Summary
Background
MPT64 is a key protein used for Mycobacterium tuberculosis (MTB) complex strain identification. We describe protracted transmission of an MPT64 negative MTB strain in Queensland, Australia, and explore genomic factors related to its successful spread.
Methods
All MPT64 negative strains identified between 2002 and 2022 by the Queensland Mycobacteria Reference Laboratory, and an additional 2 isolates from New South Wales (NSW), were whole genome sequenced. Bayesian modelling and phylogeographical analyses were used to assess their evolutionary history and transmission dynamics. Protein structural modelling to understand the putative functional effects of the mutated gene coding for MPT64 protein was performed.
Findings
Forty-three MPT64 negative isolates were sequenced, belonging to a single MTB cluster of Lineage 4.1.1.1 strains. Combined with a UK dataset of the same lineage, molecular dating estimated 1990 (95% HPD 1987–1993) as the likely time of strain introduction into Australia. Although the strain has spread over a wide geographic area and new cases linked to the cluster continue to arise, phylodynamic analysis suggest the outbreak peaked around 2003. All MPT64 negative strains had a frame shift mutation (delAT, p.Val216fs) within the MPT64 gene, which confers two major structural rearrangements at the C-terminus of the protein.
Interpretation
This study uncovered the origins of an MPT64 negative MTB outbreak in Australia, providing a richer understanding of its biology and transmission dynamics, as well as guidance for clinical diagnosis and public health action. The potential spread of MPT64 negative strains undermines the diagnostic utility of the MPT64 immunochromatographic test.
Funding
This study was funded from an operational budget provided to the Queensland Mycobacterium Reference Laboratory by Pathology Queensland, Queensland Department of Health.
Keywords: Tuberculosis, Mycobacterium tuberculosis, MPT64, Phylodynamics, Laboratory diagnostics, Transmission, Queensland, Australia
Research in context.
Evidence before this study
Multiple studies have described the accuracy of MPT64 detection as a test to differentiate non-tuberculous mycobacteria (NTM) from Mycobacterium tuberculosis complex (MTB) in positive mycobacterial cultures. Only one study highlighted that the diagnostic accuracy of the MPT64 antigen assay may be negatively influenced by the presence of a 63-bp deletion in the MPT64 gene and associated this variant with a Lineage 4.2.2 strain. No previous studies have utilised genomic data of MPT64 negative strains to examine the temporal evolution and test their ability to spread within communities. Moreover, no retrospective or prospective studies have identified any MPT64 negative strain of MTB in Australia.
Added value of this study
This is the first study to report a large MTB transmission cluster of a unique MPT64 negative Lineage 4.1.1.1 variant (p.Val216fs), with predicted structural rearrangement of the MPT64 protein impacting immune regulation. We utilised whole genome sequencing and advanced Bayesian analyses to understand the evolutionary history and transmission dynamics of the strain. Molecular dating estimated this strain was introduced into Queensland, Australia in the 1990s. Although cases mapped to large geographic areas in North and Central Queensland covering many hundreds of square kilometres, the incidence of genomic clustering was not restricted based on postcode. Our results indicate that this is an active ongoing outbreak. Phylodynamic analyses suggest an initial high reproductive number (R0 > 1.8) with reduced transmission since the strain and cluster has been identified.
Implications of all the available evidence
Molecular characterisation of a novel MPT64 negative strain with wide geographic spread, demonstrates the potential risk of using antigen specific tests, like the MPT64 immunochromatographic test, for disease diagnosis unless additional molecular tests are utilised. Misdiagnosis of such strains may facilitate diagnostic selection, leading to delayed treatment initiation. WGS can identify instances of potential misdiagnosis, clarify the underlying mechanism, provide high resolution transmission dynamics to guide control efforts and uncover the origins and historical burden of an outbreak.
Introduction
Australia is a low tuberculosis (TB) incidence country that has already achieved End TB pre-elimination targets (<10 case/million population) in the nonindigenous Australia-born population. Yet Australian Aboriginal and Torres-Strait Islander First Nations people, experience five to six times the TB disease burden compared to non-indigenous Australian born citizens.1
TB incidence rates have been sustained at 5–6/100,000 population since the late 1980s.2 However, progress has stagnated and achieving the TB elimination target of <1 case per million population remains out of reach. Variation in TB incidence rates between different States and Territories mainly reflect differences in migration patterns.1,3 The State of Queensland (QLD) has one of the lowest TB incidence rates (3–4/100,000 population),1 but cross-border TB transmission from Papua New Guinea (PNG) via the Torres Strait has been a major challenge.4
Acid-fast bacilli (AFB) microscopy has been the main TB diagnostic tool for more than a century, yet it cannot distinguish MTBC (M. tuberculosis complex) from non-tuberculous mycobacteria (NTM) or identify drug resistant disease.5 Despite the widespread use of rapid molecular diagnostic tools such as Xpert MTB/RIF assays, bacterial culture remains the most sensitive laboratory test for the diagnosis of TB and is a prerequisite for comprehensive phenotypic drug susceptibility testing (pDST), strain typing and sequencing.5
Rapid differentiation of MTBC from NTM in a positive culture is necessary for prompt and appropriate treatment initiation where a rapid molecular test has not been used or returned an initial negative direct result for MTBC detection. The MPT64 antigen test is a commonly used immunochromatographic assay that is easy to perform and rapidly differentiates MTBC from NTM in positive cultures. A false negative test may either delay diagnosis and early treatment initiation or negate the opportunity for comprehensive pDST noting rapid molecular detection assays are unable to interrogate drug resistance to many new and repurposed treatment agents. MPT64 (Rv1980c) is a 24-kDa secreted protein found in all MTBC species except some strains of M.bovis and M.bovis BCG,6 that is hypothesised to be one of the first proteins to interact with the host immune system.7 It seems important in activating and regulating the host immune response, affecting bacterial proliferation and survival.7,8 Given that it is recognised by human Th1 cells, it has been used as a TB disease marker and potential vaccine candidate.9 Although the MPT64 antigen is highly conserved, a recent study highlighted a lineage 4.2.2 strain with 63bp deletion variant in MPT6410 that impacts on the diagnostic sensitivity of the assay while lineage 5 and 6 strains are thought to have a lower diagnostic sensitivity without known MPT64 gene polymorphisms.11,12
In September 2017, a case of drug susceptible pulmonary TB was diagnosed in North Queensland, Australia, as a part of a routine contact investigation. The M. tuberculosis strain isolated tested MPT64 negative as did strains from three other close contacts also diagnosed with pulmonary TB months prior and after the above case was detected. Retrospective and prospective investigations were initiated to identify similar cases that may have formed a genotypically similar cluster. In this study, we confirmed the existence of a globally unique MPT64 negative M.tuberculosis outbreak using whole genome sequencing (WGS). We examined the evolutionary history of this strain and mapped the transmission patterns and temporal dynamics of this outbreak by using phylodynamic approaches together with relevant epidemiological information.
