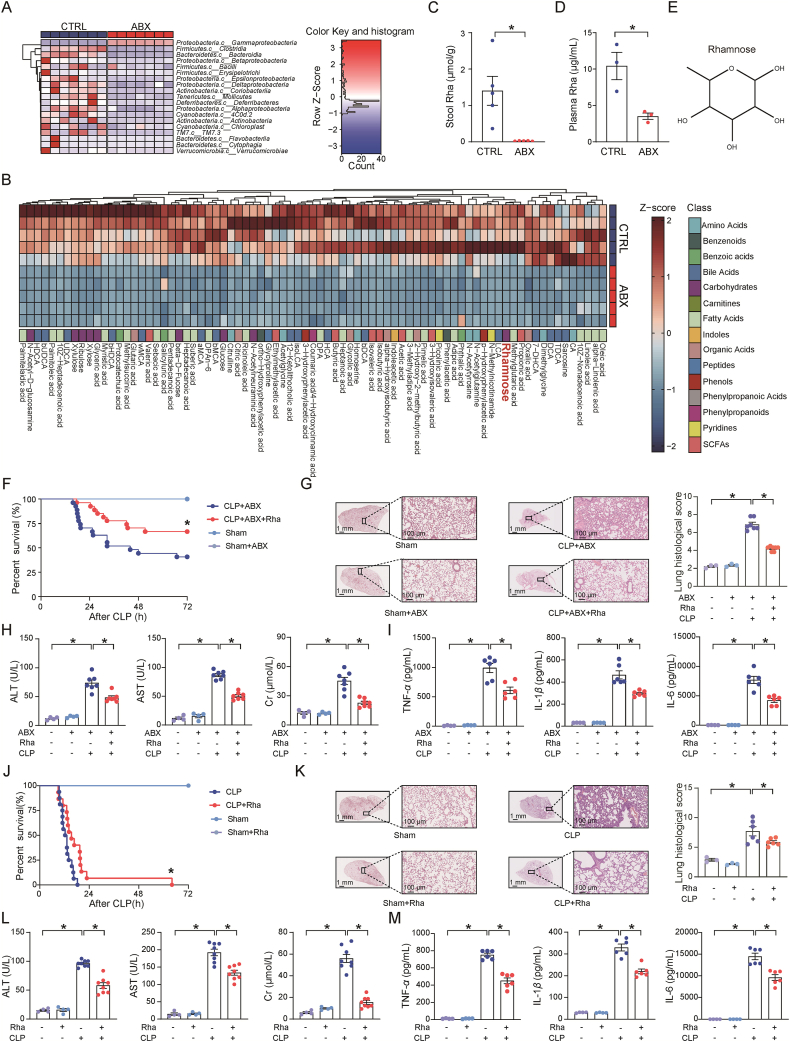
Next, we explored how commensal microbiota can protect against sepsis by performing 16S rDNA sequencing and metabolomics analysis of the feces from ABX-treated and untreated animals. As expected, ABX treatment markedly decreased the microbial diversity and depleted the majority of the bacteria in the gut (Fig. 2A, Supporting Information Fig. S2A). Moreover, antibiotic treatment shifted the microbial metabolic profile, in addition to significantly eliminating several metabolites present in the gut (Fig. 2B, Fig. S2B). Subsequently, we have investigated the contribution of metabolites lost due to ABX treatment on sepsis development. In the preliminary screening, we found the level of prokaryote-derived rhamnose from plasma and stools was decreased significantly in ABX pretreated mice (Fig. 2C–E). Meanwhile, the concentration of rhamnose in ABX pretreated group was also far less than no-ABX pretreated group in CLP model. The plasma rhamnose concentration was significantly increased in septic mice than healthy mice (Fig. S2C), possibly because sepsis could increase gut permeability and the microbial products including rhamnose could penetrate into the circulatory system. Importantly, supplementation of ABX-pretreated CLP mice with rhamnose could significantly improve the survival rate (Fig. 2F). Multiple organ damage tests including lung pathological examination, mRNA levels of pulmonary inflammatory factors, plasma cytokines, cytokine and chemokine genes in PLF cells and plasma biochemical assays, supported the protective effects of rhamnose (Fig. 2G–I, Fig. S2D and S2E). Similarly, in the ABX untreated mice, rhamnose also improved the survival rate (Fig. 2J), lung pathological damage (Fig. 2K) and suppressed the plasma concentrations of ALT, AST, and Cr (Fig. 2L). Besides, rhamnose decreased plasma cytokines concentration (Fig. 2M), the mRNA levels of inflammatory factors of PLF cells and lung tissue (Fig. S2F and S2G). We further employed another pre-clinical sepsis model to confirm the above finding. Specifically, lethal CLP surgery was performed in mice and the imipenem/cilastatin (14 mg/kg) was administrated at 6 h after surgery36. As shown in Fig. S2H and S2I, rhamnose still protected mice against organ injury in this pre-clinical sepsis model, which further validated the beneficial effect of gut-derived rhamnose in sepsis progression. Collectively, our data demonstrated that rhamnose is one of the key responsive bioactive metabolites that improves host defense against sepsis.
Figure 2.
Microbial metabolite rhamnose protects against polymicrobial sepsis. (A) The heatmap of the relative abundance of gut microbiota in the PBS (CTRL) and ABX treated mice. Stool samples were collected for metabolic analysis after PBS or ABX treatment (n = 7/group). (B) Hierarchical clustering of features identified in LC–MS analysis of stools from mice treated with PBS or ABX (n = 5/group). (C) Rhamnose (Rha) concentration present in the stools of PBS (CTRL) or ABX treated mice (n = 5/group). (D) Rhamnose concentration in plasma from mice subjected to pretreatment with PBS (CTRL) or ABX (n = 3/group). (E) The molecular structure of rhamnose. (F) Survival curve of ABX pretreated mice. Mice were pretreated with PBS or ABX for three days and then orally administrated with PBS or rhamnose 2 h prior to moderate CLP or sham surgery (Sham, n = 4/group; CLP, n = 27/group). (G) Representative H&E images and histological scores of lung tissues. Insets are low magnification images, Scale bar = 1 mm; high magnification view (200 × ), Scale bar = 100 μm. (Sham, n = 3/group, CLP, n = 7/group). Experimental design as in Fig. 2F. (H) Plasma ALT, AST, and Cr levels at 16 h (Sham, n = 4/group; CLP, n = 7/group). Experimental design as in Fig. 2F. (I) Plasma TNF-α, IL-1β and IL-6 protein levels at 16 h after CLP (Sham, n = 4/group; CLP, n = 6/group). Experimental design as in Fig. 2F. (J) Survival curve of PBS or rhamnose treated mice after sham or lethal CLP surgery. Mice were pretreatment with PBS or rhamnose for 2 h followed by lethal CLP or sham surgery (Sham, n = 4, CLP, n = 15–16/group). (K) Representative H&E images and histological scores of lung tissues at 12 h after CLP (Sham, n = 3; CLP, n = 6/group). Insets are low magnification images, Scale bar = 1 mm; high magnification view (200 × ), Scale bar = 100 μm. Experimental design as in Fig. 2J. (L) Plasma ALT, AST, and Cr levels at 12 h after CLP (Sham, n = 4; CLP, n = 8/group). Experimental design as in Fig. 2J. (M) Plasma TNF-α, IL-1β and IL-6 protein levels (sham, n = 4; CLP, n = 6/group). Experimental design as in Fig. 2J. Data are expressed as mean ± SEM. The survival curves were analyzed by the log-rank test. P values were determined by two-tailed unpaired Student's t-test for two groups, one-way analysis of variance (ANOVA) analysis was used for three or more groups. ∗P < 0.05 was considered statistically significant. n: indicates number of samples.
