Summary
Background
Current surveillance modalities of osteosarcoma relapse exhibit limited sensitivity and specificity. Although circulating tumor DNA (ctDNA) has been established as a biomarker of minimal residual disease (MRD) in many solid tumors, a sensitive ctDNA detection technique has not been thoroughly explored for longitudinal MRD detection in osteosarcoma.
Methods
From August 2019 to June 2023, 59 patients diagnosed with osteosarcoma at the First Affiliated Hospital of Sun Yat-sen University were evaluated in this study. Tumor-informed MRD panels were developed through whole exome sequencing (WES) of tumor tissues. Longitudinal blood samples were collected during treatment and subjected to multiplex PCR-based next-generation sequencing (NGS). Kaplan–Meier curves and Log-rank tests were used to compare outcomes, and Cox regression analysis was performed to identify prognostic factors.
Findings
WES analysis of 83 patients revealed substantial mutational heterogeneity, with non-recurrent mutated genes accounting for 58.1%. Tumor-informed MRD panels were successfully obtained for 85.5% of patients (71/83). Among 59 patients with successful MRD panel customization and available blood samples, 13 patients exhibited positive ctDNA detection after surgery. Patients with negative post-operative ctDNA had better event-free survival (EFS) compared to those with positive ctDNA, at 1–6 months after surgery, after adjuvant chemotherapy, and more than 6 months after surgery (p < 0.05). In both univariate and multivariate Cox regression analysis, ctDNA results emerged as a significant predictor of EFS (p < 0.05). ctDNA detection preceded positive imaging in 5 patients, with an average lead time of 92.6 days. Thirty-nine patients remained disease-free, with ctDNA results consistently negative or turning negative during follow-up.
Interpretation
Our study underscores the applicability of tumor-informed deep sequencing of ctDNA in osteosarcoma MRD surveillance and, to our knowledge, represents the largest cohort to date. ctDNA detection is a significant prognostic factor, enabling the early identification of tumor relapse and progression compared to standard imaging, thus offering valuable insights in guiding osteosarcoma patient management.
Funding
The Grants of National Natural Science Foundation of China (No. 82072964, 82072965, 82203798, 82203026), the Natural Science Foundation of Guangdong (No. 2023A1515012659, 2023A1515010302), and the Regional Combination Project of Basic and Applied Basic Research Foundation of Guangdong (No. 2020A1515110010).
Keywords: Osteosarcoma relapse, Circulating tumor DNA (ctDNA), Minimal residual disease (MRD), Next-generation sequencing (NGS), Tumor-informed MRD panel
Research in context.
Evidence before this study
Current radiological modalities lack the requisite sensitivity and specificity in identifying osteosarcoma relapse. Circulating tumor DNA (ctDNA) is increasingly being implemented in cancer screening and recurrence surveillance as a noninvasive liquid biopsy, but it remains inadequately explored whether ctDNA detection can reflect minimal residual disease (MRD) in osteosarcoma.
Added value of this study
Due to the pronounced mutational heterogeneity of osteosarcoma, we utilized a tumor-informed ctDNA panel approach to investigate its applicability in MRD surveillance in 59 osteosarcoma patients. Patients with negative post-operative ctDNA had better event-free survival (EFS) compared to those with positive ctDNA, at 1–6 months after surgery, after adjuvant chemotherapy, and more than 6 months after surgery (p all <0.05). In both univariate and multivariate Cox regression analysis, ctDNA results emerged as a significant predictor of EFS (p all <0.05).
Implications of all the available evidence
Our study highlights the clinical relevance and applicability of tumor-informed deep sequencing of ctDNA for MRD surveillance in osteosarcoma patients, presenting the largest cohort to our knowledge. The strategic application of ctDNA sequencing holds promise for improved patient monitoring, allowing for timely interventions and potentially enhancing outcomes in osteosarcoma management.
Introduction
Osteosarcoma is the most common primary bone malignancy in children and adolescents.1 Despite the deepened biological understanding of osteosarcoma and the introduction of new treatment modalities, the 5-year overall survival rate of osteosarcoma have plateaued for several decades. It remains at 60–70% for localized disease and only 20–30% for metastatic disease.2 Disease monitoring with high sensitivity and minimal harm remains challenging with current surveillance methods. Although computed tomography (CT) and magnetic resonance imaging (MRI) are widely adopted, they lack the requested sensitivity and specificity, and no universally accepted and reliable predictive biomarkers of relapse have been established for osteosarcoma.3,4
Circulating tumor DNA (ctDNA) detection based on next-generation sequencing (NGS) is increasingly employed to monitor minimal residual disease (MRD) in various cancers, including acute myeloid leukemia (AML), breast cancer, and lung cancer.5, 6, 7 As a highly sensitive liquid biopsy methodology, NGS-based ctDNA detection can enable the identification of tumor relapse earlier than traditional radiological examinations, thereby facilitating prompt treatment decisions.8, 9, 10 Currently, two major designs of ctDNA panels exist: tumor-agnostic assays and tumor-informed (bespoke) assays.11,12 Tumor-agnostic assays utilize an “off-the-shelf” uniform panel and are more widely adopted in AML and lung cancer, where patients harbor a limited set of mutated genes.13 However, due to the high heterogeneity of somatic mutations in osteosarcoma,14,15 the use of a tumor-agnostic MRD panel would compromise sensitivity, leading to reduced sequencing depth and overlooking unique somatic variants. Additionally, whole genome sequencing (WGS) has been investigated as an alternative approach for detecting ctDNA in osteosarcoma, Ewing sarcoma, and rhabdomyosarcoma.16, 17, 18 Nevertheless, the application of WGS in longitudinal MRD detection remains challenging due to its relatively high sequencing cost and lower sensitivity, compared to NGS-based ctDNA panel detection. There is no standardized strategy for MRD detection in osteosarcoma, and its implementation is still in its nascent stages.
