Abstract
Previous work has shown that spleen necrosis virus (SNV) long terminal repeats (LTRs) are associated with Rex/Rex-responsive element-independent expression of bovine leukemia virus RNA and supports the hypothesis that SNV RNA contains a cis-acting element that interacts with cellular Rex-like proteins. To test this hypothesis, the human immunodeficiency virus type 1 (HIV) Rev/RRE-dependent gag gene was used as a reporter to analyze various SNV sequences. Gag enzyme-linked immunosorbent assay and Western blot analyses reveal that HIV Gag production is enhanced at least 20,000-fold by the 5′ SNV LTR in COS, D17, and 293 cells. Furthermore, SNV RU5 in the sense but not the antisense orientation is sufficient to confer Rev/RRE-independent expression onto a cytomegalovirus-gag plasmid. In contrast, the SNV 3′ LTR and 3′ untranslated sequence between env and the LTR did not support Rev-independent gag expression. Quantitative RNase protection assays indicate that the SNV 5′ RNA terminus enhances cytoplasmic accumulation and polysome association of HIV unspliced and spliced transcripts. However, comparison of the absolute amounts of polysomal RNA indicates that polysome association is not sufficient to account for the significant increase in Gag production by the SNV sequences. Our analysis reveals that the SNV 5′ RNA terminus contains a unique cis-acting posttranscriptional control element that interacts with hypothetical cellular Rev-like proteins to facilitate HIV RNA transport and efficient translation.
Retroviruses require cytoplasmic expression of unspliced RNA to produce infectious progeny. Complex retroviruses including human immunodeficiency virus type 1 (HIV) and bovine leukemia virus (BLV) exert similar posttranscriptional control by their regulatory protein and cis-acting responsive elements, Rev/Rex and RRE/RxRE, respectively. Rev and RRE are necessary for efficient transport, stability, and translation of unspliced HIV RNAs (1–3, 9–11, 13, 15, 16, 18–20, 27). Simple retroviruses lack analogous regulatory protein. However, recent studies have identified cis-acting elements in some simple retroviruses that function in conjunction with a Rev-like cellular factor(s) to modulate cytoplasmic expression of their unspliced RNA (4, 14, 23, 29, 32, 33, 38). These elements, designated constitutive/cytoplasmic transport elements or cis-acting trans-activation elements (CTEs), have been identified in Mason-Pfizer monkey virus (MPMV) (4), the related simian retrovirus 1 (SRV-1) (38), and the avian Rous sarcoma virus (RSV) (23, 29). The CTEs are structured RNA elements positioned in the 3′ untranslated region (UTR) (12, 24, 32) and were identified by their ability to replace HIV Rev/RRE function in subgenomic HIV plasmids (4, 23, 38). They function to increase stability and nucleocytoplasmic transport of unspliced transcripts. The RSV CTE is proposed also to facilitate efficient processing of Gag precursor protein (29).
Recent characterization of BLV retroviral vector genomes that contain spleen necrosis virus (SNV) long terminal repeats (LTRs) revealed Rex/RxRE-independent expression of BLV structural gene vectors (5, 6). This observation indicates that SNV RNA may contain a cis-acting element that interacts with cellular Rex-like factors. SNV is an avian simple retrovirus that is unrelated to MPMV, SRV-1, or RSV and is instead related to murine leukemia virus (37).
The goal of this study was to test the hypothesis that SNV RNA contains a CTE that would interact with cellular Rev-like factors. Our analysis focused on two SNV regions: the 3′ UTR that corresponds to the position of the MPMV, SRV-1, and RSV CTEs; and the LTRs, because BLV structural gene vectors that contain the SNV LTRs are Rex/RxRE independent (5). Our data eliminate the possibility that the SNV 3′ UTR and 3′ LTR facilitate Rev-independent gene expression and establish that the SNV 5′ LTR functions in a position-dependent manner to facilitate Rev/RRE-independent expression of HIV gag RNA. The SNV 5′ LTR facilitates cytoplasmic accumulation and polysome association of HIV unspliced and spliced RNAs. These data identify a novel retrovirus posttranscriptional control element located at the 5′ terminus of a simple retrovirus RNA that facilitates Rev/RRE-independent expression of unspliced and spliced HIV RNAs.
MATERIALS AND METHODS
Plasmid construction.
HIV-based plasmid pSVgagpol-rre, pSVgagpol, or pBBgagpol encodes HIV Gag, contains the simian virus 40 (SV40) promoter, and either contains RRE, lacks RRE but contains a β-globin intron, or lacks RRE and a β-globin intron, respectively (30). To construct derivatives of each of these plasmids, the SNV 3′ UTR was excised from pKB477 on a BamHI/BglII fragment and subcloned into the BamHI site of pSVgagpol-rre, pSVgagpol, or pBBgagpol to construct pKB634, pKB636, or pKB637, respectively. The SNV LTR was excised from pKB404 on a BamHI fragment and subcloned into the BamHI site of pSVgagpol-rre, pSVgagpol, or pBBgagpol to construct pKB624, pKB628, or pKB632, respectively.
pKB504gagpol was constructed in five steps beginning with pKB404, which contains two copies of the SNV LTR ligated at opposite ends of the multiple cloning site in pUC19 (5). pKB404 modified by insertion of the HIV polypurine tract (PPT) (HIVBRU coordinates 8662 to 8699 [36]) on an oligonucleotide at the SphI/HindIII sites adjacent to the 3′ SNV LTR to create pKB504. HIVBRU sequences from U5 through gag (100 to 2040) were amplified by eight cycles of PCR by Taq polymerase with primers having EcoRI/XbaI termini and ligated into pUC19. Subsequently, the EcoRI fragment that encompasses HIV coordinates 100 to 2040 was subcloned at the EcoRI site of pKB504. HIV coordinates 1521 to 4655 (HIV gag-pol) were amplified with primers having XbaI termini, ligated into pUC19, and then subcloned into the preceding plasmid at ApaI (1552) and XbaI (4655) sites to construct pKB504gagpol. The MPMV CTE was PCR amplified from MPMV provirus pSHRM-15 (coordinates 8022 to 8193; gift from Eric Hunter [32]) with primers having XhoI termini and inserted into the SalI site of pKB504gagpol to create pKB504gagpolCTE.
