Abstract
Hydroxylation of prolines to 4-trans-hydroxyproline (Hyp) is mediated by prolyl-4 hydroxylases (P4Hs). In plants, Hyps occur in Hydroxyproline-rich glycoproteins (HRGPs), and are frequently O-glycosylated. While both modifications are important, e.g. for cell wall stability, they are undesired in plant-made pharmaceuticals. Sequence motifs for prolyl-hydroxylation were proposed but did not include data from mosses, such as Physcomitrella. We identified six moss P4Hs by phylogenetic reconstruction. Our analysis of 73 Hyps in 24 secretory proteins from multiple mass spectrometry datasets revealed that prolines near other prolines, alanine, serine, threonine and valine were preferentially hydroxylated. About 95 % of Hyps were predictable with combined established methods. In our data, AOV was the most frequent pattern. A combination of 443 AlphaFold models and MS data with 3000 prolines found Hyps mainly on protein surfaces in disordered regions. Moss-produced human erythropoietin (EPO) exhibited O-glycosylation with arabinose chains on two Hyps. This modification was significantly reduced in a p4h1 knock-out (KO) Physcomitrella mutant. Quantitative proteomics with different p4h mutants revealed specific changes in protein amounts, and a modified prolyl-hydroxylation pattern, suggesting a differential function of the Physcomitrella P4Hs. Quantitative RT-PCR revealed a differential effect of single p4h KOs on the expression of the other five p4h genes, suggesting a partial compensation of the mutation. AlphaFold-Multimer models for Physcomitrella P4H1 and its target EPO peptide superposed with the crystal structure of Chlamydomonas P4H1 suggested significant amino acids in the active centre of the enzyme and revealed differences between P4H1 and the other Physcomitrella P4Hs.
Keywords: Biopharmaceutical, Erythropoietin, O-glycosylation, Plant-made pharmaceutical, Posttranslational protein modification, Prolyl-hydroxylation
Graphical Abstract
Highlights
-
•
Mosses, such as Physcomitrella, encode multiple prolyl 4-hydroxylases in four subfamilies.
-
•
Computational analysis reveals a specific environment for proline hydroxylation.
-
•
AlphaFold structure models and mass spectrometry reveal features of modification.
-
•
Quantitative proteomics and mutant analysis identify relevant modifying enzyme.
-
•
Modelling suggests relevant amino acids in active centre of the modifying enzyme.
1. Introduction
Hydroxyproline-rich glycoproteins (HRGPs) are structural proteins of the plant cell wall with functions in growth, stress response, signalling and reproductive development [11], [21], [24], [34]. HRGPs possess signal peptides that mediate their entry into the endoplasmic reticulum (ER). In the ER and Golgi apparatus, proline residues of HRGPs are frequently hydroxylated by prolyl-4 hydroxylases (P4Hs) to 4-trans hydroxyproline (Hyp) that can serve as an anchor for the attachment of O-glycans. In a final step HRGPs are secreted to the apoplast [100], [72] where they cross-link with other elements of the cell wall [30].
One class of HRGPs are extensins that contain characteristic hydrophilic Ser-(Pro)n≥2 motifs in which the prolines are hydroxylated and O-glycosylated with linear chains of one to five arabinoses [39], [50]. O-glycosylated Hyps are associated with an increased stability of polyproline-II helical conformations as they exist in extensins [115], [69], [80]. Extensins undergo cross-linking mediated by hydrophobic tyrosine-rich motifs that depend on the presence of arabinosylated Hyps in vitro [12].
Another class of HRGPs are arabinogalactan proteins (AGPs) that carry large, branched, and complex glycans with variable structures which constitute up to 90 % of the glycoprotein mass. Glycans on AGPs consist mainly of galactose and arabinose, but also rhamnose, fucose and glucuronic acid [100]. Characteristic for AGPs is a high proportion of the amino acids (AAs) proline, alanine, serine and threonine (PAST), that facilitate the bioinformatic identification of AGPs using thresholds with PAST contents of 50 % and above [47], [65], [95], [97]. Target sites of O-glycosylation on AGPs are repeats of Ala-Hyp, Ser-Hyp, Thr-Hyp and Val-Hyp that, when present in synthetic peptides, showed varying degrees of prolyl-hydroxylation and O-glycosylation depending on the neighbouring AAs [99], [104]. Among other functions [27], AGPs are important for expansion of Physcomitrella protonemata [59], the tissue used for molecular farming.
Based on studies on prolyl-hydroxylation motifs in HRGPs [99], [98], Gomord et al. [31] proposed a motif for prolyl-hydroxylation and subsequent O-glycosylation, the [Ala ⁄ Ser ⁄ Thr ⁄ Val]-Pro(1,4)-X(0,10)-[Ala ⁄ Ser ⁄ Thr ⁄ Val]-Pro(1,4) glycomodule, where X can be any AA. According to the Hyp contiguity hypothesis mostly arabinose chains are attached to blocks with neighbouring Hyps, probably due to space constraints, while arabinogalactans are added more often to single non-contiguous Hyps [50]. Subsequently, Canut et al. [9] proposed an extended prolyl-hydroxylation code including data from 25 plant species taking into consideration the neighbouring AAs of Hyps. However, several exceptions from these rules are known. For example, in a Lolium multiflorum AGP only the first proline in the motif Ser-Pro-Pro-Ala was hydroxylated even though both prolines should be hydroxylated according to the extended prolyl-hydroxylation code [9]. More recently, an R package for analysis of HRGPs was developed including a function for the prediction of Hyps in plant proteins [25]. It is based on a machine-learning algorithm that was trained on plant protein sequences from UniProt, some of them containing experimentally validated Hyps.
Usually, plants have a set of P4H isoforms with diverging substrate preferences. For example, Arabidopsis thaliana (Arabidopsis) P4H1 hydroxylates mostly the second proline in the Pro-Pro-Gly motif from collagen, which is a substrate for mammalian P4Hs [33], whereas hydroxylation of such peptides is inefficient for P4H2 [106], [38]. Comparably, in Nicotiana benthamiana (Nicotiana) three of four examined P4Hs were able to hydroxylate a proline in an erythropoietin (EPO) peptide [73]. The same proline is also prolyl-hydroxylated in recombinant EPO from Physcomitrella [122], specifically by P4H1 [83].
Prolyl-hydroxylation and subsequent O-glycosylation on recombinant proteins can deteriorate plant-made pharmaceuticals (PMPs) because mammals may show an immune response against the arabinose residues of plant-specific O-glycans [2]. Knowledge of preferred prolyl-hydroxylation motifs of the different plant P4H isoforms would help to solve the issue of undesired prolyl-hydroxylation and Hyp-anchored O-glycosylation on PMPs. The production of PMPs relies on vascular plants, such as Nicotiana, and on the moss Physcomitrella. Therefore, attempts are made to eliminate single P4Hs [112], [83] without harming plant performance. While the general N-glycosylation pattern is similar between the two [18], [53], there are subtle differences [102], suggesting that the existing prediction tools for prolyl-hydroxylation and O-glycosylation might not be sufficient for the prediction in Physcomitrella.
Here, we studied prolyl-hydroxylation patterns in the model moss Physcomitrella (Physcomitrium patens; [63]) which is an attractive production system for PMPs [20], [90] and combines advantages such as high gene-targeting rates via homologous recombination, facilitating precise genome engineering [13], [41], [89], a fully sequenced genome, and a published secretome [40], [58], [57]. Our results provide a better understanding of moss prolyl-hydroxylation and may help to avoid undesired prolyl-hydroxylation and Hyp-anchored O-glycosylation of plant-made pharmaceuticals.
2. Methods
2.1. Phylogenetic reconstruction
With the v3.3 Physcomitrella P4H protein sequences as queries, BLASTp-like searches were performed with DIAMOND (version 2.1.8; [8]) in ‘ultra-sensitive’ mode against the protein sequences of 18 selected plant species: Anthoceros angustus [125], Arabidopsis thaliana [14], Calohypnum plumiforme [66], Ceratodon purpureus [10], Ceratopteris richardii [67], Funaria hygrometrica [51], Klebsormidium nitens [42], Marchantia polymorpha [7], Mesotaenium endlicherianum [15], Nicotiana benthamiana [87], Oryza sativa [79], Populus trichocarpa [111], Selaginella moellendorffii [3], Sphagnum fallax [36], Sphagnum magellanicum [36], Spirogloea muscicola [15], Takakia lepidozioides [43] and Vitis vinifera [45]. Initial hits were searched for signatures of the ‘2OG-Fe(II) oxygenase superfamily’ from Pfam (PF13640), the ’PROLYL 4-HYDROXYLASE ALPHA SUBUNIT’ from PANTHER (PTHR10869) and the ‘Fe(2 +) 2-oxoglutarate dioxygenase domain profile’ from prosite (PS51471) with InterProScan (version 5.66–98.0; [48]). Only sequences with all three signatures were kept and complemented by the P4Hs of Homo sapiens (UniProt P13674, O15460, Q7Z4N8, Q9NXG6) and Mus musculus (UniProt Q60715, Q60716, Q6W3F0, Q8BG58) as an outgroup for phylogenetic reconstruction. Several iterations of multiple sequence alignments and maximum-likelihood tree inferences were performed using MAFFT (version 7.149b; [49]) and FastTree (version 2.1.11; [85]), to filter out non-homologous sequences. The multiple sequence alignment of the resulting protein sequences was performed with UPP/SEPP (version 4.5.2; [77]) and converted into a codon-aware alignment of associated coding sequences with PAL2NAL (version 14; [103]). A maximum-likelihood tree was calculated with RAxML-NG (version 1.2.1; [55]) using the ‘GTR+G’ model and 1000 bootstrap replicates, outgroup-rooted and visualized using R (version 4.3.2; R [86]) and the ggtree package (version 3.10.0; [124]).
2.2. Mass spectrometry data sets
Hyps were selected from a collection of MS/MS measurements using samples with an enrichment of secretory Physcomitrella proteins from ER, Golgi apparatus, cell wall and extracellular space. The datasets contained: measurements of an EPO-producing line (174.16), quantitative MS data from mixtures of EPO-producing maternal line (174.16; labelled with 15N as described in [74]) with single KO lines of p4h1 – p4h6, respectively, separate secretome analysis of EPO-producing maternal line (174.16) and p4h1 KO line [109], new Mascot searches of the host cell proteome [40] using Hyp with zero to three arabinoses as variable modifications, Mascot search with no protease specificity from chemically deglycosylated protein samples digested with thermolysin, elastase or trypsin, and double KO of two galactosyltransferase genes (Δgalt2/3; [82]).
2.3. Cultivation and isotopic labelling
Physcomitrella plants were grown in axenic suspension in 500 mL flasks containing 180 mL of Knop ME medium [26] in stable conditions of 22 °C and a light cycle of 16/8 h at an intensity of 55 µmol/m2 s. The flasks were kept on a rotary shaker at 125 rpm and held on protonema stage by dispersion with an Ultra-Turrax (IKA, Staufen, Germany) at 18,000 rpm for 1 min every two weeks. After dispersion, protonemata were harvested with a 100 µm sieve and transferred to a 500 mL flask containing fresh Knop ME medium using sterile tweezers. After turraxation, sterility controls were taken directly from the Ultra-Turrax tip for each cell line to check for potential contamination according to the protocol from Heck et al. [37]. For the quantitative approach in the MS analysis, the maternal line was inoculated in a small amount of 5–10 gametophores in medium containing the heavy 15N isotope for at least 6 weeks with weekly turraxation.
For sample preparation, protonema suspensions were cultivated for one week in 400 mL BM medium [82] in a 1 L aerated round flask (modified after [56]). The supernatant of each flask was precipitated with 10 % TCA overnight on ice in a cold room (6 °C). Protein pellets were obtained by centrifugation at 15,000 x g and 2 °C for 2 h. Pellets were resuspended in ice-cold acetone, transferred into 50 mL tubes and centrifuged at 5000 x g, 4 °C for 15 min each time. Pellets were again resuspended in ice-cold acetone, transferred to 1.5 mL tubes and centrifuged at 14,000 x g, 4 °C for 15 min. Supernatants were discarded and pellets air-dried.
Protonemata were filtered using a 100 µm sieve and resuspended in extraction buffer (408 mM NaCl, 60 mM Na2HPO4 x 2 H2O, 10.56 mM KH2PO4, 60 mM EDTA, 1 % plant protease inhibitor cocktail (Sigma P9599), pH7.4, using 3 mL buffer / g protonema (fresh weight). The sample was then homogenized using an Ultra-Turrax for 10 min at 10,000 rpm on ice and subsequently centrifuged at 4500 x g for 3 min at 4 °C. The supernatant was transferred to a new tube and again centrifuged at 4500 x g for 5 min at 4 °C. The supernatant was precipitated with methanol/chloroform as described in Lang et al. [56].