Methods
Routine laboratory methods
All new MTB isolated in Queensland are characterised at the Queensland Mycobacterium Reference Laboratory (QMRL) and are derived from either primary isolation or referred from private pathology services. While methods have evolved over time, all specimens undergo acid fast smear microscopy and culture using both liquid (automated Mycobacterial Growth Indicator Tube (MGIT) system) and solid (Lowenstein Jansen with pyruvate) media. The immunochromatographic MPT64 assay (SD Bioline TB AgMPT64 Rapid) was introduced in 2008 for rapid identification of MTBC isolates, with further confirmation by a molecular method for differentiation of MTBC members performed using Taqman multiplex Real-time PCR assay targeting regions of difference (RDs; RD1, RD4, RD9, RD9ext, and RD12).13 Any isolates with morphology consistent with M. tuberculosis but with a negative MPT64 test are subjected to a molecular test that has varied in type over the two decades of this study. Xpert MTB/RIF is used selectively since 2010. Phenotypic DST is performed using the MGIT system as previously described.14 From 2001 onwards, all new TB strains were genotyped, initially by five loci variable number tandem repeat (VNTR) typing and progressively with the addition of 12 and then 24 loci mycobacterial interspersed repetitive unit (MIRU) genotyping. GeneMapper v4.0 (Applied Biosystems) was used for fragment analysis and Bionumerics v6.7 (Applied Maths, Belgium) used for clonal structure analysis and VNTR-MIRU profiles storage. As a routine, all new MTBC strains are stored for later retrieval if necessary.
Study population and specimen selection
Once the MPT64 negative cluster was suspected, investigations were carried out retrospectively and prospectively to identify all possible linked patients. Firstly, Bionumerics data were interrogated for strains with profiles consistent with the VNTR-MIRU profile of the cluster type strain identified in 2017, noting that earlier characterised isolates may have had less loci examined. Such strains were sub-cultured from storage and tested for the presence of MPT64 antigen production. All strains were subjected to full VNTR-MIRU analysis if not already performed. Secondly, review of surveillance data collected by the Queensland Communicable Diseases Branch and regional TB Control Units was done to identify any clinically diagnosed cases with strong epidemiological links to any of the identified cluster cases. Thirdly, the typing profile was shared with other Australian jurisdictions via the National Tuberculosis Advisory Committee (NTAC) and they were requested to identify any cases with the same typing profile. Fourthly, any TB patient with an MPT64 negative strain or whose isolate clustered with the outbreak were prospectively included. Some of the demographic and microbiological data for all isolates were collated for analysis. Lastly in 2023, a recent type sequence for the outbreak was submitted to the Communicable Diseases Genomic Network (CDGN), Commonwealth Department of Health and Ageing for WGS matching with jurisdictional strains contained in the national database (2015–2022).
Use of information obtained to investigate this outbreak of public health significance regarding a Notifiable Condition in Queensland is supported by the Public Health Act (2005). In additional the institutional review board of the Children’s Health Queensland Hospital and Health Service Human Research Ethics Committee and Ethics and the Forensic and Scientific Services Human Ethics Committee (FSS-HEC) granted ethical approval for the study. Ethical approval included a waiver of individual patient consent.
Whole genome sequencing and genotypic drug resistance prediction
Genomic DNA was extracted using previous described methods.14 WGS was performed at Forensic and Scientific Services (FSS, Coopers Plains, Brisbane) with NextSeq 550 Illumina technology using Nextera XT library preparation kits (Illumina, San Diego, CA). Primary analysis of generated paired-end reads was done using updated versions of previously described methods (see Supplementary Materials and Methods for additional information).4 The VCF file was used to infer MTB lineage with SNP typing according to Napier et al.15 TB-Profiler v4.4.216 and an in-house custom resistance mutation dataset was used for resistance prediction. Polymorphism within MPT64 was assessed and protein structure predicted (see Supplementary Materials and Methods for additional information).
Phylogenetic reconstruction and strain diversity
A maximum likelihood phylogenetic analysis of study strains and publicly available lineage 4 datasets,17,18 as reference genomes was undertaken using IQ-TREE v2.1.219 on a multi-sample fasta alignment. ModelFinder20 was used to identify the best substitution model to run IQ-TREE, at 1000 standard nonparametric bootstrap replicates and tree visualised using FigTree v1.4.4. We used iTol21 to annotate the phylogenies. Pairwise SNP distance was determined using the ape library in R v4.0.2.22
Time dependent phylogenetic reconstruction
We used Markov chain Monte Carlo (MCMC)-based Bayesian phylogenetic inference using Beast v2.6.723,24 to reconstruct dated phylogenetic trees and infer the time to the most recent common ancestor. To find the best model, we used both strict and relaxed molecular clock with an informative prior distribution on the mutation rate, used both Hasegawa-Kishino-Yano (HKY) and general-time-reversible (GTR) substitution models. We ran each MCMC chain for at least 100 million states with 10% burn-in (see Supplementary Methods for additional information).
Bayesian phylodynamic inference
Inference of phylogenetic topology together with epidemiological parameters, particularly effective reproductive number (R0) and non-infectious rate (δ), birth-death skyline serial (BDS) model was performed using Beast v2.6.7. This model assumes an infection event to be considered as “birth” of a newly infected individual while a recovery event (successful treatment) as a “death”, on heterochronous data (sequences sampled at different times). For this analysis, the priors were configured as previously described25 but with some adjustments; lognormal prior (μ = 2.5, σ = 0.5) on time of origin parameter (T), covering the last 40 years and longer. For reproductive number (R0), we chose a lognormal prior (μ = 0, σ = 1), which placed most weight below 5.18 (95% quartile) and estimated at 10 independent intervals with recovery rate assumed to be constant throughout the time. The sampling proportion (ρ) was set to zero before the first sample, then configured to a Beta (45,5) distribution prior. A lognormal prior was utilised to configure non-infectious rate parameter (δ) that reflected a median infectious period of 3 years (μ = 0.5, σ = 1.0), which allows for short and longer durations of infection. An informative uniform prior was defined for substitution rate (initial value 7.9 × 10−8 nucleotide change/site/year; range 1 × 10−9 to 1 × 10−6), GTR substitution model with four gamma categories, and Markov chain Monte Carlo (MCMC) chain length and burn-in were set up as described above. Examination of log files was performed (see Supplementary Materials and Methods for additional information) to ensure each model had sufficient statistical support. We ran the model with the same parameters as above using Hasegawa-Kishino-Yano (HKY) substitution model in place of GTR.
