Abstract
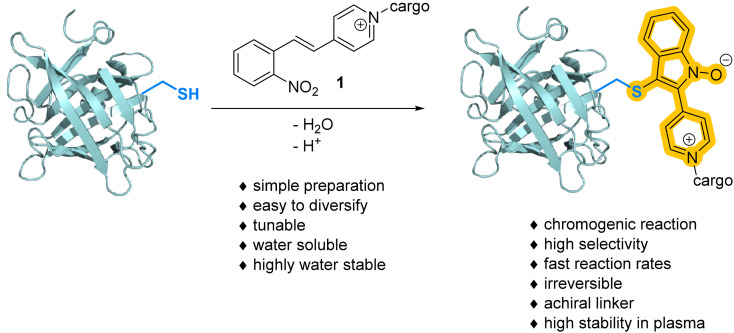
The addition of a sulfhydryl group to water-soluble N-alkyl(o-nitrostyryl)pyridinium ions (NSPs) followed by fast and irreversible cyclization and aromatization results in a stable S–C sp2-bond. The reaction sequence, termed Click & Lock, engages accessible cysteine residues under the formation of N-hydroxy indole pyridinium ions. The accompanying red shift of >70 nm to around 385 nm enables convenient monitoring of the labeling yield by UV-vis spectroscopy at extinction coefficients of ≥2 × 104 M−1 cm−1. The versatility of the linker is demonstrated in the stapling of peptides and the derivatization of proteins, including the modification of reduced trastuzumab with Val-Cit-PAB-MMAE. The high stability of the linker in human plasma, fast reaction rates (kapp up to 4.4 M−1 s−1 at 20 °C), high selectivity for cysteine, favorable solubility of the electrophilic moiety and the bathochromic properties of the Click & Lock reaction provide an appealing alternative to existing methods for cysteine conjugation.
o-Nitrostyryl-pyridinium ions (NSPs) are easily accessible electrophiles that react rapidly and irreversibly with sulfhydryl groups accompanied by a large bathochromic shift.
Protein and peptide modification by chemical means serves the study and tracking of biomolecules,1,2 the exploration of the proteome,3–6 target-selective drug delivery,7–9 the improvement of the pharmacokinetic properties of protein drugs,10,11 and the immobilization of enzymes,12 to name a few.13–23 The increasing market share of biologicals acts as an additional incentive in this industrious area of research.16,17,22,24,25
Residues targeted for protein modification are most commonly cysteines and lysines. Cysteines are particularly attractive due to their relatively low abundance, the high nucleophilicity of surface-exposed sulfhydryl groups at moderate pH values and the diverse chemistry that can be realized.13–15 Reactive cysteines can be introduced recombinantly26 or made accessible by cleavage of intramolecular disulfide bonds under mild reductive conditions. Thiols also serve as attractive nucleophiles in materials and polymer science.27–32
Traditional approaches for thiol conjugation utilize maleimides and haloacetamides as electrophilic moieties and their widespread application continues.33 For example, the majority of FDA-approved antibody-drug-conjugates are loaded with their drug cargo through a maleimide linker.34,35
Recent examples include new designs for disulfide rebridging electrophiles,36–38 tunable equilibrium constants for conjugation,28 and electrophiles that show particularly high reaction rates.39,40 Other conjugates can be cleaved41 or locked on demand,36,42 or provide additional functionality for further elaboration.43–47 High stability48 and excellent pharmacokinetic profiles49 have been demonstrated. The introduction of TM-mediated cross-coupling methods has diversified the motifs that can be directly coupled to sulfur further.22,50–52 Despite these advances, the search for valuable conjugation techniques continues since a variety of complementary properties is desired.53–56
Inspired by vinyl-pyridinium electrophiles57 and the possibility to extend the aromatic system of o-nitroarylethane derivatives and related compounds58 by intramolecular condensation,59,60 we designed N-alkyl(o-nitrostyryl)pyridinium ions 1 (Scheme 1). Here the nucleophilic attack of a thiol at an electron-deficient alkene results in an o-nitroarylethane intermediate that can subsequently form the N-hydroxyindole product under elimination of water. The reaction enables direct UV-vis spectroscopic monitoring of the conjugation progress due to the accompanying bathochromic shift. This feature can serve to conveniently estimate reaction rates, determine conjugation yields and estimate the availability of free thiols without the need for additional chromophores in the attached cargo or chromatographic analysis. In contrast to Ellman's reagent,61 the chromogenic unit doubles as the functional linker and is directly attached to the protein of interest.
