Abstract
Persistent infection with human papillomavirus type 16 (HPV-16) is strongly associated with the development of cervical cancer. Neutralizing epitopes present on the major coat protein, L1, have not been well characterized, although three neutralizing monoclonal antibodies (MAbs) had been identified by using HPV-16 pseudovirions (R. B. Roden et al., J. Virol. 71:6247–6252, 1997). Here, two of these MAbs (H16.V5 and H16.E70) were demonstrated to neutralize authentic HPV-16 in vitro, while the third (H16.U4) did not. Binding studies were conducted with the three MAbs and virus-like particles (VLPs) composed of the reference L1 sequence (114K) and three variant L1 sequences: Rochester-1k (derived from viral stock DNA), GU-1 (derived from cervical biopsy DNA), and GU-2 (derived from biopsy DNA, but containing some sequence changes likely to be artifactual). While all three MAbs bound to 114K and Rochester-1k VLPs, GU-1 VLPs were not recognized by H16.E70, and both H16.E70 and H16.V5 failed to bind to GU-2 VLPs. Site-directed mutagenesis was used to replace disparate amino acids in the GU-2 L1 with those found in the 114K L1. Alteration of the amino acid at position 50, from L to F, completely restored H16.V5 binding and partially restored H16.E70 binding, while complete restoration of H16.E70 binding occurred with GU-2 VLPs containing both L50F and T266A alterations. Immunization of mice with L1 variant VLPs revealed that GU-2 VLPs were poorly immunogenic. The L50F mutant of GU-2 L1, in which the H16.V5 epitope was restored, elicited HPV-16 antibody responses comparable to those obtained with 114K VLPs. These results demonstrate the importance of the H16.V5 epitope in the generation of potent HPV-16 neutralizing antibody responses.
There is strong epidemiological and biological evidence that infection with certain high-risk types of human papillomavirus (HPV) is the primary cause of cervical cancer, the second most common cancer in women worldwide (1, 2). Among the HPV types associated with this carcinoma, HPV-16 is the most prevalent and is present in about 50% of tumor specimens.
Recent results with animal models of papillomavirus-associated disease have suggested that development of a prophylactic vaccine against HPV-16 may be possible (4, 11, 13, 23). These animal studies demonstrated the protective effects generated by immunization with virus-like particles (VLPs) composed of the virus major coat protein, L1. In addition, passive transfer experiments provided compelling evidence that neutralizing antibody responses against the L1 protein are sufficient for protection against papillomavirus disease (4, 23).
Limited information is available about the neutralizing epitopes present on the L1 protein of HPV-16, in part due to a lack of viral stock to conduct infectivity experiments. However, using HPV-16 pseudovirions, which are recombinant capsids composed of HPV-16 structural proteins and bovine papillomavirus DNA, Roden et al. identified three monoclonal antibodies (MAbs), H16.V5 (V5), H16.E70 (E70), and H16.U4 (U4), which may be capable of neutralizing HPV-16 (18). All of these MAbs are specific for HPV-16 and require conformationally intact HPV-16 L1 for binding (6). Mapping of the epitopes recognized by these MAbs has been hampered by the complex structure of the VLPs.
A successful approach to mapping conformation-dependent neutralizing epitopes on HPV L1 has been the identification of amino acids involved in the differential binding of neutralizing MAbs to VLPs composed of natural sequence variants or site-directed mutants of L1 proteins (16, 18). Roden et al. investigated the conservation of neutralization epitopes among HPV-16 intratype variants by examining the binding profiles of V5, E70, and U4 on HPV-16 L1 VLPs composed of the reference sequence (114K isolate) and a Zairian isolate which differed from the reference L1 protein at seven amino acid positions (18). The inefficient binding of the E70 MAb to the Zairian isolate L1 VLPs enabled the identification of a critical amino acid in the binding site of this MAb.
In contrast to the E70 epitope, no information is available on the binding site of the V5 MAb. However, the V5 epitope is recognized by most human antisera following HPV-16 infection (24). Binding of the V5 MAb to HPV-16 VLPs completely blocked the reactivity of more than 75% of human antisera. Thus, identification of the V5 epitope would provide important information regarding the targeting of the humoral response against the HPV-16 major capsid protein.
In the present study, we confirm and extend previously published results by demonstrating that MAbs V5 and E70 neutralize authentic HPV-16 virions. Amino acid residues critical for the binding of these MAbs to the HPV-16 L1 sequence were identified. Additionally, the ability of HPV-16 L1 VLPs lacking one or both of these epitopes to elicit neutralizing antibody responses in outbred mice were compared. The results reveal the necessity of the V5 epitope for the induction of potent neutralizing antibody responses against HPV-16 and demonstrate the paucity of other strong neutralization sites within the major capsid protein.
MATERIALS AND METHODS
MAbs.
Ascites fluids from the hybridoma cell lines V5, E70, and U4 (6) were obtained from Chemicon International, Inc. (Temecula, Calif.). H11.F1 ascites fluid was purchased from Pennsylvania State University.
HPV-16 neutralization assay.
Anti-VLP sera and MAbs were tested for HPV-16Rochester-1k/ur3 neutralizing activity with an in vitro infectivity assay as previously described (25). Cellular β-actin spliced transcript was detected in all test samples. The neutralization titer was defined as the greatest serum dilution which inhibited the detection of the E1^E4 transcript.
Construction of recombinant baculoviruses expressing HPV-16 L1 sequence variants.
The recombinant baculovirus expressing the 114K HPV-16 L1 protein was kindly provided by Martin Müller. The origin of the Rochester-1k HPV-16 L1 sequence and the generation of a recombinant baculovirus encoding the complete L1 open reading frame of this HPV-16 variant have previously been described (25).
The GU-1 and GU-2 HPV-16 L1 sequences were derived from DNA extracted from biopsy specimens from a normal cervix showing chronic cervicitis (GU-1) and from a cervix exhibiting mild dysplasia (GU-2) (provided by Attila Lorincz). HPV-16 L1 genes were amplified from this DNA by PCR with 5′-GACCGGAGATCTGCCACCATGGCGGTTTGGCTTCCT-3′ as the forward primer and 5′-GACATGCCATGGGTGCGGTGCCGGTCAG-3′ as the reverse primer (7a). The resulting PCR products were digested with BglII and NcoI and cloned into the multiple cloning region of pBlueBacIII (Invitrogen, Carlsbad, Calif.) downstream of the polyhedrin promoter. The resulting transfer vectors, pGU-1 and pGU-2, were transfected into Sf9 cells along with linearized Autographa californica multiple nuclear polyhedrosis virus DNA (Baculo-Gold; Pharmingen, San Diego, Calif.). Recombinant baculoviruses were then taken through three rounds of plaque purification as described previously (5, 22).
