Abstract
Background
Oral lichen planus (OLP) is a common chronic oral mucosal disease with 1.4 % malignant transformation rate, and its etiology especially immune pathogenesis remains unclear. This study was aimed at investigating the immune cells related molecular underlying the pathophysiology of OLP through bioinformatics analysis.
Methods
The dataset GSE52130 obtained from the Gene Expression Omnibus (GEO) database was conducted a comprehensive analysis in this study. The CIBERSORTx was used for investigating immune cells infiltration. The gene set enrichment analysis (GSEA) and gene ontology (GO) enrichment were performed for exploring the biological functions and gene annotation. The protein-protein interactions (PPI) were constructed by STRING database and visualized by Cytoscape software. The cytohubba plugin was utilized for screening hub genes. The receiver operating characteristic (ROC) was performed for evaluating diagnostic value of hub genes. The miRNAs, lncRNAs and drugs were respectively predicted by NetworkAnalyst, miRTarbase, ENCORI, and DGIdb database.
Results
This study identified 595 differentially expressed genes (DEGs). The GSEA indicated keratinization, innate immune system and biological oxidation were involved in OLP. GO analysis showed extracellular matrix and keratinocyte were mainly enriched. And we found the activated memory CD4+ T cells were lowly infiltrated in OLP. We identified 101 activated memory CD4+ T-cells-related DEGs. Three hub genes (APP, IL1B, TF) were selected. APP and IL1B were significantly up-regulated, whereas TF was down-regulated in OLP. The three hub genes show high diagnostic value in OLP. Additionally, they were involved in MAPK signal, NF-kappaB signal and iron metabolism in OLP. What's more, NEAT1/XIST - miR - 15a - 5p/miR - 155–5p - APP/IL1B signal axis was focused in competing endogenous RNA (ceRNA) network. In addition, 35 drugs were predicted for OLP.
Conclusion
Three activated memory CD4+ T-cells-related DEGs were identified by integrative analysis. It may provide novel insight into the pathogenesis of OLP and suggest potential therapeutic targets for OLP.
Keywords: Differentially expressed genes, Oral lichen planus, Immune, The activated memory CD4+ T cells, Bioinformatics
Highlights
-
•
The activated memory CD4+ T cells were lowly infiltrated in OLP.
-
•
The activated memory CD4+ T-cells-related genes were potentially important markers.
-
•
APP, IL1B and TF had good diagnostic value in OLP.
-
•
NEAT1/XIST-miR-15a-5p/miR-155-5p-APP/IL1B signal axis might play an important role in malignant transformation of OLP.
1. Introduction
Oral lichen planus (OLP) is regarded as an oral mucosa disease manifested with chronic inflammatory, which is correlated with autoimmunity. The incidence of OLP is about 0.87 % in Asian populations [1]. Clinically, the OLP is mainly divided into two types: erosive type and non erosive type. It's reported that erosive OLP has a longer course of disease and recurrent attacks which affecting the physical and mind well-being of patients [2]. Besides, it is approximately 1.4 % OLP prone to transform to oral squamous cell carcinoma (OSCC) [3]. Despite a variety of treatments for OLP, almost no strategies of treatment are curative. Currently, misdiagnosis commonly happens in OLP which causing the overestimation of the risk of malignant transformation [4]. It without doubt makes it difficult for the treatment of OLP. Therefore, the accurate diagnosis is essential and necessary for OLP.
Recently, the biomarker in diagnosing disease is increasingly widespread. Study showed IL12RB2/TNFRSF8 ratio can be applied for diagnosis for OLP [5]. In addition, salivary cytokines may also serve as biomarker to diagnose OLP prognosis [6]. However, the biomarker for precise diagnosis of OLP are still largely limited because of the unclear pathogenesis. Consequently, exploring the molecular mechanisms of OLP is essential, and it may be helpful for the identification of accurate biomarker for the diagnosis and treatment of OLP.
Although the etiology of OLP is complex and unclear, increasing studies suggest the immune mechanisms may have important functions on OLP pathogenesis [7,8]. As known, the T cell, which recruited by cytokine, chemokine and adhesion molecule, mediated the inflammatory in OLP [8]. With the population of ChIP sequencing, bioinformatic analysis without doubt has an advantage for large-scale data analysis. To date, several studies have employed the bioinformatic analysis to investigate the underlying immune mechanisms of OLP. Lou Geng et al. [9] found that the T follicular helper cells and T regulatory cells are more infiltrated in OLP by bioinformatic analysis. Interestingly, the two subtypes of OLP classified by the CD8+ T-cell marker genes showed a difference in immune infiltration [10]. Obviously, exploring the underlying immune pathogenesis may contribute to the diagnosis and treatment for OLP. However, the immune pathogenesis is still not clear and further investigation needed.
Therefore, this study aimed to investigate the abundance of immune cell infiltration in OLP. Further, the differentially expressed activated memory CD4+ T-cells-related genes were identified. Moreover, enrichment analysis was performed. Besides, the hub genes were selected. And the diagnostic value was assessed. In addition, the competing endogenous RNA (ceRNA) and drug network were constructed. Above all, this study may provide a novel idea or direction for the diagnosis and treatment for OLP.
2. Materials and methods
2.1. Data acquisition and processing
We screened and acquired the OLP-related dataset GSE52130 [11] from the Gene Expression Omnibus (GEO) database. The dataset GSE52130 was downloaded from the GPL10558 platform (Illumina HumanHT-12 V4.0 expression beadchip), containing 7 normal oral epithelium tissues (GSM1260102-GSM1260108) and 7 oral lichen planus epithelium (OLPE) tissues (GSM1260095-GSM1260101). We used the R package limma (v3.52.2) [12] normalizeBetweenArrays to normalize the dataset GSE52130. We employed the box graph to visualize the normalization of samples by R package ggplot2 (v3.3.6) [13]. And the clusters of samples were observed by PCA graph.
2.2. Identification of differentially expressed genes (DEGs)
According to sample information of the dataset GSE52130, the samples were divided into normal and OLPE groups, then we analyzed the difference between the two groups by R (v4.2.1) package limma [12], and used Benjamini-Hochberg method to adjust p-values for multiple testing. Setting |logFC| (absolute log2 value in the fold change) > 1 and adjust p < 0.05 as the threshold. The differential analysis results were visualized using volcano map and heatmap, which plotted by R package ggplot2 (v3.3.6) [13].