3.3. Rhamnose activates immune cells and promotes bacterial phagocytosis
To further investigate the modulatory effect of rhamnose on sepsis progression, we explored the impact of rhamnose on immune cell activation by performing a single cell RNA sequencing (scRNA-seq) of peripheral blood mononuclear cells (PBMCs) in an unbiased manner by using the droplet-based single-cell RNA sequencing method. We isolated and sequenced a total of 83,525 cells from the inferior vena cava blood cell suspensions collected from three groups of septic mice (n = 3 mice/group): control (CLP), after depleting the gut microbiota (CLP + ABX) and rhamnose treatment (CLP + ABX + Rha) groups. Using stringent quality controls, we further analyzed 72,368 cells in total (Supporting Information Fig. S3A). For identification of each cell cluster, we normalized the expression levels to the representative marker genes, Itgam for myeloid cells; Cd3d, Cd3e, and Cd3g for T cells; Cd19, Cd79a and Cd79b for B cells; and Ncr1 and Klrb1c for NK cells (Fig. S3B). Clustering analysis identified 6 distinct cell clusters (Fig. S3C).
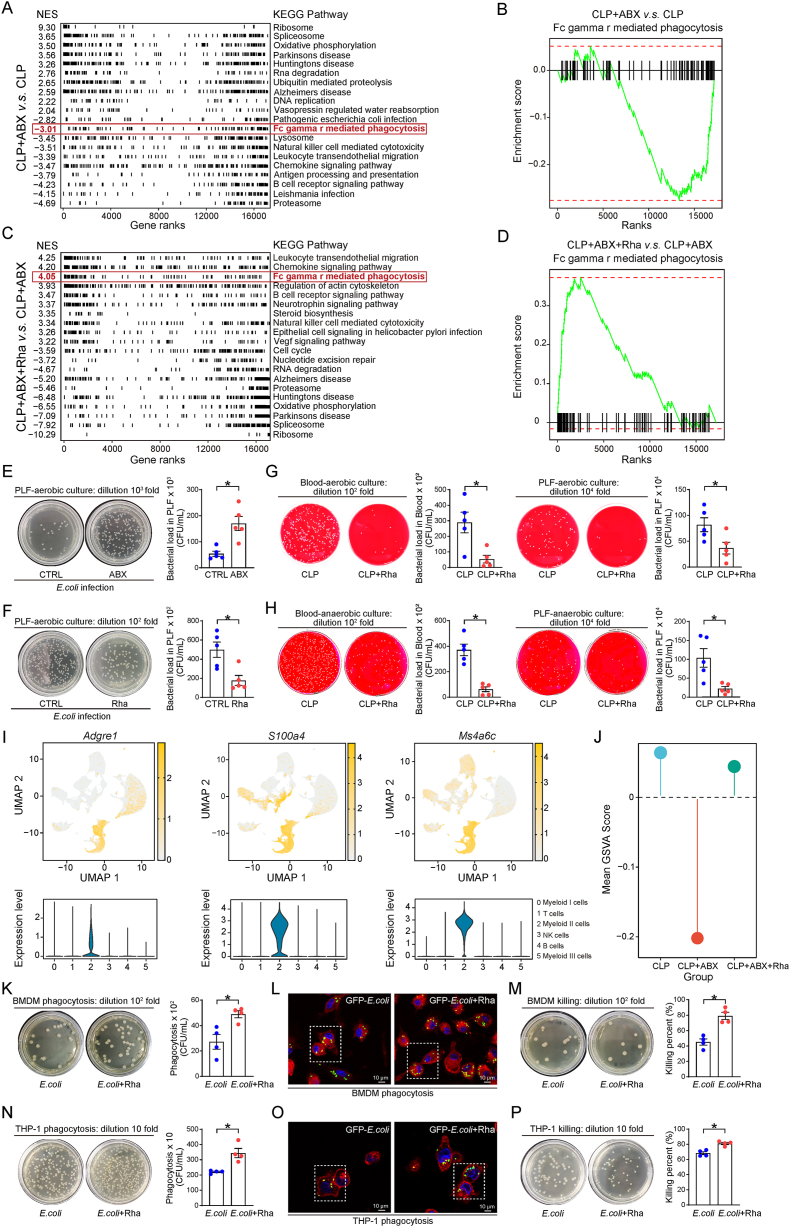
Next, we performed the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of differentially expressed genes between different groups, to find the critical biological process associated with rhamnose treatment. Interestingly, the FcγR-mediated phagocytosis pathway involved in affecting bacterial load and clearance in sepsis, was downregulated in CLP + ABX group compared to CLP group (Fig. 3A and B) in contrast to its significant upregulation in CLP + ABX + Rha group compared to CLP + ABX group (Fig. 3C and D). These results demonstrate a possibility that an increase in bacterial load after depletion of gut microbiota in septic mice. Conversely, rhamnose may protect the host from sepsis by regulating phagocytosis and decreasing the bacterial load.
Figure 3.
Rhamnose decreased the bacterial load in sepsis mice by enhancing the bacterial clearance capacity of macrophages. (A) Pathway enrichment analysis for gene sets comparing peripheral blood mononuclear cells (PBMCs) from CLP + ABX group and CLP group. Mice were pretreated with PBS or ABX for three days followed by moderate CLP surgery, and then PBMCs were isolated from whole blood samples at 16 h for further single cell RNA sequencing (scRNA-seq). NES, normalized enrichment score, (n = 3/group). (B) Gene set enrichment analysis (GSEA) enrichment plot of FcγR-mediated phagocytosis signaling of total cells in CLP + ABX versus CLP group (n = 3/group). Experimental design as in Fig. 3A. (C) Pathway enrichment analysis for gene sets comparing PBMCs from CLP + ABX + Rha group and CLP + ABX group. Mice were pretreated with ABX for three days followed by rhamnose or PBS administration and then moderate CLP model was performed to induce sepsis, PBMCs were collected at 16 h after CLP model. NES, normalized enrichment score, (n = 3/group). (D) Gene set enrichment analysis (GSEA) enrichment plot of FcγR-mediated phagocytosis signaling of total cells in CLP + ABX + Rha versus CLP + ABX group (n = 3/group). Experimental design as in Fig. 3C. (E) Quantity of E. coli in the PLF at 12 h after injecting with E. coli in ABX and no-ABX pretreated mice (n = 5/group). (F) Quantity of E. coli in the PLF at 12 h after injecting with E. coli. Mice were orally administrated with PBS or rhamnose for 2 h followed by injection with E. coli (n = 5/group). (G, H) Quantification of bacterial colonies in the blood and PLF from the CLP mice in aerobic (G) or anaerobic (H) cultures. Mice were pretreated with PBS or rhamnose for 2 h followed by lethal CLP and tissue samples were collected at 12 h for the detection of bacterial load (n = 5/group). (I) According to macrophage marker genes (Adgre1, S100a4, Ms4a6c) to label single cells on a dimensional reduction plot. Violin plot evaluated the expression levels of macrophage marker genes in each cluster. (J) Gene set variation analysis (GSVA) enrichment lollipop chart of FcγR-mediated phagocytosis signaling in macrophages. The x-axes represent different group, and y-axes represent the average GSVA score in each group (n = 3/group). (K) Phagocytosis of E. coli in BMDMs. BMDMs were pretreated with rhamnose (100 μmol/L) or PBS for 18 h and then incubated with E. coli at 37 °C in 5% CO2 for 45 min with a multiplicity of infection (MOI) of 60 (n = 4/group). (L) Representative confocal images showing phagocytosis of GFP-E. coli by BMDMs. Experimental design as in Fig. 3K. Scar bar = 10 μm. (M) Percentage of E. coli killing by BMDMs in control (CTRL) and rhamnose (Rha) groups (n = 4/group). (N) Phagocytosis of E. coli in THP-1 derived macrophages (THP-1 cells). THP-1 cells were pretreated with rhamnose (100 μmol/L) or PBS for 18 h and then incubated with E. coli at 37 °C in 5% CO2 for 45 min with a MOI of 60 (n = 4/group). (O) Representative confocal images showing phagocytosis of GFP-E. coli by THP-1 cells. Experimental design as in Fig. 3N. Scar bar = 10 μm. (P) Percentage of E. coli killing by THP-1 cells in control (CTRL) and rhamnose (Rha) groups (n = 4/group). Data are expressed as mean ± SEM and P values were determined by a two-tailed unpaired Student's t-test. ∗P < 0.05 was considered statistically significant. n: indicates number of samples.