Tumor-informed assays, based on the mutations existing in the tumor tissue, enable deep sequencing targeting known mutations and MRD detection, which has been validated across multiple solid tumors.19, 20, 21 To address the challenges of MRD surveillance in osteosarcoma, we utilized a tumor-informed ctDNA panel approach to investigate its predictive capability for tumor relapse in a prospective collection of blood and tissue samples.
Methods
Study design and patient enrollment
This study was carried out at the Department of Musculoskeletal Oncology, the First Affiliated Hospital of Sun Yat-sen University. Patients who were diagnosed with osteosarcoma between August 2019 and June 2023 and whose primary tumor samples were sent for whole exome sequencing were included for this study. Exclusion criteria comprised failure of MRD panel customization and absence of collected and sequenced blood samples.
The primary tumor tissues of patients were collected through biopsy or surgical resection. The peripheral blood samples of patients were collected at various timepoints: 1) at diagnosis before neoadjuvant chemotherapy; 2) during/after neoadjuvant chemotherapy before surgery; 3) within 1 month after surgery, before adjuvant chemotherapy; 4) during/after adjuvant chemotherapy.
Somatic mutation identification in tumor tissues and blood samples
Tumor-specific somatic single nucleotide variants (SNVs) were identified through whole exome sequencing (WES) of tumor tissue (sequencing depth: >300×) and matched germline DNA (sequencing depth: >100×), conducted by OrigiMed in Shanghai, China. The quality control protocol for WES included an assessment of tumor purity, specifying that the neoplastic fraction must constitute at least 10% of the total cellular composition. The OriSelector algorithm was employed to select 16 clonal SNVs based on PyClone,22 which informed the customization of the MRD panel. Pathogenic clonal SNVs were first enter into the panel, and if the number is less than 16, variants of unknown significance (VUS) were selected. Then the primers targeting these clonal SNVs were designed and the length of amplicon was around 100 base pairs.
For those patients with successful MRD panel customization, their peripheral blood was collected in Streck tubes and further processed for extraction of cell-free DNA (cfDNA) with the QIAamp Circulating Nuclear Acid Kit (QIAGEN, Venlo, Netherlands). Multiplex polymerase chain reaction (PCR) was performed using NEBNext Ultra II Q5 Master Mix (New England Biolabs, Inc., MA, USA), followed by library construction with the VAHTS® Universal DNA Library Prep Kit for Illumina V3 (Nanjing Vazyme Biotech Co. Ltd., Nanjing, China). Sequencing was carried out on a NovaSeq 6000 sequencer (Illumina Inc., CA, USA) at a depth of >100,000×. A noise filtration step was incorporated to systematically eliminate interference, such as germline mutations and clonal hematopoietic variations, originating from the DNA of hematopoietic cells. Then molecular tracking and variant calling for SNVs were performed. If 3 or more variants were detected in a sample, it was considered ctDNA positive, which indicated the presence of MRD. The detection limit for variant allele frequency (VAF) is as low as 0.01%, with an input range of 25–100 ng DNA from an 8–20 ml blood sample. Mean tumor molecules per ml of plasma (MTM/ml) was calculated for patients with positive ctDNA results: MTM/ml = VAF ∗ cfDNA ∗ 1000/(3.3∗ plasma volume).23
Treatment and follow-up
Patients were treated with neoadjuvant chemotherapy, surgical resection, and adjuvant therapy sequentially. Patients received 2–4 rounds of neoadjuvant chemotherapy (n = 55) and then surgery (n = 59). 2–4 weeks after surgery, the patients received adjuvant therapy if no complications were noted. AJCC 8th classification system was used for patient staging.24 Tumor necrosis score was evaluated in the tumor after surgery, based on the Huvos classification.25 Imaging follow-up (MRI on surgical site and chest CT) was performed after surgery every 3–6 months.
Ethics statement
This study was approved by the institutional ethics committee for Clinical Research and Animal Trials of the First Affiliated Hospital of Sun Yat-sen University (approval ID: [2023]568). All patients included provided written informed consent.
Statistical analysis
Event-free survival (EFS) was calculated from the date of surgical resection to the date of radiologically verified tumor relapse (recurrence/metastasis), progression (for patients with metastasis at diagnosis) or death. Censorship was defined as no tumor relapse, progression, or death at last follow-up. Kaplan–Meier curves and Log-rank tests were used to compare outcomes. Cox regression analysis was performed to identify prognostic factors among age, sex, necrosis score, tumor stage, and postoperative ctDNA status. Nomogram was constructed based on the Cox regression prognostic model. Receiver operating characteristic curve (ROC) and area under the ROC curve (AUC) value were used to evaluate the performance of the prognostic model. Sensitivity, specificity, accuracy, positive predictive value, and negative predictive value were calculated to evaluate the performance of ctDNA detection. Lead time was defined as the time from first postoperative positive ctDNA sample to tumor relapse or progression verified by radiological examination. Statistical analyses were performed using the R software (version: 4.3.1) and a two-sided p value < 0.05 was considered statistically significant.
Role of funding source
The funding source of this study had no role in study design, data collection, data analysis, or manuscript preparation. All authors read, discussed, and approved the final version of the manuscript. The co-corresponding authors (J.Y. and J.S.) had full access to all the data in the study. All authors had the final responsibility for the decision to submit the manuscript for publication.
Results
Clinical characteristics and mutation landscape
The study design and patient enrollment are illustrated in Fig. 1A. Analysis of the WES results from 83 patients revealed that 58.1% (5212/8971, Fig. 1B) of mutated genes were shared by no more than one patient, and variants in 96.4% (17821/18486) of mutated genes were VUS. Additionally, 53.4% (149/279) of pathogenic mutated genes were shared by no more than one patient (Fig. 1B), with only 1.8% shared by more than ten patients. This finding underscores the inefficiency and impracticality of designing a universal MRD detection panel targeting frequently mutated osteosarcoma pathogenic genes. Therefore, a tumor-informed individualized MRD panel was utilized in this study to enhance the sensitivity and efficiency of ctDNA detection in osteosarcoma patients.