To construct pYW100, pKB504gagpol was modified by deletion of the region between the HIV PPT and the 3′ SNV LTR and replacement with a heterologous polyadenylation signal, p(A). pKB504gagpol was digested with AflIII, treated with Klenow enzyme and digested with XbaI. pCMVglobinSPA (gift from Dan Schoenberg), which contains an optimized 47-base synthetic p(A), was digested with HindIII, treated with Klenow enzyme, and digested with XbaI, and the fragment containing p(A) was ligated with the vector backbone to make pYW100. An intermediate plasmid, pYW201, was constructed by ligation into pUC19 of the BamHI fragment of pKB504gagpol that contains the sequence from HIV U5 through the 3′ SNV LTR. Then the cytomegalovirus (CMV) immediate-early (IE) promoter of pCMVglobinSPA was excised by using SalI, treated with Klenow enzyme and ligated at the SmaI site to make pYW202. To construct pYW203, a deleted SNV LTR that lacks the 3′ 29 bases of R and 60 bases of U5 (Δ486-575) was amplified by PCR, treated with Klenow enzyme and ligated at the SmaI site of pYW201. To construct pYW209, a deleted SNV LTR that lacks the U5 (Δ512-575) was amplified by PCR, treated with Klenow enzyme, and ligated at the SmaI site of pYW201. To construct pYW99, the SphI fragment of pYW202 that contains the CMV promoter and 5′ gag gene were ligated to the SphI fragment of pYW100 that contains the 3′ region of gag and p(A). pYW204 was constructed by ligation of SphI fragments of pYW203 and pYW100. pYW205 was constructed by deletion of RU5 sequences starting at +2 (Δ436-575) by AvaI/BamHI digestion followed by treatment with Klenow enzyme and blunt-end ligation. To construct pYW207 and pYW208, SNV RU5 was excised with AvaI from pKB402 (SNV positions 435 to 599 and pUC19 positions 396 to 412), treated with Klenow enzyme, and ligated at the Klenow enzyme-treated BamHI site of pYW99. The plasmid with the antisense RU5 orientation is pYW207, and the plasmid with the sense orientation is pYW208. The sequence of each plasmid was verified by DNA sequencing of the 5′ transcriptional control region through gag and the 3′ UTR through the LTR or heterologous p(A). pGEM(140-440) was derived from pGEM(400-600) of McBride and Panganiban (22) by replacement of the HIVNL4-3 5′ UTR with the HIVBRU 5′ UTR. Plasmid pMBSVT7 was constructed by PCR amplification of the 5′ UTR regions of pSVgag-pol-rre and ligation into the SrfI site of PCR Script Cam SK+ (Stratagene).
RNA preparation.
Total, nuclear, or cytoplasmic RNA was prepared with Tri-Reagent or Tri-Reagent LS, respectively, as instructed by the manufacturer (Molecular Dynamics, Inc., Sunnyvale, Calif.). Transfected COS or 293 cells from two 10-cm-diameter plates or T150 flasks were harvested into phosphate-buffered saline, centrifuged at 2,000 × g for 5 min, and resuspended in 0.9 ml of cold cell lysis buffer (10 mM Tris [pH 8.3], 150 mM NaCl, 1.5 mM MgCl2) and 0.1 ml of 5% Nonidet P-40. After thorough mixing, incubation on ice for 10 min, and centrifugation twice at 2,000 × g for 10 min at 4°C, the nuclei were treated with 1 ml of Tri-Reagent and frozen for future extraction of nuclear RNA. Following a second centrifugation step, the cytoplasmic supernatant was mixed with 3 volumes of Tri-Reagent LS, and RNA was extracted. To prepare polysomal RNA, the clarified cytoplasmic extract was supplemented with cycloheximide (50 μg/ml), RNasin (100 U/ml), and dithiothreitol (DTT; 2 mM) and layered onto a 9-ml linear gradient of 15 to 40% sucrose in 30 mM Tris (pH 7.4)–2 mM DTT–10 mM EGTA–5 mM MgCl2 that was underlaid with 2 ml of 60% sucrose in 30 mM Tris (pH 7.4)–2 mM DTT–10 mM EDTA–5 mM MgCl2 (26). The gradient was centrifuged 225,000 × gmax for 3.5 h at 4°C in a Beckman SW41 rotor. The EDTA in the 60% sucrose pad causes free polysomes to dissociate and sediment with membrane-bound polysomes at the 60% boundary (29). Polysomal RNA was extracted from the 60% boundary (1 ml), and nonpolysomal RNA was extracted from the upper fraction (8 ml) with Tri-Reagent. All RNA preparations were treated extensively with RQ DNase (Promega), phenol extracted, and ethanol precipitated.
RPA.
Antisense runoff α-32P-labeled RNA transcripts were synthesized with MAXscript T7 RNA polymerase (Ambion) according to the manufacturer’s instructions. Template pGEM(140-440) was digested with NotI, and pGAPDH was digested with NcoI. Template from pMBSVT7 was prepared by PCR amplification. The in vitro-transcribed RNAs were isolated by gel elution, and the RNase protection assays (RPAs) were performed with RPAIII (Ambion) according to the instruction manual, with some modifications. Typically, 15 μg of RNA was ethanol precipitated with 3 × 105 cpm of HIV probe and 3 × 103 cpm of glyceraldehyde-3-phosphate dehydrogenase (gapdh) probe. Samples were resuspended in 10 μl of hybridization buffer, heated at 90°C for 3 min, and hybridized at 42°C for 16 h. An RNase digestion mixture (1:100) was added to each sample (150 μl) and incubated at 37°C for 30 min. Sodium dodecyl sulfate (SDS) and proteinase K were added to final concentrations of 1% and 0.5 mg/ml, respectively; the samples were incubated at 37°C for 30 min and then subjected to extraction with phenol-chloroform and chloroform and precipitation with ethanol in the presence of 10 μg of yeast RNA. Pellets were dissolved in 6 μl of loading buffer, heated at 90°C for 3 min, and subjected to denaturing polyacrylamide gel electrophoresis (PAGE) on 5% gels. RNase protection products were visualized by PhosphorImager (Molecular Dynamics) analysis using ImageQuaNT version 4.2 (Molecular Dynamics).
RESULTS
SNV LTR facilitates Rev/RRE-independent production of HIV Gag.
The MPMV CTE was identified originally by its ability to modulate Rev/RRE-independent expression of Gag from subgenomic HIV plasmids (4). These HIV plasmids encode gag and either contain RRE, lack RRE and contain a 3′ β-globin intron, or lack both RRE and a β-globin intron (pSVgagpol-rre, pSVgagpol, or pBBgagpol, respectively) (35). Two SNV regions were evaluated for Rev/RRE-independent Gag production in comparison to MPMV CTE: (i) the SNV 3′ UTR between env and the 3′ LTR, which is analogous to the position of the previously defined CTEs (4, 24, 25, 39); and (ii) the SNV LTR, which is associated with Rex/RxRE-independent expression of BLV structural genes (5, 6). pSVgagpol-rre-MPMV and derivatives containing SNV 3′ UTR (pSVgagpol-rre-3′UTR) or SNV LTR (pSVgagpol-rre-LTR) are shown in Fig. 1.
FIG. 1.
Structures of subgenomic HIV plasmid pSVgagpol-rreMPMV (5) and derivatives that contain the SNV 3′ UTR and SNV LTR. SV40, SV40 late promoter; black rectangle, HIV gag-pol-vif. The SNV 3′ UTR extends from the 3′ 200 bp of env through the PPT.