2.3. Identification of the activated memory CD4+ T-cells-related DEGs
We estimated the abundance of immune cells using gene expression data of the dataset GSE52130 by CIBERSORTx [14] (https://cibersortx.stanford.edu/). The LM22 matrix was selected as reference. LM22 matrix, leukocyte signature matrix, is gene expression signature data for 22 immune cells, including seven T-cell types, naive and memory B cells, plasma cells, NK cells, and bone marrow subsets. Then, we used correlation analysis to obtain the genes correlated with the abundance of the activated memory CD4+ T cells. R (correlation coefficients) ≥ 0.6 was regarded as the activated memory CD4+ T-cells-related DEGs.
2.4. Enrichment analysis
To better explore the biological functions and molecular mechanisms of the DEGs in OLP, we performed the enrichment analysis utilizing gene set enrichment analysis (GSEA) [15], gene ontology (GO) enrichment and kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis. Enrichment analysis was conducted by R package ClusterProfiler (v4.4.4) [16]. FDR (false discovery rate) < 0.25 and p < 0.05 were considered as statistical significance in GSEA. A threshold p < 0.05 was considered as significant in GO enrichment and KEGG pathway enrichment analysis. The results of enrichment analysis were visualized by R package ggplot2 (v3.3.6) [13].
2.5. Construction of protein–protein interaction (PPI) network and identification of hub genes
PPI network was predicted and constructed by the STRING database [17] (https://cn.string-db.org/). The confidence threshold was set as 0.15. PPI network was visualized using the CytoScape (v3.8.2) software [18]. The top 10 hub genes were screened through the Cytohubba plugin [19] according to MCC, MNC and Degree, respectively.
2.6. Receiver operating characteristic (ROC) curve analysis of the hub genes
The ROC curve reflects the relationship between sensitivity and specificity, and the area under the curve (AUC) can evaluate the value of diagnostic experiments. We performed the ROC analysis of the hub genes by the R package pROC (v1.18.0) [20]. p < 0.05 was regarded as significant. And we applied the R package ggplot2 (v3.3.6) [13] to visualize the results.
2.7. Construction of ceRNA network
We predicted the target miRNAs by the NetworkAnalyst 3.0 [21] (https://www.networkanalyst.ca/) and miRTarbase [22] (https://mirtarbase.cuhk.edu.cn/). The miRNAs which performed in both tools were selected. The lncRNAs were analyzed based on the ENCORI [23] (http://starbase.sysu.edu.cn/). The values were set as following: human,CLIP-Data ≥5,CLIP-Region p < 0.05. We used the CytoScape (v3.8.2) software to construct and visualize ceRNA network among lncRNAs, miRNAs and mRNAs.
2.8. Drug network construction
The DGIdb (v4.2.0) [24] (https://dgidb.org/) provides the information of drug-gene interactions and potentially druggable category. Hence, we explored the target drugs based on the DGIdb. The screening criteria were as following: FDA approved, 22 source databases, 43 gene categories, 31 interaction types. The drugs-mRNAs network was performed through the CytoScape (v3.8.2) software.
2.9. Statistical analysis
Normality analysis was assessed by Shapiro-Wilk normulity test. Levene's test was used for variance analysis. We performed the differential analysis between the two groups with a non-normal distribution by the Mann-Whitney U test. p < 0.05 was considered statistically significant. Spearman correlation was used to perform the correlation analysis.
3. Results
3.1. Identification of DEGs in OLP
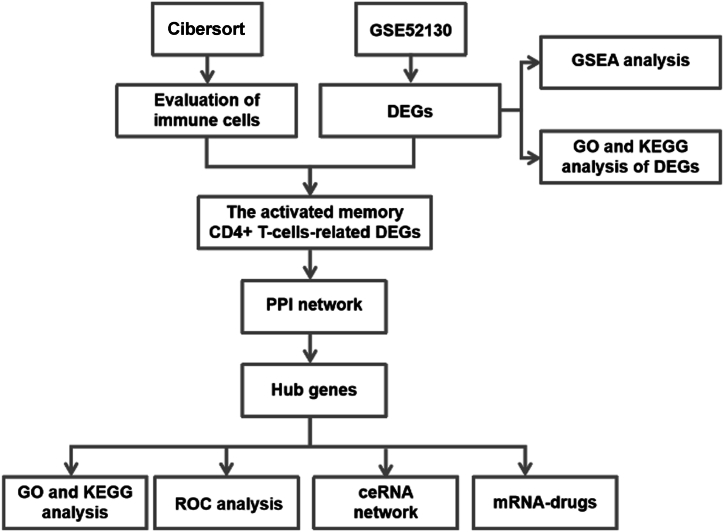
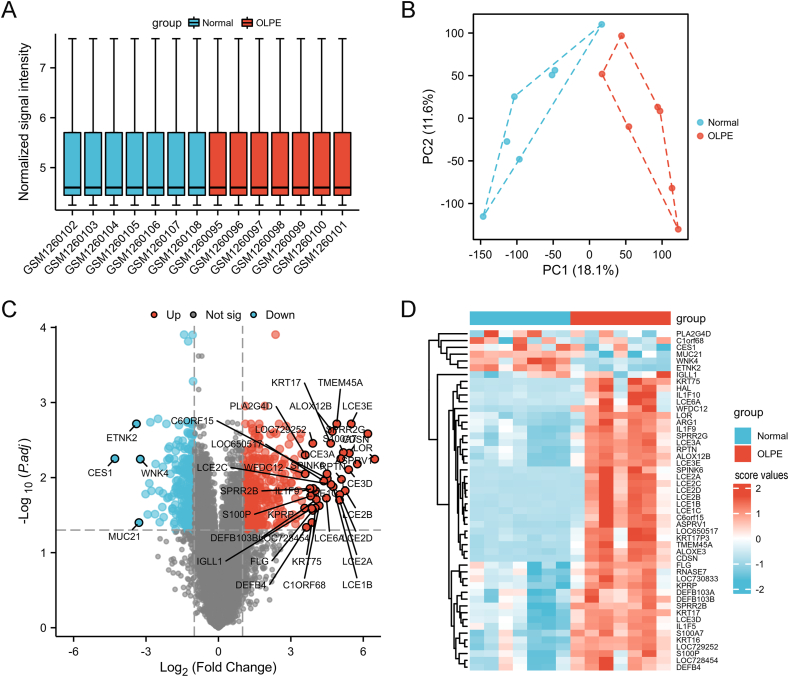
The workflow chat of this study was illustrated in Fig. 1. Firstly, we performed the standardization on the dataset GSE52130. As Fig. 2A shown, the medium intensity of each sample was consistent after normalization. The principal component analysis (PCA) plot suggested a good separation between normal and OLPE samples (Fig. 2B). We identified 595 DEGs through differential analysis between normal and OLPE groups. Additionally, there were 332 up-regulated genes and 263 down-regulated genes. The DEGs were showed in Table S1. The volcano map (Fig. 2C) and heatmap (Fig. 2D) displayed the DEGs, respectively.