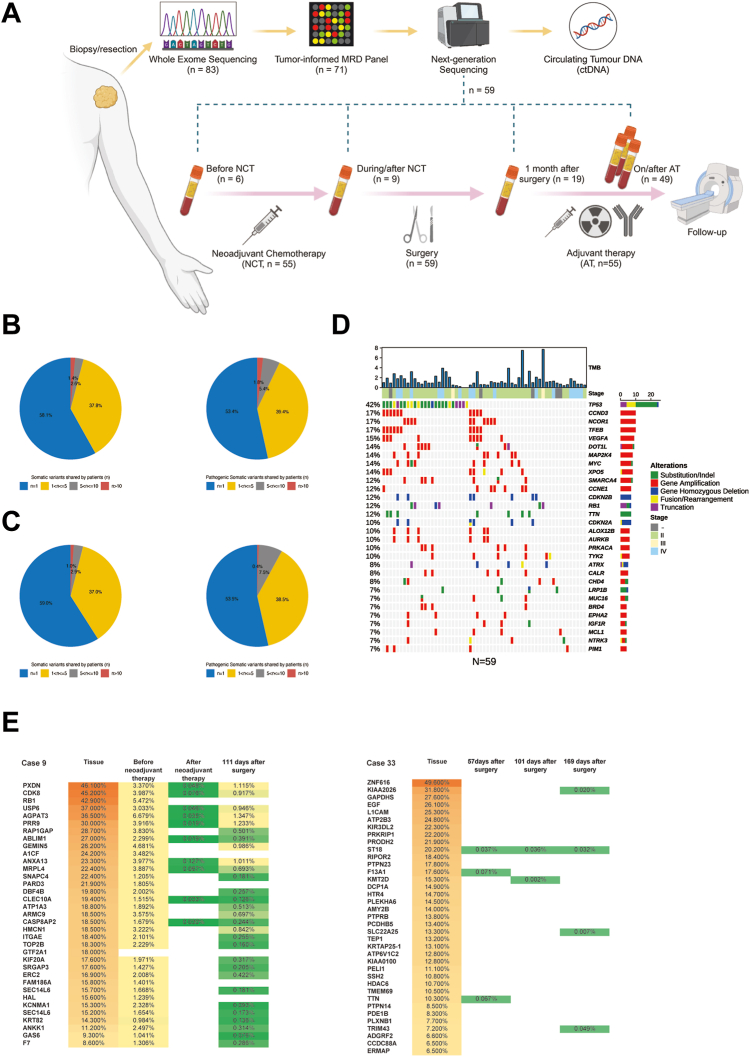
Fig. 1.
Study design and mutation distribution. (A) Study flowchart of enrollment process and treatment scheme. (B) Percentage of somatic variants/pathologic somatic variants shared among different numbers of patients in 83 osteosarcoma patients. (C) Percentage of somatic variants/pathologic somatic variants shared among different numbers of patients in 59 osteosarcoma patients included. (D) Mutation oncoplot based on WES results in 59 osteosarcoma patients included. (E) Heatmap of the variant allele frequency in WES and ctDNA sequencing for cases 9 and 33.
In 71 out of 83 patients (85.5%), a tumor-informed MRD panel was successfully generated, and 59 patients meeting both successful MRD panel customization and available blood sample detection were included in the study. Among the variants in the MRD panels of these patients, 98.0% were VUS (Table S1). Despite its uncharacterized clinical significance, VUS may reflect tumor growth and disease progression.26 To increase the sensitivity of MRD detection, VUS were not excluded during customization.
Clinicopathological characteristics are presented in Table 1. A highly sparse mutation profile was also observed among the 59 osteosarcoma patients (Fig. 1C). The majority (81.4%) of the WES samples were obtained from surgical resection. The mutational landscape for the 59 osteosarcoma patients depicted mutation profiles of top 30 mutated genes (Fig. 1D). Due to the significant heterogeneity in osteosarcoma, tumor-informed MRD panel approach increases the sensitivity of detecting tumor mutations compared to tumor-agnostic approach.
Table 1.
Clinicopathological characteristics.
| Characteristic | Number (Percentage) |
|---|---|
| Sex | |
| Male | 42 (71.2%) |
| Female | 17 (28.8%) |
| Age | |
| <18 years old | 40 (67.8%) |
| ≥18 years old | 19 (32.2%) |
| Tumor stage (AJCC 8th) at diagnosis | |
| II | 37 (62.7%) |
| III | 2 (3.4%) |
| IV | 15 (25.4%) |
| Uncertain | 5 (8.5%) |
| Sampling method for WES | |
| Surgery | 48 (81.4%) |
| Core-needle biopsy | 10 (16.9%) |
| Open biopsy | 1 (1.7%) |
| Lesion type | |
| Primary lesion | 52 (88.1%) |
| Recurrent lesion | 5 (8.5%) |
| Metastatic lesion | 2 (3.4%) |
| Adjuvant therapy | |
| Chemotherapy only | 50 (84.7%) |
| Chemotherapy + Radiotherapy | 1 (1.7%) |
| Chemotherapy + ICIs | 3 (5.1%) |
| Radiotherapy only | 1 (1.7%) |
| None | 4 (6.8%) |
WES, whole exome sequencing; ICIs, Immune checkpoint inhibitors.