2.4. Chemical deglycosylation
Chemical deglycosylation was achieved with the GlycoProfile™ Kit (Sigma Aldrich, PP0510) using 1 mg total protein, which was harvested from supernatant fractions as described above. All steps were performed as recommended by the manufacturer using the scavenging procedure. Subsequently, samples were precipitated with acetone according to Hoernstein et al. [40].
2.5. In-solution digest of protein samples
In-solution digest of proteins for MS analysis was modified after Reimann et al. [88]. In brief, one pellet each of precipitated total protein was dissolved in 100 µL 8 M urea and 50 mM ammonium bicarbonate (AmBic, Witney, UK). Protein concentration was determined either with a BCA assay (Pierce™) or via absorbance measurement at 280 nm (A280). In the latter case, values were adjusted using an empirical correction factor according to Pace et al. [81]. An amount of 20 µg protein at a concentration of 1 µg/µL was employed for trypsin digestion. Heavy labelled (15N) and corresponding light labelled (14N) protein samples were mixed 1:1. Prior to digestion, samples were reduced and alkylated at a final concentration of 5 mM TCEP (37 °C, 30 min) and 50 mM iodoacetamide (RT, 30 min in darkness). The reaction was quenched at a final concentration of 20 mM DTT and the sample solution was diluted with 50 mM AmBic to reach an urea concentration of 2 M. Trypsin (V5117; Promega, Walldorf, Germany) was added at a ratio of 1:50 and the digestion was performed over night at 37 °C. The chemically deglycosylated samples were digested with thermolysin at a ratio of 1:50 for 60 min at 45 °C or with trypsin or elastase at 37 °C respectively. Peptides were purified via C18 STAGE-Tips as described by Hoernstein et al. [40] and eluted in 30 % ACN in 0.1 % FA.
2.6. Mass spectrometry and data analysis
MS analysis of samples from the in-solution digests of the chemically deglycosylated samples was performed according to Top et al. [108]. Samples of the metabolically labelled samples were measured in the same way but using a 3 h gradient. Raw data were processed with Mascot Distiller 2.8.3.0 (Matrix Science, London, UK) and database searches were performed with Mascot Server (version 2.7.0). Processed peak lists were searched against a database containing all Physcomitrella protein models v3 [57] and the sequence of human erythropoietin (EPO; P01588). “15 N Metabolic [MD]” was specified as quantitation option and carbamidomethylation (C + 57.021464 Da) was specified as fixed modification. Variable modifications were Gln → pyro Glu (N term Q − 17.026549 Da), oxidation (M + 15.994915 Da), hydroxyproline (P + 15.994915 Da), mono-arabinosylation (P + 148.037173 Da), di-arabinosylation (P + 280.079432 Da) and tri-arabinosylation (P + 412.121691 Da), deamidation (N + 0.984016 Da) and glycosylation (S + 162.052823 Da).
2.7. MS sample preparation
Identification of hydroxyproline sites or arabinosylated hydroxyproline (Hyp) residues was performed on human EPO recombinantly produced in Physcomitrella. In brief, cell culture supernatant of an EPO producing line (174.16; IMSC 40216, www.moss-stock-center.org) [122], [82] was TCA-precipitated and dried protein pellets were subjected to SDS PAGE as described in Top et al. [108]. The band of EPO was excised and digested simultaneously with trypsin (Promega) and GluC (Thermo Fisher Scientific, Bremen, Germany). Peptides were cleaned as described in Top et al. [108]. MS measurements were performed on RSLCnano system (Dionex LC Packings/Thermo Fisher Scientific, Dreieich, Germany) coupled online to a QExactive Plus instrument (Thermo Fisher Scientific) as described in Top et al. [108] using 35 % normalized collision energy.
Raw data were processed with Mascot Distiller V2.5.1.0 and database searches on generated peak lists were performed using Mascot Server v2.6.2 and a database containing all P. patens protein models v1.6 [126], the sequence of human EPO (P01588) as well as their reversed sequences used as decoys. Simultaneously, the search was performed against a custom in-house database containing sequences of known MS contaminations such as human keratin or trypsin (267 total entries, available on request). Carbamidomethylation (C + 57.021464 Da) was set as fixed modification. Variable modifications were Gln → pyro Glu (N term Q − 17.026549 Da), oxidation (M + 15.994915 Da), acetylation (N-term + 42.010565 Da), hydroxyproline (P + 15.994915 Da), mono-arabinosylation (P + 148.037173 Da), di-arabinosylation (P + 280.079432 Da) and tri-arabinosylation (P + 412.121691 Da). The peptide mass tolerance was ± 5 ppm, and the fragment mass tolerance was ± 0.02 Da. Enzyme specificity was set to “none” and a maximum of two missed cleavages was allowed. Results were loaded in Scaffold4 software v4.11.0 using the Legacy Independent Sample Grouping Option and Legacy PeptideProphet Scoring (high mass accuracy).
2.8. Identifying secretory proteins and HRGPs
The presence of signal peptides in Physcomitrella proteins was predicted with SignalP 5 [1] via the library ragp (version 0.3.5.9000; [25]) in R (version 4.3.0; R [86]). Proteins identified by MS were filtered for the presence of a predicted signal peptide and only those proteins were further considered. HRGP classes were assigned based on Liu et al. [62] and Ma et al. [64]. Physcomitrella v1.6 IDs as used by Liu et al. [62] were translated into Physcomitrella v3.3 IDs via the PpGML DB [29] and proteins were considered as chimeric AGP if the Physcomitrella v3.3 protein sequence was identical to that of a predicted chimeric AGP from Ma et al. [64] or if the complete sequence of a predicted chimeric AGP was part of the v3.3 sequence of the measured protein.
2.9. Selecting Hyp sites and non-hydroxylated proline sites
Database search results were filtered in Scaffold 4 (or Scaffold 5 in the case of the p4h1 KO – p4h6 KO dataset; Proteome Software Inc., Portland, USA) for a protein and peptide probability > 90 % and a minimum number of unique peptides per protein of 1. Only peptides with a Mascot Ion Score > 25 were accepted. Proteins in the results from Mascot searches performed with a database containing Physcomitrella v1.6 protein sequences were translated to the major isoform of Physcomitrella v3.3 proteins using the P. patens lookup table downloaded from the PpGML DB [29]. If a protein v1.6 ID translated to two v3.3 IDs both were kept and proteins without a v3.3 counterpart were removed. Similarly, peptides that did not fit into the translated v3.3 protein sequence were removed. Translation from v1.6 to v3.3 was performed for the following datasets: host cell proteome [40], measurements of EPO-producing line (174.16), separate measurements of maternal line (174.16) and p4h1 KO line. For all other datasets the Mascot searches were directly performed with a database containing Physcomitrella v3.3 protein sequences.
Results in mzIdentML format were exported from Scaffold and converted to pepXML format with help of the OpenMS software (version 2.7.0; [91]). The probability for the localization of the hydroxylation (+15.99) at a specific proline compared to other prolines as well as methionine and tryptophan that can be hydroxylated as artefacts during electrospray ionization [101] was computed with PTMProphet from the Trans-Proteomic Pipeline (version 5.2.0; [23]). Spectra in which the hydroxylation of proline could not be distinguished from hydroxylation of carbamidomethylated cysteine, which was described to become hydroxylated in Na et al. [76], as well as histidine, tyrosine and phenylalanine, which are susceptible to oxidation [4], were not considered further. For peptides that fitted twice in the same protein, the first match was chosen as the Hyp site. If peptides were assigned to two different but very similar proteins, one representative protein was selected.
Prolyl-hydroxylation as a modification was accepted at a PTMProphet probability > 0.7 at the specific proline. Moreover, a set of prolines that were not measured to be hydroxylated was selected from the MS data of lines without p4h KO, including proline sites with a very low probability of hydroxylation (PTMProphet probability < 0.01).
2.10. Computing the degree of prolyl-hydroxylation
The degree of prolyl-hydroxylation of Hyp-containing peptides was estimated from peptide intensity values of the maternal line in the quantified MS data (data from p4h KO lines was not included). The sum of intensities from all forms of a peptide with prolyl-hydroxylation (i.e. all charges and all combinations of post-translational modifications) were divided by the sum of intensities of the peptide (prolyl-hydroxylated and not prolyl-hydroxylated version) and the mean over three technical replicates from two datasets was taken.
2.11. Hyp sequence environment
The logo showing the AA frequency in sequence windows of length 15 centred around Hyps was created with the standalone version of WebLogo (version 3.7.9; [17]) setting probability as the unit and none for composition. The two-sample logo was created with the Two Sample Logo web-based application [114] using sequence windows of length 15 centred around Hyps as positive samples and sequence windows centred around the selected set of prolines that were not measured to be hydroxylated as negative samples. The P value cut-off was set to the default value of 0.05. In the WebLogo and the two-sample logo the P of the central proline was replaced with an O using GIMP (version 2.10.18).
2.12. Predicting Hyp sites
The prediction of Hyp sites was performed for all isoforms of all secretory Physcomitrella proteins with a predicted signal peptide excluding proteins encoded by plastids or mitochondria. Identification of prolines hydroxylated according to the glycomodule and the extended prolyl-hydroxylation code was done with a Python script. Further, Hyp sites were predicted with the library ragp (version 0.3.5.9000; [25]) in R (version 4.3.0; R [86]) using the default probability threshold of 0.224.
2.13. Structural analysis
Models of proteins containing validated Hyps or prolines from the selected set of non-hydroxylated prolines were downloaded from the AlphaFold Protein Structure Database [117]. The relative accessible surface area as well as the seven secondary structure elements (3–10 helix, α-helix, π-helix, strand (participates in β ladder), isolated β-bridge, turn (hydrogen bonded), bend) or none of the previous were assigned to each residue with the DSSP module from Biopython (version 1.80; [16]).
Models of the six Physcomitrella P4Hs with the bound EPO peptide EAISPPDAASAAPLR were generated with ColabFold (version 1.3.0; [71]). The models were built with AlphaFold2-multimer-v3 [28] using no template information. The program was run with default settings and the top-ranking model of P4H1 was relaxed with molecular dynamics. Additionally, models of the six Physcomitrella P4Hs with the bound EPO peptide were generated with AlphaFold2-multimer-v2 [28] using no template information and 48 recycles. All further visualizations and analyses were performed in PyMOL (The PyMOL Molecular Graphics System, version 2.3.0, Schrödinger, LLC): the identification of hydrogen bonds between P4H1 and the substrate peptide, the alignment with the crystal structure from the Chlamydomonas reinhardtii P4H1 with a bound peptide substrate downloaded from the PDB (https://www.rcsb.org/; PDB ID: 3GZE chain C; [54]) and the computation of the root-mean-square deviation (RMSD) between the two peptide substrates. The latter was computed with the rms_cur command considering the five Cα atoms in the range of two AAs around the proline that becomes hydroxylated.
2.14. Protein and peptide abundance
Significant changes in the abundance of Hyp-containing peptides in p4h KO lines (light) compared to the 15N labelled maternal line (heavy) were computed with the light/heavy (L/H) intensity ratios for the p4h1 – p4h6 single KO dataset. The L/H ratios of each replicate were log2 transformed and normalized to a median of zero by subtraction of the median. For protein-level analysis the normalization was performed using the median value of the peptides in the replicate. For peptide-level analysis the normalization was performed with the median value of the respective protein. A one sample t-test was performed with an expected value of zero using SciPy (version 1.10.0; [120]) and peptides with P < 0.05 were accepted. Further, the mean of the normalized log2 transformed L/H ratios of the three replicates was computed and only peptides with a reduced abundance where this value was < 1 were kept. If there was a reduction in the abundance of the unmodified peptide that was comparable to the reduction in the abundance of the Hyp-containing peptide, this peptide was not further considered.
For computation of significant changes in the abundance of proteins in p4h KO lines, the same procedure to compute the t-test was applied as for the peptides but with the protein L/H intensity ratios and only proteins present in all three replicates were considered. Afterwards, multiple testing correction was performed using the Benjamini/Hochberg method via the Python module statsmodels (version 0.13.2; [96]). Proteins were filtered for an adjusted P value < 0.05 and |mean normalized log2 L/H ratio| > 1. Finally, only proteins that had a probability for correct protein identification > 90 % in Scaffold 5 (Proteome Software) were selected. Proteins for which the direction of the change in abundance was opposite in the two datasets used for this analysis were removed.