Sequencing of the HPV-16 L1 variants. (i) 114K.
DNA was extracted from recombinant baculovirus particles encoding 114K HPV-16 L1, and the sequence of the L1 gene was determined by automated DNA sequencing (Dye-Terminator; ABI, Foster City, Calif.) as previously described (25). The sequence was found to be identical to that published for the reference HPV-16 L1 gene (9), except that a G instead of an A was detected at nucleotide position 6432 (numbering is for the HPV-16 genomic sequence). This nucleotide change results in an alanine instead of a threonine at amino acid position 266 in the HPV-16 L1 protein.
(ii) GU-1 and GU-2.
The nucleotide sequence of the HPV-16 L1 genes contained in the baculovirus transfer vectors pGU-1 and pGU-2 was determined by automated DNA sequencing. The GU-1 L1 gene was found to contain seven amino acid differences from the 114K L1 sequence: arginine to glycine at position 41 (A5757G), histidine to tyrosine at position 76 (C5862T), threonine to asparagine at position 176 (C6163A), asparagine to threonine at position 181 (A6178C), serine to proline at position 282 (T6480C), threonine to proline at position 353 (A6693C), and leucine to phenylalanine at position 474 (G7058T). GU-1 L1 also contained 10 silent nucleotide changes: G5696A, T5909C, T6245C, A6314G, C6557T, G6719A, C6852T, C6863T, C6968T, and G6992A. GU-2 L1 contained three amino acid differences from the reference sequence: phenylalanine to leucine at position 50 (T5786A), alanine to threonine at position 266 (G6432A), and lysine to asparagine at position 380 (A6776C). The GU-2 L1 gene also contained a silent base change of T6281C.
Detection of the GU-2 L1 gene in biopsy specimen DNA.
Because several of the changes found in GU-2 L1 had not been previously reported in any other HPV-16 L1 variant, we sought to verify the presence of this L1 sequence in the biopsy specimen from which the gene was originally derived. DNA from this biopsy specimen was used as template for 35 cycles of PCR with 5′-TGCTGATGCAGGTGACTTTTA-3′ (located at bases 5554 to 5574 in the HPV-16 genomic sequence) as the forward primer and 5′-CAACACTAATTCAACATACATAC-3′ (located at bases 7160 to 7182 in the HPV-16 genomic sequence) as the reverse primer. The 1.6-kb PCR product was purified by agarose gel electrophoresis, extracted with the Qiagen gel extraction kit (Qiagen, Chatsworth, Calif.), and then sequenced as described above. The DNA sequence of this PCR product was found to be identical to that determined for the GU-2 L1 gene in pGU-2, except that a T was found at nucleotide position 5786 instead of an A, and an A was detected at position 6776 instead of a C. Thus, the nucleotide changes (relative to the HPV-16 L1 reference sequence) which gave rise to the leucine residue at amino acid 50 and the asparagine at amino acid 380 in the GU-2 HPV-16 L1 protein were not detected.
Oligonucleotide primers were designed to specifically amplify the GU-2 HPV16 L1 gene. The forward primer was 5′-GCAGTTGGACATCCCTATTTA-3′, with the 3′ base corresponding to the nucleotide change that resulted in a leucine at amino acid position 50, and the reverse primer was 5′-TAACGTCTGCAGTTAAGGTTATG-3′, with the 3′ base corresponding to the nucleotide change that resulted in an asparagine at position 380. Biopsy specimen DNAs, as well as the L1 gene generated by PCR from this DNA and the cloned GU-2 L1 gene, were used as a template for 40 cycles of PCR. PCR products were separated in agarose gels and stained with ethidium bromide.
Generation of GU-2 HPV-16 L1 mutants.
Single amino acid substitutions were introduced into the HPV16 L1 GU-2 protein by overlap extension PCR as described by Ho et al. with pGU-2 as the template (8). Overlapping PCR, which produced the full-length L1 gene, placed a BglII site directly 5′ to the ATG codon and a HindIII site directly 3′ to the TAA codon. Full-length PCR products were digested with BglII and HindIII and cloned into pFastBac (Gibco/BRL, Gaithersburg, Md.). Individual nucleotide substitutions were confirmed by automated DNA sequencing as described above. Upon confirmation of a mutant clone, recombinant baculoviruses were produced by using the Bac to Bac system (Gibco/BRL). HPV-16 L1 GU-2 double mutants were constructed by subcloning restriction fragments of the single mutants generated by PCR.
Expression and purification of HPV-16 L1 VLPs.
HPV-16 L1 proteins were heterologously expressed in Trichoplusia ni (High Five) cells infected with recombinant baculovirus, and VLPs were purified as described previously (17). The protein concentration was determined by the Bradford assay (3) with bovine serum albumin as the reference protein. VLPs were adsorbed to carbon-coated grids, stained with 2% phosphotungstic acid (pH 6.8), and examined under an electron microscope as described previously (17).
Size exclusion chromatography.
One hundred microliters of purified L1 protein (10 to 30 μg) was injected onto a TosoHAAS G6000PWx1 high-performance liquid chromatography column (7.8 mm by 30 cm, 13-μm particle size) (TosoHAAS, Montgomeryville, Pa.) equilibrated with phosphate-buffered saline (PBS [pH 7.4]) containing 0.5 M NaCl, and eluted at a flow rate of 0.8 ml/min. Eluted protein was detected by A215. The column had previously been calibrated with homogeneous preparations of HPV-18 L1 VLPs (∼20,000 kDa) and HPV-11 capsomeres produced by trypsinization of HPV-11 VLPs (∼230 kDa) (15), which eluted at approximately 11.5 and 14.2 min, respectively. Additional calibration standards included purified catalase (∼252 kDa) and ovalbumin (∼45-kDa), which eluted at 14.2 and 14.5 min, respectively.
HPV-16 VLP ELISA.