Interestingly, the VAF of mutations exhibited divergence between tumor and ctDNA in specific cases. The predominant mutations in tumor samples did not consistently maintain their prominence in blood samples and were occasionally absent in longitudinal blood samples (Fig. 1E). Furthermore, the percentage of each variant in longitudinal blood samples fluctuated following neoadjuvant chemotherapy and surgery (Fig. 1E).
ctDNA detection before surgery
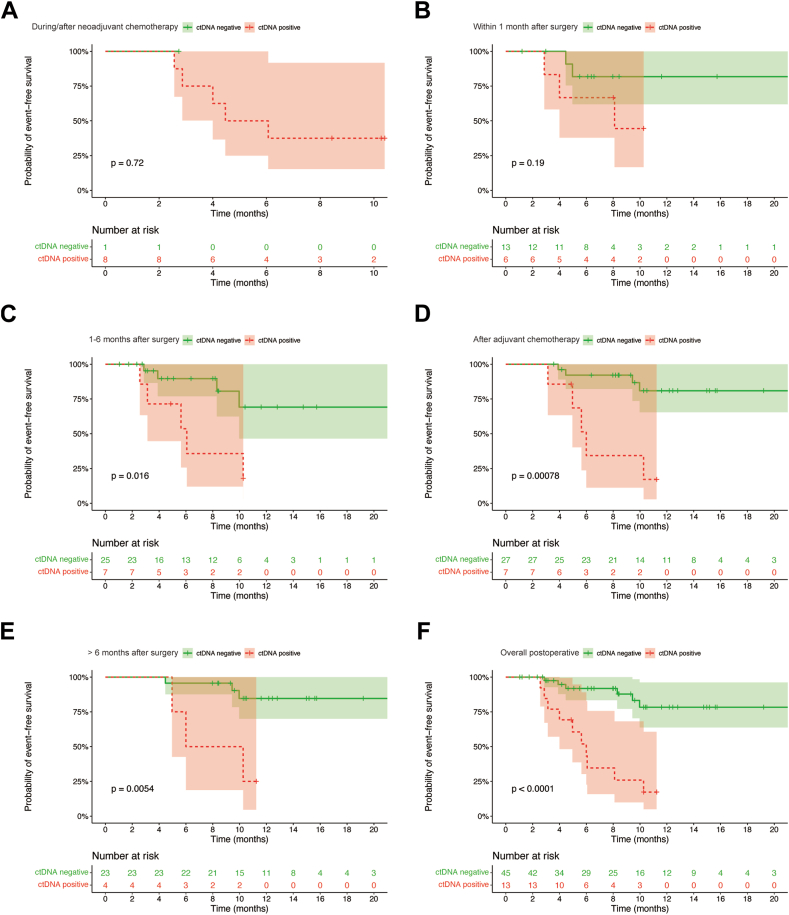
Blood samples obtained before the initial neoadjuvant chemotherapy were analyzed in 6 patients, with all 6 samples testing positive. ctDNA detection was performed during/after neoadjuvant chemotherapy in 9 patients, and only 1 patient had negative ctDNA results (positive rate: 88.9%). There was no significant difference in EFS between patients with negative ctDNA during/after neoadjuvant chemotherapy and those with positive (p = 0.72, respectively, Fig. 2A).
Fig. 2.
Longitudinal ctDNA detection through treatment. (A) EFS curve in patients with different ctDNA statuses during/after neoadjuvant chemotherapy. (B) EFS curve in patients with different ctDNA statuses within 1 month after surgery (before adjuvant chemotherapy). (C) EFS curve in patients with different ctDNA statuses in 1–6 months after surgery. (D) EFS curve in patients with different ctDNA statuses after adjuvant chemotherapy. (E) EFS curve in patients with different ctDNA statuses in >6 months after surgery. (F) EFS curve in overall patients with different ctDNA statuses after surgery.
ctDNA detection after surgery as a prognostic predictor
We hypothesized that longitudinal ctDNA detection post-surgery may provide a sensitive window into MRD status and serve as a viable surveillance strategy for tumor relapse. 58 patients provided at least one post-operative blood sample, with 29 patients contributing at least 2 samples at different timepoints (average: 1.8, range: 1–5). 13 patients exhibited positive ctDNA detection after surgery. The patients were followed up for 1.0–21.8 months (median: 8.1 months).
Survival differences between patients with distinct ctDNA statuses were assessed longitudinally. For patients who had not received adjuvant chemotherapy (within 1 month after surgery), no significant difference was observed between those patients with different ctDNA status (p = 0.19, Fig. 2B). Among 6 patients with positive ctDNA within 1 month after surgery, two patients (case 12 and 50) subsequently tested negative for ctDNA, with no observed relapse in their cases.
Patients whose ctDNA became negative or stayed negative had better EFS compared to those with positive ctDNA, at 1–6 months after surgery, after adjuvant chemotherapy, and more than 6 months after surgery (all p < 0.05. Fig. 2C–E). In total, patients with post-operative negative ctDNA demonstrated better EFS than those with positive ctDNA (p < 0.001, Fig. 2F).
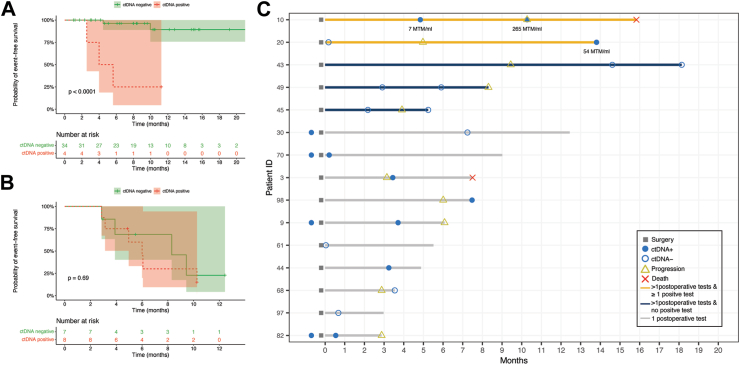
Subgroup analyses were conducted to determine whether post-operative ctDNA status could predict EFS in both localized and metastatic patients. Among localized stage I-III patients, those with positive ctDNA had significantly worse EFS compared to those with negative results (p < 0.001), while in metastatic stage IV patients, there was no significant difference observed (p = 0.69, Fig. 3A and B). Although post-operative ctDNA status did not differentiate outcomes in metastatic patients, the elevation in ctDNA levels or the transition from negative to positive ctDNA status correlated with tumor progression in one patient (case 10, Fig. 3C).
Fig. 3.