The plasmids were transfected into COS cells in the presence or absence of Rev expression plasmid pRev1 (gift from David Rekosh, University of Virginia) (30). The cells were cultured in 2 ml of Dulbecco modified Eagle medium (DMEM) with 10% fetal calf serum, and then cell-associated Gag protein was quantified by enzyme-linked immunosorbent assay (ELISA) as an endpoint for gag RNA transcription, cytoplasmic accumulation, and translation. The Gag antigen capture ELISA uses antibodies specific for the capsid domain of Gag to detect precursor Gag p55 and processed Gag p24 (Coulter Corp). The minimum detectable by the assay is 15 pg. As expected (4), Gag production from pSVgagpol-rre was Rev dependent, while Gag production from pSVgagpol-rre-MPMV was Rev independent (Table 1). Gag production from the plasmids containing SNV sequences (pSVgagpol-rre-3′UTR and pSVgagpol-rre-LTR) remained Rev dependent (Table 1). The Rev responsiveness of the plasmids indicates that they are competent for Gag production. Transfection analysis of pSVgagpol and pBBgagpol derivatives that contain SNV 3′ UTR or SNV LTR further determined that the SNV 3′ UTR or SNV LTR does not replace Rev or MPMV CTE function in COS cells even in the absence of a β-globin intron and RRE (data not shown). Therefore, the presence of RRE or a β-globin intron does not affect the hypothetical SNV CTE function.
TABLE 1.
HIV Gag production
| Plasmid | Gag concn (pg/ml)a
|
|
|---|---|---|
| COSb | D17c | |
| pSVgagpol-rre | <MDd | <MD |
| pSVgagpol-rre + pRev1 | 6,000 | 3,300 |
| pSVgagpol-rre-MPMV | 5,500 | 2,100 |
| pSVgagpol-rre-LTR | <MD | <MD |
| pSVgagpol-rre-LTR + pRev1 | 6,100 | 3,200 |
| pSVgagpol-rre-3′UTR | <MD | <MD |
| pSVgagpol-rre-3′UTR + pRev1 | 6,500 | 4,000 |
| pKB504gagpol | 600 | 600 |
| pKB504gagpol-MPMV | 2,800 | 900 |
| None (mock transfection) | <MD | <MD |
Representative data from at least three independent transfections. Three days posttransfection, cell-free supernatant medium from 3 × 105 cells was harvested, and Gag levels were quantified by Gag ELISA and normalized to transfection efficiency.
COS cells were transfected with a mixture of Lipofectamine (Gibco-BRL) and test DNA (1.5 μg and 0.3 μg of pRev1) plus 0.2 μg of pEGFPN1 reporter plasmid. After 5 h, the cells were washed and cultured in 2 ml of DMEM with 10% fetal calf serum. Transfection efficiency was determined as percentage of green fluorescent cells in 1,000 cells.
D17 cells were transfected with a mixture of Polybrene (30 μg/ml), test DNA, and pEGFPN1 or pCMVluc and then cultured in DMEM supplemented with 5% fetal calf serum.
<MD, less than the minimum detectable.
We evaluated potential cell type specificity of the SNV sequences by transfecting the various plasmids into D17 cells, a dog osteosarcoma cell line that supports SNV replication and Rex/RxRE-independent replication of hybrid SNV-BLV structural gene vectors (5, 6). As expected, Gag production from pSVgagpol-rre was Rev dependent (Table 1). The D17 cells also supported Rev-independent Gag production from pSVgagpol-rre-MPMV, consistent with the presence of appropriate MPMV CTE-interacting factors. However, Gag production from the derivatives containing the SNV sequences remained Rev dependent. In summary, in the context of the 3′ UTR of pSVgagpol-rre and derivative plasmids, neither the SNV 3′ UTR nor the SNV LTR facilitates Rev-independent Gag production in COS and D17 cells.
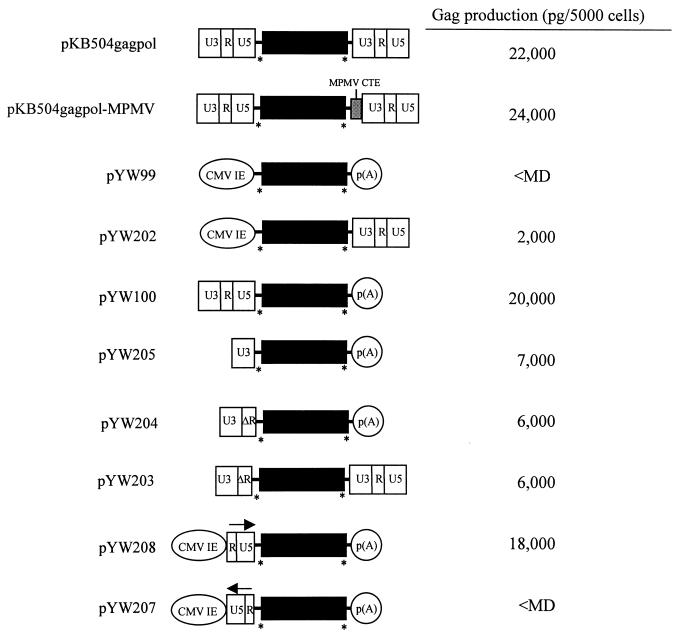
In our previous characterization of the Rex/RxRE-independent hybrid SNV-BLV structural gene vectors, the SNV LTR sequences corresponded to the 5′ and 3′ termini of the RNA (5, 6). Therefore, the position dependence of the putative SNV cis-acting element was considered by analyzing hybrid SNV-HIV structural gene vectors in which the SNV sequences comprise the 5′ and 3′ termini of the RNA (pKB504gagpol and pKB504gagpol-MPMV [Fig. 2]).
FIG. 2.
Structures of hybrid SNV-HIV plasmids and HIV Gag production. Black lines and rectangle, HIV 5′ UTR beginning at HIV U5 and extending through gag-pol, and the HIV PPT through the attL site, respectively (HIVBRU coordinates 100 to 4655 and 8662 to 8699, respectively); ∗, major HIV splice donor (left) and vif splice acceptor (right); arrows, sense and antisense orientation of SNV RU5. Shown at the right are representative data from 10 independent Gag ELISAs (Coulter Corp.), in which 105 293 cells were cotransfected with 2 μg of test plasmid and 0.2 μg of pEGFPN1 (Clontech) or pGL3 (Promega) reporter plasmid by the calcium phosphate protocol and maintained in DMEM with 10% fetal calf serum. Total cell proteins were harvested at 3 days posttransfection. Gag production was quantified by Gag ELISA (Coulter Corp.), and transfection efficiency was quantified as percentage of green fluorescent cells in 2,000 cells by UV microscopy or relative luciferase activity. Gag levels are normalized to transfection efficiency. <MD, less than the minimum detectable.
Upon transfection into COS and D17 cells, Rev/RRE-independent HIV Gag production was detected from pKB504gagpol (Table 1). The MPMV CTE in pKB504gagpol-MPMV had a stimulatory effect on Gag production in COS cells. The plasmids were also transfected into 293 human embryonic kidney cells, which consistently exhibited a higher transfection efficiency than the COS or D17 cells. pKB504gagpol and pKB504gagpol-MPMV also exhibited Rev/RRE-independent HIV Gag production in 293 cells, although a stimulatory effect of MPMV CTE was not detected (Fig. 2). Possible reasons for the increased level of Gag in 293 cells include higher transfection efficiency and/or increased availability of pertinent cellular factors. These results indicate that the SNV LTRs facilitate Rev/RRE-independent Gag expression in COS, D17, and 293 cells. Furthermore, the SNV sequences function in a position-dependent manner that corresponds to the termini of the RNA.