Fig. 1.
Workflow chart of this study.
Fig. 2.
Identification of DEGs between the normal and OLPE groups for GSE52130 dataset.
(A) The boxplot of GSE52130 dataset after normalization. (B) The PCA plot. Volcano plot (C) and heatmap (D) representing the differential expression genes, respectively. OLPE, oral lichen planus epithelium; DEGs, differentially expressed genes; PCA, principal component analysis.
3.2. Enrichment analysis for DEGs
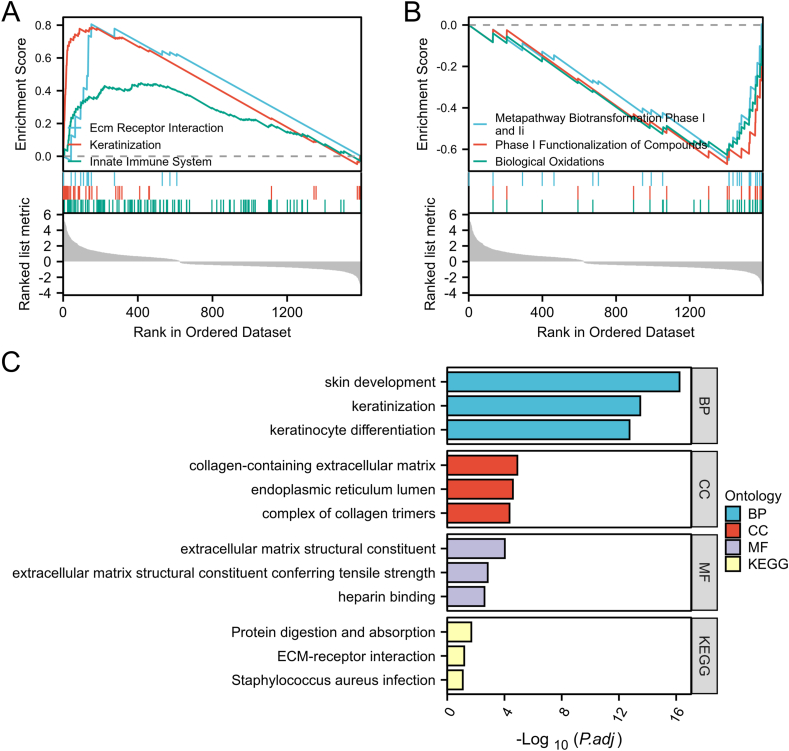
To figure out the biological functions of the DEGs, GSEA analysis was performed for all genes. The results indicated the OLP was mainly related to keratinization, receptor interaction, innate immune system, biological oxidation, metapathway biotransformation phase I and II and phase I functionalization of compounds (Fig. 3A and B). Subsequently, we applied GO enrichment and KEGG pathway enrichment analysis for 595 DEGs. GO enrichment analysis showed the 595 DEGs were mainly enriched in extracellular matrix and keratinocyte. And KEGG pathway enrichment included ECM-receptor interaction, protein digestion and absorption and so on (Fig. 3C). Taken together, enrichment analysis revealed that the DEGs mainly mediated into ECM process, an important component of the tumor microenvironment.
Fig. 3.
Enrichment analysis of DEGs.
The significantly up-regulated (A) and down-regulated (B) enrichment gene sets by GSEA. (C) The bar chart showing the significant enrichment terms through GO enrichment analysis and KEGG pathway enrichment analysis. BP, biological process; MF, molecular function; CC, cellular component.
3.3. Identification of the activated memory CD4+ T-cells-related DEGs
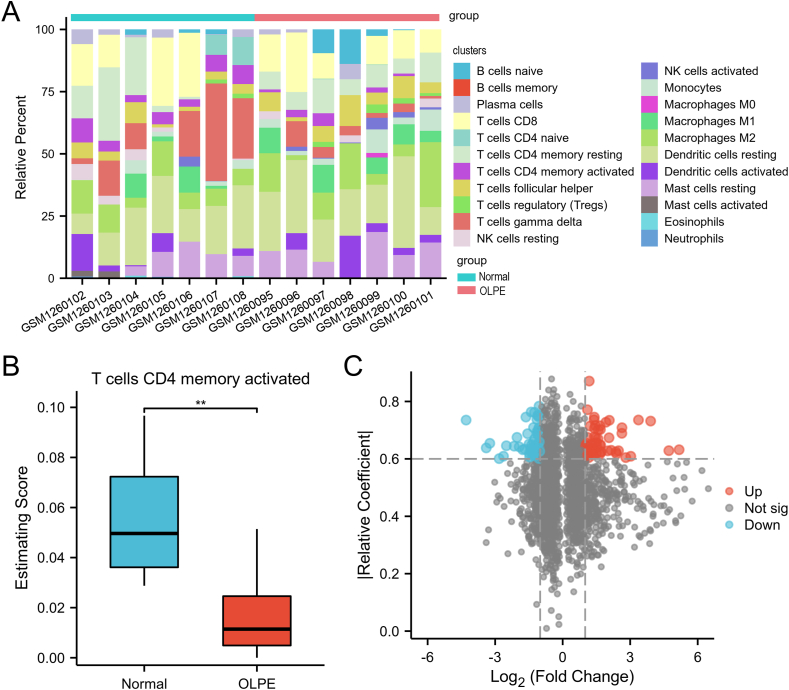
As the ECM may play important role in the immune status, we explored the immune microenvironment between normal and OLP groups. The different immune cells infiltration between the normal and OLPE groups was estimated by CIBERSORT algorithm performed based on the CIBERSORTx website. Fig. 4A displayed the composition of 22 immune cells in each sample. Further analysis indicated the OLPE group was infiltrated with lower abundance of activated memory CD4+ T cells comparing with the normal group (Fig. 4B). Then, to further explore the landscape of activated memory CD4+ T cells, the correlation analysis was performed between the DEGs and the activated memory CD4+ T cells. We identified 101 DEGs correlated with the abundance the activated memory CD4+ T cells (R ≥ 0.6, p < 0.05), regarded as the activated memory CD4+ T-cells-related DEGs (Fig. 4C). 50 up-regulated and 51 down-regulated genes were observed among the activated memory CD4+ T-cells-related DEGs, respectively. These activated memory CD4+ T-cells-related DEGs may played important role in the OLP happening.