2.15. Transcript abundance
In order to quantify the expression levels of p4h genes, RNA was isolated from 100 mg plant material of wild type and the KO lines, respectively. The RNA was first digested with DNAse I. After incubation for 1 h at 37 °C, the reaction was stopped by addition of 2 µL EDTA (25 mM) and incubation at 65 °C for 10 min. After DNAse I digest, the RNA was reverse-transcribed with TaqMan® Reverse Transcription Kit, using Multiscribe RT. RT-PCR was performed with appropriate primers (efficiency =2 ± 0.1; calculated by the Abs Quant\2nd Derivate max). The qRT-PCR amplification was performed with SensiMix™ SYBR NO-Rox Kit (Bioline) according to the manufacturer’s recommendations. Gene expression was normalized against the housekeeping genes EF1α (Pp3c2_10310V3.1) and L21 (Pp3c13_2360V3.1) [5], [123] and the relative quantification was calculated based on Advanced Relative Quantification provided by the LightCycler®480 (software release 1.5, Roche Diagnostics). Finally, the expression of the p4h genes was normalized against the maternal line and statistical analysis of the mean values of the qRT-PCR was performed using the GraphPad Prism software (version 8.0; La Jolla, California, USA) using an ANOVA with Durentt‘s test (P < 0.05).
2.16. Data analysis and visualisation
For data analysis in Python 3 (version 3.8.10, van Rossum and [116]) the libraries Pandas (version 1.3.4, [70]; The pandas development team [105]) and numpy (version 1.21.4, [35]) were applied and figures were generated with the libraries Matplotlib (version 3.2.1; [44]), Seaborn (version 0.10.1; [121]) and pyvenn (version 0.1.3, https://github.com/tctianchi/pyvenn).
3. Results
3.1. Six Physcomitrella prolyl hydroxylases in four subfamilies
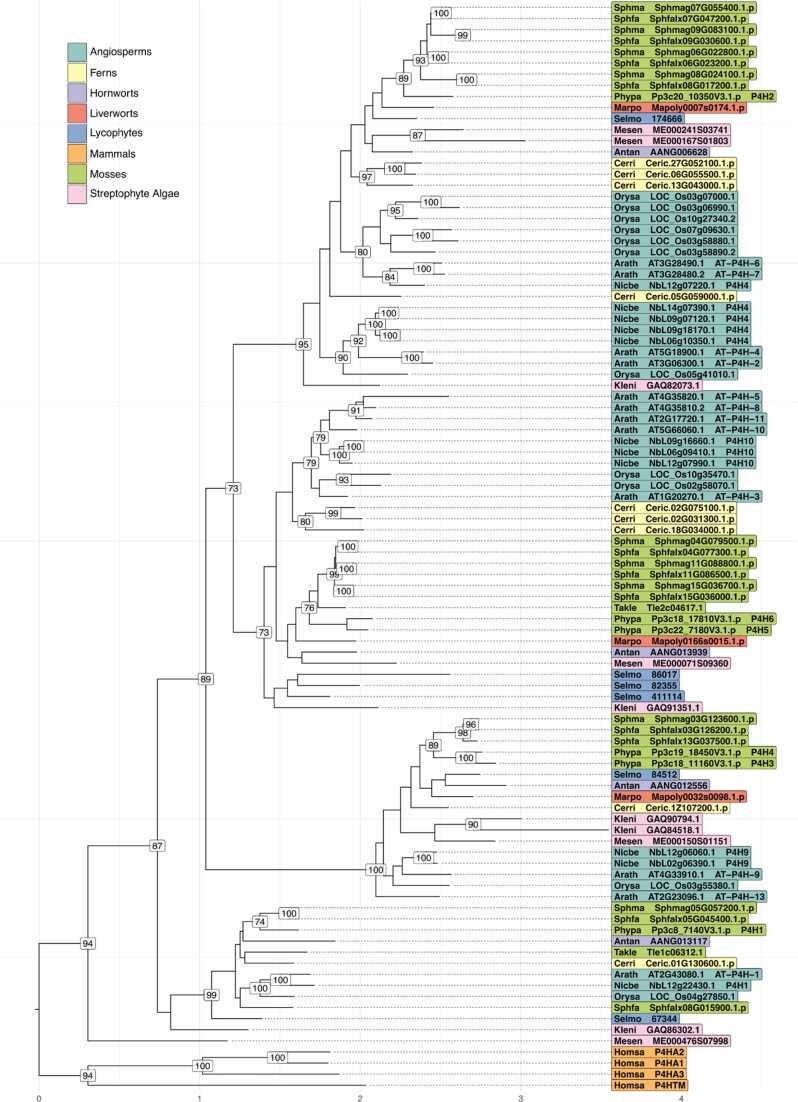
Due to sub- and neofunctionalisation, different enzyme isoforms may have different substrate specificities. Therefore, we evaluated the P4H family by phylogenetic reconstruction and identified six Physcomitrella P4Hs in four distinct clades resembling putative subfamilies in the Viridiplantae (Fig. 1, detailed in Supplementary Fig. S1). Two Physcomitrella P4Hs, namely P4H1 and P4H2, belong to a distinct subfamily whereas P4H3 and P4H4 as well as P4H5 and P4H6 group pairwise in common clades. All Physcomitrella P4Hs with the exception of P4H4 have direct orthologues in Funaria hygrometrica from the same family of mosses. While we found members of all subfamilies encoded by the mosses Calohypnum plumiforme, Ceratodon purpureus, Sphagnum fallax and Sphagnum magellanicum, the living fossil Takakia lepidozioides, sister to all other mosses, encodes only two P4Hs: one orthologue of Physcomitrella P4H1 and one single orthologue of the Physcomitrella P4H5 and P4H6. Homologues of the hornwort Anthoceros angustus are present in all clades, while we found homologues of the liverwort Marchantia polymorpha in all clades except the one containing Physcomitrella P4H1. Of the twelve Arabidopsis P4Hs, AtP4H1 is an orthologue of Physcomitrella P4H1. AtP4H2, AtP4H4, AtP4H6 and AtP4H7 are co-orthologues of Physcomitrella P4H2. AtP4H9 and AtP4H13 are co-orthologues of Physcomitrella P4H3 and P4H4, while AtP4H3, AtP4H5, AtP4H8, AtP4H10 and AtP4H11 are co-orthologues of Physcomitrella P4H5 and P4H6. In addition, we identified eleven P4Hs within the latest annotation of Nicotiana benthamiana [87], which clustered in congruence with Mócsai et al. [73] and were labelled accordingly.
Fig. 1.
Phylogenetic reconstruction of the plant P4H family. Condensed maximum-likelihood tree of a codon-aware multiple sequence alignment of P4H coding sequences annotated with internal bootstrap support values (≥ 70 %) and outgroup-rooted with mammal sequences. Tip labels are coloured by taxonomic units, reference sequences are complemented by their trivial name and species abbreviations following a five-letter code. Antan: Anthoceros angustus; Arath: Arabidopsis thaliana; Cerri: Ceratopteris richardii; Homsa: Homo sapiens; Kleni: Klebsormidium nitens; Marpo: Marchantia polymorpha; Mesen: Mesotaenium endlicherianum; Nicbe: Nicotiana benthamiana; Orysa: Oryza sativa; Phypa: Physcomitrium patens; Selmo: Selaginella moellendorfii; Sphfa: Sphagnum fallax; Sphma: Sphagnum magellanicum; Takle: Takakia lepidozioides. A detailed version of this tree containing the complete taxon set can be found in Supplementary Fig. S1.
3.2. Seventy-three Hyps in 24 secretory proteins
We identified Hyps in Physcomitrella proteins using multiple mass spectrometry (MS/MS) measurements. According to our extraction protocols the samples were enriched for secretory proteins from ER, Golgi apparatus, cell wall and extracellular space. In total, 5139 proteins were measured. From these, 602 had a predicted signal peptide that allows them to enter the secretory pathway via the ER and get in contact with the P4H enzymes which are localized there [83]. The MS data covered 23.3 % of all proteins with a predicted signal peptide from the Physcomitrella proteome (Supplementary Fig. S2a) but only 6.86 % of their proline sites were covered by identified peptides (Supplementary Fig. S2b). No signal peptide was predicted for the other 4537 proteins, so it is not certain whether they pass through the secretory compartments.
Hyps were collected exclusively from proteins with predicted signal peptide. This resulted in 73 validated Hyps (PTMProphet probability > 0.7; Supplementary Fig. S3) from 26 peptides after trypsin, elastase or thermolysin cleavage belonging to 24 proteins (Supplementary Table T1). Peptide versions with different cleavage sites were not counted additionally. Some of the 26 peptides with validated Hyps were also measured without prolyl-hydroxylation. For seven of these the degree of prolyl-hydroxylation was estimated using peptide intensities from the quantified MS data (Supplementary Fig. S4). These seven peptides had a varying degree of prolyl-hydroxylation between 0.04 % and 10.83 %. With one exception (AASILLYHIVOSOATAADLTDGQTLTTALGK) these sites with a low degree of prolyl-hydroxylation were isolated prolines that had no other proline in the neighbourhood of two AAs at each side.
Physcomitrella HRGPs were predicted by Liu et al. [62] and Ma et al. [64]. According to their classification, none of the 24 Hyp-containing proteins was an extensin but eight of them were chimeric AGPs. These were one laminin G-like AGP (Pp3c1_2420V3.1), two xylogen-like AGPs (Pp3c1_11030V3.1; Pp3c1_31020V3.1), one phytocyanin-like AGP (Pp3c16_22330V3.1), one fructose‐1,6‐bisphosphatase-like AGP (Pp3c18_4950V3.1) and three fasciclin-like AGPs (Pp3c4_16840V3.1; Pp3c7_430V3.1; Pp3c21_10620V3.1) comprising 48 of the measured Hyps (Supplementary Table T1). The protein with the highest number of validated Hyps (11) was a myosin light-chain kinase of the chimeric xylogen-like AGP family (Pp3c1_11030V3.1). Also, in several of the other chimeric AGPs a high number of Hyps was measured. These were nine Hyps in the chimeric phytocyanin-like AGP (Pp3c16_22330V3.1) as well as seven Hyps each in the chimeric xylogen-like AGP (Pp3c1_31020V3.1) and in one of the three chimeric fasciclin-like AGPs (Pp3c4_16840V3.1). From the 16 Physcomitrella proteins that were not HRGPs, only one Hyp each was identified in 11 proteins, two proteins contained two Hyps, two other proteins contained three Hyps and in one protein four Hyps were detected.
3.3. Alanine, threonine, proline, serine and valine are enriched around Hyps
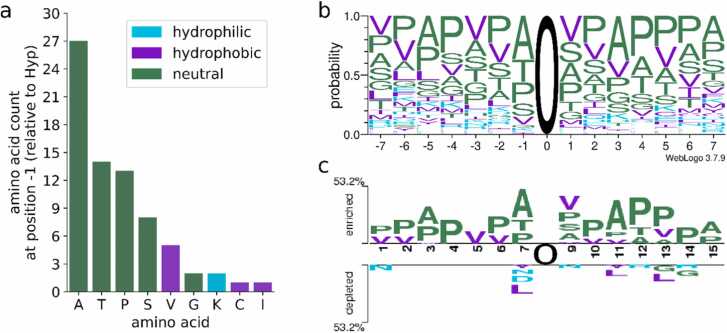
Since the AA directly before the Hyp (position −1) has a special importance according to established prolyl-hydroxylation motifs for plants [31], [9], we determined frequencies of each AA which was found immediately before the 73 measured Hyps. The most frequent AAs were in descending order alanine, threonine, proline/hydroxyproline and serine (Fig. 2). Valine was slightly more abundant than the remaining AAs that were either counted once or twice before a Hyp (twice: G, K; once: C, I) or not detected at all (D, E, F, H, L, M, N, Q, R, W, Y). Due to the applied filtering criteria, excluding all sites with ambiguous localization of the hydroxyl group on the proline, Hyps in proximity to methionine and tryptophan might be underrepresented and to a lesser extent this might also affect the easily oxidable cysteine, histidine, phenylalanine, and tyrosine (Berlett and Stadtmann, 1997).
Fig. 2.