To further confirm these sequencing data, we employed an E. coli-induced peritonitis and CLP-induced sepsis model and then collected blood and peritoneal lavage fluid (PLF) for evaluating the bacterial load in mice. In the E. coli-induced peritonitis model, the number of colonies plated from PLF of control group (treated with PBS before a single intraperitoneal injection of E. coli) was far less than the ABX-pretreated mice (Fig. 3E). The ABX cocktail used in the current study was nonabsorbable37, namely they can kill intestinal bacteria but had limited effects on circulatory bacteria. To rule out the possibility that ABX pretreatment may markedly damage E. coli in the peritonitis model, we cultured the bacteria from mice PLF after E. coli injection, the growth of E. coli from PLF of ABX pretreatment group was comparable with no-ABX pretreatment group (Fig. S3D). Therefore, it is clear that the depletion of gut microbiota could impair the ability of immune cells to clear the bacteria during an infection in mice. Next, we investigated whether rhamnose affects the bacterial load in E. coli-infected mice. We calculated the number of colonies present in the peritoneal lavage fluid of E. coli-infected mice with or without rhamnose treatment. We found that rhamnose-treated mice had fewer colonies than the control mice (Fig. 3F). To further investigate the effect of rhamnose on phagocytosis, we also employed the CLP model. As expected, rhamnose significantly decreased the bacterial load in the blood and peritoneal lavage fluid (Fig. 3G and H).
Macrophages are the key cell types involved in the clearance of bacteria following an infection38,39, such as sepsis and peritonitis. We then grouped and analyzed macrophages based on our single-cell sequencing data to explore the functional association between rhamnose and macrophages. We selected three relatively high-expression macrophage marker genes, Adgre1, S100a4, and Ms4a6c40,41, and defined myeloid II cells as macrophages (Fig. 3I). The gene set variation analysis (GSVA) was performed by using the gene set in the Molecular Signatures Database (MSigDB), and interestingly, the Fcγ R mediated phagocytosis pathway was obviously downregulated after ABX treatment, whereas it was rescued by supplementation with rhamnose (Fig. 3J). To confirm this result, we tested the effects of rhamnose on the phagocytic ability of macrophages. Interestingly, results from flow cytometry, plate colony formation assay, and confocal microscopy indicated that rhamnose-treated murine bone marrow-derived macrophages (BMDMs) and the human leukemia monocytic cell line (THP-1) derived macrophages showed an increased level of phagocytosis and bacterial clearance than the control group after 45 min incubation with E. coli (Fig. 3K–P, Fig. S3E and S3F). This finding was reproduced when the E. coli-macrophages incubation time was at 20 min (Fig. S3G–S3L). These data demonstrate that the beneficial effects of rhamnose during sepsis progression were associated with improved phagocytosis.
3.4. Rhamnose directly binds to and activates SLC12A4 in macrophages
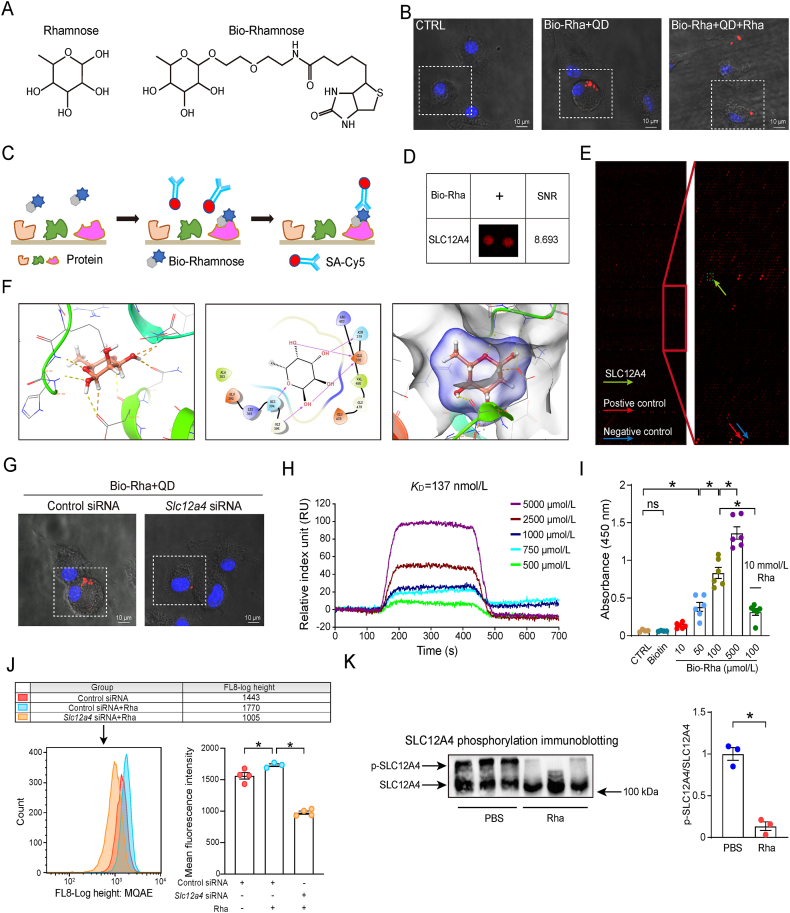
To explore the underlying mechanisms involved in the actions of rhamnose, we synthesized a biotinylated rhamnose (Bio-Rha, Bio-Rhamnose, Fig. 4A) and determined its biological effects. Indeed bio-rhamnose retained its capacity to promote phagocytosis and clearance of bacteria in BMDMs and THP-1 derived macrophages (Supporting Information Fig. S4A–S4D). To investigate the subcellular localization of rhamnose, we labeled BMDMs with biotinylated rhamnose, which binds to streptavidin quantum dots (QDs), and visualized the location of rhamnose by confocal microscopy. Bio-rhamnose-QDs were mainly localized on the cell surface, suggesting the presence of rhamnose binding proteins on the cell surface. To further corroborate our findings, we did a competitive binding assay with rhamnose and bio-rhamnose. Briefly, BMDMs were at first cultured with free rhamnose for 1 h, before the addition of biotinylated-rhamnose-QDs. Consequently, fluorescence was found to be substantially reduced on the cellular surface with rhamnose treatment (Fig. 4B), which confirmed the presence of rhamnose binding proteins on the cellular surface.
Figure 4.
Rhamnose interacts with SLC12A4 protein. (A) Chemical structures of rhamnose and biotinylated rhamnose (Bio-Rhamnose). (B) Representative confocal images of Bio-Rhamnose (Bio-Rha) binding to the surface of BMDMs. Confocal image of cells labeled with probes (red) and Hoechst 33342 stain (blue). Bio-rhamnose binding was blocked by free rhamnose (rhamnose + Bio-rhamnose). Scar bar = 10 μm. (C) A schematic diagram showing the experimental design for identifying rhamnose binding proteins using HuProt™20K Proteome Microarray Chip. (D) A magnified image showing interactions between Bio-rhamnose and SLC12A4 spots on the microarrays. Signal to noise ratio (SNR) is shown. (E) A representative image of HuProt™20K Proteome Microarray Chip showing positive (red arrow) and negative (blue arrow) spots, as well as spots for SLC12A4 (yellow spots). (F) Predicted binding modes of rhamnose targeting SLC12A4. Key residues are shown in red sticks, and hydrogen bonds are depicted by red arrows. (G) Representative confocal images of Bio-Rhamnose (Bio-Rha) binding to the surface of BMDMs pretreated with negative control or Slc12a4 siRNA. Cells were labeled with probes (red) and Hoechst 33342 stain (blue). Scar bar = 10 μm. (H) Direct interactions between rhamnose and SLC12A4 were determined by Surface plasmon resonance (SPR) analysis. (I) Rhamnose binding to SLC12A4 in ELISA. Recombinant hSLC12A4 was captured in wells pre-coated with anti-SLC12A4 antibodies. Bio-rhamnose at increasing concentrations and free rhamnose were added to perform the competition ELISA (n = 4–6/group). (J) Flow cytometry of MQAE intensity in BMDMs. Chloride ion could quench fluorescence signal, higher fluorescence intensity represents lower chloride ion concentration. Histogram of MQAE intensity (Mean Fluorescence Intensity) and the quantification (n = 3–4/group). (K) Western blot of phosphorylated SLC12A4 (p-SLC12A4) in BMDMs. Cells were treated with PBS or rhamnose for 5 min and the phosphorylation level of SLC12A4 was detected (n = 3/group). Data are expressed as mean ± SEM. One-way analysis of variance (ANOVA) analysis was used for three or more groups. Two groups were determined by a two-tailed unpaired Student's t-test, ∗P < 0.05 was considered statistically significant. ns: denotes not significant. n: indicates number of samples.