Fig. 4.
Identification of the activated memory CD4+ T-cells-related DEGs in OLP.
(A) Histogram overlay showing the abundance of 22 immune cells of GSM1260095 - GSM1260108 in OLP. (B) Abundance of activated memory CD4+ T cells between the normal and OLPE groups. (C) Volcano plot for the activated memory CD4+ T-cells-related DEGs. *p < 0.05, **p < 0.01, ***p < 0.001.
3.4. Construction of PPI network and identification of the hub genes
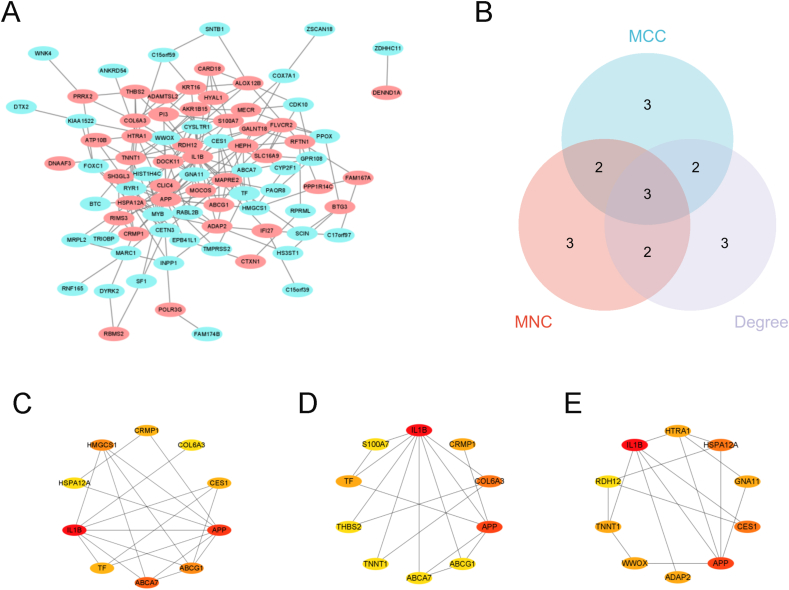
To filter out the key gene from the activated memory CD4+ T-cells-related DEGs, the PPI network was constructed. Fig. 5A was PPI network of the activated memory CD4+ T cells related DEGs. We selected the top 10 hub genes according to MCC, MNC and Degree ranks calculated by the Cytohubba plugin, respectively (Table 1) (Fig. 5C–E). Three hub genes, including APP, IL1B and TF, were acquired by taking the intersection of the genes in MCC, MNC and Degree (Table 2) (Fig. 5B). This indicated that the three hub genes might be the driven-genes in OLP happening and development.
Fig. 5.
The PPI network of the activated memory CD4+ T-cells-related DEGs and the hub genes.
(A) The PPI network of 101 genes. (B) Venn diagram showing the three hub genes of overlap among MCC, MNC and Degree. The top 10 hub genes ranked by MCC (C), MNC (D), Degree (E). Blue ellipse represented the down-regulated the activated memory CD4+ T-cells-related DEGs, pink ellipse represented the up-regulated the activated memory CD4+ T-cells-related DEGs. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
The top 10 hub genes ranked by MCC, MNC and Degree, respectively.
| MCC | MNC | Degree |
|---|---|---|
| IL1B | IL1B | IL1B |
| APP | APP | APP |
| ABCA7 | COL6A3 | HSPA12A |
| ABCG1 | TF | CES1 |
| HMGCS1 | CRMP1 | HTRA1 |
| TF | HTRA1 | TNNT1 |
| CRMP1 | RIMS3 | GNA11 |
| CES1 | SH3GL3 | ADAP2 |
| HSPA12A | TNNT1 | WWOX |
| COL6A3 | S100A7 | TF |
Table 2.
The details of the intersected hub genes.