All enzyme-linked immunosorbent assays (ELISAs) were carried out with 114K VLPs except where indicated. VLPs were diluted in PBS to 800 ng/ml, and 100-μl aliquots were dispensed into 96-well microtiter plates (Immulon II; Dynex Technologies, Inc., Chantilly, Va.). Plates were incubated at 37° for 1 h and then washed with PBS with 0.1% Tween 20. Plates were blocked at room temperature for 1 h with 5% nonfat dry milk in PBS. Following washing, serial twofold dilutions of anti-VLP sera or ascites fluid containing anti-HPV-16 MAbs in 1% nonfat dry milk in PBS were added to the wells in duplicate. Normal mouse serum and ascites fluid with an irrelevant MAb were used as negative controls. After 2 h of incubation at room temperature, the plates were washed and horseradish peroxidase-labeled, goat anti-mouse immunoglobulin G (Southern Biotechnology, Inc., Birmingham, Ala.) was added to the plates. The plates were incubated for 1 h at room temperature, washed, and developed with ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid] (Kirkegaard and Perry, Gaithersburg, Md.). Optical density readings were made at 405 nm (OD405) at the 30-min endpoint. Averages of duplicate wells were calculated as the final OD values. Titers were defined as the greatest serial dilution which yielded an OD value greater than twice that obtained with normal mouse serum.
MAb binding to 114K HPV-16 L1 VLPs: surface plasmon resonance (BIACORE).
All steps were performed at 25°C. CM5 sensor chips (BIACORE, Inc., Piscataway, N.J.) were activated with N-hydroxysuccinimide–1-ethyl-3-[3-dimethylaminopropyl]-carbodiimide as directed by the manufacturer. A 400 nM solution of purified 114K HPV-16 VLPs in 10 mM sodium acetate (pH 5) was injected over an activated chip (12). Unreacted ester groups were then blocked with 1 M ethanolamine. Binding studies were conducted with ascites fluid from the V5, E70, and U4 hybridoma cell lines diluted with HEPES-buffered saline (pH 7.4). Approximately 100 μl of dilute ascites was injected onto the VLP-coupled sensor chip at a flow rate of 10 μl/min. Binding was indicated as a change in resonance units (RU) on the chip. An anti-HPV-11 MAb was used as a control for nonspecific binding. For pairwise competitive binding studies, a saturating amount of one MAb was passed over the VLP-coupled sensor chip, followed by an injection of the same or a different MAb. Following each binding cycle, the VLP-coupled surface was regenerated with 10 μl of 100 mM phosphoric acid.
Production of antisera.
Groups of five outbred ND4 Swiss mice (Harlan Sprague-Dawley, Indianapolis, Ind.) or BALB/c ByJ mice (Jackson Laboratories, Bar Harbor, Maine) received two subcutaneous injections (100 μl), spaced 4 to 5 weeks apart, of 5, 0.5, or 0.05 μg of HPV-16 VLPs adsorbed to aluminum hydroxide (Alhydrogel, E. M. Sergeant Pulp and Chemical Co., Inc., Clifton, N.J.) at a final concentration of 1 mg of aluminum per ml. Two weeks following the last immunization, serum samples were collected, pooled, and stored at −70°C.
Nucleotide sequence accession number.
Sequence data from this article have been deposited in the GenBank data library under accession no. AF134175 (Rochester-1k L1), AF134178 (GU-1 L1), and AF134177 (GU-2 L1).
RESULTS
MAb neutralization of authentic HPV-16.
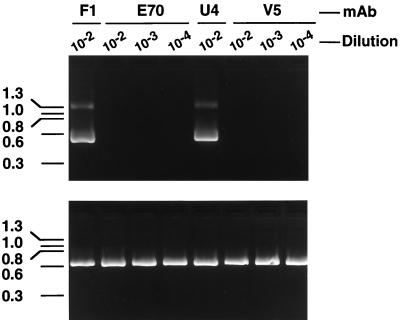
We have recently developed an in vitro infectivity assay for HPV-16 in which infection of cultured cells by authentic HPV-16 virions (generated in xenografts implanted in severe combined immunodeficiency mice) is documented by the appearance of a virus-specific spliced mRNA in cultured keratinocytes (25). To assess the ability of the HPV-16-specific MAbs V5, E70, and U4 to neutralize HPV-16, serial 10-fold dilutions of ascitic fluids were preincubated with the virus stock prior to in vitro culture. After 7 days in culture, total RNA extracted from the cells was used for cDNA synthesis. Nested primers were then used to amplify a 487-bp HPV-16 E1^E4 cDNA. As shown in Fig. 1, the V5 and E70 ascites fluids inhibited detection of the E1^E4 mRNA at dilutions of ≤1:10,000. In contrast, the U4 ascites fluid failed to neutralize virus at a 1:100 dilution. In subsequent studies, U4 was tested at a 1:25 dilution and still demonstrated no neutralizing activity against authentic HPV-16 (data not shown).
FIG. 1.
HPV-16 L1 MAbs V5 and E70 inhibit in vitro infection of HPV-16. HPV-16Rochester-1k/ur3/ur3 was preincubated with the indicated dilutions of E70, U4, or V5 ascites fluid prior to infection of cultured keratinocytes. As a control, virus was also preincubated with the HPV-11-specific MAb F1, which was previously demonstrated not to neutralize HPV-16. Seven days postinfection, cells were harvested, and the presence of the HPV-16 E1^E4 spliced message, diagnostic of HPV-16 infection, was detected by RT-PCR (top). PCR amplification of cellular β-actin was performed with all cDNA samples as an internal control (bottom).
Differential binding of MAbs to HPV-16 L1 sequence variants.
The inability of MAb U4 to neutralize authentic HPV-16 infection contrasts with results previously obtained with an HPV-16 pseudovirion infectivity assay (18). However, as shown in Table 1, the sequence of the L1 protein in our virus stock (Rochester-1k) differs at seven amino acid positions from the L1 sequence present in the HPV-16 pseudovirions (114K). Thus, the inability of the U4 MAb to neutralize HPV-16Rochester-1k/ur3 might be due to the failure of this antibody to bind to the Rochester-1k L1 protein. To address this issue and to determine whether disparate HPV-16 L1 sequences might be useful in mapping residues involved in the binding of the three MAbs, HPV-16 L1 proteins of the 114K and Rochester-1k variants were generated by using the recombinant baculovirus expression system. In addition, the L1 proteins of two other HPV-16 variants, referred to as GU-1 and GU-2, were expressed by using recombinant baculoviruses.
TABLE 1.