Amino acid distribution around Hyps from Physcomitrella proteins. Depicted is the frequency of different AAs located directly before the identified Hyps (a), the AA distribution in AA sequence windows of length 15 centred at the 73 Hyp sites (b) and a two-sample logo illustrating the AAs that were enriched or depleted around Hyps compared to 2773 proline sites not measured to be hydroxylated (c). In (b) and (c) the central Hyps are depicted as O at position 0. The AAs and corresponding bars are coloured by their properties: green = neutral AAs (A, G, H, P, S, T), purple = hydrophobic AAs (C, F, I, L, M, V, W, Y), cyan = hydrophilic AAs (D, E, K, N, Q, R).
To investigate which AAs are present in the near and more distant neighbourhood of the Hyps, AA sequence windows of length 15 centred around the Hyps were generated. This revealed that the tolerance for the presence of AAs other than proline, alanine, serine, threonine, and valine was smallest at position − 1 and increased for positions further away from the Hyp. Proline/hydroxyproline, alanine and valine (Fig. 2a, b) were the most frequent AAs at several positions of the sequence windows. Indeed, these three AAs as well as threonine and serine were significantly enriched at one or several positions around the Hyps, when using as reference the AA distribution around 2773 prolines that were non-hydroxylated (PTMProphet probability for hydroxylation <0.01, sites less than seven amino acids apart from protein termini were not considered, Supplementary Fig. S3) in secretory proteins with predicted signal peptide (Fig. 2c). In contrast, leucine, by far the most frequent AA in all Physcomitrella proteins with predicted signal peptide (Supplementary Fig. S5), as well as asparagine and aspartic acid were underrepresented at one or several positions around Hyps (Fig. 2c). Hereinafter, hydroxylated proline residues will be depicted as “O” for easier differentiation from non-hydroxylated residues.
3.4. AOV is a frequent prolyl-hydroxylation pattern
Of the 73 Hyps, 50 were not surrounded by other prolines or Hyps, while the other 23 were part of blocks of two to four AAs that included combinations of prolines or Hyps. Analysing the combination of the AA before and after a single non-contiguous Hyp, the combination of AOV was most frequent and present in total 15 times (Supplementary Table T2). AOV was found in six of the Hyp-containing proteins, two of them not being HRGPs and four being chimeric AGPs. In two of the chimeric AGPs (Pp3c4_16840V3.1, Pp3c16_22330V3.1) this combination was part of the repetitive motif (AOVV)3–4 (Supplementary Fig. S6). Other combinations were AOA, TOS and VOA (four times), as well as AOG, AOT and TOT (three times), which were most often part of the glycomodule motif in chimeric AGPs (Supplementary Fig. S6). AA combinations flanking two prolyl-hydroxylation sites were AOOM, SOOQ, SOOS, TOOD, TOOM, and TOOS. Such combinations were found in the longer motif of [A/T]OOMGSTOOS, that was identified three times in one of the chimeric AGPs (Supplementary Fig. S6). While each individual proline written as O in the previously mentioned patterns was hydroxylated at least once, in the motif QPOK the first proline was never hydroxylated. Blocks with more than two prolyl-hydroxylation sites were only identified in the peptide KOOOOSOPPK.
3.5. Arabinose on two pectinesterases
To identify which Hyps are O-glycosylated with arabinoses, modifications of Hyp with one to three arabinosyl residues were searched for in the MS data. In the two tryptic peptides TEGMGIAGTOODDGSSSOSOSTPTCIR (Pp3c5_12660V3.1) and YEAQNSESTVLDTQTLPGGDFSVEATOSOOQEATCIR (Pp3c25_760V3.1) from two different cell-wall located pectinesterases, prolines within the glycomodule motif were prolyl-hydroxylated and O-glycosylated with arabinose residues (Supplementary Figs. S7, S8). Most frequently, two arabinose residues were present in the peptides, however, in the first peptide up to five arabinoses were found.
In order to obtain data about peptides containing Hyps O-glycosylated with arabinogalactans, which normally prevent the detection of these peptides by MS, two approaches for deglycosylation were applied: One was employing a Physcomitrella double mutant of galactosyltransferase (Δgalt2/3) which might be responsible for O-glycosylation [82], and the second was chemical deglycosylation and treatment with three different proteases (trypsin, elastase, thermolysin) to increase the sequence coverage of the identified proteins. MS/MS spectra from eight of the 26 Hyp-containing peptides were exclusively obtained from the deglycosylated datasets (chemical deglycosylation: six peptides, Δgalt2/3: two peptides; Supplementary Table T1). All peptides contained at least one Hyp within a glycomodule and some contained a long glycomodule spanning many Hyps (e.g. LVAOVOAOVVKAOAOAOVIKAOTOGOA), making these suitable candidates for O-glycosylation.
3.6. Combination of three tools predicts about 95 % of the Hyps
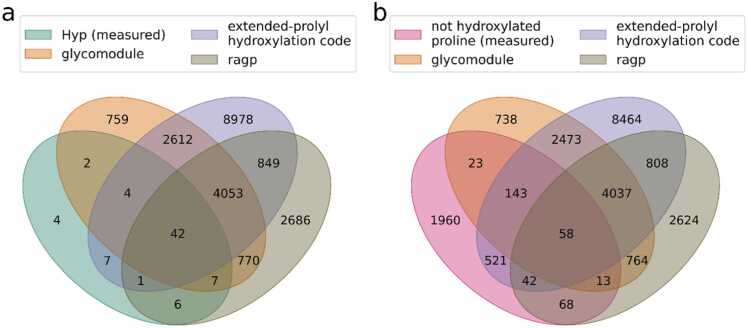
We searched for possible prolyl-hydroxylation sites in all 1920 secretory Physcomitrella proteins (major isoforms) with a predicted signal peptide (no organelles) using three methods appropriate for plant-like prolyl-hydroxylation: the glycomodule, the extended prolyl-hydroxylation code and the R package ragp using default settings [25], [31], [9]. This resulted in 8249 predicted Hyp sites from the glycomodule, 16,546 from the extended prolyl-hydroxylation code and 8414 from ragp. A high number of 4095 prolines was predicted to be hydroxylated in accordance with all three methods (Fig. 3).
Fig. 3.
Overlap between Hyps predicted by three methods and measured hydroxylation status of prolines. Hyps in all secretory Physcomitrella proteins with a signal peptide (SignalP 5) were predicted using the ragp tool, the glycomodule and the extended prolyl-hydroxylation code. Depicted is the overlap between predicted Hyps with measured Hyps (a) and prolines not measured to be hydroxylated (b).
Subsequently, data from the measured secretory proteins with a predicted signal peptide were used to check whether the three methods correctly predict the 73 MS-verified Hyps and whether they predict no hydroxylation of the 2828 prolines that we identified as non-hydroxylated. The prediction performance was assessed using the balanced accuracy score (0 =no correct prediction; 1 =all predictions correct). Here, the ragp tool and the glycomodule performed best and had a comparable performance with 56 (76.71 %) and 55 (75.34 %) correctly predicted Hyps (Fig. 3a). The ragp tool, however, predicted a possible hydroxylation for 181 additional prolines that were not hydroxylated, while 237 were predicted by the glycomodule (Fig. 3b), leading to a slightly higher balanced accuracy of the ragp tool compared to the glycomodule (0.85 and 0.83, respectively). The extended prolyl-hydroxylation code correctly predicted 54 of the Hyps (73.97 %; Fig. 3a) and a possible hydroxylation for 764 (27.02 %) further prolines that were measured to be non-hydroxylated (Fig. 3b), resulting in a balanced accuracy score of 0.73. Only four Hyps were not predicted by any of the three methods (Fig. 3a, Supplementary Fig. S9). One of them (YYPPFKOELVK) was in proximity to a proline-proline sequence segment, while another one (GHEGOSSVYTOSSDTEPFNFHDPR, underlined) was the first Hyp in a Gly-Hyp-Xaa4-Thr-Hyp motif. The third Hyp (WNSNIVVVGVDDIOLR) was from a chimeric laminin G-like AGP but it was at an isolated proline more than 140 AAs distant from the proline-rich region of the protein (Supplementary Fig. S6). The fourth was 10 AAs behind a short proline-rich segment of the protein (…SPNPPNPGPTPPSPPPPEVICDKWRTCOAENTCCCTFPVGK…, Hyp-containing peptide is underlined).
Next, we tested to what extent a combination of two or all three methods could improve the prediction performance either by a higher number of correctly predicted Hyps or an increased balanced accuracy score. When considering a proline as being hydroxylated based on any of the three methods, the vast majority of the measured Hyps (69 out of 73 = 94.52 %) were predicted with a balanced accuracy of 0.82. When a proline was only considered to be hydroxylated after prediction by all three methods, far fewer of the measured Hyps (42) were predicted, but compared to the other methods also the smallest number of prolines was incorrectly predicted as Hyps (58), making this combination the most precise. Considering a proline as being hydroxylated after prediction by either ragp or the glycomodule, the two methods with the highest accuracy, a balanced accuracy of 0.86 was achieved – better than that of ragp or the glycomodule alone – and more of the experimentally verified Hyps were predicted (62).
3.7. Hyps predominantly in disordered regions
To analyse if specific structural features favour prolyl-hydroxylation, 443 protein structure models of secretory proteins with predicted signal peptide, containing the 73 measured Hyps and 2828 prolines that were non-hydroxylated, were downloaded from the AlphaFold Protein Structure Database [117]. The pLDDT confidence score of the model (0 =minimum quality, 100 =maximum quality) was for most of the non-hydroxylated proline residues higher than 90 (2026 out of 2828 sites), indicating a high quality of the local model structure, whereas it was below 50 for 42 of the 73 Hyps (Supplementary Fig. S10). Low pLDDT scores can be an indication for disorder [110].
The DSSP tool was used to determine where the protein structure model was folded into any of the seven secondary structure elements (3–10 helix, α-helix, π-helix, strand (participates in β ladder), isolated β-bridge, turn (hydrogen bonded), bend). Additionally, the relative accessible surface area (0 =completely buried within protein structure, 1 =fully exposed to solvent) of each residue was computed. The Hyps were located mostly in well accessible protein regions with a median relative accessible surface area of 0.86, whereas non-hydroxylated prolines were often less accessible, having a median relative accessible surface area of 0.37 (Supplementary Fig. S10). Considering the secondary structure of the protein, Hyps were only present in four of the defined secondary structure elements (in bends, 3–10 helices, strands and turns; Supplementary Fig. S10), but most frequently both, Hyps and non-hydroxylated prolines, were located in regions where none of the seven secondary structure elements were assigned (Supplementary Fig. S10). With 87.67 % the proportion of Hyps in regions without assigned secondary structure was much higher than for the non-hydroxylated prolines with 43.71 %. With five exceptions, the regions without assigned secondary structure containing the Hyps were spanning more than 10 and up to 296 AAs (Fig. 4c, Supplementary Fig. S11, Supplementary Fig. S12), while they were shorter than 10 AAs for the vast majority of the non-hydroxylated prolines (1139 out of 1236 sites without secondary structure; e.g. in Fig. 4a and Supplementary Fig. S11).
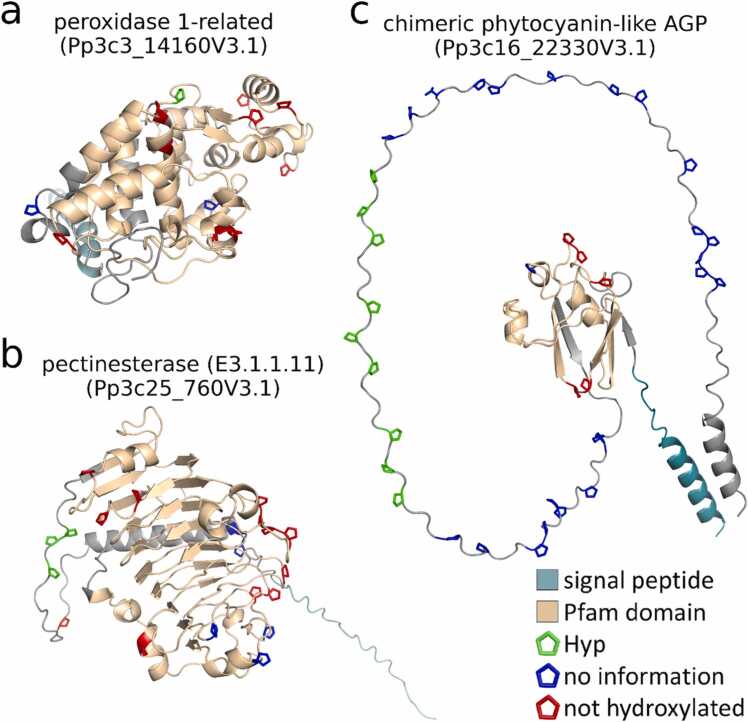
Fig. 4.