| Gene symbol | Full name | Description |
|---|---|---|
| TF | transferrin | TF is iron binding transport proteins which transports iron from sites of absorption |
| IL1B | interleukin 1 beta | IL1B is potent proinflammatory cytokine |
| APP | amyloid beta precursor protein | APP is involved in cell mobility and transcription regulation. |
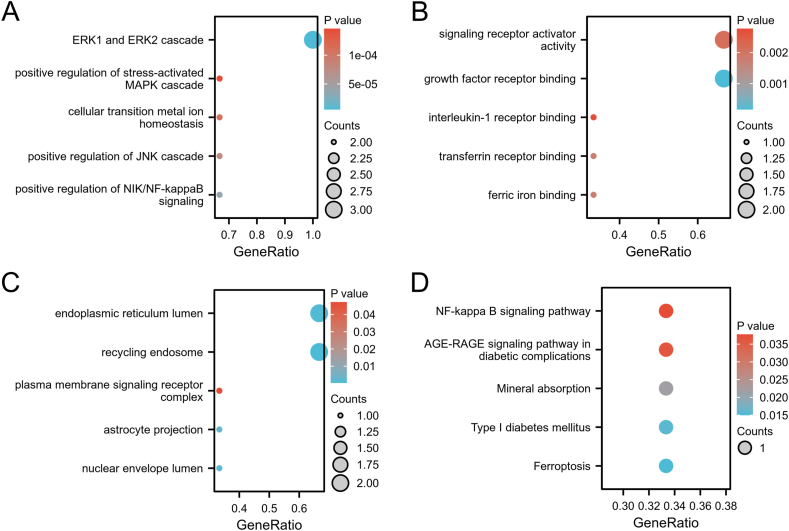
3.5. Enrichment analysis for the hub genes
In order to further undestand the functions and mechanisms of hub genes in OLP genesis, we performed the GO enrichment analysis and KEGG pathway enrichment analysis for the hub genes. The GO enrichment analysis revealed the hub genes mainly enriched in MAPK signal, NF-kappaB signal and iron metabolism (Fig. 6A–C). In addition, the KEGG pathway enrichment analysis showed NF-kappaB signal and ferroptosis were enriched (Fig. 6D). These results showed the hub genes might be involved in OLP through NF-kappaB and ferroptosis signalling pathway.
Fig. 6.
Enrichment analysis of the three hub genes.
(A) Biological process terms. (B) Molecular function terms. (C) Cellular component terms. (D) KEGG pathway enrichment terms.
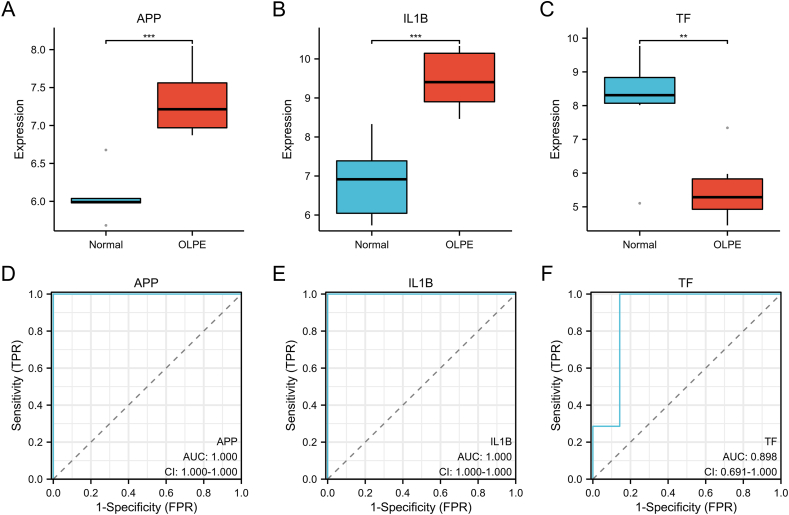
3.6. Diagnostic value of the hub genes
In GSE52130 dataset, the APP and IL1B were significantly up-regulated and TF was significantly down-regulated in the OLPE group (p < 0.05) (Fig. 7A–C). In order to estimate the diagnostic significance of the hub genes, the ROC analysis was conducted. The results suggested the APP, IL1B and TF showed the high diagnostic value with the AUC of 1.000, 1.000 and 0.898, respectively.
Fig. 7.
The diagnostic value of the three hub genes.
The box plot displayed the expression level of APP (A), IL1B (B) and TF (C) between the normal and OLPE groups, respectively. ROC curve for APP (D), IL1B (E), TF (F).
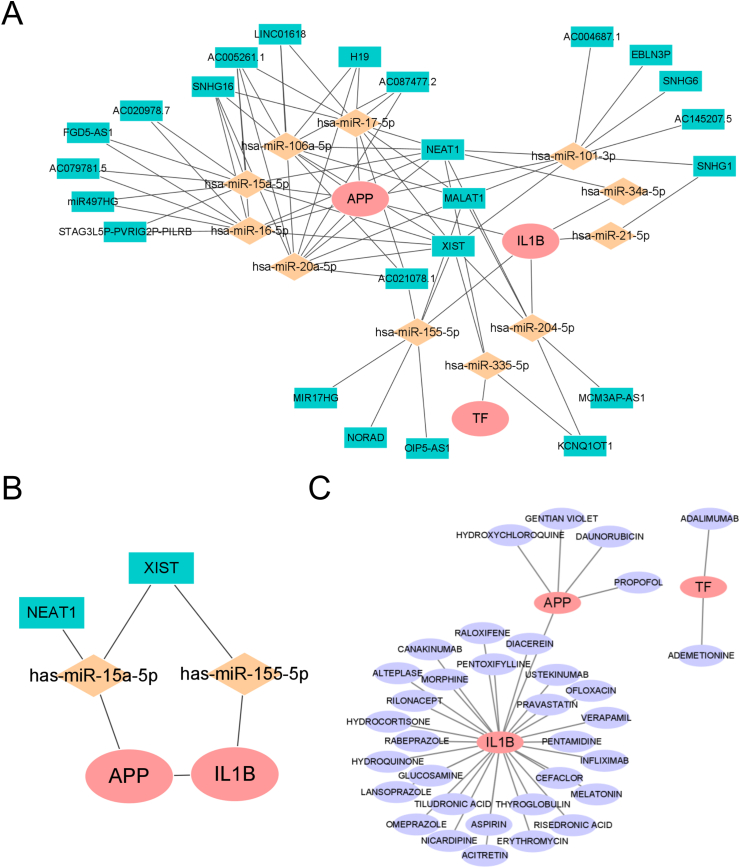
3.7. Construction of ceRNA and drug networks for the hub genes
To comprehensively understand the molecular mechanisms in OLP, we predicted the lncRNAs-miRNAs-mRNAs ceRNA network. As shown (Fig. 8A), there were 6 miRNAs and 19 lncRNAs predicted for APP. 4 miRNAs and 10 lncRNAs were selected for IL1B. One miRNA and three lncRNAs of TF were acquired. Next, we selected miRNAs (miR-15–5p and miR-155–5p) and lncRNAs (XIST and NEAT1) reported in OSCC for the further study (Fig. 8B). We speculated the NEAT1/XIST-miR-15a-5p/miR-155-5p-APP/IL1B signal axis might play an important role in the malignant transformation of OLP.