SNV RU5 RNA facilitates Rev/RRE-independent Gag production.
To evaluate the contribution of the individual SNV LTR sequences, we analyzed a panel of hybrid SNV-HIV replacement plasmids (Fig. 2). In retrovirus DNA, the LTRs are present in two copies that are segregated into three regions: U3, R, and U5. The 5′ U3 region corresponds to the promoter/enhancer, and the 3′ RU5 region contains the 3′ RNA processing signals. In retrovirus RNA, R sequences are repeated at both ends of the RNA transcript, U5 is unique to the 5′ RNA terminus, and U3 is unique to the 3′ RNA terminus. pKB504gagpol was modified by replacement of both SNV LTRs with heterologous transcriptional control sequences. The 5′ SNV LTR was replaced with the CMV IE promoter and the 3′ LTR was replaced with a synthetic p(A) signal to generate pYW99. Less than the minimum detectable level of Gag protein is exhibited in cells transfected with pYW99 (Fig. 2). When the 5′ LTR is replaced and the 3′ LTR is maintained (pYW202), low levels of Gag are observed. In contrast, when the 5′ LTR is maintained and the 3′ LTR is replaced (pYW100), Gag is produced at a level similar to that with pKB504gagpol. These results indicate that sequences within the 5′ SNV LTR modulate Rev/RRE-independent Gag production.
To determine the region of the 5′ SNV LTR necessary for Rev/RRE-independent Gag expression, we analyzed LTR deletion mutants (Fig. 2). Complete deletion of SNV RU5 (pYW205) or partial deletion of R and all of U5 (pYW204) yields low but detectable levels of Gag (Fig. 2). This defect is not complemented by concurrent addition of the 3′ SNV LTR (pYW203). These results suggest that the SNV RU5 RNA encoded by the 5′ LTR is necessary for maximal levels of Gag production. To test directly the contribution of SNV RU5 to Gag expression, SNV RU5 was inserted adjacent to the CMV IE promoter in pYW99 to make pYW207 and pYW208. The presence of RU5 in the sense orientation (pYW208) but not the antisense orientation (pYW207) correlates with Gag production (Fig. 2). These results indicate that SNV RU5 RNA is sufficient for Rev/RRE-independent expression of HIV Gag.
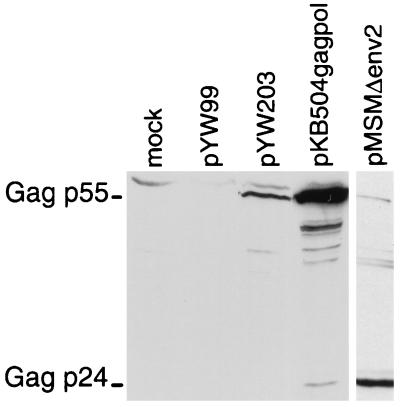
Western blot assay with Gag antibody was used to confirm that the differences observed by ELISA are not attributable to differential specificity of the Gag ELISA antibodies for precursor Gag p55 or processed Gag p24. This was important to evaluate directly because the RSV CTE has been proposed to facilitate Gag protein processing (29) and because simian immunodeficiency virus constructs containing the SRV-1 CTE exhibit impaired Gag processing in 293 cells (35). As expected, Western blot analysis does not detect Gag proteins in cells transfected with mock DNA or with pYW99 (Fig. 3). A low level of Gag p55 is observed in cells transfected with pYW203, which contains a deletion of 5′ RU5 sequence, whereas high levels of Gag p55 are observed in cells transfected with pKB504gagpol, which maintains the 5′ RU5. Thus, the Western blot data are consistent with the ELISA results. Consistent results were also observed for control cells transfected with HIV provirus (pMSMΔenv2 [22]); high levels of Gag were detected by ELISA (200,000 pg) and by Western blot analysis (Fig. 3). For the HIV control, the ratio between precursor Gag p55 and processed Gag p24 was low, consistent with high-level Gag production and efficient Gag processing. Analysis of a similar amount of Gag protein expressed from pKB504gagpol (150,000 pg) reveals both precursor Gag p55 and processed Gag p24, but the ratio between precursor Gag p55 and processed Gag p24 was high (Fig. 3). These results indicate that either the subcellular concentration of precursor Gag p55 is inadequate to drive Gag processing or Gag processing is inefficient for pKB504gagpol. Future experiments will address the relationship between the SNV element and inefficient Gag precursor processing.
FIG. 3.
Western blot immunoassay. Total cell proteins from transfected 293 cells were separated by SDS-PAGE, transferred to nitrocellulose, and reacted with polyclonal rabbit sera against HIV Gag (gift from Antonito Panganiban, University of Wisconsin). HIV Gag proteins were detected by enhanced chemiluminescence (ECL kit; Amersham). The sizes of Gag p55 and Gag p24 indicated are based on comparison with molecular weight markers (not shown).
In summary, results of both ELISA and Western blot analysis indicate that the 5′ SNV RU5 RNA facilitates maximal levels of Rev-independent Gag production. Comparison of Gag levels produced from pYW205, pYW100, and pYW208 indicates that maximal levels of Gag are yielded by the combination of the SNV RU5 with the SNV U3 promoter/enhancer rather than the combination of the SNV RU5 with the CMV IE promoter/enhancer. The apparent synergy between SNV U3 and RU5 may reflect cooperative interaction between cellular factors mediated by U3 and RU5 that together stimulate high-level Rev-independent Gag production. Consistent with this model, the R regions of murine leukemia virus and other related simple retroviruses have been shown to be important for stimulation of gene expression (9).
Unexpectedly pYW205, which encodes the SNV promoter/enhancer alone, exhibits low-level Rev/RRE-independent Gag production, whereas pYW99, which encodes the CMV promoter/enhancer alone, exhibits the expected undetectable level of Gag. One possible explanation for this difference is that the RNAs expressed from pYW205 and pYW99 have different 5′ ends and exhibit different splicing patterns. Low-level Rev/RRE-independent Gag production from pYW205 may be attributable to expression of gag transcripts that either lack a 5′ splice donor (7) or contain an excisable intron upstream of gag (17). In the following experiments, RPAs were used to evaluate the role of SNV LTR sequences in HIV RNA expression, steady-state level, cytoplasmic accumulation, and polysome loading.
Rev/RRE-independent Gag levels are not attributable to differences in steady-state RNA.