Three-dimensional structures of Hyp-containing proteins. Measured Hyps (green) and non-hydroxylated prolines (red) are highlighted in two exemplary structures from secretory proteins not predicted to be HRGPs Pp3c3_14160V3.1 in (a) and Pp3c25_760V3.1 in (b) as well as a chimeric phytocyanin-like AGP Pp3c16_22330V3.1 in (c). All remaining prolines are coloured blue. For these, no definite information about their hydroxylation status could be obtained from the MS data. The Pfam domains for peroxidase PF00141 in (a), pectinesterase PF01095 in (b) and the plastocyanin-like domain PF02298 in (c), respectively, as given by Phytozome (version 13; [32]), are coloured in light brown.
In the chimeric AGPs the long, disordered regions containing the Hyps were rich in prolines and glycomodules while non-hydroxylated prolines were found predominantly in the structured domain of the chimeric AGP (Fig. 4c, Supplementary Fig. S11). An exception was a Hyp in a chimeric AGP, a xyloglucan endo-transglycosylase (WNSNIVVVGVDDIOLR, Pp3c1_2420V3.1) where according to the NCBI Conserved Domain Search webtool (CD-Search; [68]) the single Hyp is located in the active site of the protein. In the proteins that were not HRGPs, Hyps were found in short glycomodules in the N-terminal disordered regions from two pectinesterases (Pp3c25_760V3.1 and Pp3c5_12660V3.1, Fig. 4b, Supplementary Fig. S12) where the Hyps were O-glycosylated with arabinoses.
3.8. Physcomitrella-produced EPO with plant-specific O-glycans
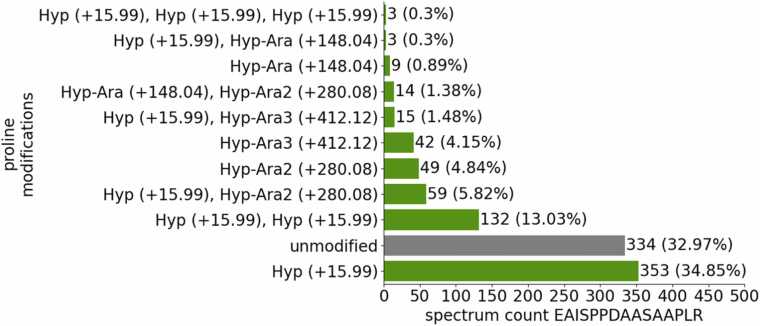
We did not only identify Hyps in native Physcomitrella proteins, but also in Physcomitrella-produced recombinant human erythropoietin (EPO). In accordance with Parsons et al. [83], we found prolines of the EPO peptide EAISPPDAASAAPLR to be hydroxylated. In addition, post-translational modifications of the peptide EAISPPDAASAAPLR included not only prolyl-hydroxylation but also plant-specific O-glycosylation of Hyps with arabinose chains on the Ser-Pro-Pro motif (Supplementary Fig. S13) as well as an additional glycosylation of the first serine with a single hexose, if neighbouring prolines were hydroxylated and O-glycosylated. From the 1013 spectra of the respective peptide, in 18.86 % (191 spectra) the peptide was glycosylated, mostly with more than one arabinose, in 48.17 % (488 spectra) the peptide was just hydroxylated (one, two or three Hyps; no arabinose) and in 32.97 % (334 spectra) the peptide was unmodified (Fig. 5). All the hydroxylated prolines in the EPO peptide fit the glycomodule, but only the second and third proline were predicted to be hydroxylated by the ragp tool with probabilities of 0.33 and 0.26, respectively. Two further Hyps in EPO were predicted by the ragp tool at positions 29 and 30 in an Ala-Pro-Pro motif. These prolines were not covered in the MS data, but Parsons et al. [83] reported these as non-hydroxylated.
Fig. 5.
Modified versions of the EAISPPDAASAAPLR peptide from recombinant EPO. Depicted are the number of spectra of the unmodified, prolyl-hydroxylated and the arabinosylated versions of the peptide, respectively. The spectra are counted over several MS measurements and replicates.
3.9. Multiple effects of single p4h knockouts
To analyse the effects of the six Physcomitrella P4Hs more deeply, we employed p4h knockout (KO) mutants. Samples enriched with secretory proteins (from ER, Golgi, cell wall and extracellular space) were obtained using two different protocols. The samples from the EPO-producing maternal line were labelled with 15N (heavy) and mixed with the single KO lines of each of the six p4hs (light), respectively, prior to MS measurements and quantification. Significant changes in protein abundance were determined over the light/heavy ratios (= intensity in p4h KO line / intensity in maternal line) in three replicates via a t-test (p adjusted < 0.05, |log2 light/heavy ratio|>1) and filtered for secretory proteins with a signal peptide. Combining results from both protocols, the p4h6 KO dataset contained the highest number of proteins with altered abundance compared to the maternal line (52 proteins), whereas the smallest number of proteins with altered abundance was 12 proteins in the p4h4 KO dataset (Supplementary Fig. S14). Considering the direction of the change, many more proteins had an increased abundance in the p4h1 KO and p4h2 KO while the number of proteins with reduced and increased abundance, respectively, was balanced in the other p4h KOs. Some proteins had an altered abundance only in one specific p4h KO while it differed for others in multiple of the p4h KOs (Supplementary Fig. S14). Only one protein, a subtilisin-like protease (Pp3c11_4360V3.1), was significantly altered (increased) in all p4h KOs. A galactose oxidase (Pp3c10_8570V3.1) and a polygalacturonase (Pp3c21_6170V3.1) had an increased abundance in five of the p4h KOs, with the latter being strongly increased in the p4h6 KO. Moreover, the abundance of four chimeric AGPs was increased in one or several of the p4h KOs (Pp3c1_2420V3.1, Pp3c4_16840V3.1, Pp3c16_22330V3.1, Pp3c26_5590V3.1, Pp3c4_3520V3.1), and the abundance of one (Pp3c4_3520V3.1) was decreased (Supplementary Table T3).
Additionally, we used these datasets to search for peptides from secretory Physcomitrella proteins with a predicted signal peptide where the abundance of peptide versions with prolyl-hydroxylation was significantly reduced in the p4h KO mutants compared to the maternal line (P < 0.05, log2(light/heavy intensity ratio)< 1), while the abundance of the corresponding protein and, if present, that of the unmodified peptide were not significantly reduced. In these datasets eight of the previously collected peptides with validated Hyps were measured in more than one replicate and hence appropriate for statistical evaluation. We identified a single peptide, GANYAITFCPTVTOVAK from a thaumatin family protein (Pp3c16_17280V3.1) in the p4h5 KO with a reduced abundance by a log2 light/heavy ratio of − 9.57 (Supplementary Table T4).
Considering the EPO peptide EAISPPDAASAAPLR (or ALGAQKEAISPPDAASAAPLR) a clear trend for reduction in the abundance of its prolyl-hydroxylated form was visible in the p4h1 KO (Supplementary Fig. S15). In all cases the reduction of Hyp-containing or arabinosylated peptides was more than 90 %, e.g. the abundance of the peptide with hydroxylation on the second proline (EAISPODAASAAPLR) was reduced by a log2 light/heavy ratio of − 4.09 (Supplementary Table T4). In none of the other p4h KOs the filtering criteria for significant reduction in the abundance of this peptide were fulfilled and no major changes in prolyl-hydroxylation of the peptide were observed (Supplementary Fig. S15). These findings further support the major role of p4h1 in the hydroxylation of EPO in Physcomitrella.
To study if the KO of a single p4h influences the expression of the five other Physcomitrella p4h genes, transcript abundances of each p4h gene were determined in the maternal line (174.16) and the six p4h single KOs. Significance in the changes of transcript abundance was computed with ANOVA and Durentt‘s test (P < 0.05). The KOs of p4h1, p4h2 and p4h4, respectively, did not significantly change the expression of any of the remaining p4h genes. However, our data showed an increase of p4h1 transcript abundance in the p4h3 KO, whereas in the p4h5 KO both p4h1 and p4h2 had an increased abundance, while the p4h6 KO led to an increased transcript abundance of p4h5 (Supplementary Fig. S16).
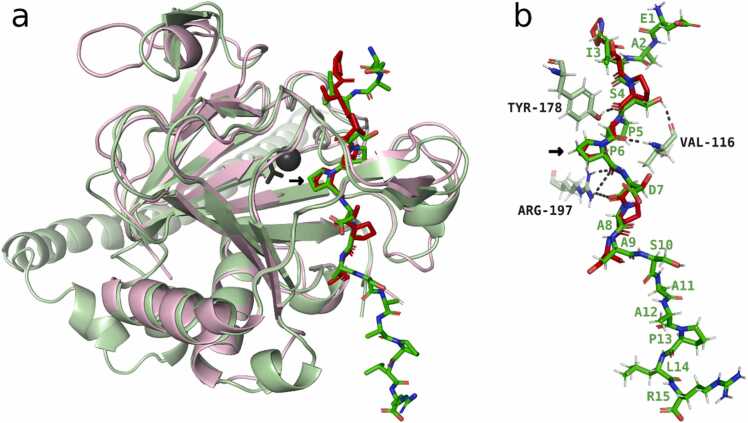
3.10. Modelling suggests peptide interactions in the active site of P4H1
Since AlphaFold-Multimer can predict the interaction between protein-peptide complexes [46], we modelled the interaction between P4H1 and its target sequence, the EPO peptide EAISPPDAASAAPLR, with AlphaFold2-multimer-v3 and no template information. The top-ranking model structure was superposed with the experimentally solved crystal structure of Chlamydomonas reinhardtii (Chlamydomonas) P4H1, which has a PSPSPSPS peptide bound in its active site (PDB ID 3GZE chain C; [54]). The third proline in this peptide (bold) is located at the catalytic active position within the active site of Chlamydomonas P4H1, where the prolyl-hydroxylation reaction takes place and is buried under two loops of the enzyme. In the superimposed model of Physcomitrella P4H1, the second proline from the EPO peptide (EAISPPDAASAAPLR) is located at this position (marked with an arrow in Fig. 6a, b). The root-mean-square deviation (RMSD) of the Cα atoms from the five residues of the two substrate peptides centred within the active site (PSPSPSPS and EAISPPDAASAAPLR) is 0.4 Å, indicating a highly similar fold between the backbones of two substrate peptides within the active site. The EPO peptide interacts over eight hydrogen bonds with residues of the P4H1 protein (Fig. 6b). Five of these are also present in the interaction between Chlamydomonas P4H1 and its peptide. Two are located between the central proline that becomes hydroxylated and the ARG197 from P4H1 (corresponding to ARG161 in Chlamydomonas P4H1), two are located between residues of the peptide with VAL116 and one with TYR178 of P4H1 (corresponding to VAL80 and TYR140 in Chlamydomonas P4H1).
Fig. 6.
Interaction between P4H1 and EPO peptide EAISPPDAASAAPLR modelled with AlphaFold2-multimer-v3 without template information. (a) The top-ranking model (light green) with the bound peptide EAISPPDAASAAPLR (green) superimposed on the experimentally solved crystal structure of Chlamydomonas reinhardtii P4H1 (light red) having a bound (Pro-Ser)4 peptide (red) in its active site (PDB ID 3GZE chain C; [54]). (b) Conformation of the two substrate peptides in the superposed structures with hydrogen bonds (dashed lines) between the EPO peptide and P4H1 to the residues VAL80, TYR140 and ARG197.
To test if EPO is a substrate for any of the other five Physcomitrella P4Hs, those were modelled with the EPO peptide using versions 2 and 3 of AlphaFold2-Multimer. While the peptide fits into the P4H1 models computed with both versions, it was not modelled into any of the other Physcomitrella P4H proteins with AlphaFold2-Multimer-v2. In contrast, AlphaFold2-Multimer-v3 computed models for P4H2, P4H5 and P4H6 with either the first (P4H2, P4H5) or the second proline (P4H2, P4H5, P4H6) of the peptide in the catalytic active position. For P4H3 and P4H4 a part of the peptide was modelled at the active site, but other AAs than proline were placed at the catalytic active position (Supplementary Fig. S17). Thus, AlphaFold2-Multimer-v3 also computed P4H-EPO interactions less favourable for the other five Physcomitrella P4Hs than for P4H1. We consider this as further indication for different substrate specificities of the moss P4Hs, and as support of our experimental findings.