Next, we employed a human proteome microarray consisting of 20,000 affinity-purified GST-tagged proteins to identify the potential rhamnose-binding proteins. We incubated bio-rhamnose with this proteome microarray, and the binding signals were identified by using Cy5-conjugated streptavidin (Cy5-SA, Fig. 4C). The signal to noise ratio (SNR) was calculated as the ratio between the foreground and background values. Interestingly, our data pointed out that rhamnose potentially interacted with SLC12A4 with a robust SNR of 8.693 (Fig. 4D and E). Molecular docking analysis identified the four key resides present in the active pocket (Glu-378 Asn-379 His-394 Gly-395) of SLC12A4 protein that could possibly be interacting with the rhamnose molecule (Fig. 4F). Next, we transfected BMDMs with Slc12a4 or control siRNA, and cells were incubated with bio-rhamnose-QDs. Interestingly, SLC12A4-knockdown cells had an obvious reduction in the red fluorescence on the cellular surface (Fig. 4G), which further confirmed SLC12A4 in macrophages as a target for rhamnose.
To validate the binding nature between SLC12A4 and rhamnose, a surface plasmon resonance (SPR) experiment was used. The data suggested positive affinity interactions of rhamnose (KD = 137 nmol/L) with the recombinant human SLC12A4 (rh SLC12A4) protein (Fig. 4H). Furthermore, we used a competitive ELISA-based system to consolidate our conclusion in which free rhamnose was found to inhibit bio-rhamnose binding with SLC12A4 (Fig. 4I).
SLC12A4 is a well-known ion channel that governs the exportation of intracellular chloride upon activation. We then explored if the binding between rhamnose and SLC12A4 exerts biological activity by monitoring the chloride ion level alteration in macrophages. As shown in Fig. 4J, rhamnose significantly decreased the intracellular chloride level while SLC12A4 knockdown rescued the chloride concentration in macrophages. Importantly, phosphorylated SLC12A4 (p-SLC12A4) is recognized as the inactive status of this ion channel, so the detection of the level of p-SLC12A4 may be important. We found that p-SLC12A4 was decreased in CLP group than sham groups (possibly because bacteria may active SLC12A4), while rhamnose could further significantly decrease the level of p-SLC12A4. Importantly, p-SLC12A4 was increased in CLP + ABX group than CLP groups, while rhamnose could significantly decrease the level of p-SLC12A4 in CLP + ABX + Rha group compared to CLP + ABX group (Fig. S4E and S4F). Besides, we found that rhamnose was able to decrease p-SLC12A4 in BMDMs (Fig. 4K), further indicating rhamnose could activate SLC12A4. Collectively, the above results demonstrated that rhamnose directly binds to and activates SLC12A4 in macrophages.
3.5. SLC12A4 in macrophages is required for bacterial phagocytosis
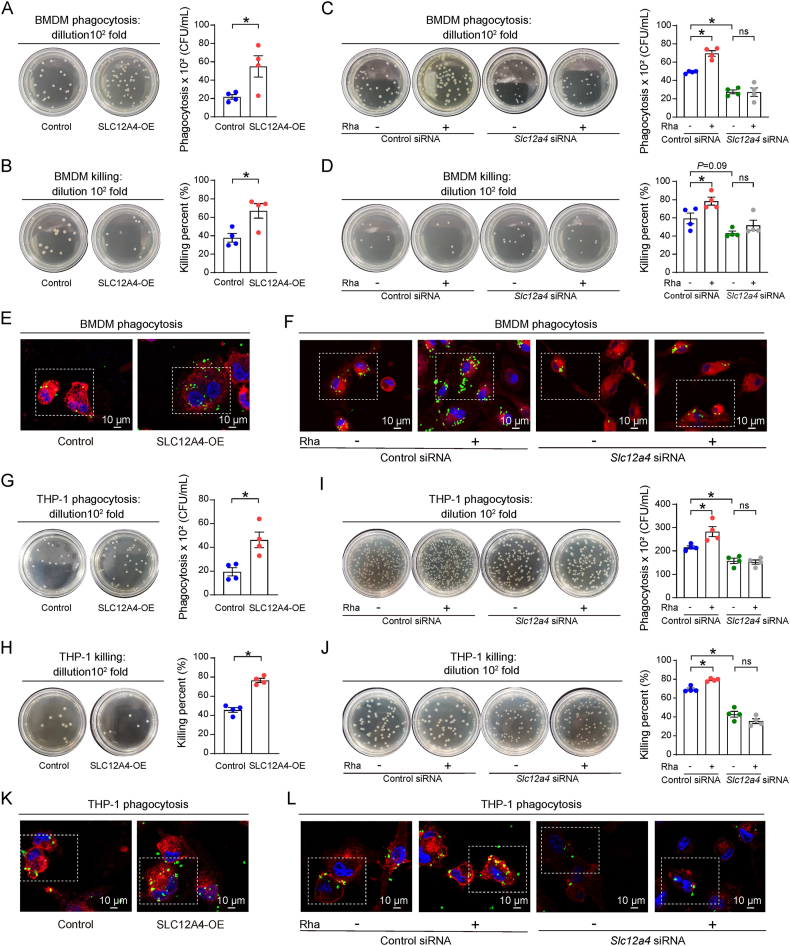
We next evaluated whether and how SLC12A4 participated in macrophage bacterial phagocytosis by using SLC12A4 overexpression plasmid and siRNA in macrophages (Supporting Information Fig. S5A–S5D). E. coli was added to the macrophage cultures to assay its phagocytosis and bacterial clearance properties. Interestingly, SLC12A4 overexpression in BMDMs has significantly enhanced their phagocytotic activity and bacterial killing (Fig. 5A and B), while transfection with Slc12a4 siRNA showed a significantly decreased phagocytosis and bacterial killing (Fig. 5C and D). These results were confirmed by confocal microscopy (Fig. 5E and F). To confirm our results, we designed the same experiments in THP-1 derived macrophages. As expected, similar results were obtained with phorbol-12-myristate-13-acetate (PMA)-differentiated THP-1 cells as well (Fig. 5G–L). These data indicated a requirement for SLC12A4 in the macrophage phagocytosis function.
Figure 5.
Defective E. coli phagocytosis in SLC12A4-defecient cells. (A) Phagocytosis of E. coli by BMDMs transfected with control or SLC12A4-overexpression (SLC12A4-OE) plasmids (n = 4/group). (B) Percentage of E. coli killing by BMDMs transfected with control or SLC12A4-overpression (SLC12A4-OE) plasmids (n = 4/group). (C) Phagocytosis of E. coli by BMDMs transfected with Slc12a4 siRNA or control siRNA for 72 h in the presence or absence of rhamnose (100 μmol/L) (n = 4/group). (D) Percentage of E. coli killing by BMDMs transfected with Slc12a4 siRNA or control siRNA for 72 h in the presence or absence of rhamnose (100 μmol/L) (n = 4/group). (E) Representative confocal images showing phagocytosis of GFP-E. coli in BMDMs transfected with SLC12A4 overexpression or control plasmids for 48 h in the presence or absence of rhamnose (100 μmol/L). Scale bar = 10 μm. (F) Representative confocal images showing phagocytosis of GFP-E. coli in BMDMs transfected with Slc12a4 siRNA or control siRNA for 72 h in the presence or absence of rhamnose. Scale bar = 10 μm. (G) Phagocytosis of E. coli in THP-1 cells transfected with control or SLC12A4-overpression (SLC12A4-OE) plasmids (n = 4/group). (H) Percentage of E. coli killing in THP-1 cells transfected with control or SLC12A4-overpression (SLC12A4-OE) plasmids (n = 4/group). (I) Phagocytosis of E. coli in THP-1 cells transfected with Slc12a4 or control siRNA for 48 h in the presence or absence of rhamnose (100 μmol/L) (n = 4/group). (J) Percentage of E. coli killing in THP-1 cells transfected with Slc12a4 or control siRNA for 48 h in the presence or absence of rhamnose (100 μmol/L) (n = 4/group). (K) Representative confocal images showing phagocytosis of GFP-E. coli in THP-1 cells transfected with SLC12A4 overexpression or control plasmids for 48 h in the presence or absence of rhamnose (100 μmol/L). Scale bar = 10 μm. (L) Representative confocal images showing phagocytosis of GFP-E. coli in THP-1 cells transfected with Slc12a4 or control siRNA for 48 h in the presence or absence of rhamnose. Scale bar = 10 μm. Data are expressed as mean ± SEM. One-way analysis of variance (ANOVA) analysis was used for three or more groups. Two groups were determined by a two-tailed unpaired Student's t-test. ∗P < 0.05 was considered statistically significant. ns, denotes not significant. n: indicates number of samples.