EFS curve in localized stage I-III osteosarcoma patients (A) and metastatic stage IV osteosarcoma patients (B). (C) Swimmer plot depicting the MRD surveillance and tumor events in metastatic stage IV patients.
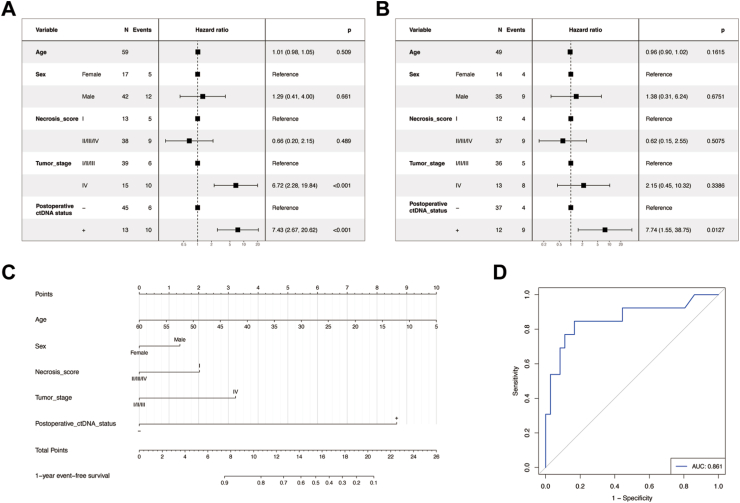
Both univariable and multivariable Cox regression analyses identified postoperative ctDNA status as a statistically significant variable (Fig. 4A and B). To be specific, the hazard ratio (HR) for positive ctDNA results were 7.43 (95% CI: 2.67–20.62) and 7.74 (95% CI: 1.55–38.75) in univariate and multivariate Cox regression analyses, respectively, indicating that a positive ctDNA result was a significant predictor of poor prognosis.
Fig. 4.
Univariate (A) and multivariate (B) Cox regression analysis of event-free survival by clinicopathological variables. (C) Nomogram of prognostic model comprised of clinicopathological factors and post-operative ctDNA. (D) The ROC of prognostic model.
Additionally, a nomogram was developed for the multivariable Cox regression prognostic model, encompassing age, sex, necrosis score, tumor stage, and postoperative ctDNA status (Fig. 4C), with an AUC of 0.861 (Fig. 4D).
ctDNA detection preceded imaging for recurrence detection
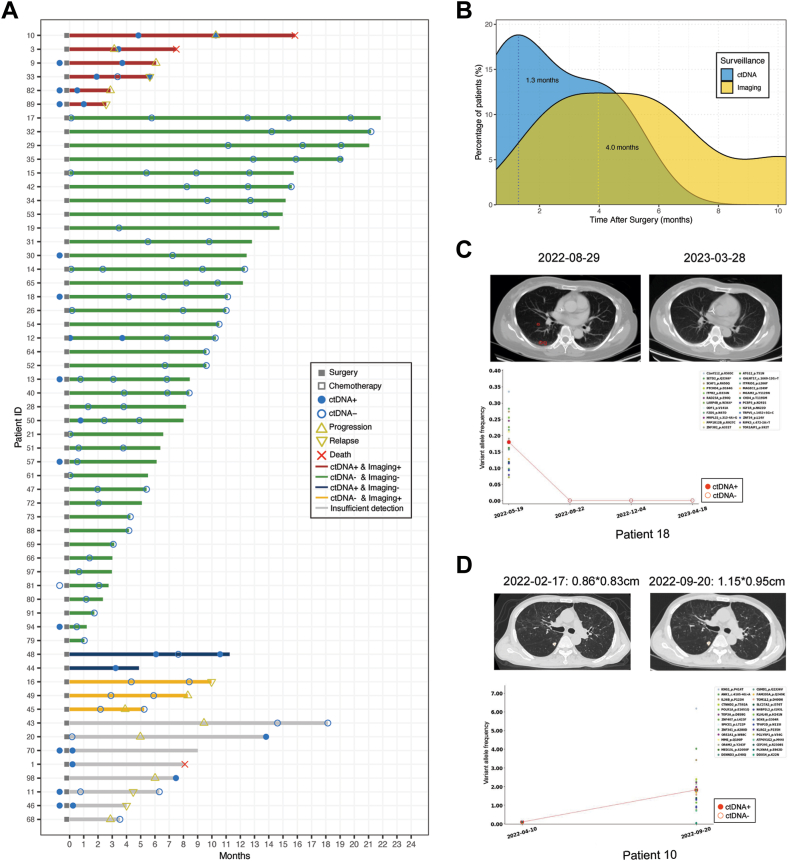
To assess the predictive performance of ctDNA detection, patients were categorized into different groups based on the relationship between tumor relapse/progression and ctDNA positivity (Fig. 5A). Six patients had positive ctDNA results within 3 months prior to tumor relapse/progression or around the same time, while two patients had positive ctDNA results but experienced no tumor relapse/progression after three months. Thirty-nine patients remained disease-free, with ctDNA results consistently negative or turning negative during follow-up. Three patients experienced tumor relapse/progression, but their ctDNA results within three months before relapse/progression were negative. Patients without ctDNA detection within three months before tumor relapse/progression were classified as having insufficient detection and were not included in sensitivity and specificity calculations (n = 8). Consequently, the sensitivity and specificity rates of ctDNA in predicting tumor relapse were 66.7% and 95.1%, respectively, with an accuracy rate of 90.0%. The positive predictive value and negative predictive value were 75.0% and 92.9%, respectively.
Fig. 5.
MRD surveillance precedes imaging. (A) Swimmer plot depicting MRD surveillance and tumor events. (B) Density curve in 5 patients indicating the peak of positive ctDNA occurred 2.7 months earlier than that of imaging examinations. (C) Negative ctDNA results helped differentiate pulmonary nodules (red circle) in case 18. (D) ctDNA quantification in case 10 reflected treatment efficacy.
ctDNA detection identified relapse/progression events earlier than imaging in 5 patients (Fig. 5A), and average lead time was 92.6 days. The peak for positive ctDNA occurred 2.7 months earlier than the peak for relapse/progression verified by imaging (Fig. 5B).