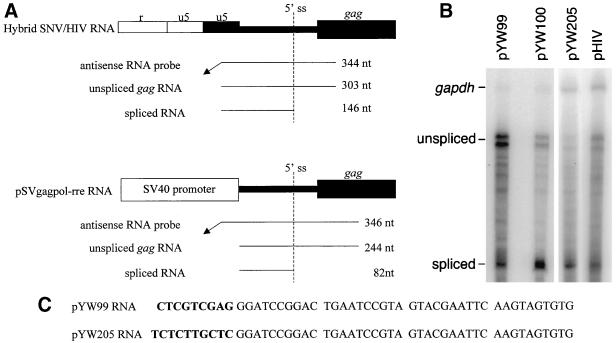
Steady-state RNAs from pYW99, pYW100, pYW205, and HIV provirus were subjected to quantitative RPAs with an antisense RNA probe that extends across the HIV major splice donor and distinguishes unspliced and spliced HIV transcripts (Fig. 4A) (22). A gapdh probe (gift from Ing-Ming Chiu, Ohio State University) was used to normalize differences in RNA loading. Control RNA expressed from HIVNL4-3 exhibited the HIV unspliced RNA and spliced RNAs previously characterized by McBride and Panganiban (22) (Fig. 4B). These HIV unspliced and spliced RNAs were also expressed from pYW100, pYW99, and pYW205. Interestingly, pYW100 exhibits an increased amount of spliced RNA. The size of spliced transcripts corresponds to pre-mRNAs spliced at the HIV major 5′ splice donor upstream of gag and the vif splice acceptor. Results from four independent experiments indicate that steady-state gag RNA levels are 3.5 ± 1.6-fold higher for pYW99 than pYW100 and indicate that Rev/RRE-independent Gag production is not attributable to increased promoter activity or RNA stability. While a difference in splicing pattern is not observed among the RNAs, experiments were performed to more completely evaluate the possibility that the low-level Gag production from pYW205 is attributable to altered RNA splicing. The 5′ RNA terminus was characterized by primer extension analysis with an antisense primer in the HIV 5′ UTR. Compared to pYW99 RNA, pYW205 RNA was 1 nucleotide (nt) longer and differed in sequence at the 5′ terminal 9 nt (Fig. 4C). RPAs were performed with an antisense pYW205 RNA probe that extends across SNV U3 and the HIV 5′ UTR splice donor. Again similar unspliced and spliced HIV transcripts were observed from pYW205 and pYW99 (data not shown). Comparison of the protected RNAs against DNA sequence ladders confirmed that, as expected, the protected pYW99 RNAs are 10 nt shorter than the pYW205 RNAs. These data eliminate the possibility that the low level of Gag production from pYW205 is attributable to altered RNA splicing that yields new gag transcripts that either lack a 5′ splice site (7) or contain an excisable intron positioned upstream of gag (17). Further experiments are necessary to explain the low-level Gag production from pYW205.
FIG. 4.
RPA. (A) Regions of complementarity between hybrid SNV-HIV sequence and the antisense HIV RNA probes, and the protected unspliced and spliced transcripts. 5′ ss, 5′ splice site. SNV R and U5 RNA regions are shown in white. HIV U5 and 5′ UTR (narrow line) and gag are shown in black. (B) Quantification of steady-state RNA levels by RPA. Two days posttransfection, total cellular RNA was harvested and subjected to DNase treatment. Aliquots of 15 μg were subjected to RPA with uniformly labeled antisense HIV RNA and gapdh RNA probes, PAGE, and PhosphorImager analysis. The protected RNAs are labeled. (C) Sequence comparison of the 5′ termini of pYW99 and pYW205 RNA. Primer extension analysis was performed on total cell RNA from transfected cells with murine leukemia virus reverse transcriptase and primer complementary to the HIV 5′ UTR. The extension products were approximately 100 bases in length and were analyzed by electrophoresis in parallel with homologous DNA sequencing reactions and PhosphorImager analysis. Sequence differences are indicated in boldface.
SNV sequences facilitate cytoplasmic accumulation of HIV RNA.
To begin to address the role of SNV RU5 in the cytoplasmic accumulation of HIV RNAs, RPAs were used to analyze total and cytoplasmic RNAs from cells transfected with pYW99, pYW100, pYW205, or pYW208. The presence of the SNV RU5 correlates with a two- to fourfold increase in nucleocytoplasmic transport of both unspliced and spliced RNA (Table 2; compare pYW99 with pYW208 and pYW205 with pYW100). Moreover, the presence of the SNV LTR in pYW100 also increased the relative amount of spliced RNA significantly. The modest increase in nucleocytoplasmic transport by SNV RU5 is not sufficient to account for the significant increase in Gag production in the presence of the element. Therefore, we performed experiments to test the hypothesis that SNV RU5 enhances the polysome association of the HIV RNAs. Previous research has shown that Rev increases polysome association of Rev-dependent mRNAs (2, 10).
TABLE 2.
Comparison of Gag protein production and cytoplasmic accumulation of HIV RNAa
| Plasmid
|
PhosphorImager units (105) normalized to gapdh RNA signal
|
||||||
|---|---|---|---|---|---|---|---|
| Total
|
Cytoplasmic
|
Foldc
|
|||||
| Name | Gagb (pg) | Unspliced | Spliced | Unspliced | Spliced | Unspliced | Spliced |
| pYW99 | <MDd | 16.6 (1.0e) | 51.3 (3.1) | 2.3 (1.0) | 14.1 (6.1) | 1.0 | 1.0 |
| pYW208 | 8,000 | 6.4 (0.4) | 23.0 (1.4) | 3.0 (1.3) | 23.4 (10.2) | 3.3 | 4.3 |
| pYW205 | 12,000 | 10.4 (0.6) | 43.8 (2.6) | 1.8 (0.8) | 14.6 (6.3) | 1.3 | 1.1 |
| pYW100 | 81,000 | 9.1 (0.5) | 56.0 (3.4) | 2.4 (1.0) | 35.6 (15.5) | 2.0 | 2.3 |
Total cellular or cytoplasmic RNAs were harvested from duplicate cultures of 106 293 cells at 2 days posttransfection and subjected to DNase treatment. Fifteen-microgram aliquots were subjected to RPA with uniformly labeled antisense HIV and gapdh RNA probes, PAGE, and PhosphorImager analysis.
Cell-associated Gag protein measured by Gag ELISA.
Cytoplasmic RNA/total RNA level relative to cytoplasmic RNA/total RNA level of pYW99.
<MD, less than the minimum detectable.
RNA level presented relative to level of pYW99 unspliced RNA.
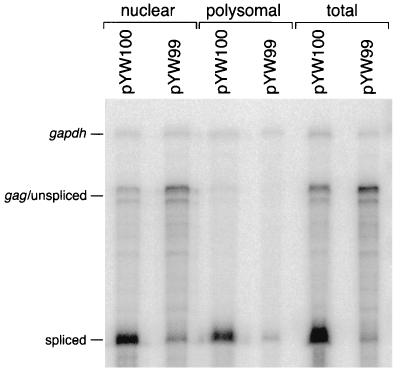
RPAs were performed on total RNA and on nuclear and polysomal RNAs from duplicate cell cultures transfected with pYW99 or pYW100. Data from three replicate experiments are summarized in Table 3, and results of a representative RPA are shown in Fig. 5. Consistent with our previous results, total steady-state gag RNA levels were lower for pYW100 than pYW99 by a factor of 3, and the amount of spliced RNA from pYW100 was increased. Comparison of gag RNA levels in polysomal and nuclear RNA indicates that pYW100 RNA exhibits an average 3.8-fold increase in polysome association compared to pYW99 RNA (Table 3). Spliced HIV transcripts from pYW100 increased an average 2.4-fold for pYW100.
TABLE 3.