To explore whether rhamnose exerts its functions through SLC12A4, we next investigated the effect of rhamnose on phagocytosis in the presence of Slc12a4 siRNA treatment. In agreement with our previous data, rhamnose alone promoted phagocytosis and clearance of bacteria but had negligible effects in the Slc12a4-silenced BMDMs and THP-1 derived macrophages (Fig. 5C, D, F, I, J and L). Thus, these data demonstrate a key role played by SLC12A4 in rhamnose-mediated phagocytosis and bacterial clearance by macrophages.
3.6. Rhamnose by regulating Rac1 and Cdc42 activities enhances phagocytosis
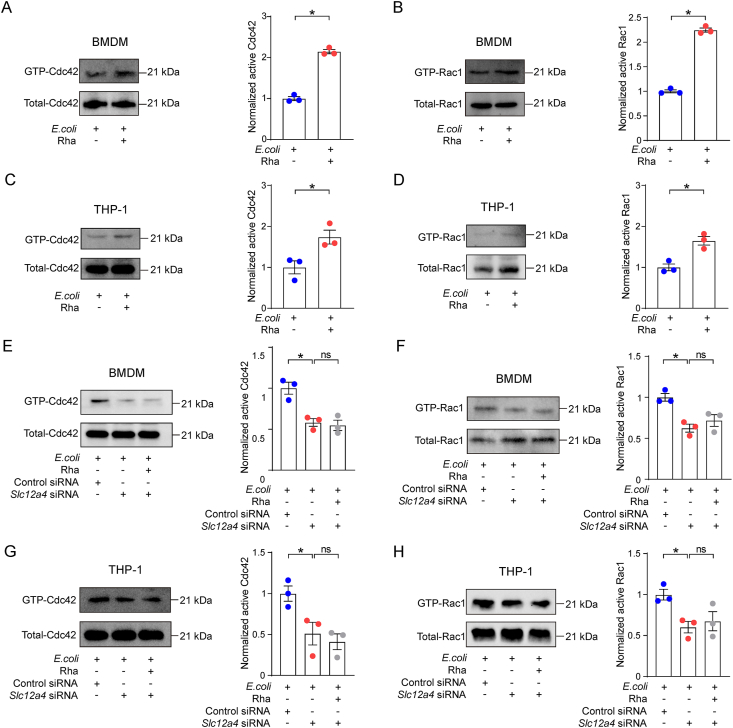
To further understand rhamnose-mediated regulatory mechanisms in phagocytosis, we next identified the proteins influenced by rhamnose. By analyzing the genes involved in the phagocytosis pathway, we found the Rho family GTPases, Cdc42 and Rac1 were downregulated in CLP + ABX group while Rha supplementation could rescue its abundance (Supporting Information Fig. S6A). It is well established that these Rho GTPases could regulate the actin cytoskeleton and phagocytosis by switching between an inactive GDP-bound state and an active GTP-bound state42. Since the Rho GTPases interact with the effectors only in their GTP-bound active state, we investigated their activities in vivo and in vitro. We found ABX treatment could significantly decrease the levels of GTP-Cdc42 and GTP-Rac1 compared to no-ABX pretreatment mice in CLP model and rhamnose supplementation could rescue these Rho GTPases levels (Fig. S6B and S6C). Besides, GTP-Cdc42 and GTP-Rac1 were also significantly elevated in lethal septic mice than sham mice, which may be attributed to the direct stimulation by bacteria in the peritoneal cavity of septic mice and rhamnose could significantly increase the levels of GTP-Cdc42 and GTP-Rac1 (Fig. S6D and S6E). Besides, Fig. 6A–D shows that activated Cdc42 and Rac1 expression was significantly elevated in rhamnose supplemented group than control, both in BMDMs and THP-1 derived macrophages during E. coli infection.
Figure 6.
Rhamnose promotes phagocytosis by activating Cdc42 and Rac1. (A, B) Representative Western blot image and quantification of GTP-cell division control protein 42 homolog (GTP-Cdc42) and GTP-Ras-related C3 botulinum toxin substrate1(GTP-Rac1) levels in BMDMs. Cells were incubated with E. coli for 5 min in the presence or absence of rhamnose (100 μmol/L) (n = 3/group). (C, D) Representative Western blot image and quantification of GTP-Cdc42 and GTP-Rac1 levels in THP-1 derived macrophages. Cells were incubated with E. coli for 5 min in the presence or absence of rhamnose (100 μmol/L) (n = 3/group). (E, F) Representative Western blot image and quantification of GTP-Cdc42 and GTP-Rac1 levels in BMDMs. BMDMs were transfected with Slc12a4, or control siRNA for 72 h and then incubated with E. coli for 5 min in the presence or absence of rhamnose (100 μmol/L) (n = 3/group). (G, H) Representative Western blot image and quantification of GTP-Cdc42 and GTP-Rac1 levels in THP-1 derived macrophages. THP-1 cells transfected with Slc12a4, or control siRNA for 48 h and then incubated with E. coli for 5 min in the presence or absence of rhamnose (100 μmol/L) (n = 3/group). Data are expressed as mean ± SEM. One-way analysis of variance (ANOVA) analysis was used for three or more groups. Two groups were determined by a two-tailed unpaired Student's t-test. ∗P < 0.05 was considered statistically significant. n: indicates number of samples. ns, denotes not significant.
We next evaluated the role of SLC12A4 in the regulation of rhamnose-mediated Cdc42 and Rac1 activity. Both Cdc42 and Rac1 activation was markedly decreased in the SLC12A4 knockdown group, indicating the key status of SLC12A4 in regulating the activation of Rac1 and Cdc42 in macrophages. Importantly, rhamnose had a negligible effect in the SLC12A4-silenced groups (Fig. 6E–H). Thus, rhamnose could possibly regulate Cdc42 and Rac1 activities through SLC12A4 in macrophages.
3.7. Rhamnose enhanced bacterial phagocytosis by human blood monocyte-derived macrophages
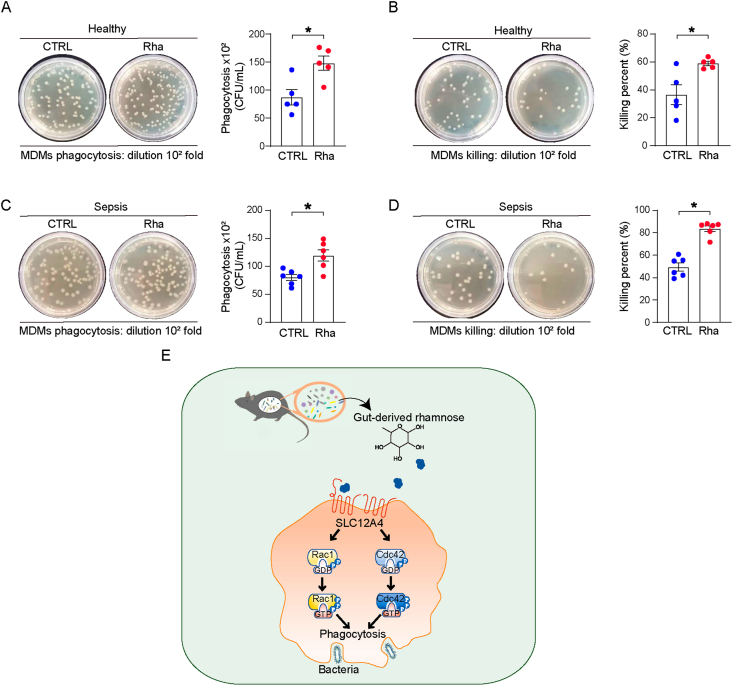
Finally, to assess whether a similar function of rhamnose in enhancing bacterial phagocytosis exists in human blood monocyte-derived macrophages (MDMs), we isolated and incubated MDMs from healthy volunteers and sepsis patients. Notably, rhamnose has significantly enhanced the phagocytosis and bacterial clearance capacity of MDMs from healthy volunteers (Fig. 7A and B) and sepsis patients (Fig. 7C and D). Thus, rhamnose may be used as a novel therapeutic agent for promoting bacterial phagocytosis in sepsis.