Transmission inference
We utilised TransPhylo26 in R package to infer number of secondary cases for all isolates, using dated-labelled phylogeny and MCMC approach. In TransPhylo, transmission is modelled as a stochastic branching process, with a negative binomial distribution of secondary cases. The times between transmission events (generation times) and the times between infection and sampling (sampling times) are drawn from a Gamma distribution, with individuals infected right before the end of the sampling period having a lower probability of being sampled. The sampling proportion reflects the proportion of sampled cases since the onset of the outbreak. A coalescent model with constant population size Ne is used to model within-host evolution and a complete transmission bottleneck is assumed at transmission.
The generation time was configured using a long-tailed gamma distribution with parameters (shape = 0.8, scale = 7), which caters for long latent TB infections before activation and transmission. The sampling proportion was set at 80%, considering the well-resourced setting with effective case-finding strategies. Within-host and reproductive number were set at default setting, and the model was run at 100,000 MCMC chain of length, with states sampled every 10 steps. The first 20% of samples from each chain were discarded as burn-in before samples from the different chains were pooled. Convergence was assessed using trace plots and by confirming that the effective sample size (ESS) was at least 200 for each of the inferred parameters. R (cluster package) was used for construction of genomic clusters.
Phylogeographic analysis
To test for geospatial genetic structure, a mantel test (geosphere R package) was utilised to evaluate the relationship between genetic and geographic distances for all Queensland isolates. The statistical significance was evaluated by permuting matrices at 10,000, using pairwise SNP distance and geographic distance between isolates as measured by haversine great-circle (between relevant Queensland post codes) for paired strains.27
To assess the historical process that might be responsible for the past to present geographical distribution of the strain, we utilised PASTML,28 which uses a rooted timed tree and geographical locations of the strains as in-put. We assigned each strain the post code of residence (according to when the first diagnostic specimen was collected) and used PASTML to reconstruct the geographical ranges and changes, using MPPA as prediction method (standard settings) and added the character predicted by the joint reconstruction even if it was not selected by the Brier score (option -forced_joint).
Role of the funding source
The funder had no role in the study design, data analysis, interpretation or report writing.
Results
Laboratory isolate characteristics
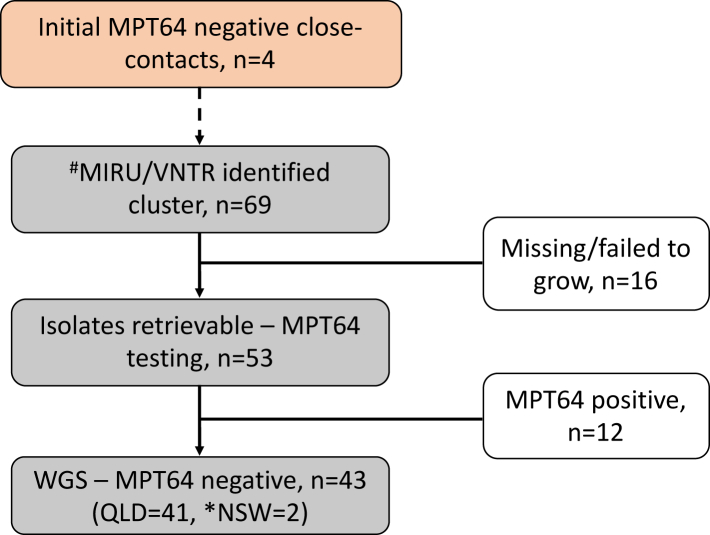
As of 31st July 2023, 69 cases from Queensland residents were identified as possible outbreak cases while 2 cases were identified from New South Wales (NSW) through the NTAC and CDGN search process (Fig. 1). Of the 53 Queensland suspected isolates that were retrieved and successfully cultured, 41 tested MPT64 negative and were all confirmed to be M. tuberculosis (Supplementary Table S1). When entire MIRU-24 profiles for all isolates (including those that initially had 5,12 and 15 loci assessed) were re-analysed, 23/24 loci were identical among all isolates (Supplementary Table S2).
Fig. 1.
Flow diagram of strain identification (2002–2022) and genomic analysis. #Bionumerics analysis of 5, 15 or 24 loci MIRU typing that was performed at different times of the study period. ∗MPT64 untested strains with similar MIRU profile.
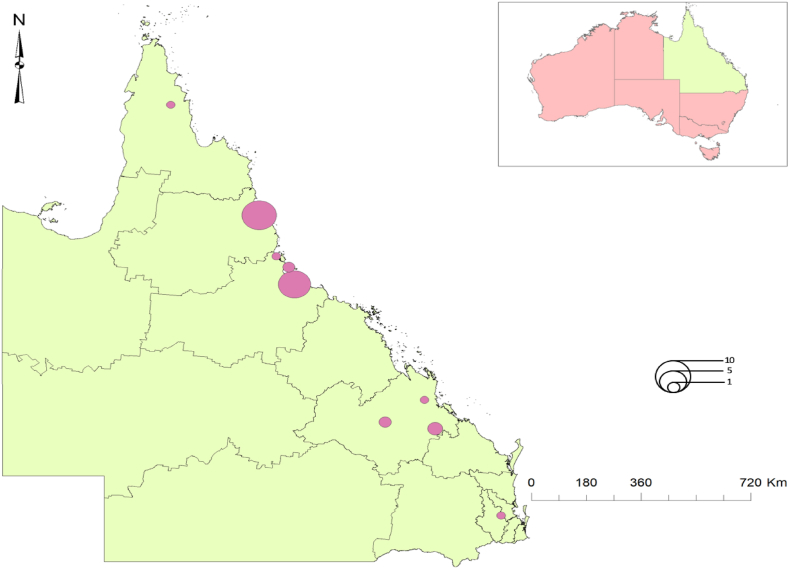
Cases were generally centred around Cairns (n = 13), Townsville (n = 12) and the Tablelands (n = 3) in North Queensland and less so, Central Queensland, Australia (Fig. 2). As previously reported,29 most cases identified were First Nations people. Five additional cases lacking culture confirmation (and thus unavailable for genomic analysis), were considered to likely be part the outbreak on consideration of epidemiological links.