Divergent amino acids in the L1 protein of HPV-16 variants
| Variant | Variationa at amino acid position:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 41 | 50 | 76 | 176 | 181 | 206 | 266 | 282 | 353 | 380 | 389 | 474 | |
| 114K | R | F | H | T | N | G | Ab | S | T | K | T | L |
| Rochester-1k | R | F | Y | N | T | S | A | S | P | K | S | F |
| GU-1 | G | F | Y | N | T | G | A | P | P | K | T | F |
| GU-2 | R | L | H | T | N | G | T | S | T | N | T | L |
The boldfaced amino acids are different from those present in the 114K sequence.
Originally reported to be a T at this position.
The GU-1 and GU-2 sequences were originally generated by PCR from biopsy specimens of cytologically normal and dysplastic tissues, respectively. Sequencing of the GU-1 L1 gene revealed differences at seven amino acid positions from 114K L1 and differences at four positions from Rochester-1k L1 (Table 1). The GU-1 sequence is highly homologous to the previously reported 1194 sequence, differing only at amino acid residue 41, where the GU-1 sequence encodes a G and the 1194 sequence contains an R (10).
The GU-2 L1 sequence contained amino acid residues at three positions which were divergent from those found in the 114K L1 protein (Table 1). Two of the changes found in GU-2 L1, the L at amino acid 50 and the D at position 380, had not been previously reported in any other HPV-16 L1 variant (9). Due to the unique nature of the GU-2 L1, we sought to verify the presence of this L1 sequence in the biopsy specimen from which the gene was originally derived. Using DNA from the original biopsy specimen as a template, PCR was carried out with both HPV-16 L1-specific primers, capable of amplifying most HPV-16 L1 sequence variants, and GU-2-specific primers, which could only amplify HPV-16 L1 sequences having the unique residues identified in GU-2 L1. Whereas a PCR product of the expected size was obtained with the HPV-16 L1-specific primers, no product was detected with the GU-2 primers. The DNA sequence of the PCR product obtained with the HPV-16 L1-specific primers was found to be identical to that determined for the cloned GU-2 L1 gene, except that the nucleotide changes which gave rise to the L at amino acid 50 and the D at position 380 were not found. When this PCR product was used as a template with GU-2-specific primers for additional rounds of PCR, no amplification was detected. Thus, we were unable to confirm the existence of the GU-2 sequence in the biopsy specimen DNA. We concluded, therefore, that either GU-2 represented a very minor component of a mixed infection in the biopsy specimen or this sequence arose as a PCR artifact. We reasoned, however, that even if the GU-2 sequence was artifactual, the unique changes in the gene relative to the reference sequence might be useful in identifying amino acid residues critical for the binding of neutralizing MAbs.
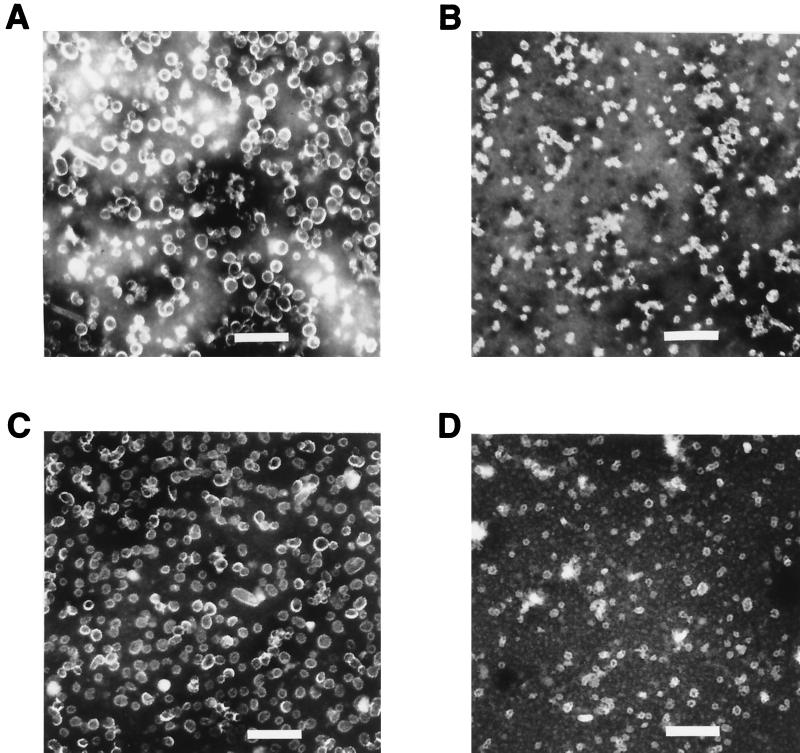
The 114K, Rochester-1k, GU-1, and GU-2 proteins were purified from recombinant baculovirus-infected insect cells by CsCl and sucrose gradient centrifugation. Since prior studies had demonstrated that a single amino acid difference could result in inefficient production of VLPs (14) and that the binding of the E70, V5 and U4 MAbs requires structurally intact HPV-16 L1 (6), the equivalency of the material produced by recombinant expression of the different HPV-16 L1 variants was assessed prior to their use in antibody binding studies. All of the purified L1 preparations exhibited retention times on a size exclusion column consistent with those of a previously documented HPV VLP control, indicating that the proteins were in large assemblies similar to VLPs (data not shown). Analysis of the different purified HPV-16 L1 variants by electron microscopy showed a mixture of particles of different sizes. Preparations of GU-2 L1 and 114K L1 contained a large proportion of full-size, 50-nm-diameter VLPs, while GU-1 L1 and Rochester L1 preparations appeared to contain predominantly smaller particles of ∼30 nm in diameter (Fig. 2). The cause of the particle size variability among the different VLP preparations is unknown but may be related to differences in the L1 sequences or differences in the expression levels of the L1 proteins and the resultant effects on VLP assembly. However, variations in the size of particles have previously been reported for isolated stocks of authentic papillomaviruses and VLP preparations (7, 17).
FIG. 2.
Electron micrographs of HPV-16 L1 variant VLPs. Purified VLPs were stained with 2% phosphotungstic acid, applied to grids, and photographed at a magnification of ×20,000. (A) 114K VLPs. (B) GU-1 VLPs. (C) GU-2 VLPs. (D) Rochester-1k VLPs. Scale bar, 230 nm.