Summary of Gag production and subcellular localization of HIV RNAa
| Expt | Plasmid
|
PhosphorImager units (105) normalized to gapdh RNA level
|
||||||
|---|---|---|---|---|---|---|---|---|
| Total
|
Nuclear
|
Polysomal
|
||||||
| Name | Gagb (pg) | Unspliced | Spliced | Unspliced | Spliced | Unspliced [foldc] | Spliced [fold] | |
| 1 | pYW99 | <MDd | 49.5 (1.0)e | 104.5 (2.1) | 45.0 (1.0) | 96.0 (2.1) | 4.2 (1.0) [1.0] | 5.4 (1.3) [1.0] |
| pYW100 | 40,000 | 14.4 (0.3) | 87.4 (1.8) | 17.1 (0.4) | 81.9 (1.8) | 5.9 (1.4) [3.5] | 15.7 (3.7) [3.2] | |
| 2 | pYW99 | <MD | 7.6 (1.0) | 4.0 (0.5) | 5.2 (1.0) | 4.4 (0.8) | 0.2 (1.0) [1.0] | 1.2 (6.0) [1.0] |
| pYW100 | 11,000 | 1.8 (0.2) | 7.7 (1.0) | 2.8 (0.5) | 17.4 (3.3) | 0.4 (2.0) [4.0] | 6.9 (34.5) [1.5] | |
| 3 | pYW99 | <MD | 8.1 (1.0) | 11.7 (1.4) | NDf | ND | 2.1 (1.0) [ND] | 19.6 (9.3) [ND] |
| pYW100 | 32,000 | 2.6 (0.3) | 10.0 (1.2) | ND | ND | 3.2 (1.5) [ND] | 42.0 (20.0) [ND] | |
Total RNA and nuclear or polysomal RNA were harvested 2 days after transfection of duplicate cultures of 2.5 × 106 293 cells and then subjected to DNase treatment and RPA with uniformly labeled antisense HIV and gapdh RNA probes, PAGE, and PhosphorImager analysis.
Cell-associated Gag protein measured by Gag ELISA.
Polysomal RNA/nuclear RNA level relative to polysomal RNA/nuclear RNA level of pYW99.
<MD, less than the minimum detectable.
RNA level presented relative to level of pYW99 unspliced RNA.
ND, not done.
FIG. 5.
Polysomal RNA accumulation. RPA of nuclear, polysomal, and total RNAs. RNAs were harvested 2 days posttransfection and subjected to DNase treatment, and 5- to 10-μg aliquots were subjected to RPA with uniformly labeled antisense HIV and gapdh RNA probes, PAGE, and PhosphorImager analysis. Labels indicate the protected RNAs.
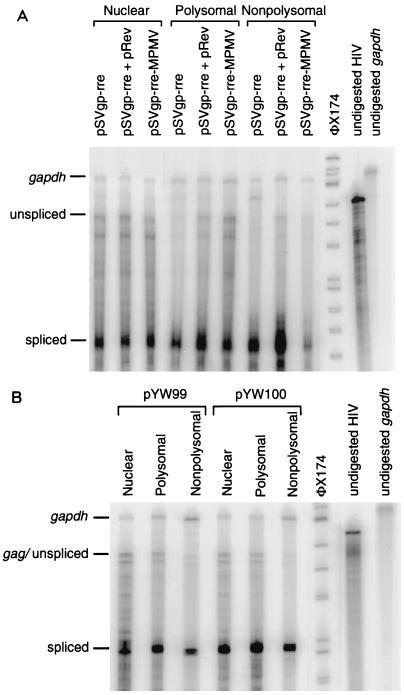
To further evaluate the cytoplasmic localization of the HIV transcripts, RPAs were also performed on nuclear, polysomal, and nonpolysomal RNAs from transfected cells. As a control, expression of Rev-dependent HIV RNAs was evaluated from pSVgagpol-rre in the absence and presence of Rev and from pSVgagpol-rre-MPMV. Similar antisense RNA probes were used for the SNV plasmids and pSVgagpol-rre-based plasmids (Fig. 4A). In the absence of Rev, gag transcripts from pSVgagpol-rre are readily observed in nuclear RNA, while levels in polysomal and nonpolysomal cytoplasmic RNA are significantly lower (Fig. 6A; Table 4). In the presence of Rev in trans or MPMV CTE in cis (pSVgagpol-rre-MPMV), polysomal gag RNA levels increase 3.4- and 6.1-fold, respectively. Nonpolysomal gag RNA levels increase 3.4- and 2.7-fold, respectively. For spliced HIV transcripts, the level in polysomal increased 1.5-fold in the presence of Rev and was not increased by CTE. Consistent with the previous RPAs, polysomal gag RNA levels expressed by pYW100 were increased compared to pYW99; the polysomal gag RNA level was increased 2.4-fold, while nonpolysomal gag RNA levels differed by 1.4-fold compared to pYW99 (Fig. 6; Table 4). In addition, polysomal and nonpolysomal levels of spliced HIV transcripts increased twofold. The results summarized in Tables 3 and 4 confirm that the SNV LTR facilitates cytoplasmic accumulation and polysome association of both HIV unspliced RNA (3.3 ± 0.8-fold) and spliced RNAs (2.3 ± 0.9-fold). By comparison, MPMV CTE selectively facilitates cytoplasmic accumulation and polysome association of the HIV unspliced transcripts (sixfold). Still, comparison of the absolute amounts of polysomal gag RNA indicates that enhanced polysome association of gag RNA is not sufficient to account for the large increases in Gag production by the SNV LTR. These results imply that enhanced translation efficiency may account for the increase in Gag production by SNV sequences.
FIG. 6.
Comparison of polysomal and nonpolysomal RNA localization. RPA of nuclear, polysomal, and nonpolysomal RNAs. RNAs were harvested 2 days posttransfection and subjected to DNase treatment, and 5- to 15-μg aliquots were subjected to RPA with uniformly labeled antisense HIV and gapdh RNA probes, PAGE, and PhosphorImager analysis. Labels indicate the protected RNAs. (A) RPA of RNA from cells transfected with pSVgagpol-rre without and with pCMVRev and pSVgagpol-rre-MPMV. (B) RPA of RNA from cells transfected with pYW99 and pYW100.
TABLE 4.