Fig. 2.
Map of Queensland, Australia (inset), showing local government areas where outbreak cluster members were identified.
All isolates were found to be fully susceptible to all first-line agents with no drug resistance conferring mutations detected on WGS.
MPT64 gene polymorphism and protein structure
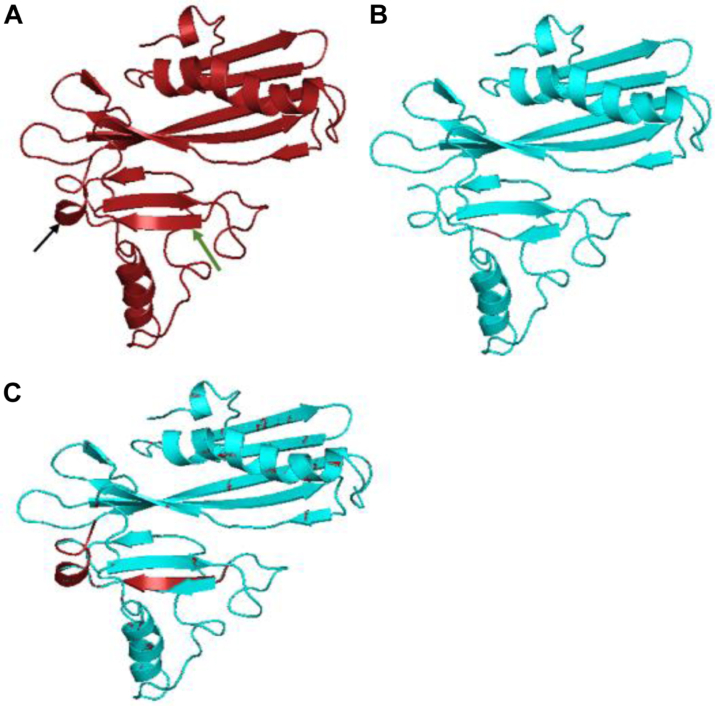
A total of forty-three sequences were analysed (including 2 from NSW) with an average read depth of 122X and average reference coverage >99%. The indel variant calling identified a deletion mutation (delAT, p.Val216fs) within the MPT64 gene among all study isolates; visual inspection of alignment reads confirmed the deletion (Supplementary Fig. S1). Fig. 3 shows the predicted tertiary structure of MPT64 protein with and without the frame-shift deletion. Of the 223 MPT64 amino acids of the mutant, 201 (90%) were modelled with 100% confidence, resulting in two major structural rearrangements at the C-terminus, when compared to the wild-type protein. There is loss of three amino acids on the last beta-strand of the protein (pos. 217–219) and rearrangement of the only transmembrane helix part of the protein (pos. 204–206) to a longer alpha-helix structure (pos. 203–207) in the mutant protein, losing the transmembrane signal.
Fig. 3.
Predicted tertiary structure of mutated MPT64 protein∗. A–unmutated MPT64 sequence (green arrow—beta strand, black arrow—transmembrane helix); B–mutated MPT64 sequence (delAT, p.Val216); C–overlap of A and B to emphasise key differences∗∗. ∗Predicted by Phyre2 software v2.0 (1). ∗∗Red arrow overlapping the turquoise arrow–loss of three amino acids on the last beta-strand of the protein (pos. 217–219). Red alpha helix structure overlapping a turquoise flat helix highlight rearrangement at pos. 203–207 in the mutant protein.
Global phylogeny of lineage 4 strains
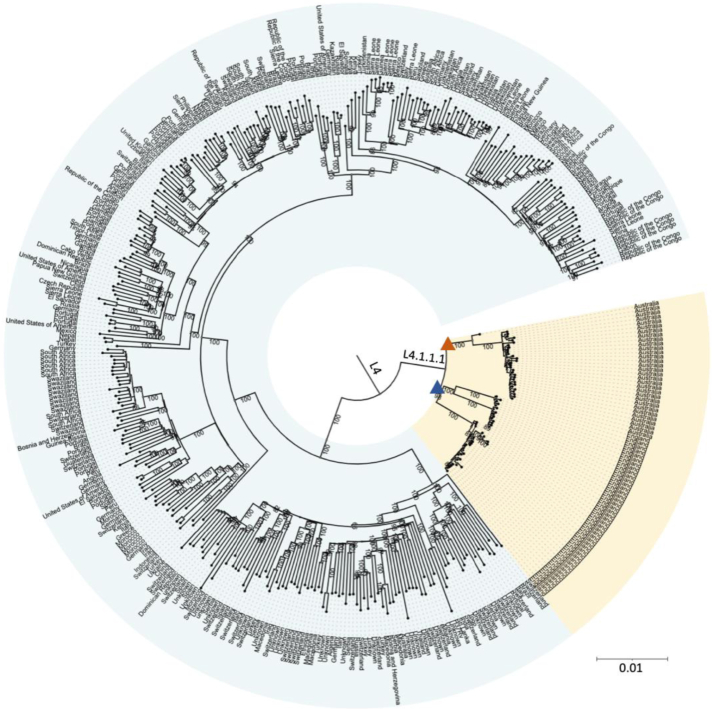
All MPT64 negative strains were identified as belonging to MTBC sub-lineage 4.1.1.1; as per Napier et al. classification.15 To investigate the origins and dispersal of these strains, we compiled a dataset comprising our 43 Australian genomes and 383 lineage 4 genomes from previous studies, representing 100 different countries.17,18 Using a high-quality variant alignment (26304 SNPs) to infer lineage 4 phylogeny, we demonstrated that the Australian strains formed a paraphyletic clade with strains from the United Kingdom (UK) Midlands (also by coincidence n = 43) (Fig. 4).
Fig. 4.
Maximum likelihood phylogeny of global Lineage 4 genomes∗. The Orange triangle marks the tree branch with the Australian genomes∗∗ and the blue triangle the tree branch with ancestral UK genomes. Single Nucleotide Polymorphism (SNP); United Kingdom (UK). ∗SNP Phylogeny of 383 Lineage 4 isolates from a Stucki et al., including UK genomes (2). ∗∗Total number of Lineage 4.1.1.1 strains sequenced in Queensland to date (41) plus two strains sequenced in NSW.