Scheme 1. The reaction of a cysteine sulhydrylgroup with a nitrostyryl electrophile results in an irreversible bionconjugation that is accompanied by a large bathochromic shift.
Results and discussion
In the initial set of experiments we employed glutathione as the nucleophile and a simple methyl group as the ‘cargo’ in electrophile 1a. Glutathione conjugates were formed quantitatively within 1 hour as revealed by LC-MS (2 mM 1a, 1.2 eq. of GSH, 37 °C, 100 mM [NH4][HCO3] (pH 8.0), buffer:DMF = 9 : 1). The reaction proceeded cleanly and was accompanied by a red-shift of 75 nm. A range of substituents at the nitroaryl moiety were tolerated; while electron-withdrawing substituents in the 4-position (1b–1f), accelerated the reaction, it was slowed down by electron-donating substituents (1g, 1j and 1l, Table 1). The products showed in all cases a strong red shift (>70 nm) relative to the N-alkyl(o-nitrostyryl)pyridinium ions allowing product formation to be detected by eye as a yellow hue of the solution.
Click & Lock reaction between glutathione (GSH) and various 4-(2-nitrostyryl)pyridinium ions 1a–1na.

| |||||
|---|---|---|---|---|---|
| ID | Conversionb | λ max (nm) Substrate (1)c | λ max (nm) Product (3)c | ε (×104 M−1 cm−1) Product (3)c | k app d (M−1 s−1) |
| 3a | >99% | 310 | 385 | 2.2 | 2.2 |
| 3b | >99% | 309 | 385 | 2.2 | 2.6 |
| 3c | >99% | 311 | 387 | 2.2 | 3.1 |
| 3d | >99% | 313 | 387 | 2.2 | 3.1 |
| 3e | >99% | 308 | 379 | 2.1 | 3.9 |
| 3f | >99% | 299 | 374 | 2.0 | 4.2 |
| 3g | 90% (4 h)ef | 312 | 422 | 2.2 | — |
| 3h | >99% | 316 | 391 | 2.0 | 4.4 |
| 3i | >99% | 324 | 395 | 2.3 | 4.1 |
| 3j | Trace (24 h) | — | — | — | — |
| 3k | >99e% | 320 | 405 | 2.3 | — |
| 3l | 90% (4 h)ef | 332 | 413 | 2.1 | — |
| 3m | >99e% | 309 | 387 | 2.3 | 1.1 |
| 3n | 53% (4 h)g | 525 | 524 | 6.1 | — |
0.40 mM (1a–1n), 1.2 equiv. glutathione, 0.1 M NH4HCO3 buffer (pH 8.0)/DMF = 9 : 1, 1 h, 37 °C.
Determined by LC-MS and LC-UV. Consistent with UV-vis conversions.
In 0.1 M NH4HCO3 buffer (pH 8.0).
k app was determined at 40 μM 1a–1i, 1k–1m in the presence of 4.0 mM glutathione at 20 °C in 0.1 M NH4HCO3 buffer (pH 8.0) containing 1% DMF.
0.20 mM 1g, 1l, 1m, 2.4 equiv. glutathione.
Conversion determined by UV-vis.
Isolated yield; DMF was replaced by MeOH, bodipy is the dominant chromophore. Isolated yields for all compounds (except 3j) are reported in the ESI. Preparative reactions were carried at room temperature, at higher concentrations (2–4 mM) in buffer/DMF = 5 : 1 (except 3n, see ESI for further details).
Cargo can be attached to the pyridine ring directly by alkylation as shown for the bodipy chromophore in 3n or via additional linker moieties that have alternative functional groups, such as an alkyne in 3h or a primary amine displayed by a hydrophilic PEG-linker in 3i. Intriguingly, the nature of the linker can also affect reaction rates: i.e., the reaction of 1i to 3i carrying a triazole-PEG-amine cargo was significantly faster than the reaction of 1m to 3m having a lipophilic hydrocarbon chain and a terminal carboxylate. Possibly, limited solubility and mass transfer play a role for the lower reaction rate observed with 3m.
We next probed the bathochromic properties of the reaction for the estimation of reaction rate constants under pseudo-first-order conditions. The UV-spectrum of a solution containing initial concentrations of 1a (40 μM) and GSH (4.0 mM) in 100 mM NH4HCO3 buffer (pH 8.0) was recorded in the wavelength range from 270–470 nm every 3 seconds at 20 °C (Fig. 1). The decreasing absorption of 1a and the increasing absorption of 3a in the course of the reaction resulted in a well-defined isosbestic point at 355 nm. Its sharp definition indicates that the cyclization (the ‘lock’) is indeed very fast since the presence of underlying intermediates in significant concentrations can be excluded. The lower limits for the apparent second order rate constants obtained by the method of initial rates range from 1.1 M−1 s−1 for 3m to 4.4 M−1 s−1 for 3i (Table 1 and Fig. 1).