Twenty-seven patients (45.8%) had blood samples tested 6 months after surgery. ctDNA was detected as positive in case 10 and case 33 almost concurrently with recurrences, because blood samples were taken along with imaging follow-up. But in these two cases, patients also have previous positive predictive results (Fig. 5A).
Representative cases were selected to highlight the clinical significance of MRD surveillance in osteosarcoma. In case 18 (Fig. 5C), a 53-year-old female diagnosed with osteosarcoma, a chest CT performed 3 months after surgery revealed multiple pulmonary nodules with uncertain nature, necessitating imaging follow-up to differentiate between inflammatory nodules and neoplasms. However, all post-operative ctDNA results were negative, and 10 months after surgery, pulmonary nodules disappeared on chest CT. In case 10 (Fig. 5D), a 58-year-old male diagnosed with osteosarcoma, ctDNA quantification increased during adjuvant chemotherapy, and a chest CT confirmed the enlargement of pulmonary metastasis. These cases demonstrate the utility of ctDNA in differentiating suspicious metastasis, identifying early relapse, and monitoring treatment efficacy.
Discussion
ctDNA is increasingly being implemented in clinical practice, as the capacity of this technique has been successfully shown in a variety of settings of the clinical cancer care continuum from cancer screening, evaluation of treatment efficacy, detection of MRD to recurrence surveillance.27 In the context of recurrence surveillance, positive ctDNA serves as a sensitive early indicator of tumor recurrence, while serial negative ctDNA in longitudinal testing suggests a lower risk of recurrence or metastasis, which could guide the selection of adjuvant therapy. It remains inadequately explored whether ctDNA detection can reflect MRD and identify early tumor relapse in osteosarcoma patients.
Barris et al. demonstrated tumor specific somatic mutations through an NGS-based 7-gene-panel, confirming the presence of ctDNA in 8 osteosarcoma patients. However, the prognostic significance was not explored extensively due to the small sample size and low depth of coverage (mean: 698.4.1×).28 Low passage whole genome sequencing (lpWGS) has been explored as an alternative strategy for ctDNA detection in osteosarcoma.16,29 Audinot et al. conducted lpWGS in 183 osteosarcoma patients for copy number alteration detection, revealing that ctDNA quantification at diagnosis is a major prognostic factor.16 However, ctDNA quantification by lpWGS at the time of surgery and at the end of treatment failed to be prognostic factors in this study due to the low sensitivity of lpWGS, rendering it unsuitable for longitudinal MRD detection.16
It has been validated in multiple solid tumors that tumor-informed strategy can be used for MRD detection. Therefore, we opted for a tumor-informed strategy in designing the MRD panel and conducted MRD surveillance in 59 osteosarcoma patients, constituting the largest cohort for MRD surveillance in osteosarcoma to our knowledge.
In our study, the WES results in tumor samples of 83 osteosarcoma patients illustrated a high degree of mutational heterogeneity (Fig. 1B). Given this pronounced heterogeneity, we hypothesized that the adoption of a tumor-informed MRD panel would be a more practical and cost-effective approach with heightened sensitivity and specificity, as the panel incorporates only 16–40 tumor-specific variants and allows deep sequencing. Conversely, a universal MRD panel would necessitate the inclusion of hundreds of targets, leading to elevated costs, particularly in the context of longitudinal multiple testing, and compromised sensitivity due to a lower sequencing depth.
Our results indicate that clonal somatic variants in osteosarcoma are predominantly VUS, and VUS accounted for 98.0% of the variants in the MRD panel. This observation underscores the unmet need to comprehend the potential role of VUS in osteosarcoma tumorigenesis and suggests that VUS should be considered when designing MRD panels and exploring potential therapeutic targets.
We successfully generated a tumor informed MRD panel in 85.5% of osteosarcoma patients (71/83), confirming the feasibility of tumor-informed MRD panel customization for the majority of osteosarcoma patients. 12 patients failed in the customization of MRD, primarily due to reasons such as having fewer than 16 clonal mutations or the presence of alternative structural variants, including fusion, rearrangement, and amplification.
ctDNA is a valuable tool for tracking the dynamic change of tumor-specific variants. The variants with highest fraction in the primary tumor didn't consistently rank as most frequent during longitudinal ctDNA detection, whereas the fraction of those variants with low VAF in the primary tumor occasionally increased when tumor relapse occurred, potentially revealing tumor evolution in response to treatment (Fig. 1E).
In this study, we demonstrate that post-operative ctDNA status serves as a significant predictor of EFS and acts as a sensitive and specific biomarker for MRD surveillance in osteosarcoma patients. For metastatic osteosarcoma patients, the rise in ctDNA levels or the transition from negative to positive ctDNA status could potentially serve as a marker of tumor progression. As tumor-informed ctDNA detection is highly sensitive, ctDNA quantification may reflect tumor burden when the residual disease is known to exist. Importantly, post-operative ctDNA identified tumor relapse earlier than radiological examinations, with an average lead time of 92.6 days. A nomogram prognostic model was developed to predict patient survival based on clinicopathological characteristics and ctDNA status.
Audinot et al. and Shulman et al. utilized WGS to quantify ctDNA in osteosarcoma.16,29 Both found that ctDNA detection by WGS at diagnosis was a significant predictor of patient outcome, but they did not monitor MRD in those patients due to the low sensitivity of WGS. Our findings demonstrates that tumor-informed ctDNA detection is more suitable for MRD surveillance than WGS-based ctDNA detection, with the advantages of high sensitivity and relatively low cost for longitudinal, repeated detection. Nevertheless, this technique may miss newly acquired mutations due to tumor evolution.11 Moreover, three patients were observed with tumor relapse or progression, yet their ctDNA stayed negative. The negative ctDNA results could be attributed to poor representation of the MRD panel, the acquisition of new mutations during treatment or evolution, and low quality of ctDNA samples. Longitudinal ctDNA detection can reflect patients’ response to adjuvant chemotherapy. Two patients (case 12 and 50) with ctDNA clearance during adjuvant chemotherapy remained disease-free until last follow-up, and thus, ctDNA clearance may indicate a good response to therapy. Conversely, persistent positive ctDNA with stable or increased VAF strongly suggests the presence of MRD and resistance to current therapy. MRD surveillance through longitudinal ctDNA detection could significantly contribute to monitoring treatment efficacy and guiding decisions regarding de-escalation or escalation of chemotherapy.