Temporal evolution of the MPT64 negative strain
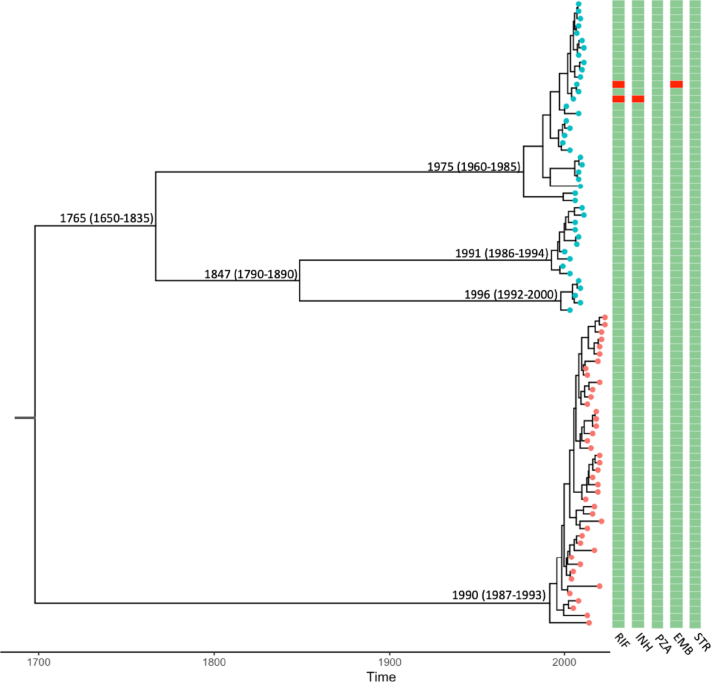
We explored the shared ancestral origin and temporal evolution of the Australian and UK L4.1.1.1 strains using Beast2 (with an alignment of 611 SNPs). Date randomisation test for temporal signal within the combined data showed sufficient temporal signal for analysis (Supplementary Fig. S2). Bayesian model comparison using path sampling determined coalescent exponential demographic model, with HKY nucleotide substitution rate as the best model (Supplementary Table S3). Under this model, we estimated a mean substitution rate of 0.37 SNPs per genome per year (95% HPD 0.26–0.5), which is similar to studies using contemporary L4 and mixed lineage MTBC genomes.30, 31, 32, 33 The phylogenetic tree of all L4.1.1.1 strains (Fig. 5) estimated time of the most recent common ancestor (TMRCA) in the year 1683 (95% HPD 1472–1876), with 1765 (95% HPD 1650–1835) estimated for the UK strains. The TMRCA for the Australian strain was estimated to be 1990 (95% HPD 1987–1993), indicating relatively recent introduction or clonal expansion from a previously introduced unsampled L4.1.1.1 strain that has attained local adaptations.
Fig. 5.
Phylogenetic reconstruction∗ of closely related Lineage 4.1.1.1 M. tuberculosis genomes. UK genomes marked in blue and Australian genomes marked in orange. Nodes labelled with estimated time of divergence (95% highest probability density intervals). Columns show predicted drug resistance in red. RIF-Rifampicin, INH-Isoniazid, PZA-Pyrazinamide, ETB-Ethambutol and STR-Streptomycin. ∗Bayesian phylogenetic reconstruction using the best evolutionary model (relaxed lognormal clock, Hasegawa–Kishino–Yano substitution and exponential demographic).
Phylodynamic inference
MPT64 negative L4.1.1.1 strain, Australia
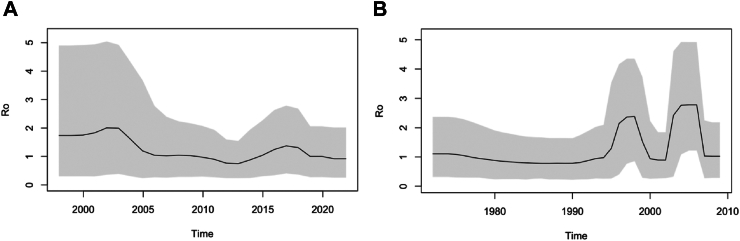
Path sampling revealed a relaxed molecular clock with HKY nucleotide substitution rate as the best BDS model (Supplementary Table S4), facilitating estimation of posterior distributions from the model for parameters such as the effective reproductive number (R0) and non-infectiousness (Supplementary Fig. S3). The skyline plot (Fig. 6A) of Ro revealed an estimated 1.8 (95% HPD 0.3–4.9) at the start, which increased to 2 (its maximum) by 2004, and gradually declined (with some expected variability) to levels below 1 at the present time. The median rate to become non-infectious was estimated to be 0.14 (95% HPD 0.09–0.19), which implies an infectious period of roughly 7 years. The medium sampling proportion was estimated to be 90% (95% HPD 82–97%), with an estimated mean substitution rate of 0.33 SNPs per genome per year (95% HPD 0.24–0.45).
Fig. 6.
Birth-death skyline plots of effective reproductive number (R0) for Australian (A) and UK (B) L4.1.1.1 genomes. The grey region is the 95% highest probability density intervals (HPD), while the black line is the median.
MPT64 positive L4.1.1.1 strain, UK
For the UK strain (Fig. 6B), the same BDS model indicated a R0 estimate of slightly above 1 at the start, with a sharp increase peaking at 2.2 around 1997, before declining and peaking again at 2.4 around 2005. Since 2008 it has declined to levels below 1 (declining epidemic), where it has remained ever since. The median rate to become non-infectious was estimated to be 0.18, suggesting an infected period of around 5 years. The median sampling proportion was estimated to be 89% (95% HPD 81–97%), with an estimated mean substitution rate of 0.52 SNPs per genome per year (95% HPD 0.35–0.61).
Comparison of transmission fitness
The mean pairwise SNP distance among the 43 Australian genomes (MPT64 negative) was 6 SNPs (IQR 4–10), illustrating limited diversity among the isolates, while a mean of 147 SNPs (IQR 10–242) was identified among the UK genomes (MPT64 positive). To assess the relative transmission dynamics between the two outbreaks, we used ≤5 and ≤ 12 SNP distance thresholds for cluster identification. With a 5 SNP cut-off, 81% (35/43) of MPT64 negative isolates formed 5 genomic clusters, whereas 83% (36/43) of the UK MPT64 positive isolates produced 9 genomic clusters (Table 1). At the 12 SNP threshold, MPT64 negative strains formed a single genomic cluster compared to 5 genomic clusters for MPT64 positive strains. TransPhylo (getOffspringDist) was used to estimate the number of secondary cases from each isolate (representing a single patient). We calculated the posterior probability that each patient generated at least one secondary case and labelled those with a posterior probability >0.9 as a ‘transmitter’.34 We identified 7 transmitters among the UK strains, while 3 transmitters were identified among the MPT64 negative outbreak.