Fig. 8.
Construction of ceRNA network and drug network for the three hub genes.
(A) The lncRNAs - miRNAs - mRNAs ceRNA network. (B) NEAT1/XIST - miR - 15a - 5p/miR - 155–5p - APP/IL1B signal axis. C The drug network. Pink ellipses indicated the hub genes, orange diamonds indicated miRNAs, blue rectangles indicated lncRNAs, purple ellipses indicated drugs. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
What’ s more, we predicted the potential target drugs of the hub genes by the DGIdb database. 35 drugs were selected for the hub genes. There were 4, 28 and 2 drugs targeted on APP, IL1B and TF, respectively (Fig. 8C).
4. Discussion
The OLP is one of the most common chronic inflammatory diseases of oral mucosa, which is easy to recurrent attacks [1]. It is considered as a potential malignant oral disease by the WHO [3]. The pathogenesis of OLP is still not clear. Several studies have found that immune factors participated in the pathogenesis of OLP [7]. Even so, how the immune mechanism responds in OLP remains unclear. Therefore, it is necessary to further elucidate the exact role of immune factors in the pathogenesis of OLP.
Hence, to explore the underlying immune mechanism of OLP, we performed an integrated analysis on GSE52130 dataset with 7 OLPE samples and 7 normal samples. We identified 595 DEGs between normal and OLPE groups. Further, the GSEA analysis and GO and KEGG enrichment analysis indicated these genes were mainly involved in extracellular matrix (ECM) and keratinocyte. As known, the ECM is an important component of the tumor microenvironment [25]. ECM structures influence immune cell localization and function [26]. In addition, ECM remodeling regulated immune cells and resulted in immune escape in tumor progression [27]. Consistently, previous study found ECM proteins significantly increased might be involved in self-driving mechanism in OLP [28]. Taken together, the ECM may play a key role in the immune status in OLP.
To investigate the immune cell infiltration level, the 22 immune cells were estimated by CIBERSORTx. Of note, the activated memory CD4+ T cells were low infiltrated in OLP tissue. As known, memory CD4+ T cells have several notable characteristics, including long-term survival, low-threshold activation, enhanced ability to migrate to lymph nodes [29,30]. An increased number of activated memory CD4+ T cells and differentiation into certain T cells subset was found after secondary antigenic stimulation [31,32]. It has been reported the memory CD4+ T cells from immunological non-responders of HIV might weaken metabolic activity of lymphocyte, thus reduced T cell ability to divide [33]. Consequently, we speculate that the low activated memory CD4+ T cells may be associated with the immune restoration in OLP. Next, in order to explore the role of the activated memory CD4+ T cells, we analyzed its associated genes in OLP. 101 genes were identified.
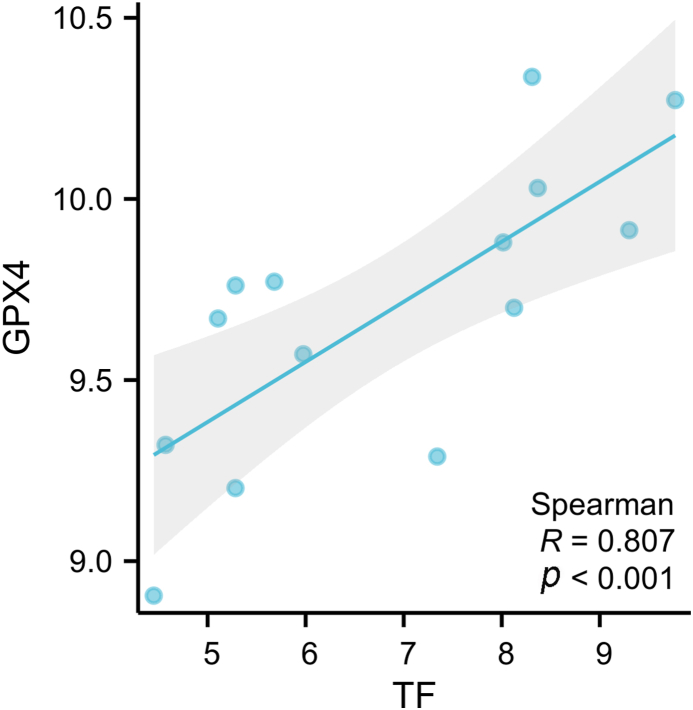
In the PPI network constructed in this study, the top 10 genes with multiple interactions were highlighted as the most significant hub genes by the cytohubba plugin. What's more, to further investigate the roles of these genes in OLP, three hub genes including APP, IL1B and TF were selected by taking the intersection of different algorithms. Among them, APP and IL1B were significantly up-regulated in OLP, yet TF was significantly down-regulated. As for the diagnostic value, APP and IL1B showed high diagnostic accuracy with both the AUC 1.000, and TF have a moderate diagnostic accuracy (AUC = 0.898) [34]. It indicated the three genes may be promising diagnostic targets for OLP. APP, namely amyloid beta precursor protein, was known to participate in the progression, proliferation, and migration of cancer cells [35]. It was reported APP was up-regulated in multiple tumors, including breast, pancreatic and OSCC [36,37]. Obviously, this study investigated the role of APP in OLP, providing the potential immune molecular mechanism, firstly. Consistent with our results, de Lanna CA et al. [38] reported immunity-related gene IL1B was overexpressed in OLP. Similarly, a previous study has demonstrated expression of IL1B in keratinocytes was increased in OLP [39]. As known, TF is an important molecule binding and transporting iron. Especially, we found that TF was positively associated with the expression of the key ferroptosis-regulated protein glutathione peroxidase (GPX4) in OLP (R = 0.807, p < 0.001) (Fig. 9). Consistently, TF was mainly involved in iron metabolism and ferroptosis by enrichment analysis (Fig. 6) in this study. In addition, another proteomic analysis showed salivary TF might serve as a biomarker for early detection in oral cancer [40]. Therefore, it's necessary to explore whether TF influenced the progress of OLP through ferroptosis. Taken together, investigating the molecular mechanism of the three hub genes may improve our understanding of the etiology of OLP.