Fig. 1. (a) Electrophiles of type 1 react with thiol nucleophiles under fast addition and even faster cyclization (b) time-resolved UV-vis of the reaction mixture: 1a (40 μM) and glutathione (4.0 mM), 0.1 M NH4HCO3 buffer (pH 8.0), scan from 270 nm to 470 nm every 3 s; (c) apparent second order rate constants for transformation of 1a–1f, 1h, 1i, 1m (40 μM) with glutathione (4.0 mM), 20 °C obtained by a linear fit of the early data-points of the UV-vis data under pseudo-first order conditions; the lines show a first order fit to guide the eye. (d) Reversibility of thiol-Michael addition for slow reacting electrophile 1m; 2m was isolated and re-exposed to the reaction buffer with or without cysteine (e) LC-MS (m/z = 100–1000) of 2m (0.40 mM) in NH4HCO3 buffer (100 mM, pH 8.0), 37 °C, 2 h incubation; (f) LC-MS (m/z = 100–1000) of a mixture of 2m (0.40 mM) and cysteine (4.0 mM) in NH4HCO3 buffer (100 mM, pH 8.0), 37 °C, 4 h incubation.
No sharp isosbestic point was observed for the reactions of electrophiles 1g–1l to products 3g–3l, respectively, possibly due to a reduced electrophilicity of the nitro group. When the reactions for slow reacting substrates 1g and 1m with GSH (1.2 eq.) were performed at higher substrate concentrations (4.0 mM) for preparative purposes, the intermediates 2g and 2m, respectively, could be detected by LC-MS. A larger scale reaction under these conditions allowed the isolation of intermediate 2m by preparative HPLC in the presence of TFA. When 2m was exposed to the reaction buffer, a mixture of 1m, 2m and 3m was observed by LC-MS after 2 hours (Fig. 1e). The addition of cysteine to 2m led to competitive formation of the cysteine addition and cyclization product, 2m(cys) and 3m(cys), respectively (Fig. 1f), confirming that the aromatization is indeed necessary to render the reaction irreversible.
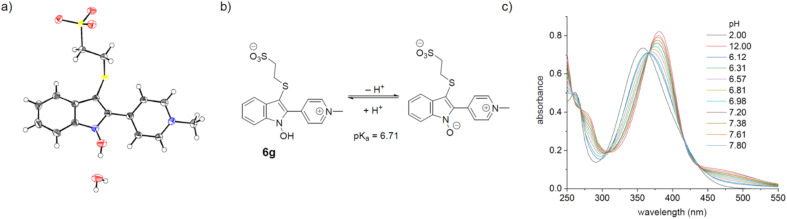
The X-ray structure of the cyclized conjugate 6g revealed the presence of a water molecule associated with the N-hydroxy group, hinting at a potential acidity (Fig. 2).62,63 Since the OH-group is part of the chromophore, the absorption spectrum of the cyclized conjugates was expected to be pH-dependent. A red-shift of 24 nm was observed for 6g between pH 2 and 12 and the pKa-value (pKa = 6.71) could be conveniently determined by UV-vis-titration (see ESI†). In the reaction buffer (pH 8.0) 95% of the formed hydroxyindole 6g is deprotonated.
Fig. 2. (a) The X-ray structure of conjugate 6g reveals an associated water molecule; CCDC ID 2278235 (b) protonation equilibrium of conjugate 6g (c) pH-dependence of the absorption spectrum of conjugate 6g.
To test the applicability of the NSP-motif, first a small set of peptides (4–16 AA) with a single cysteine residue were reacted with electrophile 1a (Fig. 3). The conjugates were isolated in good yields after preparative HPLC (75–80%) and had formed quantitatively after 30 minutes (6c, 4 amino acids; 6d, 8 amino acids) or after 2 hours for longer peptides (6e, 6f, 12 and 16 amino acids respectively). Importantly, only singly modified products were observed, confirming that a potential cross-reactivity with lysine is of no concern under the conditions employed. We further scrutinized this conclusion by an attempted reaction of 3a with lysine itself where no reaction was observed (see ESI†).