Table 1.
Genomic clusters for Australian and UK L4.1.1.1 genomes using different SNP cut-offs.
| Lineage 4.1.1.1 strain | Year | Total No. | 5 SNPs cut-off |
12 SNPs cut-off |
||
|---|---|---|---|---|---|---|
| No. in clusters | No. of clusters | No. in clusters | No. of clusters | |||
| MPT64-Neg (Aus) | 2002–2022 | 43 | 35 | 5 | 42 | 1 |
| MPT64-Pos (UK) | 1998–2010 | 43 | 36 | 9 | 43 | 5 |
SNP clustering performed using cluster package in R.
Geospatial and phylogeographical analysis
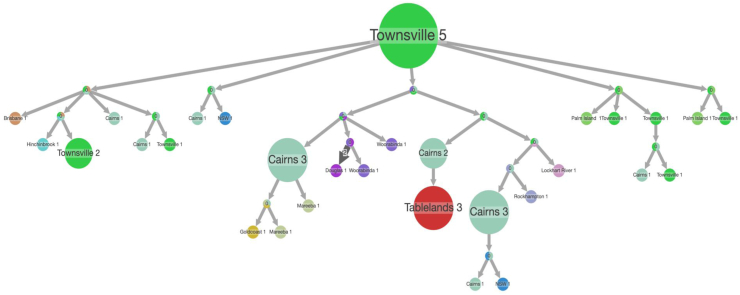
Comparing the 41 MPT64 negative Queensland isolates using Mantel test for pairwise SNP difference and geospatial distance showed poor correlation (r = 0.184, p = 0.690) (Supplementary Fig. S4). This illustrates poor geospatial clustering at the postcode level, suggesting high patient mobility. Using the time-rooted tree for pastml analysis, we found that Townsville was predicted to be the geographic origin (Fig. 7), with subsequent spread to surrounding towns and beyond. Five back migration events, where the strain was re-introduced into Townsville, illustrates the mobility of the affected population in and out of Townsville. Although there were lots of uncertain nodes, Cairns was identified as the likely intermediary to spread further north (i.e. Mareeba and the Tablelands).
Fig. 7.
Ancestral reconstruction of MPT64 negative strains with likely geographic spread∗. The size of the circles is proportional to the number of patients, arrows depict the likely direction of spread. The size of the arrows reflects migration pattern as seen on the tree, the dark arrow represents independent introductions in the same location. Different circle colours correspond to different post codes of the strains. ∗From PastML analysis showing a compressed tree for all locations.
Discussion
We describe the largest and longest standing TB outbreak ever investigated in Australia, caused by an MPT64 negative strain introduced into Queensland around 1990. The MPT64 protein stimulates a strong T-cell mediated immune response,35 and the frame-shift mutation observed in this MPT64 negative cluster may have caused reduced T-cell immune responses resulting in an attenuated phenotype potentially facilitating the sustained cryptic transmission. The observed mutation induced rearrangement of the C-terminus, which is required for localisation within the endoreticulum of macrophages, would impair protein response.35
Our geospatial phylogenetic structure analysis revealed that these MPT64 negative strains were spread by internal migration and people movement within Queensland. Mobility over long distances and at times homelessness were noted in this group of predominantly First Nations people (detailed epidemiological data to be published separately) and similar to displaced people and internal migrants elsewhere are more likely than geographically stable residents to share crowded living conditions and be of low socio-economic status, all of which are associated with an increased risk of tuberculosis development and spread.36 Although Townsville was recognised as the most likely origin and epicentre of the outbreak, the cluster analysis indicated repeated reintroduction events demonstrating dynamic interaction with external communities and emphasising the need for vigilant surveillance and ongoing disease control efforts.
Lineage 4 (Euro-American) is genetically diverse and its global spread has been attributed to European migration and colonisation.17 Although the closely related UK strain was MPT64 positive, it allowed us to perform comparative genomic analysis gaining valuable evolutionary insight. Our dating analysis estimated 1683 as the Lineage 4.1.1.1 TMRCA, which coincides with major waves of European migration out of Europe to Asia.31 However, the MPT64 negative strain identified in Queensland, Australia was only introduced around 1990. The vast majority of TB notifications in Australia are attributed to people movement and recent importation from high TB incidence settings.4,37 Despite the fact that sampling dates range between 2002 and 2022, with majority of the Australian cases reported in 2012 (7 cases), our R0 skyline plot indicated that the outbreak peaked around 2003. The fact that the peak of the outbreak occurred several years before it was detected, shows that this strain may have a long latency period. People have tried to infer latency periods from such data, but multiple factors contribute to periods of more effective spread. Kühnert et al. inferred long latency periods after phylodynamic analysis of a Swiss outbreak,25 but the affected population included people with homelessness and substance abuse, suggesting that social factors were likely central to observed transmission dynamics.
Interestingly, the estimated mutation rate for UK strains was found to be slightly higher than that for Australian strains (0.52 vs 0.33 SNP/genome/year). A study analysing two independent sub-lineage 4.1.2 datasets, one from Argentina (drug resistant) and the other from Canada (drug susceptible), found mutation rates to be broadly similar (0.22 vs 0.39 SNP/genome/year), suggesting that the evolution of antibiotic resistance did not have a large impact on the rate of molecular evolution of MTBC.38 We noted less genetic diversity among the Australian strains and high clustering using a 5 SNP threshold compared to UK strains, which is not unexpected given its recent introduction. A 5 SNP threshold has been used to infer recent local transmission, while a 12 SNP threshold may identify older transmission events or transmission of more diverse minority variants.39,40 This suggests more recent and ongoing transmission in Australia, but other factors such as length of latency, the rate of the molecular clock, sub-population transmission and sampling period may all influence the clustering signal.41 The use of a Bayesian-based approach (TransPhylo) showed more plausible secondary cases inferred from the UK strains than Australian strains, which suggests a lower transmissibility among the Australian strains.
Our study has some limitations. Transmission inferences based on genomics as yet have not been fully corroborated with detailed epidemiological data. Although the reference laboratory provides comprehensive passive surveillance, some historical isolates may have been missed or misidentified, although the practice of using a second molecular test to confirm MTBC makes misidentification unlikely. Inclusion criteria included the use of MIRU-24 profiling or MPT64 test results, which were in place from 2000 to 2007, respectively, but the outbreak was predicted to have begun earlier. It was not feasible to filter isolates prior to 2000 as to likelihood of cluster membership and retrieval of isolates from the 1990's is often unsuccessful in our laboratory. Nevertheless, we were able to gain deep insight from comparative genomic analyses and our retrospective sampling should have identified the vast majority of historical isolates. Although all Australian jurisdictions examined their own VNTR-MIRU databases for matching strains, the degree of rigor and date ranges were determined by the jurisdictions themselves. The request for interjurisdictional WGS data from the national CDGN database was limited by the degree of completeness and date range of the banked sequences.