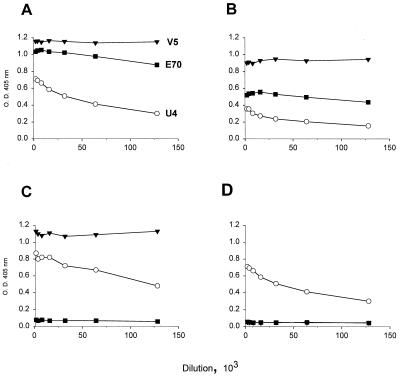
The binding of the V5, E70, and U4 MAbs to the different VLPs was assessed by ELISA (Fig. 3). Each of the MAbs was found to bind the 114K and Rochester-1k VLPs. Whereas the binding of the MAbs to the Rochester-1k VLPs was generally lower than that observed with the 114K VLPs, the overall binding patterns to these two L1 variants were similar. Thus, the ability of U4 to neutralize HPV-16 114K pseudovirions, but not authentic HPV-16Rochester-1k/ur3 virus, cannot be explained by the failure of this MAb to recognize the Rochester-1k L1 protein.
FIG. 3.
MAb binding to HPV-16 L1 variant VLPs. Serial twofold dilutions of V5 (▾), E70 (■), and U4 (○) ascites fluids were reacted with 114K VLPs (A), Rochester-1k VLPs (B), GU-1 VLPs (C), or GU-2 VLPs (D) in an ELISA.
Figure 3 also shows that while the U4 and V5 MAbs bound efficiently to the GU-1 L1 VLPs, E70 failed to recognize this L1 variant (Fig. 3C). The inability of E70 to bind to GU-1 VLPs was not unexpected, since this L1 variant contains a P at amino acid 282. Roden et al. had previously demonstrated that an S at position 282 in the HPV-16 L1 sequence was critical for E70 binding and showed that a P residue at this position abrogated E70 interaction with L1 (18). However, the data presented in Fig. 3 suggest additional residues, distant in the linear L1 sequence from position 282, may also constitute part of the E70 epitope. Figure 3D shows that E70 fails to bind GU-2 VLPs even at very low dilutions. Like the 114K and Rochester-1k L1 variants which are efficiently bound by E70, the GU-2 L1 sequence has an S at position 282. However, GU-2 contains residues at three amino acid positions (L at position 50, T at position 266, and N at position 380) which are distinct from those found in the 114K and Rochester-1k L1 proteins. Interestingly, V5 also exhibits a complete inability to bind the GU-2 VLPs, whereas the pattern of U4 binding to the GU-2 VLPs is indistinguishable from that observed with the other variant VLPs. Thus, the disparate residues in GU-2 L1 may define critical amino acids in the epitopes of two HPV-16 neutralizing MAbs, E70 and V5.
Identification of an amino acid residue in HPV-16 L1 which constitutes part of the E70 and V5 neutralizing epitopes.
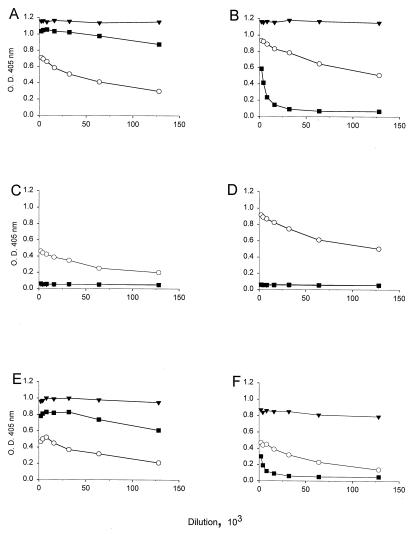
To identify the residue(s) in the HPV-16 GU-2 L1 sequence critical for E70 and V5 binding, site-directed mutagenesis was used to introduce single amino acid substitutions into GU-2 L1, replacing the divergent amino acids with those found in the other HPV-16 L1 variants. Recombinant baculoviruses were generated and used to express the mutated GU-2 L1 sequences in insect cells. The resultant purified L1 products were demonstrated to be in VLP form by electron microscopy and size exclusion chromatography (data not shown). The binding of the three MAbs to the mutated GU-2 L1 VLPs was tested by ELISA (Fig. 4). The results demonstrate that amino acid 50 in the HPV-16 L1 sequence is important for both V5 and E70 binding (Fig. 4B). Alteration of the amino acid at this position from L to F completely restored V5 binding and partially restored E70 binding. In contrast, independent mutations of the T to A at position 266 or the N to K at position 380 in GU-2 L1 failed to restore the binding of either of these two MAbs (Fig. 4C and D). As with the original GU-2 sequence, all of the mutated GU-2 L1 VLPs bound MAb U4.
FIG. 4.
Binding of MAbs to site-directed mutants of GU-2 L1 VLPs. The results represent ELISA reactivity of V5 (▾), E70 (■), and U4 (○) ascites fluids with VLPs composed of 114K L1 (A), GU-2 L1 containing an L-to-F mutation at amino acid 50 (B), GU-2 L1 containing a T-to-A change at position 266 (C), GU-2 L1 in which the N at position 380 was changed to K (D), GU-2 L1 containing an L-to-F change at position 50 and a T-to-A change at position 266 (E), or GU-2 L1 in which both the L at position 50 is changed to F and the N at position 380 is changed to K (F).
These results showed that position 50 in the amino acid sequence of HPV-16 L1 is a critical residue in both the V5 and E70 epitopes. However, other divergent amino acids in the GU-2 L1 sequence may comprise part of the E70 epitope, since mutation at position 50 only partially restored binding of E70 to GU-2 L1. To explore the possibility that an alteration at either amino acid 266 or 380 combined with the change at position 50 could completely restore E70 reactivity, GU-2 L1 double mutants were generated and purified from recombinant baculovirus-infected insect cells. The mutant GU-2 L1 proteins were demonstrated to be in VLP form as described above (data not shown). The data presented in Fig. 4 show that the binding efficiency of E70 to the GU-2 L1 double mutant containing alterations at positions 50 and 380 (GU-2 L1 L50F, N380K) was similar to that observed with the GU-2 L1 L50F single mutant (compare Fig. 4B and F). However, a GU-2 L1 double mutant containing changes at both amino acids 50 and 266 (GU-2 L1 L50F, T266A) bound E70 as efficiently as 114K VLPs (compare Fig. 4A and E). The binding pattern of V5 and U4 to both of the GU-2 double mutants was similar to that observed with the GU-2 L1 containing a single amino acid substitution at position 50. Thus, while amino acid 50 in the HPV-16 L1 sequence is critical for both V5 and E70 binding, the residue at position 266 is also important for reactivity with E70.
Competitive binding studies with the HPV-16-specific MAbs.