Fig. 3. Selective modification of one, two or three sulfhydryl groups in peptides with mono-, bi- and trifunctional 4-(2-nitrostyryl)pyridinium linkers. Conditions for the coupling reactions: peptides with a single cysteine residue: 2.0 mM 1a, 1.5 equiv. peptide 5a–5f, 100 mM NH4HCO3 buffer (pH 8.0), 37 °C, 0.5–2 h; peptides with two cysteine residues: 0.50 mM 4a, 1.0 equiv. peptide 5g, 5h, 100 mM NH4HCO3 buffer (pH 8.0), rt, 1 hour; peptides with three cysteine residues: 0.20 mM 4b, 1.0 equiv. peptide 5i, 5j, 100 mM NH4HCO3 buffer (pH 8.0), 0 °C→rt, 1 hour. Isolated yields; products purified by prep. HPLC.
Next, bi- and trifunctional linkers (4a, 4b) for peptide stapling were prepared by reacting dibromopropane or 1,3,5-tri(bromomethyl)benzene with (E)-4-(2-nitrostyryl)pyridine, respectively. Peptides with two or three cysteine residues could be conveniently constrained into cyclic or bicyclic structures under moderately dilute conditions. The monocyclic constructs with ring sizes of 7 and 8 amino acids were isolated in good yields (≥60%). Bicyclic structures were prepared with 6,7- and 7,8-membered rings respectively in ≥25% yield.
We further tested the modification of bovine aprotinin (58 AA) where the three disulfide bonds had been reduced by addition of 6 eq. of TCEP. Direct addition of 20 eq. of 1a to the reduced mixture without workup resulted in the hexa-modified polypeptide (see ESI†). The excess of 1a served to compensate for competing addition of TCEP to 1a.
Next, to probe NSPs for the selective modification of an unpaired cysteine on a protein, β-lactoglobulin A (BLG-A, 162 AA) that contains two disulfide bridges and an unpaired buried cysteine was selected. Despite being buried, this single cysteine can be modified under mild conditions. Reagent 1a, alkynyl-functionalized 5 and bifunctional 4a were tested for modification at 37 °C and at pH 8.0: all reactions resulted in singly modified BLG-A within 2 h (Fig. 4).
Fig. 4. Modification of BLG-A with different NSPs (20 μM BLG-A, 10 eq. 1a and 5, 5 eq. 4a, respectively). Deconvoluted mass spectra are shown.
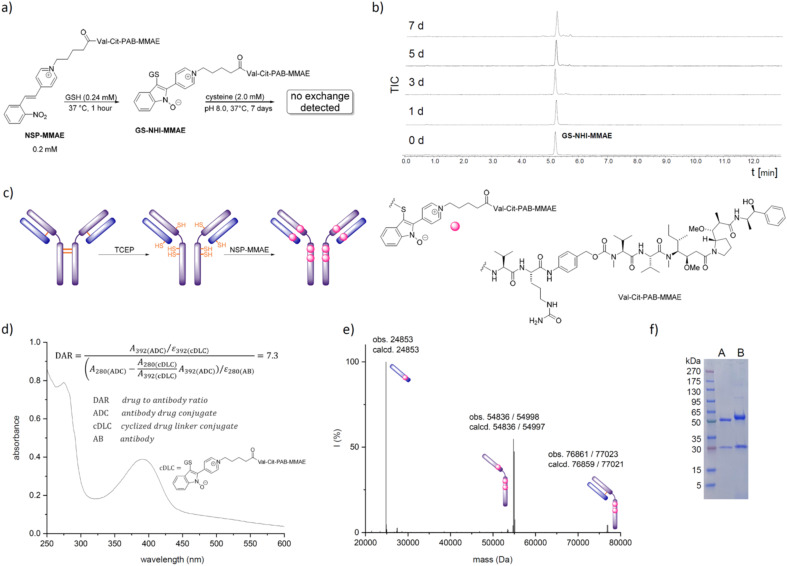
Toward a pharmacologically relevant application, we equipped the cytotoxic Val-Cit-PAB-MMAE with the electrophilic NSP moiety. The resulting reactive drug-linker NSP-Val-Cit-PAB-MMAE was first tested in the reaction with glutathione. A similar reaction rate to 1a (kapp = 1.9 M−1 s−1) indicates that the larger payload does not impede the reaction rate (Val-Cit-PAB-MMAE vs. methyl in 1a). Importantly the GSH conjugate (0.20 mM) showed only minimal decomposition after 7 days of incubation at 37 °C and pH 8.0 in the presence of 2.0 mM cysteine according to LC-MS and LC-UV analysis (Fig. 5b). This contrasts with observations made with the commercial maleimide drug-linker MC-Val-Cit-PAB-MMAE (see ESI†).64
Fig. 5. (a) The GSH conjugate of the NSP-MMAE linker does not exchange with cysteine and shows superior stability. (b) Stability test of incubated GS-NHI-MMAE by LC-MS. (c) Conjugation of NSP-MMAE with trastuzumab. (d) Determination of DAR by UV-vis. (e) Deconvoluted HRMS of the trastuzumab NSP-MMAE conjugates. (f) SDS-PAGE of reduced trastuzumab after reaction with NSP-MMAE, A: no NSP-MMAE; B: 20 eq. NSP-MMAE; 3 h incubation; 3 days of dialysis at room temperature; samples were heated to 95 °C for 10 minutes before they were applied to the gel.