Figure 7.
Rhamnose promotes phagocytosis of human macrophages. (A, B) Phagocytosis and percentage of killing of E. coli by human blood monocyte-derived macrophages (MDMs) from healthy volunteers in control (CTRL) and rhamnose (Rha) groups (n = 5/group). (C, D) Phagocytosis and percentage of killing of E. coli by human blood monocyte-derived macrophages (MDMs) from sepsis patients in control (CTRL) and rhamnose (Rha) groups (n = 6/group). (E) A schematic diagram depicting gut microbiota derived rhamnose promoted bacterial phagocytosis by activating Cdc42 and Rac1 in macrophages. Data are expressed as mean ± SEM. Two groups were determined by a two-tailed unpaired Student's t-test. ∗P < 0.05 was considered statistically significant. n: indicates number of samples.
4. Discussion
The gut commensal microbiota is widely acknowledged to be an important upstream regulator of sepsis progression. Previous research has mainly focused on the association between commensals and gut barrier function15. However, recent studies demonstrated more fundamental roles played by gut commensals in sepsis development. For example, in the context of Streptococcus pneumoniae infection, depletion of intestinal microbiota disturbed alveolar macrophage functions, which augmented infection-induced multiple organ damage43. Similarly, the absence of gut commensals impaired the pathogen clearance capacity of Kupffer cells causing an increase in the Staphylococcus aureus infection-associated mortality44,45. Moreover, gut microbiota stimulated the host immune cells to generate IgA antibodies, which inhibited bacterial sepsis in an animal model46,47. These observations increased our understanding of the basic role of gut commensals in the pathogenesis of sepsis. However, specific interactions between the gut microbiota and the host immune system are still largely unknown. In addition, the complex nature of gut microbiota and its products, emphasizes the need for identifying specific functional factors for a better understanding the modulatory landscape between the gut microbiota and sepsis. In the present work, we found depletion of gut commensals increased the bacterial load, resulting in higher levels of inflammation with aggravated polymicrobial sepsis in mice, and antibiotic-treated mice exhibited impaired phagocytosis signaling pathways, thereby implicating the important contribution of microbiota in promoting phagocytosis. The depletion of gut microbiota caused the loss of some key responsive bioactive metabolites which affect the progress of sepsis. We originally selected more commercialized metabolites with an average concentration higher than 100 nmol/g in feces according to the fold change and significance (P < 0.05) (Control compared to ABX), including rhamnose, N-acetyl-d-glucosamine, xylose, citrulline, etc. Interestingly, we identified a specific and functional microbial metabolite, rhamnose, which was the most effective compound to protect against sepsis among these metabolites. Moreover, the impaired phagocytosis signal pathways could be restored by supplying rhamnose in the antibiotic-treated mice. Together, this study demonstrates the importance of gut microbiota in promoting an effective immune system by enhancing the phagocytic capacity of innate immune cells, particularly macrophages. However, we cannot rule out the beneficial contribution of other metabolites and the detailed functions of these compounds require further investigation.
Since the precursor for rhamnose synthesis (TDP/UDP-rhamnose) is found only in the microbiome and plants, it can only be generated by the gut bacteria in mammals. Although rhamnose is an integral component of the bacterial membrane48, unlike other cell membrane components (e.g., LPS), it is not identified as a PAMP. On the other hand, rhamnose and polysaccharides containing rhamnose residues were reported to exert many beneficial effects on the host. For instance, rhamnose suppressed tumor growth and prolonged the survival of tumor-bearing mice49,50. In addition, many natural polysaccharides containing rhamnose have antioxidant activity, which could alleviate metabolic disorders51. These beneficial effects are likely associated with the immunomodulatory properties of rhamnose. This functional sugar could suppress endotoxin-induced splenocyte proliferation52 and also stimulate the immune system in clearing the cancer cells53. To our knowledge, the role of rhamnose in bacterial clearance remains poorly understood. Our present work explicitly demonstrated rhamnose-induced enhancement in phagocytosis and bacterial killing by macrophages, causing reduction in the bacterial load both within the blood and peritoneal cavity.
Our study also evaluated the mechanisms involved in the rhamnose-mediated regulation of macrophage phagocytosis. Earlier it was speculated about the presence of a mammalian cell membrane protein that can directly interact with the rhamnose54. However, so far, such a molecule has not been identified. In this study, for the first time, we identified SLC12A4 as a potential receptor of rhamnose, and we used SPR and ELISA to confirm the direct binding of rhamnose to recombinant SLC12A4 protein. Functionally, when the expression of SLC12A4 was knocked down in macrophages by using siRNA, the beneficial effect of rhamnose on phagocytosis was found to have disappeared. Thus, gut-derived rhamnose acts through its potential “receptor” SLC12A4 to protect the host from polymicrobial sepsis-induced organ injury.
SLC12A4 belongs to the SLC12 family that was initially identified in fish and then in mammals55. Since members of this protein family have been found in humans and mice, their pathophysiological effects are increasingly better appreciated. This gene family includes two major branches, one with two bumetanide-sensitive Na+–K+–2Cl cotransporters and the other with thiazide-sensitive Na+:Cl– cotransporters, including SLC12A1, SLC12A2 and SLC12A3. The second branch encodes the electroneutral K+–Cl– cotransporters, including the four genes, SLC12A4–7. SLC12A4 is a ubiquitously expressed protein, known to play a key role in regulating the trans-epithelial transport, cell volume regulation, and intracellular chloride concentration56. However, the contribution of SLC12A4 to sepsis is still largely unknown. A recent study pointed out that SLC12A4 is associated with the process of apoptotic cell clearance14. However, investigations on the modulatory mechanisms of SLC12A4 and in the bacterial clearance leading to the resolution of infection, especially in sepsis, are scarce. Herein, knocking down of SLC12A4 expression in macrophages led to an impaired bacterial clearance, but the paradox of how SLC12A4 can affect the macrophage phagocytic capacity remains to be explored. Of note, the Rho GTPases Rac1 and Cdc42 were shown to be required for the uptake of apoptotic cell phagocytosis by macrophages57. Both Rac1 and Cdc42 are important for F-actin polymerization, a critical step in phagocytosis and the activation of Cdc42 and Rac1 is a pre-requisite for the initiation of FcγR-mediated phagocytosis10,58. Therefore, as a next step, we investigated whether rhamnose could promote Rac1/Cdc42 activity in macrophages after exposing them to E. coli and cause an increase in the level of phagocytosis. Interestingly, activating SLC12A4 could enhance while inhibiting the SLC12A4 expression could decrease the activity of Rac1/Cdc42, which suggests SLC12A4 could be a new target to develop a novel bacterial clearance strategy.
The immune responses during sepsis development are quite complicated; a variety of diverse immune cells participate, and also communicate with each other to contribute together in clearing pathogens, or leading to severe immune defects causing adverse events59. The protective effects of rhamnose may also be complex, although we showed that at the onset of sepsis, rhamnose mainly targets macrophages to promote bacterial clearance. Neutrophils also play a major role in bacterial clearance in sepsis progression60. We found that supplementation with rhamnose could also enhance the mouse bone marrow-derived neutrophils (BMDNs) phagocytosis and bacteria killing (Supporting Information Fig. S7A–S7C). The reasons for the effect of rhamnose on phagocytosis and bacteria killing observed in neutrophils may be complex, and the detailed mechanism should be investigated in the future. The effect of rhamnose on the interaction and cooperation between macrophages and neutrophils in the elimination of pathogens may be important for the protective effects observed in the animal study. Besides, how rhamnose affects adaptive immune cell functions during sepsis is still unknown. This knowledge gap is important to be filled and undoubtedly it requires further investigations.