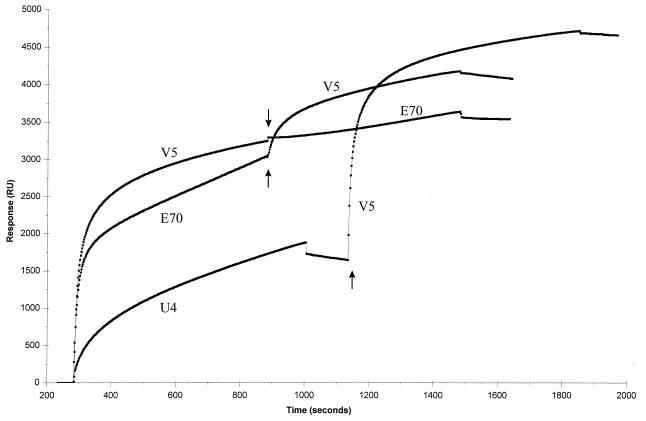
The results obtained with the HPV-16 L1 variant VLPs provided evidence that MAbs V5 and E70 recognized distinct but overlapping epitopes, different from the site recognized by U4. These results were corroborated by pairwise competitive binding studies conducted with Biosensor technology (Fig. 5). For these studies, HPV-16 114K VLPs were covalently attached to a sensor chip, and MAb binding to the immobilized VLPs was measured as a mass increase on the chip expressed as RU. Independent interaction with U4, E70, or V5 resulted in a significant increase in RU, indicating that all three MAbs were capable of binding the immobilized antigen. However, saturation of the VLPs with V5 almost completely blocked subsequent binding with E70. The ΔRU calculated for E70 in the presence and absence of saturating quantities of V5 were 392 and 3,038, respectively. When the VLPs were saturated with E70, V5 binding was significantly reduced. The ΔRU measured for V5 in the absence of E70 was 3,247, and that in the presence of saturating amounts of E70 was 1,117. In contrast, U4 was completely incapable of blocking V5 binding. The ΔRU for V5 in the presence and absence of U4 were nearly identical (3,037 and 3,247, respectively). These results are consistent with partially overlapping binding sites for V5 and E70 and a distinct U4 binding site on the HPV-16 VLPs.
FIG. 5.
Pairwise binding studies of HPV-16-specific MAbs to HPV-16 VLPs with Biosensor technology. A saturating amount of one MAb, either V5, E70, or U4, was bound to a sensor chip covalently coupled with 114K VLPs prior to the injection of the second MAb. Arrows indicate the time of injection of the second MAb.
Immunization of mice with HPV-16 L1 variant VLPs.
We have demonstrated that MAbs V5 and E70 recognize overlapping neutralizing epitopes on HPV-16. However, HPV-16 VLPs may possess additional neutralizing sites, distinct from those defined by V5 and E70. To investigate this, we tested the ability of HPV-16 VLPs lacking one or both of these epitopes to elicit HPV-16 neutralizing antibody responses. Swiss mice were immunized with either GU-1 VLPs, which contain the V5 epitope but lack the E70 binding site, or GU-2 VLPs, which lack both the E70 and V5 epitopes. Additional mice were immunized with either 114K or Rochester-1k VLPs, which contain both the E70 and V5 epitopes. Dosages of 5, 0.5, or 0.05 μg of the HPV-16 L1 intratype variants were administered following formulation with aluminum hydroxide adjuvant. ELISA titers and HPV-16 neutralization titers were determined with serum samples obtained after two immunizations. The results presented in Table 2 show that the 114K, Rochester-1k, and GU-1 VLPs elicited comparable antibody responses, as measured by ELISA with 114K VLPs as the coating antigen. In contrast, the ELISA titers generated by the GU-2 VLPs were at least fourfold lower. The anti-GU-2 VLP titers remained identical when the GU-2 VLPs were used as the coating antigen. However, antisera against the three other variant VLPs exhibited lower titers when screened on GU-2 VLPs. These data suggested that there was a difference in the epitopes displayed by GU-2 VLPs compared to those of the other variants.
TABLE 2.
Immunogenicity of HPV-16 variant VLPs
| HPV-16 variant VLP | L1 concn (ng/dose) | ELISA titer
|
Neutralization titer | |
|---|---|---|---|---|
| 114K VLPs | GU-2 VLPs | |||
| 114K | 5,000 | 2,048,000 | 512,000 | 10−5 |
| 500 | 256,000 | 128,000 | 10−4 | |
| 50 | 128,000 | 64,000 | 10−4 | |
| Rochester-1k | 5,000 | 512,000 | 256,000 | 10−5 |
| 500 | 256,000 | 128,000 | 10−4 | |
| 50 | 128,000 | 32,000 | 10−3 | |
| GU-1 | 5,000 | 1,024,000 | 64,000 | 10−5 |
| 500 | 512,000 | 64,000 | 10−5 | |
| 50 | 128,000 | 32,000 | 10−4 | |
| GU-2 | 5,000 | 32,000 | 32,000 | <10−2 |
| 500 | 64,000 | 64,000 | <10−2 | |
| 50 | 2,000 | 2,000 | <10−2 | |
Table 2 also shows that the GU-2 VLPs were very poor at eliciting HPV-16 neutralizing antibodies in the outbred Swiss mice. In contrast to the high level of neutralizing activity elicited by the 114K, Rochester-1k, and GU-1 VLPs, no HPV-16-neutralizing activity was detected with the anti-GU-2 VLP serum. Similar results were obtained in a repeat experiment conducted with BALB/c mice (data not shown).
When the V5 epitope was completely restored in GU-2 L1, by mutation of the amino acid at position 50 from L to F, the ability to elicit potent neutralizing antibody responses was also regenerated. Thus, immunization of mice with VLPs composed of GU-2 L1 L50F resulted in serum ELISA and HPV-16 neutralization titers similar to those generated by immunization with 114K VLPs (Table 3). These results demonstrate that the epitope defined by MAb V5 is necessary for the induction of potent neutralizing antibody responses against HPV-16.
TABLE 3.
Restoration of the ability to elicit potent HPV-16 neutralizing antibody responses by alteration of amino acid 50 in the GU-2 L1 protein
| HPV-16 variant VLP | Titer
|
|
|---|---|---|
| ELISA | Neutralization | |
| 114K | 1,024,000 | 10−5 |
| GU-2 | 128,000 | 10−2 |
| GU-2 L50F | 2,048,000 | 10−5 |
DISCUSSION
The primary objective of a prophylactic HPV-16 VLP-based vaccine is the induction of protective immune responses which, in large part, are associated with virus-neutralizing antibodies. However, the number of neutralization sites on HPV-16 is unknown, and the neutralization epitopes which have been identified on the viral capsid have not been thoroughly characterized. Identification of the neutralization sites present on the major capsid protein may be useful in characterizing the immune responses in serum samples from both vaccine recipients and naturally exposed individuals, as well as providing insight into virus structure-function relationships.