We further tested the stability of the glutathione conjugates 3a, 3f, 3g in human plasma (pooled human plasma (blood-derived) K2 EDTA, Loxo GmbH). Within 48 hours more than half of the species with an intact conjugated-N-hydroxy indole pyridinium unit remained. Identified decomposition products stemed from cleavage of the glutamate–cysteine amide bond in the glutathione moiety (49% for 3a, 56% for 3f, and 12% for 3g, see ESI†).
The high stability of the conjugated N-hydroxy indole pyridinium unit against thiol exchange and in human plasma illustrates the superior applicability of the conjugation method.
Encouraged by these results, we modified the widely applied anti-HER2 antibody trastuzumab. Interchain disulfide bonds were reduced with TCEP and the mixture subsequently treated with 2.5 eq. of NSP-MMAE per free thiol (4 μM trastuzumab, 40 μM TCEP, 37°, 90 min, followed by 80 μM NSP-MMAE, 3 h; 3 days dialysis at room temperature). HRMS showed, as expected, the singly modified light chain and the triply modified heavy chain. Additional signals of the heavy chain are due to heterogeneous glycosylation commonly observed in commercial trastuzumab.65
The red-shifted absorption spectrum of the conjugate allows for a simple estimation of the drug to antibody ratio by comparing the absorption at 280 nm and 392 nm resulting in a value of DAR = 7.3 under not further optimized conditions. A small signal for the doubly modified half-antibody indicates that the DAR < 8.0 might be caused by incomplete TCEP reduction (Fig. 5e).
Conclusion
In summary we present a versatile and readily accessible electrophilic moiety for aqueous cysteine conjugation that allows facile monitoring of the reaction progress by virtue of its bathochromic properties and whose conjugates show superior stability to thiol exchange. The thiol N-hydroxy indole pyridinium unit furthermore showed half-lifes of >48 hours in human plasma. The chromogenic reaction proved to be an effective method for the determination of the reaction rate constants, the concentration of free sulfhydryl residues in the reaction mixture and the conjugation yield as exemplified in the drug-to-antibody ratio (DAR) of an antibody-drug conjugate (ADC). Moreover, a synthesized drug conjugate (GS-NHI-MMAE) displayed excellent stability, even when confronted with an excess of free thiol groups. These features collectively make Click & Lock an appealing alternative to existing methods for cysteine conjugation and open new possibilities in the field of protein modification and bioconjugation strategies.
Data availability
The data is included in the ESI.†
Author contributions
Conceptualization: Y. Hua; formal analysis: Y. Hua, V. Köhler; funding acquisition: M. Mayor, V. Köhler, T. R. Ward; investigation: Y. Hua, Z. Zou, A. Prescimone; methodology: Y. Hua; project administration: M. Mayor, V. Köhler; resources: M. Mayor, T. R. Ward, V. Köhler; supervision: V. Köhler, M. Mayor, T. R. Ward; validation: Y. Hua, V. Köhler; visualization: Y. Hua, V. Köhler, Z. Zou, A. Prescimone; writing – orginal draft: Y. Hua, V. Köhler; writing – review & editing: Y. Hua, V. Köhler, T. R. Ward, M. Mayor.
Conflicts of interest
The authors declare no competing financial interest.
Supplementary Material
Acknowledgments
This project has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreements #860713. We acknowledge funding by the Gordon & Betty Moore foundation under grant agreement #10771 and the Swiss National Science Foundation #200020_207744. We thank Michael Pfeffer for the HRMS analysis and Daniel Häussinger for advice on NMR spectroscopy.