Considering the predilection of osteosarcoma in children and adolescents, the significant side effects of radiological surveillance cannot be overlooked. Radiation exposure from CT scans elevates the risk of brain tumors and hematological malignancies.30,31 Adopting ctDNA-based MRD detection has the potential to mitigate the risk of secondary malignancies by reducing the frequency of CT scans in persistently ctDNA-negative patients.
Therefore, the potential clinical applications of ctDNA detection in osteosarcoma can be envisioned as follows: 1) early identification of tumor relapse, enabling timely management; 2) escalation and/or extension of adjuvant chemotherapy in patients with persistently positive ctDNA; 3) de-escalation of adjuvant chemotherapy and reduction of radiological examinations in patients with persistently negative ctDNA; 4) guiding the management of osteosarcoma patients who have undergone unplanned tumor resection. Prospective studies should be conducted to further delineate the role of ctDNA-guided management in the clinical setting of osteosarcoma treatment.
The limitation of our study is primarily that not all patients underwent regular sequential blood withdrawal, resulting in a reduced effective sample size for survival analysis. Second, the relatively short follow-up duration precludes the evaluation of ctDNA's predictive value in overall survival. Third, a validation cohort is needed to confirm the findings and facilitate clinical application.
In summary, our study highlights the clinical relevance and applicability of tumor-informed deep sequencing of ctDNA for MRD surveillance in osteosarcoma patients, presenting the largest cohort to our knowledge. Notably, our findings suggest that ctDNA has the potential to enable earlier detection of tumor recurrence and disease progression compared to conventional imaging modalities. The strategic application of ctDNA sequencing holds promise for improved patient monitoring, allowing for timely interventions and potentially enhancing outcomes in osteosarcoma management. However, these observations are still indicative rather than definitive and require further validation in larger and more diverse populations to establish their clinical utility.
Contributors
J.Y. and J.S. conceived and implemented the study. G.H., C.Z., X.X., J.S., and J.Y. supervised the sample collection and assessment. Y.F., Y.X., W.L., J.Z., and F.W. collected clinical information. Q.J., F.P., F.L. and K.W. performed experiment and data curation. Y.F. and Q.J. performed data analysis and visualization. The manuscript was prepared and revised by Y.F., Y.X., W.L., J.Z., A.H.K., D.M., and J.Y. with the input from all co-authors.
Data sharing statement
The data presented in this study are available on request from the corresponding author.
Declaration of interests
Q.J., F.P., F.L. and K.W. are employees of OrigiMed. A.H.K. is a consultant for Monteris Medical and received a research grant on dural substitute from Stryker. The other authors declare no competing interests.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82072964, 82072965, 82203798, 82203026), the Natural Science Foundation of Guangdong (No. 2023A1515012659, 2023A1515010302), and the Regional Combination Project of Basic and Applied Basic Research Foundation of Guangdong (No. 2020A1515110010).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2024.102697.
Contributor Information
Jingnan Shen, Email: shenjn@mail.sysu.edu.cn.
Junqiang Yin, Email: yinjunq@mail.sysu.edu.cn.
Appendix A. Supplementary data
References
- 1.Beird H.C., Bielack S.S., Flanagan A.M., et al. Osteosarcoma. Nat Rev Dis Primers. 2022;8(1):1–19. doi: 10.1038/s41572-022-00409-y. [DOI] [PubMed] [Google Scholar]
- 2.Durfee R.A., Mohammed M., Luu H.H. Review of osteosarcoma and current management. Rheumatol Ther. 2016;3(2):221–243. doi: 10.1007/s40744-016-0046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallander K., Öfverholm I., Boye K., et al. Sarcoma care in the era of precision medicine. J Intern Med. 2023;294(6):690–707. doi: 10.1111/joim.13717. [DOI] [PubMed] [Google Scholar]
- 4.Aran V., Devalle S., Meohas W., et al. Osteosarcoma, chondrosarcoma and Ewing sarcoma: clinical aspects, biomarker discovery and liquid biopsy. Crit Rev Oncol Hematol. 2021;162 doi: 10.1016/j.critrevonc.2021.103340. [DOI] [PubMed] [Google Scholar]
- 5.Yu S., Lin T., Nie D., et al. Dynamic assessment of measurable residual disease in favorable-risk acute myeloid leukemia in first remission, treatment, and outcomes. Blood Cancer J. 2021;11(12):195. doi: 10.1038/s41408-021-00591-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipsyc-Sharf M., de Bruin E.C., Santos K., et al. Circulating tumor DNA and late recurrence in high-risk hormone receptor-positive, human epidermal growth factor receptor 2-negative breast cancer. J Clin Oncol. 2022;40(22):2408–2419. doi: 10.1200/JCO.22.00908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J.T., Liu S.Y., Gao W., et al. Longitudinal undetectable molecular residual disease defines potentially cured population in localized non–small cell lung cancer. Cancer Discov. 2022;12(7):1690–1701. doi: 10.1158/2159-8290.CD-21-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Underwood J.J., Quadri R.S., Kalva S.P., et al. Liquid biopsy for cancer: review and implications for the radiologist. Radiology. 2020;294(1):5–17. doi: 10.1148/radiol.2019182584. [DOI] [PubMed] [Google Scholar]
- 9.Tivey A., Church M., Rothwell D., Dive C., Cook N. Circulating tumour DNA — looking beyond the blood. Nat Rev Clin Oncol. 2022;19(9):600–612. doi: 10.1038/s41571-022-00660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pantel K., Alix-Panabières C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol. 2019;16(7):409–424. doi: 10.1038/s41571-019-0187-3. [DOI] [PubMed] [Google Scholar]