Summary of Gag production and subcellular localization of HIV RNAa
| Plasmid
|
Phosphorimager units (105) normalized to gapdh RNA level
|
||||||
|---|---|---|---|---|---|---|---|
| Nuclear
|
Polysomal
|
Nonpolysomal
|
|||||
| Name | Gagb (pg) | Unspliced | Spliced | Unspliced [foldc] | Spliced [fold] | Unspliced [foldd] | Spliced [fold] |
| pSVgagpol-rre | |||||||
| −pRev1 | <MDe | 23.6 (1.0f) | 136.3 (5.8) | 2.6 (1.0) [1.0] | 167.5 (64.4) [1.0] | 1.1 (1.0) [1.0] | 302.4 (274.9) [1.0] |
| +pRev1 | 347,000 | 34.2 (1.4) | 137.7 (5.8) | 12.6 (4.8) [3.4] | 243.0 (93.5) [1.5] | 5.2 (4.7) [3.4] | 357.1 (324.6) [1.2] |
| +MPMV | 3,000 | 29.4 (1.2) | 212.7 (9.0) | 18.9 (7.3) [6.1] | 115.1 (44.3) [0.4] | 3.5 (3.2) [2.7] | 91.6 (83.3) [0.2] |
| pYW99 | <MD | 57.7 (1.0g) | 110.9 (1.9) | 40.4 (1.0) [1.0] | 122.0 (3.0) [1.0] | 19.3 (1.0) [1.0] | 88.0 (4.6) [1.0] |
| pYW100 | 81,000 | 28.4 (0.5) | 101.3 (1.8) | 49.6 (1.2) [2.4] | 248.4 (6.1) [2.2] | 13.6 (0.7) [1.4] | 170.1 (8.8) [2.1] |
Nuclear RNA and polysomal or nonpolysomal RNA were harvested 2 days after transfection of 2.5 × 106 293 cells and then subjected to DNase treatment and RPA with uniformly labeled antisense HIV and gapdh RNA probes, PAGE, and PhosphorImager analysis.
Cell-associated Gag protein normalized to transfection efficiency which was determined by cotransfection of pGL-3 and luciferase assay.
Polysomal RNA/nuclear RNA level relative to polysomal RNA/nuclear RNA level of pSVgagpol-rre without pRev1 or pYW99.
Nonpolysomal RNA/nuclear RNA level relative to nonpolysomal RNA/nuclear RNA level of pSVgagpol-rre without pRev1 or pYW99.
<MD, less than the minimum detectable.
RNA level presented relative to unspliced RNA from pSVgagpol-rre without pRev1.
RNA level presented relative to unspliced RNA from pYW99.
DISCUSSION
The goal of this study was to test the hypothesis that SNV sequences contain a cis-acting element that facilitates Rev/RRE-independent expression of HIV gag RNA. SNV 3′ UTR and LTR regions were analyzed in the context of HIV-based plasmids that were used previously for identification of the MPMV CTE (4). Exchange of the MPMV CTE with the SNV 3′ UTR or SNV LTR indicated that these SNV regions do not function in the 3′ UTR of pSVgagpol-rre to replace the function of MPMV CTE, even in the absence of RRE or a β-globin intron (Table 1). Previous observation of BLV Rex/RxRE-independent gene expression in the context of hybrid SNV-BLV retrovirus vectors suggested that the SNV LTRs possess a position-dependent CTE-like function (5, 6). Consistent with this prediction, the SNV LTRs facilitated HIV Rev/RRE-independent Gag expression in the context of a hybrid SNV-HIV retrovirus vector (Table 1; Fig. 2). The observation of Rev/RRE-independent gene expression in COS, D17, and 293 cells indicates that putative cellular factors necessary for function of the SNV element are expressed in each of these cell lines (Table 1; Fig. 2). Analysis of Gag production from a panel of LTR deletion mutants indicates that the SNV element functions in a position- and orientation-dependent manner that corresponds to the 5′ LTR (Fig. 2 and 3). Specifically, the SNV RU5 RNA is necessary and sufficient for efficient Rev/RRE-independent Gag production. Quantitative RPAs were used to evaluate the contributions of transcription, RNA stability, nucleocytoplasmic transport, and translation efficiency to Rev/RRE-independent Gag production. Analysis of steady-state RNA indicates that the effect of SNV RU5 is not attributable to increased steady-state gag RNA level or changes in splicing pattern, although SNV sequences do increase the amount of spliced HIV transcripts (Fig. 4B, 5, and 6B). RPAs of nuclear and cytoplasmic gag RNA indicate that SNV RU5 RNA facilitates cytoplasmic accumulation of both unspliced and spliced HIV transcripts (Table 3). Because the two- to fourfold increase in transport is insufficient to account for the significant increase in Gag production, a translational effect of the SNV sequence was investigated. Analysis of cytoplasmic localization of the RNAs indicates that the SNV 5′ LTR enhances polysome association of HIV unspliced and spliced RNAs (Fig. 5 and 6; Tables 3 and 4). Control experiments with the Rev-dependent RNAs indicate that the MPMV CTE selectively facilitates polysome association of HIV unspliced transcripts (Fig. 6; Table 4).
Unexpectedly, low-level Rev/RRE-independent Gag production is detected from pYW205, which encodes the SNV U3 promoter/enhancer region alone. As expected, Gag production is not detectable from pYW99, which encodes the CMV IE promoter/enhancer alone. Comparative analysis of pYW205 and pYW99 by RPA and primer extension eliminated the possibility that pYW205 RNA lacks a 5′ splice site (7) or contains an excisable intron positioned upstream of gag (17). Further experiments are necessary to understand the low-level Gag production from pYW205.
Comparison of gag RNA and protein levels from pYW205, pYW100, pYW99, and pYW208 indicates that the combination of SNV RU5 with the SNV U3 promoter/enhancer, rather than combination of SNV RU5 with the CMV promoter/enhancer, produces maximal levels of Gag. A possible explanation for the apparent synergy between the SNV U3 promoter/enhancer and RU5 is cooperative interaction between cellular factors mediated by U3 and RU5 that stimulates high-level Rev-independent Gag production. The R regions of related simple retroviruses (i.e., murine leukemia virus and chicken syncytial virus) have been shown to be important for stimulation of gene expression and the mechanisms involved in RNA processing (9).
trans activation of Gag production by Rev/RRE involves derepression of cis-acting translational repressive sequences in HIV RNA (8, 21, 25, 28) that bind cytoplasmic poly(A)-binding protein 1 (1); release of this protein is proposed to enhance polysome association and efficient translation of gag RNA by facilitating interaction between the 3′ poly(A) tail and 5′ 7-methylguanosine cap (1, 34). Future experiments will consider whether the SNV sequences neutralize these translational repressive sequences in the HIV RNA or supply stimulatory sequences.
In summary, SNV encodes a position- and orientation-dependent posttranscriptional control element that is distinct in location and function from the MPMV CTE. The possibility of the existence of a CTE elsewhere in the SNV genome remains. It is also possible that the MPMV LTR contains a posttranscriptional control element similar to that of SNV. The SNV element corresponds to the 5′ terminus of the SNV RNA and increases cytoplasmic accumulation and polysome association of both unspliced and spliced HIV transcripts. In contrast, the MPMV CTE selectively stimulates cytoplasmic accumulation and polysome association of unspliced viral transcripts. Importantly, comparison of the absolute amounts of polysomal RNA indicates that polysome association is not sufficient to account for Rev/RRE-independent Gag production by SNV sequences or MPMV CTE. The data imply that a significant effect of the SNV element is enhancement of the translation efficiency of HIV gag RNA. Elucidation of SNV primary sequence and RNA structure that are necessary and sufficient for Rev/RRE-independent transport and translation will be important for identification of cellular Rev-like factors that modulate the function of this unique posttranscriptional control element.
ACKNOWLEDGMENTS
We thank Ing-Ming Chiu, Marie-Louise Hammarskjöld, Eric Hunter, Scott McBride, Nito Panganiban, David Rekosh, and Dan Schoenberg for gifts of plasmids and Gag antiserum, and we thank Patrick Green, Michael Lairmore, and Dan Schoenberg for critical comments on the manuscript.