Electronic supplementary information (ESI) available. CCDC 2278235. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4sc01710b
References
- Giepmans B. N. G. Adams S. R. Ellisman M. H. Tsien R. Y. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- Wang L. Frei M. S. Salim A. Johnsson K. J. Am. Chem. Soc. 2019;141:2770–2781. doi: 10.1021/jacs.8b11134. [DOI] [PubMed] [Google Scholar]
- Weerapana E. Wang C. Simon G. M. Richter F. Khare S. Dillon M. B. Bachovchin D. A. Mowen K. Baker D. Cravatt B. F. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backus K. M. Correia B. E. Lum K. M. Forli S. Horning B. D. González-Páez G. E. Chatterjee S. Lanning B. R. Teijaro J. R. Olson A. J. Wolan D. W. Cravatt B. F. Nature. 2016;534:570–574. doi: 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y. Chin Chan S. Welsh E. A. Fang B. Sun L. Schonbrunn E. Koomen J. M. Duckett D. R. Haura E. B. Monastyrskyi A. Rix U. ACS Chem. Biol. 2023;18:251–264. doi: 10.1021/acschembio.2c00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K. C. Maddox S. M. Backus K. M. Raj M. Chem. Sci. 2022;13:763–774. doi: 10.1039/D1SC04139H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck A. Goetsch L. Dumontet C. Corvaia N. Nat. Rev. Drug Discovery. 2017;16:315–337. doi: 10.1038/nrd.2016.268. [DOI] [PubMed] [Google Scholar]
- Fani M. Maecke H. R. Okarvi S. M. Theranostics. 2012;2:481–501. doi: 10.7150/thno.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh S. J. Bargh J. D. Dannheim F. M. Hanby A. R. Seki H. Counsell A. J. Ou X. Fowler E. Ashman N. Takada Y. Isidro-Llobet A. Parker J. S. Carroll J. S. Spring D. R. Chem. Soc. Rev. 2021;50:1305–1353. doi: 10.1039/D0CS00310G. [DOI] [PubMed] [Google Scholar]
- Horn J. M. Obermeyer A. C. Biomacromolecules. 2021;22:4883–4904. doi: 10.1021/acs.biomac.1c00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailon P. Won C.-Y. Expert Opin. Drug Delivery. 2009;6:1–16. doi: 10.1517/17425240802650568. [DOI] [PubMed] [Google Scholar]
- Mateo C. Palomo J. M. Fernandez-Lorente G. Guisan J. M. Fernandez-Lafuente R. Enzyme Microb. Technol. 2007;40:1451–1463. doi: 10.1016/j.enzmictec.2007.01.018. [DOI] [Google Scholar]
- Chalker J. M. Bernardes G. J. Lin Y. A. Davis B. G. Chem.–Asian J. 2009;4:630–640. doi: 10.1002/asia.200800427. [DOI] [PubMed] [Google Scholar]
- Gunnoo S. B. Madder A. ChemBioChem. 2016;17:529–553. doi: 10.1002/cbic.201500667. [DOI] [PubMed] [Google Scholar]
- Ochtrop P. Hackenberger C. P. R. Curr. Opin. Chem. Biol. 2020;58:28–36. doi: 10.1016/j.cbpa.2020.04.017. [DOI] [PubMed] [Google Scholar]
- Boutureira O. Bernardes G. J. L. Chem. Rev. 2015;115:2174–2195. doi: 10.1021/cr500399p. [DOI] [PubMed] [Google Scholar]
- Krall N. da Cruz F. P. Boutureira O. Bernardes G. J. L. Nat. Chem. 2016;8:103–113. doi: 10.1038/nchem.2393. [DOI] [PubMed] [Google Scholar]
- Stephanopoulos N. Francis M. B. Nat. Chem. Biol. 2011;7:876–884. doi: 10.1038/nchembio.720. [DOI] [PubMed] [Google Scholar]
- Taiariol L. Chaix C. Farre C. Moreau E. Chem. Rev. 2022;122:340–384. doi: 10.1021/acs.chemrev.1c00484. [DOI] [PubMed] [Google Scholar]
- Holz E. D. Zou M. Tesar D. B. Shatz-Binder W. Pharmaceutics. 2023;15:1–54. doi: 10.3390/pharmaceutics15020600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenegger P. G. Davis B. G. J. Am. Chem. Soc. 2019;141:8005–8013. doi: 10.1021/jacs.8b13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohata J. Martin S. C. Ball Z. T. Angew. Chem., Int. Ed. 2019;58:6176–6199. doi: 10.1002/anie.201807536. [DOI] [PubMed] [Google Scholar]