4. Discussion
Plants are gaining increasing importance for the production of valuable compounds, such as pharmaceuticals. While most production hosts are vascular plants, such as Nicotiana benthamiana (Nicotiana) and Daucus carota (carrot), the non-vascular moss Physcomitrella has a proven track-record for the production of pharmaceuticals and bioactive ingredients [119], [20], [75]. In our attempts to constantly optimize this moss for molecular pharming [90], [92], we concentrate on gene expression [107], [78], bioproduction [93], and glycoengineering [19], [6]. In the latter field, plant-typical glyco-structures have been abolished [82], and stable in-vivo protein sialylation has been achieved [5]. In contrast to the well-studied N-glycosylation of recombinant proteins, O-glycosylation is still underexplored in plants although it might deteriorate product quality. While mosses, such as Physcomitrella, and vascular plants, such as Nicotiana, share similar N-glycosylation patterns [53], they may differ in their O-glycosylation pattern. A gene responsible for prolyl-hydroxylation of recombinant erythropoietin (EPO) from Physcomitrella has been identified [83], but a reliable bioinformatic tool to predict this protein modification was not available.
Based on genome information we made a phylogenetic reconstruction of plant P4Hs, the enzymes responsible for prolyl-hydroxylation. The Physcomitrella genome encodes six P4Hs in four subfamilies, indicating neofunctionalisation during evolution. P4H1 is the only homologue of Physcomitrella within its subfamily and clusters with P4H1 from Arabidopsis. Arabidopsis P4H1 hydroxylates poly-proline and proline-rich motifs in plant proteins, and motifs of the human hypoxia-inducible factor and collagen-like peptides [38] that are substrates for mammalian P4Hs [33]. Arabidopsis P4H2 [106] clusters with Physcomitrella P4H2 and preferentially hydroxylates substrates with three neighbouring prolines. Arabidopsis P4H5 clusters with Physcomitrella P4H5 and P4H6 and hydroxylates all except the last proline in Ser-(Pro)4 extensin motifs [118]. Another P4H from this cluster is Arabidopsis P4H3, which plays a role in the response to low oxygen [52].
To gain a better understanding of the favoured targets for prolyl-hydroxylation by the six Physcomitrella P4Hs, we collected Hyps from a set of MS/MS measurements. Since the P4Hs are located in secretory compartments, we focused on secretory proteins with predicted signal peptide. With 48 out of the 73 identified Hyps, the majority originated from chimeric arabinogalactan proteins (AGPs) and were mostly located in disordered regions of the protein. AGPs have domains with a high content of proline, alanine, serine and threonine (PAST), and non-contiguous prolines in repetitive motifs preceded by alanine, serine and threonine are frequently hydroxylated [60]. Accordingly, in our study these AAs and additionally valine dominated over a window of 15 AAs centred around the Hyps.
In agreement with MS measurements of 114 Hyps from 62 glycoproteins in rice [61], we found alanine most often before Hyps. Valine was the second most frequent AA before Hyp in rice. In contrast, it was the fifth most abundant in our data. Leucine, which was significantly depleted before the Hyps compared to non-hydroxylated prolines in Physcomitrella, was the third most frequent AA preceding Hyps in rice. Other AAs such as aspartic acid, glutamic acid and glutamine were not identified in our MS data, although peptides with prolyl-hydroxylation after these AAs occur in vascular plants, e.g. in Zea mays and Echinacea purpurea [9].
Most Hyps were non-contiguous and thus not directly surrounded by other prolines. Combinations of AOV, AOA, TOS, VOA, AOG, AOT and TOT were most frequent before and after a non-contiguous Hyp. These combinations were found particularly often in chimeric AGPs and lay within long glycomodule motifs, spanning multiple Hyps. Most of these peptides were from data where deglycosylation was performed to allow identification of peptides that were likely O-glycosylated with large arabinogalactan chains, and which cannot be identified by MS. Taken together, these peptides are suitable candidates for O-glycosylation in Physcomitrella. Interestingly, two peptides that contain several prolines in the glycomodule with arabinose residues were not HRGPs but two pectinesterases.
Some of the Physcomitrella mutants used in this study were producing recombinant human EPO. In the EPO peptide EAISPPDAASAAPLR we found not only the previously reported prolyl-hydroxylation of the first two prolines [83], but in rare cases also hydroxylation of the third proline as well as O-glycosylation with up to three arabinose residues. In agreement with the Hyp contiguity hypothesis that predicts addition of arabinogalactans to single non-contiguous Hyps and arabinose chains to neighbouring contiguous Hyps [50], the arabinose chains were assigned to the segment with two neighbouring contiguous prolines. Further, in a small fraction of the peptides, the serine of the Ser-Pro-Pro motif was glycosylated with a hexose, resembling the O-glycosylation pattern in extensins where the hexose attached to the serine is a galactose [94].
Three methods to predict Hyps in plants developed with data from various plant species, but not mosses, were used to predict the hydroxylation status of the prolines in secretory Physcomitrella proteins: the glycomodule, the extended prolyl-hydroxylation code and the prediction tool ragp. More than 4000 candidate sites for prolyl-hydroxylation were predicted in accordance with all three methods, indicating that our MS data represents only a small fraction of the total hydroxylation pattern in Physcomitrella. Comparing the predictions by ragp and the glycomodule with MS data, both methods performed comparably well. A combination of the predicted Hyps by ragp and the glycomodule yielded an even higher number of correctly predicted Hyps with a better accuracy, making these methods well suited for the prediction of prolyl-hydroxylation in Physcomitrella. All except four of the identified Hyps were correctly predicted by at least one method. For two of these peptides the degree of hydroxylation was determined, and these peptides were only very rarely prolyl-hydroxylated. The peptide EVQLINIINAPLQGFK contained a Hyp only in 0.09 % and WNSNIVVVGVDDIPLR was prolyl-hydroxylated in 2.11 %, indicating that these two peptides are not preferred targets of P4Hs in moss.
Since different P4Hs can act on the same peptide and to some extent on the same prolines, but with diverging preferences [73], we investigated the effect of the knockout (KO) of single p4h genes on the expression of the remaining p4h genes. Mostly the expression level of the five remaining p4hs was not significantly altered by the KO of a single p4h, but we found hints for a possible compensation in the single KO of p4h3, p4h5 and p4h6 by an upregulation of one or two other p4h genes. However, functional compensation apart from transcriptional upregulation is also possible since all p4h genes are expressed in protonema under standard growth conditions. A possible rebalancing effect by the remaining P4Hs was also reported after a quadruple KO of the Nicotiana p4h4 subset, where the KO led to a reduced abundance of the unmodified version of the hinge region from a recombinant IgA1 antibody and to an increased fraction of peptides O-glycosylated with pentoses [113]. In our data, the abundance of the prolyl-hydroxylated form of the peptide GANYAITFCPTVTPVAK was strongly reduced in the p4h5 KO, indicating that in this case loss of P4H5 is hardly compensated. In addition, we confirmed the findings of Parsons et al. [83] that P4H1 plays the major role in the prolyl-hydroxylation of recombinant EPO.
The KO of single p4hs not only affected prolyl-hydroxylation but also resulted in altered abundance of secretory proteins. Among these was a polygalacturonase with strongly increased abundance in several KO datasets. Furthermore, five HRGPs showed increased abundance in at least one p4h KO mutant, except for one whose abundance was decreased in the p4h6 KO. Increased abundances of HRGPs in p4h KO mutants might be partially caused by a reduction in O-glycosylation with large arabinogalactan trees of some peptides, which prevent sufficient solubilization by our extraction method. This indicates that the deleted P4H contributes considerably to the prolyl-hydroxylation of the respective HRGP. An effect of a p4h KO on cell wall protein expression was also observed in Arabidopsis where AGP12 was downregulated in a p4h3 mutant, indicating that the presence of P4Hs is linked with transcriptional regulation of AGPs [52].
By combining 443 AlphaFold structural protein models with our MS data of peptides with nearly 3000 proline sites, we identified Hyps predominantly on accessible protein surfaces in disordered regions of the protein. AlphaFold-Multimer models of Physcomitrella P4Hs with an EPO peptide as substrate suggested a highly accurate structure and identified relevant amino acids in the active centre of P4H1 that form H-bonds with the peptide substrate. In contrast, these models were far less clear about substrate binding for the other Physcomitrella P4Hs, further supporting the differential prolyl-hydroxylation by the six moss enzymes.
5. Conclusions
We provide a comprehensive analysis of prolyl-hydroxylation in the secretome of the moss Physcomitrella, an established production host for pharmaceuticals. We confirmed that general rules for prolyl-hydroxylation derived from vascular plants also apply to the majority of Hyps in moss. Nevertheless, some Hyps had an amino acid environment diverging from common motifs and were not predictable by existing methods, demonstrating specific differences in the prolyl-hydroxylation capacity between Physcomitrella and vascular plants. The substrate specificity of the different P4Hs is still scarcely known in any plant species. While we demonstrate that some prolines are mainly hydroxylated by a single P4H, there is also evidence for a compensation of such a p4h KO by increased expression of the other p4h genes. To what extent an interplay between the P4H enzymes, such as hetero-dimerization, as observed in Arabidopsis [118], or overlapping substrate specificities, as reported for Nicotiana [73], play a role for hydroxylation in Physcomitrella has to be determined. An exact understanding of the conditions for the hydroxylation of a proline by one or several P4Hs will facilitate the modification of prolyl-hydroxylation and O-glycosylation and can enhance quality and human compatibility of plant-produced pharmaceuticals.
CRediT authorship contribution statement
Sebastian N.W. Hoernstein: Investigation. Nico van Gessel: Investigation. Andreas W. Graf: Investigation. Roxane P. Spiegelhalder: Investigation. Eva L. Decker: Writing, Supervision, Funding acquisition. Ralf Reski: Writing – review & editing, Supervision, Funding acquisition. Christine Rempfer: Writing, Investigation. Anne Bertolini: Investigation. Lennard L. Bohlender: Investigation. Juliana Parsons: Investigation.
Declaration of Competing Interest
All authors declare no conflict of interest.
Acknowledgements
This study was supported by the Ministry of Science, Research and Arts Baden-Württemberg, the German Research Foundation (DFG) under Germany’s Excellence Strategy (CIBSS – EXC-2189 – Project ID 390939984), and the Wissenschaftliche Gesellschaft Freiburg. We gratefully acknowledge language editing by Anne Katrin Prowse.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2024.06.014.
Appendix A. Supplementary material
Supplementary material
.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [22], [84] partner repository with the dataset identifier PXD051497 and 10.6019/PXD051497.