Fig. 9.
The correlation analysis between TF and GPX4.
In the lncRNAs-miRNAs-mRNAs ceRNA network, there were 6, 4 and 1 miRNAs predicted for APP, IL1B and TF, respectively. And 19, 10 and 3 lncRNAs were also separately selected for APP, IL1B and TF. A cross-sectional study found the expression of miRNA-15a-5p was increasing in OSCC [41]. In addition, it reported miR-155–5p promoted progression of oral cancer [42]. Interestingly, two predicted lncRNAs including XIST and NEAT1 were both reported to be up-regulated in OSCC and promote the malignancy [43,44]. Based on this, we speculated the NEAT1/XIST - miR-15a-5p/miR-155-5p- APP/IL1B signal axis might play an important role in the malignant transformation of OLP. Of course, further analysis and verification were necessary for clarifying the effect of these genes on OLP.
This study still has some limitations. First, in order to clarify the mechanism more comprehensively in OLP, integrated analysis of blood and oral tissue samples is necessary. However, this study only conducted analysis based on oral tissue, due to lack of microarray data from blood samples of OLP. Second, the small sample size may limit the robustness of our statistical analysis. Although the relatively small sample size was adequate to detect differences in gene expression, it may have limited our ability to detect fold change in DEGs, which may deviate from the actual value. And it may lead to low diagnostic accuracy of the three hub genes associated with OLP. Hence, expanding the sample size for further external validation is warranted. Third, high false-positive rate may resulted from single microarray analysis, thus, to improve the accuracy, analysis of integrating multiple data is needed in the future study. Finally, further experiment validation is necessary to fully identify the function of hub genes and the underlying mechanisms of OLP.
In summary, this study investigated the underlying immune molecular mechanisms of OLP by a comprehensive bioinformatic analysis. We identified the genes correlated with the abundance the activated memory CD4+ T cells in OLP. In addition, 3 hub genes were selected as potential candidate targets through a comprehensive analysis, including the function enrichment analysis, PPI network analysis, and ROC curve analysis. Nevertheless, more experiments are required for further verifying the findings in this study.
Ethics statement
The present study adhered to public database guidelines and data access policies, and approval by the local ethics committees was not required.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not for profit sectors.
Data availability statement
The datasets generated and analyzed in this study are available GEO (https://www.ncbi.nlm.nih.gov/geo/) databases. All data mentioned in the manuscript can be provided by the corresponding author upon reasonable request.
Declarations
No additional ethical approval or informed consent was required as all data were publicly available.
CRediT authorship contribution statement
Hui Zhu: Writing – original draft, Visualization, Data curation. Huanping Lu: Visualization, Data curation. Tianyou Li: Visualization, Methodology. Jing Chen: Writing – review & editing, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank GEO, and all the databases we used in the paper and contributors for uploading their datasets.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e33305.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Li C., Tang X., Zheng X., et al. Global prevalence and incidence estimates of oral lichen planus: a systematic review and meta-analysis. Jama Dermatol. 2020;156(2):172–181. doi: 10.1001/jamadermatol.2019.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iocca O., Sollecito T.P., Alawi F., et al. Potentially malignant disorders of the oral cavity and oral dysplasia: a systematic review and meta-analysis of malignant transformation rate by subtype, Head. Neck. 2020;42(3):539–555. doi: 10.1002/hed.26006. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez-Moles M.A., Ruiz-Avila I., Gonzalez-Ruiz L., et al. Malignant transformation risk of oral lichen planus: a systematic review and comprehensive meta-analysis. Oral Oncol. 2019;96:121–130. doi: 10.1016/j.oraloncology.2019.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Cai X., Zhang J., Zhang H., et al. Overestimated risk of transformation in oral lichen planus. Oral Oncol. 2022;133 doi: 10.1016/j.oraloncology.2022.106025. [DOI] [PubMed] [Google Scholar]
- 5.Jeon S.H., Jeon E.H., Lee J.Y., et al. The potential of interleukin 12 receptor beta 2 (IL12RB2) and tumor necrosis factor receptor superfamily member 8 (TNFRSF8) gene as diagnostic biomarkers of oral lichen planus (OLP) Acta Odontol. Scand. 2015;73(8):588–594. doi: 10.3109/00016357.2014.967719. [DOI] [PubMed] [Google Scholar]
- 6.Zhu Z.D., Ren X.M., Zhou M.M., et al. Salivary cytokine profile in patients with oral lichen planus. J. Dent. Sci. 2022;17(1):100–105. doi: 10.1016/j.jds.2021.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng X., Wang Y., Jiang L., et al. Updates on immunological mechanistic insights and targeting of the oral lichen planus microenvironment. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1023213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Howati A., Thornhill M.H., Colley H.E., et al. Immune mechanisms in oral lichen planus. Oral Dis. 2023;29(4):1400–1415. doi: 10.1111/odi.14142. [DOI] [PubMed] [Google Scholar]
- 9.Geng L., Zhang X., Tang Y., et al. Identification of potential key biomarkers and immune infiltration in oral lichen planus. Dis. Markers. 2022;2022 doi: 10.1155/2022/7386895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J., Peng G., Chi H., et al. CD8 + T-cell marker genes reveal different immune subtypes of oral lichen planus by integrating single-cell RNA-seq and bulk RNA-sequencing. BMC Oral Health. 2023;23(1):464. doi: 10.1186/s12903-023-03138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danielsson K., Coates P.J., Ebrahimi M., et al. Genes involved in epithelial differentiation and development are differentially expressed in oral and genital lichen planus epithelium compared to normal epithelium. Acta Derm. Venereol. 2014;94(5):526–530. doi: 10.2340/00015555-1803. [DOI] [PubMed] [Google Scholar]
- 12.Ritchie M.E., Phipson B., Wu D., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkinson L. ggplot2: elegant graphics for data analysis by WICKHAM, H. Biometrics. 2011;67(2):678–679. doi: 10.1111/j.1541-0420.2011.01616.x. [DOI] [Google Scholar]