- Wu C. S. Cheng L. ChemBioChem. 2023;24:e202200468. doi: 10.1002/cbic.202200468. [DOI] [PubMed] [Google Scholar]
- Kjaersgaard N. L. Nielsen T. B. Gothelf K. V. ChemBioChem. 2022;23:e202200245. doi: 10.1002/cbic.202200245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sornay C. Vaur V. Wagner A. Chaubet G. R. Soc. Open Sci. 2022;9:211563. doi: 10.1098/rsos.211563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohri R. Bhakta S. Fourie-O'Donohue A. dela Cruz-Chuh J. Tsai S. P. Cook R. Wei B. Ng C. Wong A. W. Bos A. B. Farahi F. Bhakta J. Pillow T. H. Raab H. Vandlen R. Polakis P. Liu Y. Erickson H. Junutula J. R. Kozak K. R. Bioconjugate Chem. 2018;29:473–485. doi: 10.1021/acs.bioconjchem.7b00791. [DOI] [PubMed] [Google Scholar]
- Bednarek C. Schepers U. Thomas F. Bräse S. Adv. Funct. Mater. 2024;34:2303613. doi: 10.1002/adfm.202303613. [DOI] [Google Scholar]
- Crolais A. E. Dolinski N. D. Boynton N. R. Radhakrishnan J. M. Snyder S. A. Rowan S. J. J. Am. Chem. Soc. 2023;145:14427–14434. doi: 10.1021/jacs.3c03643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe A. B. Polym. Chem. 2010;1:17–36. doi: 10.1039/B9PY00216B. [DOI] [Google Scholar]
- Stenzel M. H. ACS Macro Lett. 2013;2:14–18. doi: 10.1021/mz3005814. [DOI] [PubMed] [Google Scholar]
- Nair D. P. Podgórski M. Chatani S. Gong T. Xi W. Fenoli C. R. Bowman C. N. Chem. Mater. 2014;26:724–744. doi: 10.1021/cm402180t. [DOI] [Google Scholar]
- Bahou C. Spears R. J. Ramírez Rosales A. M. Rochet L. N. C. Barber L. J. Stankevich K. S. Miranda J. F. Butcher T. C. Kerrigan A. M. Lazarov V. K. Grey W. Chudasama V. Spicer C. D. Biomacromolecules. 2023;24:4646–4652. doi: 10.1021/acs.biomac.3c00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson G. T., in Bioconjugate Techniques, ed. G. T. Hermanson, Academic Press, Boston, 3rd edn, 2013, pp. 229–258 [Google Scholar]
- Chia C. S. B. ChemistryOpen. 2022;17:e202200032. doi: 10.1002/cmdc.202200032. [DOI] [PubMed] [Google Scholar]
- You J. Zhang J. Wang J. Jin M. Bioconjugate Chem. 2021;32:1525–1534. doi: 10.1021/acs.bioconjchem.1c00213. [DOI] [PubMed] [Google Scholar]
- Tallon A. M. Xu Y. West G. M. am Ende C. W. Fox J. M. J. Am. Chem. Soc. 2023;145:16069–16080. doi: 10.1021/jacs.3c04444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannheim F. M. Walsh S. J. Orozco C. T. Hansen A. H. Bargh J. D. Jackson S. E. Bond N. J. Parker J. S. Carroll J. S. Spring D. R. Chem. Sci. 2022;13:8781–8790. doi: 10.1039/D2SC02198F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. Silva M. J. S. A. Gois P. M. P. Kuan S. L. Weil T. Chem. Sci. 2021;12:13321–13330. doi: 10.1039/D1SC03250J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka B. M. Honeycutt D. S. Bassett G. M. Kowal T. N. Adamczyk M. Cartnick Z. C. Betti V. M. Goldberg J. M. Wang F. J. Am. Chem. Soc. 2023;145:23427–23432. doi: 10.1021/jacs.3c10334. [DOI] [PubMed] [Google Scholar]
- Chen F. J. Gao J. Chem.–Eur. J. 2022;28:e202201843. doi: 10.1002/chem.202201843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity S. Bingham C. Sheng W. Ehyaei N. Chakraborty D. Tahmasebi-Nick S. Kimmel T. E. Vasileiou C. Geiger J. H. Borhan B. Analyst. 2023;148:1085–1092. doi: 10.1039/D2AN01395A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavriel K. van Doeselaar D. C. A. Geers D. W. T. Neumann K. RSC Chem. Biol. 2023;4:685–691. doi: 10.1039/D3CB00062A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsopetras I. Mishra A. K. Benazza R. Hernandez-Alba O. Cianférani S. Chaubet G. Nicolai S. Waser J. Chem.–Eur. J. 2023;29:e202302689. doi: 10.1002/chem.202302689. [DOI] [PubMed] [Google Scholar]