5. Conclusions
Our findings demonstrate that a novel interaction occurs between gut commensals and host immune functions during sepsis. The depletion of gut microbiota leads to increased mortality in the CLP model. Gut-derived rhamnose is a monosaccharide with a potential pathophysiological function that could significantly enhance macrophage phagocytic activity and protect against sepsis-induced organ damage and death. More importantly, our study identified solute carrier family 12 member 4 (SLC12A4) as the potential rhamnose interacting protein in the macrophages, and we showed that SLC12A4 is indispensable for phagocytosis during infection. The schematic diagram (Fig. 7E) represents rhamnose/SLC12A4/Cdc42 and Rac1 axis present in macrophages that could plausibly be an important contributor in host defense against polymicrobial sepsis-induced bacterial dissemination and death. Therefore, rhamnose or other therapeutics targeting SLC12A4 could plausibly be translated to clinics for promoting macrophages phagocytosis during sepsis.
Acknowledgments
This study was supported by the National Key R&D Program of China (2022YFA0806400), and the National Natural Science Foundation of China (32071124, 32271230) to Peng Chen, and National Natural Science Foundation of China (82130063), Special Support Plan for Outstanding Talents of Guangdong Province (2019JC05Y340, China) to Yong Jiang.
Author contributions
Dongping Li, Rongjuan Wei, Xianglong Zhang, Shenhai Gong, Meijuan Wan, Fangzhao Wang, Meiling Chen, Yantong Wan, Yinghao Hong, Ruofan Liu and Peng Gu performed the experiments and analyzed the data; Zhang Wang participated in the bioinformatics analysis; Jiaxin Li and Zhenhua Zeng were responsible for clinical data analysis; Kutty Selva Nandakumar revised the manuscript; Yong Jiang, Hongwei Zhou and Peng Chen designed the study, interpreted the data, drafted and edit the manuscript, and supervised the study.
Conflicts of interest
The authors declare no conflicts of interest.
Data and code availability
The raw sequencing data from this study have been deposited in the Genome Sequence Archive (GSA) in BIG DATA Center (http://bigd.big.ac.cn/). All RNA-seq data for this experiment have been submitted to the GSA under accession number CRA008013. All of the 16S rDNAseq data were deposited in the GSA that under accession number CRA008012. All of the scRNAseq data were available from the GSA under accession number CRA008014. Other data are available from the corresponding author upon reasonable request.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2024.03.025.
Contributor Information
Hongwei Zhou, Email: biodegradation@gmail.com.
Peng Chen, Email: perchen@smu.edu.cn.
Appendix A. Supporting information
The following is the Supporting information to this article:
References
- 1.Deutschman C.S., Tracey K.J. Sepsis: current dogma and new perspectives. Immunity. 2014;40:463–475. doi: 10.1016/j.immuni.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Prescott H.C., Angus D.C. Enhancing recovery from sepsis: a review. JAMA. 2018;319:62–75. doi: 10.1001/jama.2017.17687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venet F., Monneret G. Advances in the understanding and treatment of sepsis-induced immunosuppression. Nat Rev Nephrol. 2018;14:121–137. doi: 10.1038/nrneph.2017.165. [DOI] [PubMed] [Google Scholar]
- 4.Akoumianaki T., Vaporidi K., Diamantaki E., Pene F., Beau R., Gresnigt M.S., et al. Uncoupling of IL-6 signaling and LC3-associated phagocytosis drives immunoparalysis during sepsis. Cell Host Microbe. 2021;29 doi: 10.1016/j.chom.2021.06.002. 1277–93.e6. [DOI] [PubMed] [Google Scholar]
- 5.Murray P.J., Wynn T.A. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serezani C.H., Aronoff D.M., Sitrin R.G., Peters-Golden M. FcγRI ligation leads to a complex with BLT1 in lipid rafts that enhances rat lung macrophage antimicrobial functions. Blood. 2009;114:3316–3324. doi: 10.1182/blood-2009-01-199919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L., Hong W., Zhu H., He Q., Yang B., Wang J., et al. Macrophage senescence in health and diseases. Acta Pharm Sin B. 2024;14:1508–1524. doi: 10.1016/j.apsb.2024.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park J.B. Phagocytosis induces superoxide formation and apoptosis in macrophages. Exp Mol Med. 2003;35:325–335. doi: 10.1038/emm.2003.44. [DOI] [PubMed] [Google Scholar]
- 9.Swanson J.A. Shaping cups into phagosomes and macropinosomes. Nat Rev Mol Cell Biol. 2008;9:639–649. doi: 10.1038/nrm2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J., Guo J., Dzhagalov I., He Y.W. An essential function for the calcium-promoted Ras inactivator in Fcγ receptor-mediated phagocytosis. Nat Immunol. 2005;6:911–919. doi: 10.1038/ni1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S., Chang S., Han B., Xu L., Zhang M., Zhao C., et al. Cryo-EM structures of the human cation-chloride cotransporter KCC1. Science. 2019;366:505–508. doi: 10.1126/science.aay3129. [DOI] [PubMed] [Google Scholar]
- 12.Garneau A.P., Slimani S., Tremblay L.E., Fiola M.J., Marcoux A.A., Isenring P. K+–Cl– cotransporter 1 (KCC1): a housekeeping membrane protein that plays key supplemental roles in hematopoietic and cancer cells. J Hematol Oncol. 2019;12:74. doi: 10.1186/s13045-019-0766-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Y., Chang S., Zhao C., Wang F., Liu S., Wang J., et al. Structures and an activation mechanism of human potassium–chloride cotransporters. Sci Adv. 2020;6 doi: 10.1126/sciadv.abc5883. eabc5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perry J.S.A., Morioka S., Medina C.B., Iker Etchegaray J., Barron B., Raymond M.H., et al. Interpreting an apoptotic corpse as anti-inflammatory involves a chloride sensing pathway. Nat Cell Biol. 2019;21:1532–1543. doi: 10.1038/s41556-019-0431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haak B.W., Wiersinga W.J. The role of the gut microbiota in sepsis. Lancet Gastroenterol Hepatol. 2017;2:135–143. doi: 10.1016/S2468-1253(16)30119-4. [DOI] [PubMed] [Google Scholar]
- 16.Ho J., Chan H., Liang Y., Liu X., Zhang L., Li Q., et al. Cathelicidin preserves intestinal barrier function in polymicrobial sepsis. Crit Care. 2020;24:47. doi: 10.1186/s13054-020-2754-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z., Li N., Fang H., Chen X., Guo Y., Gong S., et al. Enteric dysbiosis is associated with sepsis in patients. FASEB J. 2019;33:12299–12310. doi: 10.1096/fj.201900398RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rooks M.G., Garrett W.S. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J., d'Aigle J., Atadja L., Quaicoe V., Honarpisheh P., Ganesh B.P., et al. Gut microbiota-derived short-chain fatty acids promote poststroke recovery in aged mice. Circ Res. 2020;127:453–465. doi: 10.1161/CIRCRESAHA.119.316448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan L.L., Ren Z.N., Yang J., Li B.B., Huang Y.W., Song D.X., et al. Gut microbiota controls the development of chronic pancreatitis: a critical role of short-chain fatty acids-producing Gram-positive bacteria. Acta Pharm Sin B. 2023;13:4202–4216. doi: 10.1016/j.apsb.2023.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong S., Yan Z., Liu Z., Niu M., Fang H., Li N., et al. Intestinal microbiota mediates the susceptibility to polymicrobial sepsis-induced liver injury by granisetron generation in mice. Hepatology. 2019;69:1751–1767. doi: 10.1002/hep.30361. [DOI] [PubMed] [Google Scholar]
- 22.Siempos I.I., Lam H.C., Ding Y., Choi M.E., Choi A.M., Ryter S.W. Cecal ligation and puncture-induced sepsis as a model to study autophagy in mice. J Vis Exp. 2014 doi: 10.3791/51066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young M.D., Behjati S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience. 2020;9 doi: 10.1093/gigascience/giaa151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8 doi: 10.1016/j.cels.2019.03.003. 329–37.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184 doi: 10.1016/j.cell.2021.04.048. 3573–87.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu G. Using meshes for MeSH term enrichment and semantic analyses. Bioinformatics. 2018;34:3766–3767. doi: 10.1093/bioinformatics/bty410. [DOI] [PubMed] [Google Scholar]