- 11.Yoest J.M., Shirai C.L., Duncavage E.J. Sequencing-based measurable residual disease testing in acute myeloid leukemia. Front Cell Dev Biol. 2020;8:249. doi: 10.3389/fcell.2020.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malla M., Loree J.M., Kasi P.M., Parikh A.R. Using circulating tumor DNA in colorectal cancer: current and evolving practices. J Clin Oncol. 2022;40(24):2846–2857. doi: 10.1200/JCO.21.02615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santini D., Botticelli A., Galvano A., et al. Network approach in liquidomics landscape. J Exp Clin Cancer Res. 2023;42:193. doi: 10.1186/s13046-023-02743-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nacev B.A., Sanchez-Vega F., Smith S.A., et al. Clinical sequencing of soft tissue and bone sarcomas delineates diverse genomic landscapes and potential therapeutic targets. Nat Commun. 2022;13(1):3405. doi: 10.1038/s41467-022-30453-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu C.C., Beird H.C., Andrew Livingston J., et al. Immuno-genomic landscape of osteosarcoma. Nat Commun. 2020;11:1008. doi: 10.1038/s41467-020-14646-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Audinot B., Drubay D., Gaspar N., et al. ctDNA quantification improves estimation of outcomes in patients with high-grade osteosarcoma: a translational study from the OS2006 trial. Ann Oncol. 2024;35(6):559–568. doi: 10.1016/j.annonc.2023.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Peneder P., Stütz A.M., Surdez D., et al. Multimodal analysis of cell-free DNA whole-genome sequencing for pediatric cancers with low mutational burden. Nat Commun. 2021;12:3230. doi: 10.1038/s41467-021-23445-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abbou S., Klega K., Tsuji J., et al. Circulating tumor DNA is prognostic in intermediate-risk rhabdomyosarcoma: a report from the children's Oncology group. J Clin Oncol. 2023;41(13):2382–2393. doi: 10.1200/JCO.22.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santonja A., Cooper W.N., Eldridge M.D., et al. Comparison of tumor-informed and tumor-naïve sequencing assays for ctDNA detection in breast cancer. EMBO Mol Med. 2023;15(6) doi: 10.15252/emmm.202216505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abbosh C., Frankell A.M., Harrison T., et al. Tracking early lung cancer metastatic dissemination in TRACERx using ctDNA. Nature. 2023;616(7957):553–562. doi: 10.1038/s41586-023-05776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhiman A., Kothary V., Witmer H.D.D., et al. Role of tumor-informed personalized circulating tumor DNA assay in informing recurrence in patients with peritoneal metastases from colorectal and high-grade appendix cancer undergoing curative-intent surgery. Ann Surg. 2023;278(6):925–931. doi: 10.1097/SLA.0000000000005856. [DOI] [PubMed] [Google Scholar]
- 22.Roth A., Khattra J., Yap D., et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014;11(4):396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalashnikova E., Aushev V.N., Malashevich A.K., et al. Correlation between variant allele frequency and mean tumor molecules with tumor burden in patients with solid tumors. Mol Oncol. 2023;23:1878. doi: 10.1002/1878-0261.13557. 0261.13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amin MB, American Joint Committee on Cancer, American Cancer Society, editors. AJCC cancer staging manual. Eight edition/editor-in-chief, Mahul B. Amin, MD, FCAP; editors, Stephen B. Edge, MD, FACS [and 16 others]; Donna M. Gress, RHIT, CTR-Technical editor ; Laura R. Meyer, CAPM-Managing editor. American joint committee on cancer, Springer; Chicago IL: 2017. p. 1024. [Google Scholar]
- 25.Rosen G., Caparros B., Huvos A.G., et al. Preoperative chemotherapy for osteogenic sarcoma: selection of postoperative adjuvant chemotherapy based on the response of the primary tumor to preoperative chemotherapy. Cancer. 1982;49(6):1221–1230. doi: 10.1002/1097-0142(19820315)49:6<1221::aid-cncr2820490625>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 26.Baumgartner J.M., Riviere P., Lanman R.B., et al. Prognostic utility of pre- and postoperative circulating tumor DNA liquid biopsies in patients with peritoneal metastases. Ann Surg Oncol. 2020;27(9):3259–3267. doi: 10.1245/s10434-020-08331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krebs M.G., Malapelle U., André F., et al. Practical considerations for the use of circulating tumor DNA in the treatment of patients with cancer: a narrative review. JAMA Oncol. 2022;8(12):1830–1839. doi: 10.1001/jamaoncol.2022.4457. [DOI] [PubMed] [Google Scholar]
- 28.Barris D.M., Weiner S.B., Dubin R.A., et al. Detection of circulating tumor DNA in patients with osteosarcoma. Oncotarget. 2018;9(16):12695–12704. doi: 10.18632/oncotarget.24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shulman D.S., Klega K., Imamovic-Tuco A., et al. Detection of circulating tumour DNA is associated with inferior outcomes in Ewing sarcoma and osteosarcoma: a report from the children's oncology group. Br J Cancer. 2018;119(5):615–621. doi: 10.1038/s41416-018-0212-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meulepas J.M., Ronckers C.M., Smets A.M.J.B., et al. Radiation exposure from pediatric CT scans and subsequent cancer risk in The Netherlands. J Natl Cancer Inst. 2018;111(3):256–263. doi: 10.1093/jnci/djy104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosch de Basea Gomez M., Thierry-Chef I., Harbron R., et al. Risk of hematological malignancies from CT radiation exposure in children, adolescents and young adults. Nat Med. 2023;29(12):3111–3119. doi: 10.1038/s41591-023-02620-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.