References
- 1.Almagro Armenteros J.J., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 2.Altmann F. The role of protein glycosylation in allergy. Int Arch Allergy Immunol. 2007;142:99–115. doi: 10.1159/000096114. [DOI] [PubMed] [Google Scholar]
- 3.Banks J.A., Nishiyama T., Hasebe M., Bowman J.L., Gribskov M., et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlett B.S., Stadtman E.R. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 5.Bohlender L.L., Parsons J., Hoernstein S.N.W., Rempfer C., Ruiz-Molina N., et al. Stable protein sialylation in Physcomitrella. Front Plant Sci. 2020;11 doi: 10.3389/fpls.2020.610032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohlender L.L., Parsons J., Hoernstein S.N.W., Bangert N., Rodríguez-Jahnke F., et al. Unexpected arabinosylation after humanization of plant protein N-glycosylation. Front Bioeng Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.838365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowman J.L., Kohchi T., Yamato K.T., Jenkins J., Shu S., et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 2017;171:287–304.e15. doi: 10.1016/j.cell.2017.09.030. [DOI] [PubMed] [Google Scholar]
- 8.Buchfink B., Reuter K., Drost H.G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 2021;18:366–368. doi: 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canut H., Albenne C., Jamet E. Post-translational modifications of plant cell wall proteins and peptides: a survey from a proteomics point of view. Biochim Et Biophys Acta (BBA) - Proteins Proteom. 2016;1864:983–990. doi: 10.1016/j.bbapap.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 10.Carey S.B., Jenkins J., Lovell J.T., Maumus F., Sreedasyam A., et al. Gene-rich UV sex chromosomes harbor conserved regulators of sexual development. Sci Adv. 2021;7 doi: 10.1126/sciadv.abh2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cassab G.I., Varner J.E. Cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:321–353. doi: 10.1146/annurev.pp.39.060188.001541. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y., Dong W., Tan L., Held M.A., Kieliszewski M.J. Arabinosylation plays a crucial role in extensin cross-linking in vitro. Biochem Insights. 2015;8:1–13. doi: 10.4137/BCI.S31353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L.-G., Lan T., Zhang S., Zhao M., Luo G., et al. A designer synthetic chromosome fragment functions in moss. Nat Plants. 2024;10:228–239. doi: 10.1038/s41477-023-01595-7. [DOI] [PubMed] [Google Scholar]
- 14.Cheng C., Krishnakumar V., Chan A.P., Thibaud‐Nissen F., Schobel S., Town C.D. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017;89:789–804. doi: 10.1111/tpj.13415. [DOI] [PubMed] [Google Scholar]
- 15.Cheng S., Xian W., Fu Y., Marin B., Keller J., et al. Genomes of subaerial Zygnematophyceae provide insights into land plant evolution. Cell. 2019;179:1057–1067. doi: 10.1016/j.cell.2019.10.019. [DOI] [PubMed] [Google Scholar]
- 16.Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Decker E.L., Parsons J., Reski R. Glyco-engineering for biopharmaceutical production in moss bioreactors. Front Plant Sci. 2014;5:346. doi: 10.3389/fpls.2014.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Decker E.L., Reski R. Glycoprotein production in moss bioreactors. Plant Cell Rep. 2012;31:453–460. doi: 10.1007/s00299-011-1152-5. [DOI] [PubMed] [Google Scholar]
- 20.Decker E.L., Reski R. Mosses in biotechnology. Curr Opin Biotechnol. 2020;61:21–27. doi: 10.1016/j.copbio.2019.09.021. [DOI] [PubMed] [Google Scholar]
- 21.Deepak S., Shailasree S., Kini R.K., Muck A., Mithöfer A., Shetty S.H. Hydroxyproline-rich glycoproteins and plant defence. J Phytopathol. 2010;158:585–593. doi: 10.1111/j.1439-0434.2010.01669.x. [DOI] [Google Scholar]
- 22.Deutsch E.W., Bandeira N., Perez-Riverol Y., Sharma V., Carver J., et al. The ProteomeXchange Consortium at 10 years: 2023 update. Nucleic Acids Res. 2023;51:D1539–D1548. doi: 10.1093/nar/gkac1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deutsch E.W., Mendoza L., Shteynberg D., Slagel J., Sun Z., Moritz R.L. Trans‐Proteomic Pipeline, a standardized data processing pipeline for large‐scale reproducible proteomics informatics. Proteom Clin Appl. 2015;9:745–754. doi: 10.1002/prca.201400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Draeger C., Ndinyanka Fabrice T., Gineau E., Mouille G., Kuhn B.M., et al. Arabidopsis leucine-rich repeat extensin (LRX) proteins modify cell wall composition and influence plant growth. BMC Plant Biol. 2015;15:155. doi: 10.1186/s12870-015-0548-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dragićević M.B., Paunović D.M., Bogdanović M.D., Todorović S.I., Simonović A.D. ragp: Pipeline for mining of plant hydroxyproline-rich glycoproteins with implementation in R. Glycobiology. 2020;30:19–35. doi: 10.1093/glycob/cwz072. [DOI] [PubMed] [Google Scholar]
- 26.Egener T., Granado J., Guitton M.-C., Hohe A., Holtorf H., et al. High frequency of phenotypic deviations in Physcomitrella patens plants transformed with a gene-disruption library. BMC Plant Biol. 2002;2:6. doi: 10.1186/1471-2229-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellis M., Egelund J., Schultz C.J., Bacic A. Arabinogalactan-proteins: Key regulators at the cell surface? Plant Physiol. 2010;153:403–419. doi: 10.1104/pp.110.156000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans, R., O’Neill, M., Pritzel, A., Antropova, N., Senior, A. et al. (2022). Protein complex prediction with AlphaFold-Multimer. BioRxiv. 10.1101/2021.10.04.463034. [DOI]
- 29.Fernandez‐Pozo N., Haas F.B., Meyberg R., Ullrich K.K., Hiss M., et al. PEATmoss (Physcomitrella Expression Atlas Tool): a unified gene expression atlas for the model plant Physcomitrella patens. Plant J. 2020;102:165–177. doi: 10.1111/tpj.14607. [DOI] [PubMed] [Google Scholar]
- 30.Fruleux A., Verger S., Boudaoud A. Feeling stressed or strained? A biophysical model for cell wall mechanosensing in plants. Front Plant Sci. 2019;10:757. doi: 10.3389/fpls.2019.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomord V., Fitchette A., Menu‐Bouaouiche L., Saint‐Jore‐Dupas C., Plasson C., et al. Plant‐specific glycosylation patterns in the context of therapeutic protein production. Plant Biotechnol J. 2010;8:564–587. doi: 10.1111/j.1467-7652.2009.00497.x. [DOI] [PubMed] [Google Scholar]
- 32.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorres K.L., Raines R.T. Prolyl 4-hydroxylase. Crit Rev Biochem Mol Biol. 2010;45:106–124. doi: 10.3109/10409231003627991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Y., Gómez-Vásquez R., Reilly K., Li H., Tohme J., et al. Hydroxyproline-rich glycoproteins expressed during stress responses in cassava. Euphytica. 2001;120:59–70. doi: 10.1023/A:1017547419332. [DOI] [Google Scholar]
- 35.Harris, R C., Millman, van der Walt K.J., Gommers S.J., R, et al. Array programming with NumPy. Nature. 2020;585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Healey A.L., Piatkowski B., Lovell J.T., Sreedasyam A., Carey S.B., et al. Newly identified sex chromosomes in the Sphagnum (peat moss) genome alter carbon sequestration and ecosystem dynamics. Nat Plants. 2023;9:238–254. doi: 10.1038/s41477-022-01333-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heck M.A., Lüth V.M., van Gessel N., Krebs M., Kohl M., et al. Axenic in vitro cultivation of 19 peat moss (Sphagnum L.) species as a resource for basic biology, biotechnology, and paludiculture. N Phytol. 2021;229:861–876. doi: 10.1111/nph.16922. [DOI] [PubMed] [Google Scholar]
- 38.Hieta R., Myllyharju J. Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. J Biol Chem. 2002;277:23965–23971. doi: 10.1074/jbc.M201865200. [DOI] [PubMed] [Google Scholar]
- 39.Hijazi M., Velasquez S.M., Jamet E., Estevez J.M., Albenne C. An update on post-translational modifications of hydroxyproline-rich glycoproteins: toward a model highlighting their contribution to plant cell wall architecture. Front Plant Sci. 2014;5:395. doi: 10.3389/fpls.2014.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoernstein S.N.W., Fode B., Wiedemann G., Lang D., Niederkrüger H., et al. Host cell proteome of Physcomitrella patens harbors proteases and protease inhibitors under bioproduction conditions. J Proteome Res. 2018;17:3749–3760. doi: 10.1021/acs.jproteome.8b00423. [DOI] [PubMed] [Google Scholar]
- 41.Hohe A., Egener T., Lucht J.M., Holtorf H., Reinhard C., et al. An improved and highly standardised transformation procedure allows efficient production of single and multiple targeted gene-knockouts in a moss, Physcomitrella patens. Curr Genet. 2004;44:339–347. doi: 10.1007/s00294-003-0458-4. [DOI] [PubMed] [Google Scholar]
- 42.Hori K., Maruyama F., Fujisawa T., Togashi T., Yamamoto N., et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun. 2014;5:3978. doi: 10.1038/ncomms4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu R., Li X., Hu Y., Zhang R., Lv Q., et al. Adaptive evolution of the enigmatic Takakia now facing climate change in Tibet. Cell. 2023;186:3558–3576. doi: 10.1016/j.cell.2023.07.003. e17. [DOI] [PubMed] [Google Scholar]
- 44.Hunter J.D. Matplotlib: A 2D graphics environment. Comput Sci Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 45.Jaillon O., Aury J.M., Noel B., Policriti A., Clepet C., et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 46.Johansson-Åkhe I., Wallner B. Improving peptide-protein docking with AlphaFold-Multimer using forced sampling. Front Bioinforma. 2022;2 doi: 10.3389/fbinf.2022.959160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson K.L., Cassin A.M., Lonsdale A., Bacic A., Doblin M.S., Schultz C.J. Pipeline to identify hydroxyproline-rich glycoproteins. Plant Physiol. 2017;174:886–903. doi: 10.1104/pp.17.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones P., Binns D., Chang H.Y., Fraser M., Li W., et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kieliszewski M.J., Lamport D.T.A. Extensin: repetitive motifs, functional sites, post‐translational codes, and phylogeny. Plant J. 1994;5:157–172. doi: 10.1046/j.1365-313X.1994.05020157.x. [DOI] [PubMed] [Google Scholar]
- 51.Kirbis A., Waller M., Ricca M., Bont Z., Neubauer A., et al. Transcriptional landscapes of divergent sporophyte development in two mosses, Physcomitrium (Physcomitrella) patens and Funaria hygrometrica. Front Plant Sci. 2020;11:747. doi: 10.3389/fpls.2020.00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konkina A., Klepadlo M., Lakehal A., Zein Z.El, Krokida A., et al. An Arabidopsis prolyl 4 hydroxylase is involved in the low oxygen response. Front Plant Sci. 2021;12 doi: 10.3389/fpls.2021.637352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koprivova A., Altmann F., Gorr G., Kopriva S., Reski R., Decker E.L. N‐glycosylation in the moss Physcomitrella patens is organized similarly to that in higher plants. Plant Biol. 2003;5:582–591. doi: 10.1055/s-2003-44721. [DOI] [Google Scholar]
- 54.Koski M.K., Hieta R., Hirsilä M., Rönkä A., Myllyharju J., Wierenga R.K. The crystal structure of an algal prolyl 4-hydroxylase complexed with a proline-rich peptide reveals a novel buried tripeptide binding motif. J Biol Chem. 2009;284:25290–25301. doi: 10.1074/jbc.M109.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lang E.G.E., Mueller S.J., Hoernstein S.N.W., Porankiewicz-Asplund J., Vervliet-Scheebaum M., Reski R. Simultaneous isolation of pure and intact chloroplasts and mitochondria from moss as the basis for sub-cellular proteomics. Plant Cell Rep. 2011;30:205–215. doi: 10.1007/s00299-010-0935-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lang D., Ullrich K.K., Murat F., Fuchs J., Jenkins J., et al. The Physcomitrella patens chromosome‐scale assembly reveals moss genome structure and evolution. Plant J. 2018;93:515–533. doi: 10.1111/tpj.13801. [DOI] [PubMed] [Google Scholar]
- 58.Lang D., van Gessel N., Ullrich K.K., Reski R. The genome of the model moss Physcomitrella patens. Adv Bot Res. 2016;78:97–140. doi: 10.1016/bs.abr.2016.01.004. [DOI] [Google Scholar]
- 59.Lee K.J.D., Sakata Y., Mau S.-L., Pettolino F., Bacic A., et al. Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens. Plant Cell. 2005;17:3051–3065. doi: 10.1105/tpc.105.034413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leszczuk A., Kalaitzis P., Kulik J., Zdunek A. Review: structure and modifications of arabinogalactan proteins (AGPs) BMC Plant Biol. 2023;23:45. doi: 10.1186/s12870-023-04066-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang R., You L., Dong F., Zhao X., Zhao J. Identification of hydroxyproline-containing proteins and hydroxylation of proline residues in rice. Front Plant Sci. 2020;11:1207. doi: 10.3389/fpls.2020.01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X., Wolfe R., Welch L.R., Domozych D.S., Popper Z.A., Showalter A.M. Bioinformatic identification and analysis of extensins in the plant kingdom. PLOS ONE. 2016;11 doi: 10.1371/journal.pone.0150177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lueth V.M., Reski R. Mosses. Curr Biol. 2023;33:R1175–R1181. doi: 10.1016/j.cub.2023.09.042. [DOI] [PubMed] [Google Scholar]
- 64.Ma Y., Yan C., Li H., Wu W., Liu Y., et al. Bioinformatics prediction and evolution analysis of arabinogalactan proteins in the plant kingdom. Front Plant Sci. 2017;8:66. doi: 10.3389/fpls.2017.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma H., Zhao J. Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.) J Exp Bot. 2010;61:2647–2668. doi: 10.1093/jxb/erq104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mao L., Kawaide H., Higuchi T., Chen M., Miyamoto K., et al. Genomic evidence for convergent evolution of gene clusters for momilactone biosynthesis in land plants. Proc Natl Acad Sci USA. 2020;117:12472–12480. doi: 10.1073/pnas.1914373117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marchant D.B., Chen G., Cai S., Chen F., Schafran P., et al. Dynamic genome evolution in a model fern. Nat Plants. 2022;8:1038–1051. doi: 10.1038/s41477-022-01226-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marchler-Bauer A., Bryant S.H. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marzol E., Borassi C., Bringas M., Sede A., Rodríguez Garcia D.R., et al. Filling the gaps to solve the extensin puzzle. Mol Plant. 2018;11:645–658. doi: 10.1016/j.molp.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 70.McKinney W. Data structures for statistical computing in python. Proc 9th Phyton Sci Conf. 2010:56–61. doi: 10.25080/Majora-92bf1922-00a. [DOI] [Google Scholar]
- 71.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mishler-Elmore J.W., Zhou Y., Sukul A., Oblak M., Tan L., et al. Extensins: self-assembly, crosslinking, and the role of peroxidases. Front Plant Sci. 2021;12 doi: 10.3389/fpls.2021.664738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mócsai R., Göritzer K., Stenitzer D., Maresch D., Strasser R., Altmann F. Prolyl hydroxylase paralogs in Nicotiana benthamiana show high similarity with regard to substrate specificity. Front Plant Sci. 2021;12 doi: 10.3389/fpls.2021.636597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mueller S.J., Lang D., Hoernstein S.N.W., Lang E.G.E., Schuessele C., et al. Quantitative analysis of the mitochondrial and plastid proteomes of the moss Physcomitrella patens reveals protein macrocompartmentation and microcompartmentation. Plant Physiol. 2014;164:2081–2095. doi: 10.1104/pp.114.235754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Munoz C., Schröder K., Henes B., Hubert J., Leblong S., et al. Phytochemical exploration of ceruchinol in moss: a multidisciplinary study on biotechnological cultivation of Physcomitrium patens (Hedw.) Mitt. Appl Sci. 2024;14:1274. doi: 10.3390/app14031274. [DOI] [Google Scholar]
- 76.Na S., Bandeira N., Paek E. Fast multi-blind modification search through tandem mass spectrometry. Mol Cell Proteom. 2012;11 doi: 10.1074/mcp.M111.010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nguyen N.D., Mirarab S., Kumar K., Warnow T. Ultra-large alignments using phylogeny-aware profiles. Genome Biol. 2015;16:124. doi: 10.1186/s13059-015-0688-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Niederau P.A., Eglé P., Willig S., Parsons J., Hoernstein S.N.W., et al. Multifactorial analysis of terminator performance on heterologous gene expression in Physcomitrella. Plant Cell Rep. 2024;43:43. doi: 10.1007/s00299-023-03088-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ouyang S., Zhu W., Hamilton J., Lin H., Campbell M., et al. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Owens N.W., Stetefeld J., Lattová E., Schweizer F. Contiguous O -galactosylation of 4(R)-hydroxy-L-proline residues forms very stable polyproline II helices. J Am Chem Soc. 2010;132:5036–5042. doi: 10.1021/ja905724d. [DOI] [PubMed] [Google Scholar]
- 81.Pace C.N., Vajdos F., Fee L., Grimsley G., Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parsons J., Altmann F., Arrenberg C.K., Koprivova A., Beike A.K., et al. Moss‐based production of asialo‐erythropoietin devoid of Lewis A and other plant‐typical carbohydrate determinants. Plant Biotechnol J. 2012;10:851–861. doi: 10.1111/j.1467-7652.2012.00704.x. [DOI] [PubMed] [Google Scholar]
- 83.Parsons J., Altmann F., Graf M., Stadlmann J., Reski R., Decker E.L. A gene responsible for prolyl-hydroxylation of moss-produced recombinant human erythropoietin. Sci Rep. 2013;3:3019. doi: 10.1038/srep03019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perez-Riverol Y., Bai J., Bandla C., Hewapathirana S., García-Seisdedos D., et al. The PRIDE database resources in 2022: A Hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Price M.N., Dehal P.S., Arkin A.P. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.R Core Team (2024). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria.