- Ahangarpour M. Kavianinia I. Hume P. A. Harris P. W. R. Brimble M. A. J. Am. Chem. Soc. 2022;144:13652–13662. doi: 10.1021/jacs.2c04146. [DOI] [PubMed] [Google Scholar]
- Afonso C. F. Marques M. C. Antonio J. P. M. Cordeiro C. Gois P. M. P. Cal P. M. S. D. Bernardes G. J. L. Angew. Chem., Int. Ed. 2022;61:e202208543. doi: 10.1002/anie.202208543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. Kuan S. L. Weil T. Angew. Chem., Int. Ed. 2021;60:13757–13777. doi: 10.1002/anie.202012034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S. Zhang Z. Li B. Li L. Tan M. C. L. Jia Z. Loh T.-P. Angew. Chem., Int. Ed. 2023;62:e202311906. doi: 10.1002/anie.202311906. [DOI] [PubMed] [Google Scholar]
- Gober I. N. Sharan R. Villain M. J. Pept. Sci. 2023;29:e3495. doi: 10.1002/psc.3495. [DOI] [PubMed] [Google Scholar]
- Ochtrop P. Jahzerah J. Machui P. Mai I. Schumacher D. Helma J. Kasper M.-A. Hackenberger C. P. R. Chem. Sci. 2023;14:2259–2266. doi: 10.1039/D2SC05678J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A. J. Wolfe J. M. Dhanjee H. H. Buslov I. Truex N. L. Liu R. Y. Massefski W. Pentelute B. L. Buchwald S. L. Chem. Sci. 2022;13:11891–11895. doi: 10.1039/D2SC04074C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery H. R. Messina M. S. Doud E. A. Spokoyny A. M. Maynard H. D. Bioconjugate Chem. 2022;33:1536–1542. doi: 10.1021/acs.bioconjchem.2c00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J. Dhanjee H. H. Pentelute B. L. Buchwald S. L. J. Am. Chem. Soc. 2022;144:11706–11712. doi: 10.1021/jacs.2c03492. [DOI] [PubMed] [Google Scholar]
- Zhang Z. Li L. Xu H. Lee C. K. Jia Z. Loh T. P. J. Am. Chem. Soc. 2024;146:1776–1782. doi: 10.1021/jacs.3c12050. [DOI] [PubMed] [Google Scholar]
- Spears R. J. Chudasama V. Curr. Opin. Chem. Biol. 2023;75:102306. doi: 10.1016/j.cbpa.2023.102306. [DOI] [PubMed] [Google Scholar]
- O W.-Y. Cui J.-F. Yu Q. Kung K. K.-Y. Chung S.-F. Leung Y.-C. Wong M.-K. Angew. Chem., Int. Ed. 2023;62:e202218038. doi: 10.1002/anie.202218038. [DOI] [PubMed] [Google Scholar]
- Seki H. Walsh S. J. Bargh J. D. Parker J. S. Carroll J. Spring D. R. Chem. Sci. 2021;12:9060–9068. doi: 10.1039/D1SC02722K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos M. J. Navo C. D. Hakala T. Ferhati X. Guerreiro A. Hartmann D. Bernardim B. Saar K. L. Compañón I. Corzana F. Knowles T. P. J. Jiménez-Osés G. Bernardes G. J. L. Angew. Chem., Int. Ed. 2019;58:6640–6644. doi: 10.1002/anie.201901405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanusek J. Machacek V. Collect. Czech. Chem. Commun. 2009;74:811–833. doi: 10.1135/cccc2008216. [DOI] [Google Scholar]
- Wróbel Z. Mąkosza M. Synlett. 1993:597–598. doi: 10.1055/s-1993-22544. [DOI] [Google Scholar]
- Wrobel Z. Makosza M. Tetrahedron. 1997;53:5501–5514. doi: 10.1016/S0040-4020(97)00208-1. [DOI] [Google Scholar]
- Ellman G. L. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- Somei M., in Advances in Heterocyclic Chemistry, Academic Press, 2002, vol. 82, pp. 101–155 [Google Scholar]
- Bykov E. E. Lavrenov S. N. Preobrazhenskaya M. N. Chem. Heterocycl. Compd. 2006;42:42–44. doi: 10.1007/s10593-006-0044-z. [DOI] [Google Scholar]
- Lyon R. P. Setter J. R. Bovee T. D. Doronina S. O. Hunter J. H. Anderson M. E. Balasubramanian C. L. Duniho S. M. Leiske C. I. Li F. Senter P. D. Nat. Biotechnol. 2014;32:1059–1062. doi: 10.1038/nbt.2968. [DOI] [PubMed] [Google Scholar]
- Li T. Tong X. Yang Q. Giddens J. P. Wang L.-X. J. Biol. Chem. 2016;291:16508–16518. doi: 10.1074/jbc.M116.738765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data is included in the ESI.†