- 31.Korotkevich G., Sukhov V., Budin N., Shpak B., Artyomov M.N., Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2016 doi: 10.1101/060012. Available from: [DOI] [Google Scholar]
- 32.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geng J., Sun X., Wang P., Zhang S., Wang X., Wu H., et al. Kinases Mst1 and Mst2 positively regulate phagocytic induction of reactive oxygen species and bactericidal activity. Nat Immunol. 2015;16:1142–1152. doi: 10.1038/ni.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Q., Chen T., Xu Y., Zhu J., Jiang Y., Zhao Y., et al. Steroid receptor coactivator 3 is required for clearing bacteria and repressing inflammatory response in Escherichia coli-induced septic peritonitis. J Immunol. 2010;185:5444–5452. doi: 10.4049/jimmunol.0903802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuijt T.J., van der Poll T., de Vos W.M., Wiersinga W.J. The intestinal microbiota and host immune interactions in the critically ill. Trends Microbiol. 2013;21:221–229. doi: 10.1016/j.tim.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Doi K., Hu X., Yuen P.S., Leelahavanichkul A., Yasuda H., Kim S.M., et al. AP214, an analogue of α-melanocyte-stimulating hormone, ameliorates sepsis-induced acute kidney injury and mortality. Kidney Int. 2008;73:1266–1274. doi: 10.1038/ki.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du J., Zhang P., Luo J., Shen L., Zhang S., Gu H., et al. Dietary betaine prevents obesity through gut microbiota-drived microRNA-378a family. Gut Microb. 2021;13:1–19. doi: 10.1080/19490976.2020.1862612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price J.V., Vance R.E. The macrophage paradox. Immunity. 2014;41:685–693. doi: 10.1016/j.immuni.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 39.Huang X., Venet F., Wang Y.L., Lepape A., Yuan Z., Chen Y., et al. PD-1 expression by macrophages plays a pathologic role in altering microbial clearance and the innate inflammatory response to sepsis. Proc Natl Acad Sci U S A. 2009;106:6303–6308. doi: 10.1073/pnas.0809422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X., Lan Y., Xu J., Quan F., Zhao E., Deng C., et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47:D721–D728. doi: 10.1093/nar/gky900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Summers K.M., Bush S.J., Hume D.A. Network analysis of transcriptomic diversity amongst resident tissue macrophages and dendritic cells in the mouse mononuclear phagocyte system. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H., Bradbury J.A., Edin M.L., Graves J.P., Gruzdev A., Cheng J., et al. sEH promotes macrophage phagocytosis and lung clearance of Streptococcus pneumoniae. J Clin Invest. 2021;131 doi: 10.1172/JCI129679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuijt T.J., Lankelma J.M., Scicluna B.P., de Sousa e Melo F., Roelofs J.J., de Boer J.D., et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. 2016;65:575–583. doi: 10.1136/gutjnl-2015-309728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng Z., Surewaard B.G., Wong C.H., Geoghegan J.A., Jenne C.N., Kubes P. CRIg functions as a macrophage pattern recognition receptor to directly bind and capture blood–borne Gram-positive bacteria. Cell Host Microbe. 2016;20:99–106. doi: 10.1016/j.chom.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 45.McDonald B., Zucoloto A.Z., Yu I.L., Burkhard R., Brown K., Geuking M.B., et al. Programing of an intravascular immune firewall by the gut microbiota protects against pathogen dissemination during infection. Cell Host Microbe. 2020;28 doi: 10.1016/j.chom.2020.07.014. 660–8.e4. [DOI] [PubMed] [Google Scholar]
- 46.Wilmore J.R., Gaudette B.T., Gomez Atria D., Hashemi T., Jones D.D., Gardner C.A., et al. Commensal microbes induce serum IgA responses that protect against polymicrobial sepsis. Cell Host Microbe. 2018;23 doi: 10.1016/j.chom.2018.01.005. 302–11.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huus K.E., Petersen C., Finlay B.B. Diversity and dynamism of IgA-microbiota interactions. Nat Rev Immunol. 2021;21:514–525. doi: 10.1038/s41577-021-00506-1. [DOI] [PubMed] [Google Scholar]
- 48.Reinhardt A., Johnsen U., Schonheit P. l-Rhamnose catabolism in archaea. Mol Microbiol. 2019;111:1093–1108. doi: 10.1111/mmi.14213. [DOI] [PubMed] [Google Scholar]
- 49.Chen W., Gu L., Zhang W., Motari E., Cai L., Styslinger T.J., et al. l-Rhamnose antigen: a promising alternative to α-gal for cancer immunotherapies. ACS Chem Biol. 2011;6:185–191. doi: 10.1021/cb100318z. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H., Wang B., Ma Z., Wei M., Liu J., Li D., et al. l-Rhamnose enhances the immunogenicity of melanoma-associated antigen A3 for stimulating antitumor immune responses. Bioconjugate Chem. 2016;27:1112–1118. doi: 10.1021/acs.bioconjchem.6b00081. [DOI] [PubMed] [Google Scholar]
- 51.Seedevi P. Antioxidant and anticoagulant activity of crude polysaccharide and α-l-rhamnose from Grateloupia lithophila. Biomass Conversion and Biorefinery. 2022 doi: 10.1007/s13399-022-03708-2. Available from: [DOI] [Google Scholar]
- 52.Biswas T., Roy S., Banerjee K.K. l-Rhamnose inhibits proliferation of murine splenocytes by the lipopolysaccharide and polysaccharide moiety of Shigella dysenteriae type 1 lipopolysaccharide. Immunology. 1995;84:322–325. [PMC free article] [PubMed] [Google Scholar]
- 53.Sheridan R.T., Hudon J., Hank J.A., Sondel P.M., Kiessling L.L. Rhamnose glycoconjugates for the recruitment of endogenous anti-carbohydrate antibodies to tumor cells. Chembiochem. 2014;15:1393–1398. doi: 10.1002/cbic.201402019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y., Gao J., Gu G., Li G., Cui C., Sun B., et al. In situ RBL receptor visualization and its mediated anticancer activity for solasodine rhamnosides. Chembiochem. 2011;12:2418–2420. doi: 10.1002/cbic.201100551. [DOI] [PubMed] [Google Scholar]
- 55.Hebert S.C., Mount D.B., Gamba G. Molecular physiology of cation-coupled Cl– cotransport: the SLC12 family. Pflügers Archiv. 2004;447:580–593. doi: 10.1007/s00424-003-1066-3. [DOI] [PubMed] [Google Scholar]
- 56.Zhao Y., Shen J., Wang Q., Ruiz Munevar M.J., Vidossich P., De Vivo M., et al. Structure of the human cation-chloride cotransport KCC1 in an outward-open state. Proc Natl Acad Sci U S A. 2022;119 doi: 10.1073/pnas.2109083119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leverrier Y., Ridley A.J. Requirement for Rho GTPases and PI 3-kinases during apoptotic cell phagocytosis by macrophages. Curr Biol. 2001;11:195–199. doi: 10.1016/s0960-9822(01)00047-1. [DOI] [PubMed] [Google Scholar]
- 58.Greenberg S. Modular components of phagocytosis. J Leukoc Biol. 1999;66:712–717. doi: 10.1002/jlb.66.5.712. [DOI] [PubMed] [Google Scholar]
- 59.Cinel I., Opal S.M. Molecular biology of inflammation and sepsis: a primer. Crit Care Med. 2009;37:291–304. doi: 10.1097/CCM.0b013e31819267fb. [DOI] [PubMed] [Google Scholar]
- 60.Mantovani A., Cassatella M.A., Costantini C., Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data from this study have been deposited in the Genome Sequence Archive (GSA) in BIG DATA Center (http://bigd.big.ac.cn/). All RNA-seq data for this experiment have been submitted to the GSA under accession number CRA008013. All of the 16S rDNAseq data were deposited in the GSA that under accession number CRA008012. All of the scRNAseq data were available from the GSA under accession number CRA008014. Other data are available from the corresponding author upon reasonable request.