- 87.Ranawaka B., An J., Lorenc M.T., Jung H., Sulli M., et al. A multi-omic Nicotiana benthamiana resource for fundamental research and biotechnology. Nat Plants. 2023;9:1558–1571. doi: 10.1038/s41477-023-01489-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reimann L., Schwäble A.N., Fricke A.L., Mühlhäuser W.W.D., Leber Y., et al. Phosphoproteomics identifies dual-site phosphorylation in an extended basophilic motif regulating FILIP1-mediated degradation of filamin-C. Commun Biol. 2020;3:253. doi: 10.1038/s42003-020-0982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reski R., Bae H., Simonsen H.T. Physcomitrella patens, a versatile synthetic biology chassis. Plant Cell Rep. 2018;37:1409–1417. doi: 10.1007/s00299-018-2293-6. [DOI] [PubMed] [Google Scholar]
- 90.Reski R., Parsons J., Decker E.L. Moss‐made pharmaceuticals: from bench to bedside. Plant Biotechnol J. 2015;13:1191–1198. doi: 10.1111/pbi.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Röst H.L., Sachsenberg T., Aiche S., Bielow C., Weisser H., et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nat Methods. 2016;13:741–748. doi: 10.1038/nmeth.3959. [DOI] [PubMed] [Google Scholar]
- 92.Ruiz-Molina N., Parsons J., Decker E.L., Reski R. Structural modelling of human complement FHR1 and two of its synthetic derivatives provides insight into their in-vivo functions. Comput Struct Biotechnol J. 2023;21:1473–1486. doi: 10.1016/j.csbj.2023.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ruiz-Molina N., Parsons J., Schroeder S., Posten C., Reski R., Decker E.L. Process engineering of biopharmaceutical production in moss bioreactors via model-based description and evaluation of phytohormone impact. Front Bioeng Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.837965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saito F., Suyama A., Oka T., Yoko-o T., Matsuoka K., et al. Identification of novel peptidyl serine α-galactosyltransferase gene family in plants. J Biol Chem. 2014;289:20405–20420. doi: 10.1074/jbc.M114.553933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schultz C.J., Rumsewicz M.P., Johnson K.L., Jones B.J., Gaspar Y.M., Bacic A. Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiol. 2002;129:1448–1463. doi: 10.1104/pp.003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Seabold S., Perktold J. Statsmodels: Econometric and statistical modeling with python. Proc 9th Python Sci Conf. 2010:92–96. doi: 10.25080/Majora-92bf1922-011. [DOI] [Google Scholar]
- 97.Showalter A.M., Keppler B., Lichtenberg J., Gu D., Welch L.R. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 2010;153:485–513. doi: 10.1104/pp.110.156554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shpak E., Barbar E., Leykam J.F., Kieliszewski M.J. Contiguous hydroxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. J Biol Chem. 2001;276:11272–11278. doi: 10.1074/jbc.M011323200. [DOI] [PubMed] [Google Scholar]
- 99.Shpak E., Leykam J.F., Kieliszewski M.J. Synthetic genes for glycoprotein design and the elucidation of hydroxyproline- O -glycosylation codes. Proc Natl Acad Sci USA. 1999;96:14736–14741. doi: 10.1073/pnas.96.26.14736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Silva J., Ferraz R., Dupree P., Showalter A.M., Coimbra S. Three decades of advances in arabinogalactan-protein biosynthesis. Front Plant Sci. 2020;11 doi: 10.3389/fpls.2020.610377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Silva A.M.N., Vitorino R., Domingues M.R.M., Spickett C.M., Domingues P. Post-translational modifications and mass spectrometry detection. Free Radic Biol Med. 2013;65:925–941. doi: 10.1016/j.freeradbiomed.2013.08.184. [DOI] [PubMed] [Google Scholar]
- 102.Stenitzer D., Mócsai R., Zechmeister H., Reski R., Decker E.L., Altmann F. O-methylated N-glycans distinguish mosses from vascular plants. Biomolecules. 2022;12:136. doi: 10.3390/biom12010136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Suyama M., Torrents D., Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tan L., Leykam J.F., Kieliszewski M.J. Glycosylation motifs that direct arabinogalactan addition to arabinogalactan-proteins. Plant Physiol. 2003;132:1362–1369. doi: 10.1104/pp.103.021766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.The pandas development team (2020). pandas-dev/pandas: Pandas 1.3.4. Zenodo. 10.5281/zenodo.5574486. [DOI]
- 106.Tiainen P., Myllyharju J., Koivunen P. Characterization of a second Arabidopsis thaliana prolyl 4-hydroxylase with distinct substrate specificity. J Biol Chem. 2005;280:1142–1148. doi: 10.1074/jbc.M411109200. [DOI] [PubMed] [Google Scholar]
- 107.Top O., Milferstaedt S.W.L., van Gessel N., Hoernstein S.N.W., Özdemir B., et al. Expression of a human cDNA in moss results in spliced mRNAs and fragmentary protein isoforms. Commun Biol. 2021;4:964. doi: 10.1038/s42003-021-02486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Top O., Parsons J., Bohlender L.L., Michelfelder S., Kopp P., et al. Recombinant production of MFHR1, a novel synthetic multitarget complement inhibitor, in moss bioreactors. Front Plant Sci. 2019;10:260. doi: 10.3389/fpls.2019.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Toplak M., Wiedemann G., Ulicevic J., Daniel B., Hoernstein S.N.W., et al. The single berberine bridge enzyme homolog of Physcomitrella patens is a cellobiose oxidase. FEBS J. 2018;285:1923–1943. doi: 10.1111/febs.14458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tuskan G.A., DiFazio S., Jansson S., Bohlmann J., Grigoriev I., et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 112.Uetz P., Göritzer K., Vergara E., Melnik S., Gruenwald-Gruber C., et al. Implications of O-glycan modifications in the hinge region of a plant-produced SARS-CoV-2-IgA antibody on functionality. Front Bioeng Biotechnol. 2024;12:1329018. doi: 10.3389/fbioe.2024.1329018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Uetz P., Melnik S., Grünwald‐Gruber C., Strasser R., Stoger E. CRISPR/Cas9‐mediated knockout of a prolyl‐4‐hydroxylase subfamily in Nicotiana benthamiana using DsRed2 for plant selection. Biotechnol J. 2022;17 doi: 10.1002/biot.202100698. [DOI] [PubMed] [Google Scholar]
- 114.Vacic V., Iakoucheva L.M., Radivojac P. Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics. 2006;22:1536–1537. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]
- 115.van Holst G.-J., Varner J.E. Reinforced polyproline II conformation in a hydroxyproline-rich cell wall glycoprotein from carrot root. Plant Physiol. 1984;74:247–251. doi: 10.1104/pp.74.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van Rossum G., Drake F.L. CreateSpace; 2009. Python 3 Reference Manual. [Google Scholar]
- 117.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Velasquez S.M., Ricardi M.M., Poulsen C.P., Oikawa A., Dilokpimol A., et al. Complex regulation of prolyl-4-hydroxylases impacts root hair expansion. Mol Plant. 2015;8:734–746. doi: 10.1016/j.molp.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 119.Verdú-Navarro F., Moreno-Cid J.A., Weiss J., Egea-Cortines M. The advent of plant cells in bioreactors. Front Plant Sci. 2023;14 doi: 10.3389/fpls.2023.1310405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Waskom M. seaborn: statistical data visualization. J Open Source Softw. 2021;6:3021. doi: 10.21105/joss.03021. [DOI] [Google Scholar]
- 122.Weise A., Altmann F., Rodriguez‐Franco M., Sjoberg E.R., Bäumer W., et al. High‐level expression of secreted complex glycosylated recombinant human erythropoietin in the Physcomitrella Δ‐fuc‐t Δ‐xyl‐t mutant. Plant Biotechnol J. 2007;5:389–401. doi: 10.1111/j.1467-7652.2007.00248.x. [DOI] [PubMed] [Google Scholar]
- 123.Wiedemann G., van Gessel N., Köchl F., Hunn L., Schulze K., et al. RecQ helicases function in development, DNA repair, and gene targeting in Physcomitrella patens. Plant Cell. 2018;30:717–736. doi: 10.1105/tpc.17.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yu G., Smith D.K., Zhu H., Guan Y., Lam T.T.Y. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 125.Zhang J., Fu X.-X., Li R.-Q., Zhao X., Liu Y., et al. The hornwort genome and early land plant evolution. Nat Plants. 2020;6:107–118. doi: 10.1038/s41477-019-0588-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zimmer A.D., Lang D., Buchta K., Rombauts S., Nishiyama T., et al. Reannotation and extended community resources for the genome of the non-seed plant Physcomitrella patens provide insights into the evolution of plant gene structures and functions. BMC Genom. 2013;14:498. doi: 10.1186/1471-2164-14-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [22], [84] partner repository with the dataset identifier PXD051497 and 10.6019/PXD051497.