This work was supported by grants from the National Institute Allergy and Infectious Diseases (R29AI40851), National Cancer Institute, Bethesda, Md. (P30CA16058), and American Cancer Society, Ohio Division.
REFERENCES
- 1.Afonina E, Neumann M, Pavlakis G N. Preferential binding of poly (A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J Biol Chem. 1997;272:2307–2311. doi: 10.1074/jbc.272.4.2307. [DOI] [PubMed] [Google Scholar]
- 2.Arrigo S J, Chen I S Y. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu-2 RNAs. Genes Dev. 1991;5:808–819. doi: 10.1101/gad.5.5.808. [DOI] [PubMed] [Google Scholar]
- 3.Benko D M, Robinson R, Solomin L, Melinni M, Felber B K, Pavlakis G N. Binding of trans-dominant mutant Rev-responsive element does not affect the fate of viral messenger RNA. New Biol. 1990;2:1111–1122. [PubMed] [Google Scholar]
- 4.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K-T, Rekosh D, Hammarskjöld M-L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boris-Lawrie K, Temin H M. Genetically simple derivatives of bovine leukemia virus can replicate independently of Tax and Rex. J Virol. 1995;69:1920–1924. doi: 10.1128/jvi.69.3.1920-1924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boris-Lawrie K, Altanerova V, Altaner C, Janglova L, Temin H M. Genetically simple BLV derivatives are infectious in vivo and induce anti-viral antibodies. J Virol. 1997;71:1514–1520. doi: 10.1128/jvi.71.2.1514-1520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang D D, Sharp P A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 8.Cochrane A W, Jones K S, Beidas S, Dillion P J, Skalka A M, Rosen C A. Identification and characterization of intragenic sequences with repress human immunodeficiency virus structural gene expression. J Virol. 1991;65:5305–5313. doi: 10.1128/jvi.65.10.5305-5313.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cupelli L, Okenquist S A, Trubetskoy A, Lenz J. The secondary structure of the R region of a murine leukemia virus is important for stimulation of long terminal repeat-driven gene expression. J Virol. 1998;72:7807–7814. doi: 10.1128/jvi.72.10.7807-7814.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Agostino D M, Felber B K, Harrison J E, Pavlakis G N. The rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol Cell Biol. 1992;12:1375–1386. doi: 10.1128/mcb.12.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerman M, Vazeux R, Peden K. The rev gene product of the human immunodeficiency virus affects envelope-specific RNA localization. Cell. 1989;57:1155–1165. doi: 10.1016/0092-8674(89)90053-6. [DOI] [PubMed] [Google Scholar]
- 12.Ernest R K, Bray M, Rekosh D, Hammarskjöld M-L. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol Cell Biol. 1997;17:135–144. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felber B K, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis G N. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of viral mRNA. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 15.Hadzopoulou-Cladaras M, Felber B K, Cladaras C, Athanassopoulos A, Tse A, Pavlakis G N. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J Virol. 1989;63:1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammarskjöld M L, Heimer J, Hammarskjöld B, Sangwan I, Albert L, Rekosh D. Regulation of human immunodeficiency virus env expression by the rev gene product. J Virol. 1989;63:1959–1966. doi: 10.1128/jvi.63.5.1959-1966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammarskjöld M L, Li H, Rekosh D, Prasad S. Human immunodeficiency virus env expression becomes Rev-independent if the env region is not defined as an intron. J Virol. 1994;68:951–958. doi: 10.1128/jvi.68.2.951-958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence J B, Cochrane A W, Johnson C V, Perkins A, Rosen C A. The HIV-1 Rev protein: a model system for coupled RNA transport and translation. New Biol. 1991;3:1220–1232. [PubMed] [Google Scholar]
- 19.Malim M H, Hauber J, Le S-Y, Maizel J V, Cullen B R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;339:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 20.Malim M H, Cullen B R. Rev and the fate of pre-mRNA in the nucleus: implications for the regulation of RNA processing in eukaryotes. Mol Cell Biol. 1993;13:6180–6189. doi: 10.1128/mcb.13.10.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maldarelli F, Martin M A, Strebel K. Identification of post-transcriptionally active inhibitory sequences in human immunodeficiency virus type 1 RNA: novel level of gene regulation. J Virol. 1991;65:5732–5743. doi: 10.1128/jvi.65.11.5732-5743.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogert R A, Lee L H, Beemon K L. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J Virol. 1996;70:3834–3843. doi: 10.1128/jvi.70.6.3834-3843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizvi T A, Schmidt R D, Lew K A. Mason-Pfizer monkey virus (MPMV) constitutive transport element (CTE) functions in a position-dependent manner. Virology. 1997;236:118–129. doi: 10.1006/viro.1997.8728. [DOI] [PubMed] [Google Scholar]
- 25.Schneider R, Campbell M, Nasioulas G, Felber B K, Pavlakis G N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoenberg D R, Cunningham K S. Characterization of mRNA endonucleases. Methods Companion Methods Enzymol. 1999;17:60–73. doi: 10.1006/meth.1998.0708. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz S, Felber B K, Pavlakis G N. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J Virol. 1992;66:150–159. doi: 10.1128/jvi.66.1.150-159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber B K, Pavlakis G N. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J Virol. 1992;66:7176–7182. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson S B, Zhang L, Craven R C, Stoltzfus C M. Rous sarcoma virus direct repeat cis elements exert effects at several points in the virus life cycle. J Virol. 1997;71:9150–9156. doi: 10.1128/jvi.71.12.9150-9156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith A J, Cho M-I, Hammarskjöld M-L, Rekosh D. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J Virol. 1990;64:2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonigo P, Barker C, Hunter E, Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986;45:375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- 32.Tabernero C, Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. The posttranscriptional control element of the simian retrovirus type 1 forms an extensive RNA secondary structure necessary for its function. J Virol. 1996;70:5998–6011. doi: 10.1128/jvi.70.9.5998-6011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang H, Gaietta G M, Fischer W H, Ellisman M H, Wong-Staal F. A cellular cofactor for the constitutive transport element of type D retrovirus. Science. 1997;276:1412–1415. doi: 10.1126/science.276.5317.1412. [DOI] [PubMed] [Google Scholar]
- 34.Tarun S Z, Jr, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 35.von Gegerfelt A, Felber B K. Replacement of posttranscriptional regulation in SIVmac239 generated a Rev-independent infectious virus able to propagate in rhesus peripheral blood mononuclear cells. Virology. 1997;232:291–299. doi: 10.1006/viro.1997.8567. [DOI] [PubMed] [Google Scholar]
- 36.Wain-Hobson S, Sonigo P, Danos O, Cole S, Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985;40:9–17. doi: 10.1016/0092-8674(85)90303-4. [DOI] [PubMed] [Google Scholar]
- 37.Wilhemsen K C, Eggleton K, Temin H M. Nucleic acid sequences of the oncogene v-rel in reticuloendotheliosis virus stain T and its cellular homolog, the proto-oncogene c-rel. J Virol. 1984;52:172–182. doi: 10.1128/jvi.52.1.172-182.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian virus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]