In conclusion, our findings call for caution when interpreting negative MPT64 test results in a patient clinically suspected of having TB disease. A molecular assay should always be used to confirm or exclude the presence of MTB.42 This will minimise the risk of misdiagnosing MPT64 negative strains, with delayed or inappropriate treatment supporting ongoing transmission and ‘diagnostic selection”, as demonstrated by rifampicin resistant strains in eSwatini missed by XpertMTB/RIF and XpertUltra.43 Integration of routine WGS into public health surveillance will aid early cluster identification, highlight sub-lineages of concern and where relevant, identify antimicrobial resistance.44 The study also shows that MPT64 status may influence transmission success and persistence within a population, and highlights the potential benefits of routine WGS and value of jurisdictional data sharing, as encouraged by Australia’s National Tuberculosis Advisory Committee (NTAC), for enhanced national transmission surveillance45 and better targeted public health responses.
Contributors
CC, SP, BJM, LJMC, AB and VS designed the study and guided the data analysis. Data collection was done by AB, SP, BO and CC. AB performed bioinformatic analysis and drafted the manuscript. All authors participated in manuscript revision and approved the final version.
Data sharing statement
Sequencing data for this project is available in the NCBI Sequence Read Archive (BioProject accession no. PRJNA1095921). Bayesian analysis xml files can be found here https://github.com/arnoldbain/MPT64_Neg-MTB.
Editor note
The Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
Declaration of interests
DW is supported by a Queensland Advancing Clinical Research Fellowship from the Queensland Government, and also reports research funding from SpeedDx Pty Ltd and the Australian Research Council for unrelated research. Other co-authors have no conflict of interest.
Acknowledgements
We thank the staff of Queensland Mycobacterium Reference Laboratory, TB control units in Queensland, NSW Tuberculosis and Rheumatic Heart Disease Program and Forensic and Scientific Services (FSS) and Australian jurisdictional TB programmes for all their assistance. We would like to thank everyone at the Communicable Disease Branch, especially Denise Jenkins and Gemma Devlin, for their help. Dr. Brian Forde (UQCCR and IMB) and Thom Cuddihy (IMB) provided invaluable computational support.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lanwpc.2024.101105.
Contributor Information
Arnold Bainomugisa, Email: arnold.bainomugisa@health.qld.gov.au.
Christopher Coulter, Email: chris.coulter@health.qld.gov.au.
Appendix ASupplementary data
References
- 1.Bright Amy, Denholm Justin T., Coulter Chris, Waring Justin, Stapledon Rick, on behalf of the National Tuberculosis Advisory Committee for the CDNA and the AMRLN Tuberculosis notifications in Australia, 2015–2018. Commun Dis Intell. 2020;44:88. doi: 10.33321/cdi.2020.44.88. [DOI] [PubMed] [Google Scholar]
- 2.Toms C., Stapledon R., Waring J.D.P., National Tuberculosis Advisory Committee. Communicable Diseases Network Australia. Australian Mycobacterium Reference Laboratory Network Tuberculosis notifications in Australia, 2012 and 2013. Commun Dis Intell Q Rep. 2015;39:E217–E235. [PubMed] [Google Scholar]
- 3.Norton S., Bag S.K., Cho J.G., et al. Detailed characterisation of the tuberculosis epidemic in western Sydney: a descriptive epidemiological study. ERJ Open Res. 2019;5(3) doi: 10.1183/23120541.00211-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bainomugisa A., Pandey S., Donnan E., et al. Cross-border movement of highly drug-resistant Mycobacterium tuberculosis from Papua New Guinea to Australia through torres strait protected zone, 2010-2015. Emerg Infect Dis. 2019;25(3):406–415. doi: 10.3201/eid2503.181003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization (WHO) Module 3: diagnosis - Rapid diagnostics for tuberculosis detection 2021 update. 2021. WHO operational handbook on tuberculosis. [Google Scholar]
- 6.Brent A.J., Mugo D., Musyimi R., et al. Performance of the MGIT TBc identification test and meta-analysis of MPT64 assays for identification of the Mycobacterium tuberculosis complex in liquid culture. J Clin Microbiol. 2011;49(12):4343–4346. doi: 10.1128/JCM.05995-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coombes B.K., Valdez Y., Finlay B.B. Evasive maneuvers by secreted bacterial proteins to avoid innate immune responses. Curr Biol. 2004 doi: 10.1016/j.cub.2004.09.043. [DOI] [PubMed] [Google Scholar]
- 8.Mustafa A.S., Shaban F. Mapping of Th1-cell epitope regions of mycobacterium tuberculosis protein MPT64 (Rv1980c) using synthetic peptides and t-cell lines from M. tuberculosis-infected healthy humans. Med Princ Pract. 2010 doi: 10.1159/000273073. [DOI] [PubMed] [Google Scholar]
- 9.Li Q., Yu H., Zhang Y., et al. Immunogenicity and protective efficacy of a fusion protein vaccine consisting of antigen Ag85B and HspX against Mycobacterium tuberculosis infection in mice. Scand J Immunol. 2011 doi: 10.1111/j.1365-3083.2011.02531.x. [DOI] [PubMed] [Google Scholar]
- 10.Zexuan S., Wencong H., Shaojun P., et al. Association of lineage 4.2.2 of Mycobacterium tuberculosis with the 63-bp deletion variant of the mpt64 gene. Microbiol Spectr. 2023;11 doi: 10.1128/spectrum.01842-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.N’Dira Sanoussi C., de Jong B.C., Odoun M., et al. Low sensitivity of the MPT64 identification test to detect lineage 5 of the Mycobacterium tuberculosis complex. J Med Microbiol. 2018 doi: 10.1099/jmm.0.000846. [DOI] [PubMed] [Google Scholar]
- 12.Ofori-Anyinam B., Kanuteh F., Agbla S.C., et al. Impact of the mycobaterium africanum west africa 2 lineage on TB diagnostics in west africa. PLoS Negl Trop Dis. 2016;10(7) doi: 10.1371/journal.pntd.0004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halse T.A., Escuyer V.E., Musser K.A. Evaluation of a single-tube multiplex real-time PCR for differentiation of members of the Mycobacterium tuberculosis complex in clinical specimens. J Clin Microbiol. 2011 doi: 10.1128/JCM.00467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bainomugisa A., Lavu E., Hiashiri S., et al. Multi-clonal evolution of multi-drug-resistant/extensively drug-resistant Mycobacterium tuberculosis in a high-prevalence setting of Papua New Guinea for over three decades. Microb Genom. 