- 14.Newman A.M., Steen C.B., Liu C.L., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37(7):773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu G., Wang L.G., Han Y., et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szklarczyk D., Gable A.L., Nastou K.C., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chin C.H., Chen S.H., Wu H.H., et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. Suppl 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robin X., Turck N., Hainard A., et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou G., Soufan O., Ewald J., et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47(W1):W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H.Y., Lin Y.C., Cui S., et al. miRTarBase update 2022: an informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022;50(D1):D222–D230. doi: 10.1093/nar/gkab1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J.H., Liu S., Zhou H., et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freshour S.L., Kiwala S., Cotto K.C., et al. Integration of the drug-gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49(D1):D1144–D1151. doi: 10.1093/nar/gkaa1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J., Zhang L., Wan D., et al. Extracellular matrix and its therapeutic potential for cancer treatment. Signal Transduct. Targeted Ther. 2021;6(1):153. doi: 10.1038/s41392-021-00544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutherland T.E., Dyer D.P., Allen J.E. The extracellular matrix and the immune system: a mutually dependent relationship. Science. 2023;379(6633) doi: 10.1126/science.abp8964. [DOI] [PubMed] [Google Scholar]
- 27.Yuan Z., Li Y., Zhang S., et al. Extracellular matrix remodeling in tumor progression and immune escape: from mechanisms to treatments. Mol. Cancer. 2023;22(1):48. doi: 10.1186/s12943-023-01744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loncar-Brzak B., Klobucar M., Veliki-Dalic I., et al. Expression of small leucine-rich extracellular matrix proteoglycans biglycan and lumican reveals oral lichen planus malignant potential, Clin. Oral Investig. 2018;22(2):1071–1082. doi: 10.1007/s00784-017-2190-3. [DOI] [PubMed] [Google Scholar]
- 29.Liu Q., Sun Z., Chen L. Memory T cells: strategies for optimizing tumor immunotherapy. Protein Cell. 2020;11(8):549–564. doi: 10.1007/s13238-020-00707-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J.R., Hou P., Wang X.J., et al. TNFRSF11B suppresses memory CD4+ T cell infiltration in the colon cancer microenvironment: a multiomics integrative analysis. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.742358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins M.K., Khoruts A., Ingulli E., et al. In vivo activation of antigen-specific CD4 T cells. Annu. Rev. Immunol. 2001;19:23–45. doi: 10.1146/annurev.immunol.19.1.23. [DOI] [PubMed] [Google Scholar]
- 32.Li M., Zhao J., Yang R., et al. CENPF as an independent prognostic and metastasis biomarker corresponding to CD4+ memory T cells in cutaneous melanoma. Cancer Sci. 2022;113(4):1220–1234. doi: 10.1111/cas.15303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vlasova V.V., Saidakova E.V., Korolevskaya L.B., et al. Metabolic features of activated memory CD4(+) T-cells derived from HIV-infected immunological non-responders to highly active antiretroviral therapy. Dokl. Biol. Sci. 2021;501(1):206–209. doi: 10.1134/S0012496621060090. [DOI] [PubMed] [Google Scholar]
- 34.Akobeng A.K. Understanding diagnostic tests 3: receiver operating characteristic curves. Acta Paediatr. 2007;96(5):644–647. doi: 10.1111/j.1651-2227.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- 35.Tsang J., Lee M.A., Chan T.H., et al. Proteolytic cleavage of amyloid precursor protein by ADAM10 mediates proliferation and migration in breast cancer. EBioMedicine. 2018;38:89–99. doi: 10.1016/j.ebiom.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ko S.Y., Lin S.C., Chang K.W., et al. Increased expression of amyloid precursor protein in oral squamous cell carcinoma. Int. J. Cancer. 2004;111(5):727–732. doi: 10.1002/ijc.20328. [DOI] [PubMed] [Google Scholar]
- 37.Pandey P., Sliker B., Peters H.L., et al. Amyloid precursor protein and amyloid precursor-like protein 2 in cancer. Oncotarget. 2016;7(15):19430–19444. doi: 10.18632/oncotarget.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Lanna C.A., Da S.B., de Melo A.C., et al. Oral lichen planus and oral squamous cell carcinoma share key oncogenic signatures. Sci. Rep. 2022;12(1) doi: 10.1038/s41598-022-24801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y., Du G., Shi L., et al. Altered expression of CCN1 in oral lichen planus associated with keratinocyte activation and IL-1beta, ICAM1, and CCL5 up-regulation. J. Oral Pathol. Med. 2020;49(9):920–925. doi: 10.1111/jop.13087. [DOI] [PubMed] [Google Scholar]
- 40.Jou Y.J., Lin C.D., Lai C.H., et al. Proteomic identification of salivary transferrin as a biomarker for early detection of oral cancer. Anal. Chim. Acta. 2010;681(1–2):41–48. doi: 10.1016/j.aca.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 41.Saravana M.P., Kannan A., Ganesan A., et al. Evaluating the expression of microRNA-15a-5p and YAP1 gene in oral squamous cell carcinoma in comparison with normal tissue: a cross-sectional study. J. Oral Pathol. Med. 2023;52(7):593–600. doi: 10.1111/jop.13451. [DOI] [PubMed] [Google Scholar]
- 42.Wu M., Duan Q., Liu X., et al. MiR-155-5p promotes oral cancer progression by targeting chromatin remodeling gene ARID2. Biomed. Pharmacother. 2020;122 doi: 10.1016/j.biopha.2019.109696. [DOI] [PubMed] [Google Scholar]
- 43.Wu K., Wu W., Wu M., et al. Long non-coding RNA XIST promotes the malignant features of oral squamous cell carcinoma (OSCC) cells through regulating miR-133a-5p/VEGFB. Histol. Histopathol. 2023;38(1):113–126. doi: 10.14670/HH-18-504. [DOI] [PubMed] [Google Scholar]
- 44.Liu X., Shang W., Zheng F. Long non-coding RNA NEAT1 promotes migration and invasion of oral squamous cell carcinoma cells by sponging microRNA-365. Exp. Ther. Med. 2018;16(3):2243–2250. doi: 10.3892/etm.2018.6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed in this study are available GEO (https://www.ncbi.nlm.nih.gov/geo/) databases. All data mentioned in the manuscript can be provided by the corresponding author upon reasonable request.