Using a reverse transcription (RT)-PCR-based in vitro infectivity assay, we demonstrated that MAbs V5 and E70 neutralize authentic HPV-16. These results corroborate previously published findings with HPV-16 pseudovirions (18). In contrast, MAb U4, which neutralized the 114K HPV-16 pseudovirion infection, did not neutralize HPV-16Rochester-1k/ur3. The failure of MAb U4 to neutralize HPV-16Rochester-1k/ur3 was not due to an inability to bind HPV-16Rochester-1k/ur3 since this MAb was shown to recognize HPV-16 Rochester-1k VLPs (Fig. 3). However, the avidity of the U4 MAb for Rochester-1k L1 may be too low to promote neutralization. Wang et al. had previously calculated the binding constant for U4 to be 10-fold lower than those of E70 and V5 (24). It is also possible that binding to VLPs may not accurately reflect the ability of a MAb to bind authentic virus. Thus, the inability of MAb U4 to neutralize HPV-16Rochester-1k/ur3 might relate to subtle differences in the structure of authentic virions, which are composed of both viral structural proteins L1 and L2 and contain viral DNA, in contrast to the empty L1 VLPs.
We found that MAb V5 failed to bind VLPs composed of HPV-16 L1 protein corresponding to the GU-2 sequence variant. Site-directed mutagenesis of the GU-2 L1 protein demonstrated that the F at position 50 in the HPV-16 L1 sequence plays a critical role in V5 binding. The binding patterns of the E70 and V5 MAbs showed some similarity, because the E70 MAb also failed to bind to the GU-2 VLPs. Substitution of the L at position 50 in the GU-2 L1 sequence with an F, which completely restored V5 binding, partially restored the binding of the E70 MAb. However, complete restoration of E70 binding to GU-2 L1 required substitution of amino acids at both positions 50 and 266. In addition, amino acid position 282 in the HPV-16 L1 sequence was previously demonstrated by Roden et al. to be important for E70 binding (18). Taken together, these binding data provide evidence that distinct, yet overlapping epitopes are recognized by the V5 and E70 MAbs. The relatedness of the V5 and E70 epitopes was confirmed by competitive binding data which demonstrated the ability of bound V5 MAb to completely inhibit subsequent binding of the E70 MAb to HPV-16 VLPs. In addition, Roden et al. demonstrated that both E70 and V5 inhibit HPV-16 VLP-mediated hemagglutination, suggesting that both of these MAbs neutralize by blocking the binding of HPV-16 to target cells (19).
The amino acids identified as critical for E70 binding are distantly located on the linear L1 sequence (F at position 50, A at position 266, and S at position 282). For these amino acids to be within the E70 epitope, L1 would have to fold and assemble into the viral capsid so as to juxtapose these residues and make them accessible on the capsid surface. Given the complex structure of the capsid, the amino acids required for E70 binding could be located within a single L1 polypeptide or reside on adjacent intra- or intercapsomeric L1 molecules. Using capsomeres produced by reduction or trypsinization of HPV-11 VLPs, we have previously shown that the minimal structural unit required for binding of two neutralizing MAbs was the capsomere and not the VLP (17, 20). Production of recombinant capsomeres from a mucosotropic HPV has recently been reported (21). It will be of interest to determine the structural requirements (i.e., VLP or capsomere) for E70 and V5 binding.
Our results suggest that the F at position 50 in the HPV-16 L1 sequence forms part of the neutralization epitopes recognized by both the E70 and V5 MAbs. Currently, no other HPV neutralization sites have been mapped to this N-terminus region of the L1 protein. However, an alternate interpretation of the results is that amino acid 50 lies outside the E70 and V5 binding sites and that substitutions at this amino acid position disrupt the epitopes indirectly. An alignment of mammalian PV L1 amino acid sequences revealed that most L1 proteins contain an aromatic amino acid, either F or Y, at approximately the same position, suggesting that this residue may be part of a common structural motif. However, if substitutions at position 50 in HPV-16 L1 induce structural changes in the protein, these would have to be subtle, because GU-2 L1 still assembled into VLPs which were bound by the structure-specific U4 antibody. In addition, gross analysis of the GU-2 VLPs revealed no obvious size or structural differences relative to 114K VLPs.
The integrity of the E70 epitope was compromised in HPV-16 L1 proteins containing a T at amino acid position 266. Approximately 20% of the HPV-16 sequence variants reported to date contain an L1 protein with a T at this amino acid position (9), suggesting these HPV-16 variants lack an intact E70 epitope. However, we have demonstrated that VLPs composed of HPV-16 L1 proteins in which the E70 epitope is disrupted were still capable of eliciting high-titer HPV-16 neutralizing antibody responses. In contrast, GU-2 VLPs, which lacked both the V5 and E70 epitopes, were poorly immunogenic and induced little to no neutralizing activity. GU-2 L1 mutants in which the V5 epitope was restored elicited potent HPV-16-neutralizing responses. These results demonstrate that the V5 epitope is most critical for the induction of HPV-16-neutralizing antibody responses in animals and that, in the absence of the V5 epitope, few additional potent neutralization sites are present on the HPV-16 L1 VLP.
The V5 epitope was previously shown to be predominantly recognized by antibodies in serum samples from people infected with HPV-16 (24). Whether this epitope is also immunodominant in people immunized with an HPV-16 VLP-based vaccine awaits the results of future clinical studies.
ACKNOWLEDGMENTS
We thank Attila Lorincz for tracking and providing biopsy specimen DNA. We also thank the members of MedImmune’s Process Cell Culture Group for the generation of baculovirus-infected T. ni paste and Eileen Rusnock for her assistance with electron microscopy. We also acknowledge Neil Christensen for the original description of the MAbs.