2018 doi: 10.1099/mgen.0.000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Napier G., Campino S., Merid Y., et al. Robust barcoding and identification of Mycobacterium tuberculosis lineages for epidemiological and clinical studies. Genome Med. 2020 doi: 10.1186/s13073-020-00817-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phelan J.E., O’Sullivan D.M., Machado D., et al. Integrating informatics tools and portable sequencing technology for rapid detection of resistance to anti-tuberculous drugs. Genome Med. 2019 doi: 10.1186/s13073-019-0650-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stucki D., Brites D., Jeljeli L., et al. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat Genet. 2016 doi: 10.1038/ng.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coll F., McNerney R., Guerra-Assunção J.A., et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun. 2014 doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen L.T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015 doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., Von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017 doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letunic I., Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paradis E., Schliep K. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019 doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 23.Ayres D.L., Cummings M.P., Baele G., et al. Beagle 3: improved performance, scaling, and usability for a high-performance computing library for statistical phylogenetics. Syst Biol. 2019 doi: 10.1093/sysbio/syz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouckaert R., Vaughan T.G., Barido-Sottani J., et al. Beast 2.5: an advanced software platform for bayesian evolutionary analysis. bioRxiv. 2018 doi: 10.1101/474296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kühnert D., Coscolla M., Brites D., et al. Tuberculosis outbreak investigation using phylodynamic analysis. Epidemics. 2018 doi: 10.1016/j.epidem.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Didelot X., Kendall M., Xu Y., White P.J., McCarthy N. Genomic epidemiology analysis of infectious disease outbreaks using TransPhylo. Curr Protoc. 2021 doi: 10.1002/cpz1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James I. 1835. Navigation and nautical astronomy: for the use of British seamen. London, UK. [Google Scholar]
- 28.Ishikawa S.A., Zhukova A., Iwasaki W., Gascuel O., Pupko T. A fast likelihood method to reconstruct and visualize ancestral scenarios. Mol Biol Evol. 2019 doi: 10.1093/molbev/msz131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Queensland Health . 2021. Tuberculosis in Queensland 2017-2019. Brisbane.https://www.health.qld.gov.au/__data/assets/pdf_file/0020/1130555/report-tb-qld-2017-2019.pdf [Google Scholar]
- 30.Menardo F., Duchêne S., Brites D., Gagneux S. The molecular clock of Mycobacterium tuberculosis. bioRxiv. 2019 doi: 10.1101/532390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brynildsrud O.B., Pepperell C.S., Suffys P., et al. Global expansion of Mycobacterium tuberculosis lineage 4 shaped by colonial migration and local adaptation. Sci Adv. 2018 doi: 10.1126/sciadv.aat5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eldholm V., Monteserin J., Rieux A., et al. Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis outbreak strain. Nat Commun. 2015 doi: 10.1038/ncomms8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulholland C.V., Shockey A.C., Aung H.L., et al. Dispersal of Mycobacterium tuberculosis driven by historical European trade in the South pacific. Front Microbiol. 2019 doi: 10.3389/fmicb.2019.02778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loiseau C., Windels E.M., Gygli S.M., et al. The relative transmission fitness of multidrug-resistant Mycobacterium tuberculosis in a drug resistance hotspot. Nat Commun. 2023 doi: 10.1038/s41467-023-37719-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stamm C.E., Pasko B.L., Chaisavaneeyakorn S., et al. Screening Mycobacterium tuberculosis secreted proteins identifies Mpt64 as a eukaryotic membrane-binding bacterial effector. mSphere. 2019 doi: 10.1128/msphere.00354-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang C., Lu L., Warren J.L., et al. Internal migration and transmission dynamics of tuberculosis in Shanghai, China: an epidemiological, spatial, genomic analysis. Lancet Infect Dis. 2018 doi: 10.1016/S1473-3099(18)30218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lönnroth K., Migliori G.B., Abubakar I., et al. Towards tuberculosis elimination: an action framework for low-incidence countries. Eur Respir J. 2015 doi: 10.1183/09031936.00214014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menardo F., Duchêne S., Brites D., Gagneux S. The molecular clock of mycobacterium tuberculosis. PLoS Pathog. 2019 doi: 10.1371/journal.ppat.1008067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cancino-Muñoz I., López M.G., Torres-Puente M., et al. Population-based sequencing of Mycobacterium tuberculosis reveals how current population dynamics are shaped by past epidemics. Elife. 2022 doi: 10.7554/ELIFE.76605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X., Martinez E., Lam C., et al. Exploring programmatic indicators of tuberculosis control that incorporate routine Mycobacterium tuberculosis sequencing in low incidence settings: a comprehensive (2017–2021) patient cohort analysis. Lancet Reg Health West Pacific. 2023 doi: 10.1016/j.lanwpc.2023.100910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menardo F. Understanding drivers of phylogenetic clustering and terminal branch lengths distribution in epidemics of Mycobacterium tuberculosis. Elife. 2022 doi: 10.7554/eLife.76780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bastian I., Shephard L., Lumb R. Revised guidelines for Australian laboratories performing mycobacteriology testing. Commun Dis Intell. 2021 doi: 10.33321/cdi.2020.44.2. [DOI] [PubMed] [Google Scholar]
- 43.Beckert P., Sanchez-Padilla E., Merker M., et al. MDR M. tuberculosis outbreak clone in Eswatini missed by Xpert has elevated bedaquiline resistance dated to the pre-treatment era. Genome Med. 2020 doi: 10.1186/s13073-020-00793-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denholm J.T., Behr M.A., de Vries G., et al. Developing best practice public health standards for whole genome sequencing of Mycobacterium tuberculosis. Lancet Reg Health West Pacific. 2024;0 doi: 10.1016/j.lanwpc.2024.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donnan E.J., Marais B.J., Coulter C., et al. The use of whole genome sequencing for tuberculosis public health activities in Australia: a joint statement of the National Tuberculosis Advisory Committee and Communicable Diseases Genomics Network. Commun Dis Intell. 2023 doi: 10.33321/cdi.2023.47.8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.