REFERENCES
- 1.Bosch F X, Manos M M, Munoz N, Sherman M, Jansen A M, Petro J, Schifman M H, Moreno V, Kurman R, Shah K V. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 2.Bosch F X, de Sanjose S, Castellsague S, Munoz N. Geographic and social patterns of cervical cancer incidence. In: Franco E, Monsonego J, editors. New developments in cervical cancer screening and prevention. Oxford, England: Blackwell Science, Ltd.; 1997. pp. 23–33. [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 4.Breitburd F, Kirnbauer R, Hubbert N L, Nonnenmacher B, Trin-Dinh-Desmarquet C, Orth G, Schiller J T, Lowy D R. Immunization with viruslike particles from cottontail rabbit papillomavirus (CRPV) can protect against experimental CRPV infection. J Virol. 1995;69:3959–3963. doi: 10.1128/jvi.69.6.3959-3963.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown M, Faulkner P. A plaque assay for nuclear polyhedrosis viruses using a solid overlay. J Gen Virol. 1977;36:361–364. [Google Scholar]
- 6.Christensen N D, Dillner J, Eklund C, Carter J J, Wipf G C, Reed C A, Cladel N M, Galloway D A. Surface conformational and linear epitopes on HPV-16 and HPV-18 virus-like particles as defined by monoclonal antibodies. Virology. 1996;223:174–184. doi: 10.1006/viro.1996.0466. [DOI] [PubMed] [Google Scholar]
- 7.Finch J T, Klug A. The structure of viruses of the papilloma-polyoma type. J Mol Biol. 1965;13:1–12. doi: 10.1016/s0022-2836(65)80075-4. [DOI] [PubMed] [Google Scholar]
- 7a.Ghim, S.-j. Unpublished observations.
- 8.Ho S N, Hunt H N, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 9.Human Papillomavirus Compendium. Markov protein alignment of supergroup A genomes II-L1-5. Los Alamos, N.Mex: Theoretical Biology and Biophysics, Los Alamos National Laboratory; 1996. [Google Scholar]
- 10.Icenogle J P, Clancy K A, Lin S Y. Sequence variation in the capsid protein genes of human papillomavirus type 16 and type 31. Virology. 1995;214:664–669. doi: 10.1006/viro.1995.0082. [DOI] [PubMed] [Google Scholar]
- 11.Jansen K U, Rosolowsky M, Schultz L D, Markus H Z, Cook J C, Donnelly J J, Martinez D, Ellis R W, Shaw A R. Vaccination with yeast expressed cottontail rabbit papillomavirus (CRPV) virus-like particles protects rabbits from CRPV-induced papilloma formation. Vaccine. 1995;13:1509–1514. doi: 10.1016/0264-410x(95)00103-8. [DOI] [PubMed] [Google Scholar]
- 12.Karlsson R, Fält A. Experimental design for the kinetic analysis of protein-protein interactions with surface plasmon resonance biosensors. J Immunol Methods. 1997;200:121–133. doi: 10.1016/s0022-1759(96)00195-0. [DOI] [PubMed] [Google Scholar]
- 13.Kirnbauer R, Chandrachud L M, O’Neil B W, Wagner E R, Grindlay G J, Armstrong A, McGarvie G M, Schiller J T, Lowy D R, Campo M S. Virus-like particles of bovine papillomavirus type 4 in prophylactic and therapeutic immunization. Virology. 1996;219:37–44. doi: 10.1006/viro.1996.0220. [DOI] [PubMed] [Google Scholar]
- 14.Kirnbauer R, Taub J, Greenstone H, Roden R, Dürst M, Gissman L, Lowy D R, Schiller J T. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J Virol. 1993;67:6929–6936. doi: 10.1128/jvi.67.12.6929-6936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M, Cripe T P, Estes P A, Lyon M K, Rose R C, Garcea R L. Expression of the human papillomavirus type 11 L1 capsid protein in Escherichia coli: characterization of protein domains involved in DNA binding and capsid assembly. J Virol. 1997;71:2988–2995. doi: 10.1128/jvi.71.4.2988-2995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludmerer S W, Benincasa D, Mark G E., III Two amino acid residues confer type specificity to a neutralizing, conformationally dependent epitope on human papillomavirus type 11. J Virol. 1996;70:4791–4794. doi: 10.1128/jvi.70.7.4791-4794.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarthy M P, White W I, Palmer-Hill F, Koenig S, Suzich J A. Quantitative disassembly and reassembly of human papillomavirus type 11 viruslike particles in vitro. J Virol. 1998;72:32–41. doi: 10.1128/jvi.72.1.32-41.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roden R B S, Armstrong A, Haderer P, Christensen N D, Hubbert N L, Lowy D R, Schiller J T, Kirnbauer R. Characterization of a human papillomavirus type 16 variant-dependent neutralizing epitope. J Virol. 1997;71:6247–6252. doi: 10.1128/jvi.71.8.6247-6252.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roden R B S, Hubbert N L, Kirnbauer R, Christensen N D, Lowy D R, Schiller J T. Assessment of the serological relatedness of genital human papillomaviruses by hemagglutination inhibition. J Virol. 1996;70:3298–3301. doi: 10.1128/jvi.70.5.3298-3301.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose R C, White W I, Li M, Suzich J A, Lane C, Garcea R L. Human papillomavirus type 11 recombinant L1 capsomeres induce virus-neutralizing antibodies. J Virol. 1998;72:6151–6154. doi: 10.1128/jvi.72.7.6151-6154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sapp M, Fligge C, Petzak I, Harris J R, Streek R E. Papillomavirus assembly requires trimerization of the major capsid protein by disulfides between two highly conserved cysteines. J Virol. 1998;72:6186–6189. doi: 10.1128/jvi.72.7.6186-6189.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Summers M D, Smith G E. A manual of methods for baculovirus vectors and insect cell culture procedures. Tex Agric Exp Stn Bull. 1987;1555:26–33. [Google Scholar]
- 23.Suzich J A, Ghim S-J, Palmer-Hill F J, White W I, Tamura J K, Bell J A, Newsome J A, Bennett Jenson A, Schlegel R. Systemic immunization with papillomavirus L1 protein completely prevents the development of viral mucosal papillomas. Proc Natl Acad Sci USA. 1995;92:11553–11557. doi: 10.1073/pnas.92.25.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Christensen N, Schiller J T, Dillner J. A monoclonal antibody against human papillomavirus type 16 capsids blocks the serological reactivity of most human sera. J Gen Virol. 1997;78:2209–2215. doi: 10.1099/0022-1317-78-9-2209. [DOI] [PubMed] [Google Scholar]
- 25.White W I, Wilson S D, Bonnez W, Rose R C, Koenig S, Suzich J A. In vitro infection and type-restricted antibody-mediated neutralization of authentic human papillomavirus type 16. J Virol. 1998;72:959–964. doi: 10.1128/jvi.72.2.959-964.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]