Abstract
We previously showed that the 93-bp region between the enhancer and promoter (named DEN for downstream of enhancer) of the long terminal repeat (LTR) of the MCF13 murine leukemia virus is an important determinant of the ability of this virus to induce thymic lymphoma. In this study we observed that DEN plays a role in the regulation of virus replication in the thymus during the preleukemic period. A NF-κB site in the DEN region partially contributes to the effect of DEN on both lymphomagenicity and virus replication. To further study the effects of DEN and the NF-κB site on viral pathogenicity during the preleukemic period, we examined replication of wild-type and mutant viruses with a deletion of the NF-κB site or the entire DEN region in the thymus. Thymic lymphocytes which were infected with wild-type and mutant viruses were predominantly the CD3− CD4+ CD8+ and CD3+ CD4+ CD8+ cells. The increase in infection by wild-type virus and both mutant viruses of these two subpopulations during the preleukemic period ranged from 9- to 84-fold, depending upon the time point and virus. The major difference between the wild-type and both mutant viruses was the lower rate and lower level of mutant virus replication in these thymic subpopulations. Significant differences in replication between wild-type and both mutant viruses were seen in the CD3− CD4+ CD8+ and CD3− CD4− CD8− subpopulations, suggesting that these thymic cell types are important targets for viral transformation.
An important determinant of pathogenicity of murine leukemia viruses (MLVs) is the long terminal repeat (LTR) region of the viral genome (15–17, 20, 28, 43). In particular, sequences in the enhancer region of the LTR have been shown to regulate both cell type-specific transcription and pathogenicity of these viruses (4, 15–17, 20, 25, 28, 30, 43–46). Additional sequences downstream of the enhancer region also contribute to viral pathogenicity and the regulation of transcription (8, 17, 20, 28, 46). We have shown that the 93-bp region between the enhancer and promoter (named DEN for downstream of enhancer) of the LTR of the MCF13 MLV is important for the ability of this murine retrovirus to induce thymic lymphoma (46). Further studies of the ability of this region to regulate transcription have demonstrated that DEN is the only region of the LTR which potentiates transcription in activated T cells (8). We observed that the NF-κB-binding site in DEN was mainly responsible for the control of transcription in activated T cells (50). The question of whether this protein-binding site also plays a role in MCF13 pathogenicity is addressed in this report.
Although the roles of the different regions in the LTR in tumorigenesis have been extensively studied, less is known regarding their role in the early stages of disease progression, i.e., the preleukemic period. This period of thymic lymphoma development in AKR mice was best described by O’Donnell and coworkers, who defined various stages in the development of mink cell focus-forming virus (MCF virus)-accelerated leukemia (37). By flow cytometric analysis, they observed three stages of leukemogenesis before the appearance of frank leukemia. At the earliest stage, thymic lymphocytes, which were infected by MCF virus but lacked changes in both light scatter properties and the expression of differentiation alloantigens, were detectable. The appearance of a clonal population of cells, which could be resolved from normal thymic lymphocytes by light scatter and expression of the viral envelope gp70 and differentiation antigens, characterized an intermediate period of preleukemogenesis. During the final stage, which was observed at around 10 weeks postinjection, the outgrowth of fully transformed cells was apparent. In this same study they further concluded that MCF MLV pathogenesis involved the immature small cortisone-sensitive thymic cell population.
It has been observed that efficient virus replication in the thymus is essential for tumor development (22, 35, 36). However, MLVs with either a single copy of the enhancer or with certain mutations in enhancers composed of two copies, which were significantly attenuated in their ability to induce thymic lymphoma, were able to replicate as efficiently as wild-type (WT) virus in the thymus (16, 19). Thus, it appeared that there was a lack of correlation between LTR sequences which regulate virus replication in the thymus and those which play a pivotal role in tumorigenesis. It is presently unknown which LTR sequences are responsible for the regulation of virus replication in the thymus during the early stages of tumorigenesis.
To examine whether the DEN region has a role in the regulation of MCF virus replication in the thymus, we exploited our observation that MCF MLVs with mutations in their DEN region displayed different tumor incidences and latencies. We determined whether differences in the ability of these mutant viruses to replicate in the thymus during the early stages of disease and in different thymic cell types could account for their different pathogenic phenotypes.
MATERIALS AND METHODS
Construction and isolation of the ΔNF-κB and ΔDEN mutant viruses.
A plasmid containing a clone of the WT MCF13 MLV (PMSL) (46, 49) was mutated at the NF-κB site in the DEN region of the LTR by site-directed mutagenesis (Amersham). The BamHI recognition sequence was substituted for the NF-κB-binding site, which is shown in Fig. 1. The ΔNF-κB mutation was verified by DNA sequencing (Center for Molecular Medicine and Genetics DNA Sequencing Facility at Wayne State University). Construction of the ΔDEN mutant virus has been described elsewhere (46). Twenty micrograms of plasmid DNA and 33 μg of Lipofectin reagent (Gibco BRL) were transfected into Mus dunni fibroblasts, which do not contain genomic sequences related to known MLVs (26). Transfected cells were passaged at high density until 100% of the cells were infected by virus as determined by an immunofluorescence focus assay (42, 46). For this assay, we used a monoclonal antibody, MAb 514, which reacts specifically with MCF-type glycoproteins (13). This monoclonal antibody was a generous gift of L. H. Evans, Rocky Mountain Laboratories, Hamilton, Mont. Cells on the culture dish were stained with 0.5 ml of MAb 514 supernatant, followed with 1 ml of fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG) (Sigma). Both incubations were performed for 30 min at 37°C. Fluorescent foci were detected with a Nikon phase-contrast 2 fluorescence microscope.
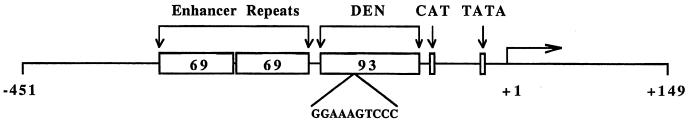
FIG. 1.
LTR of the MCF13 MLV. The CAT and TATA boxes of the basal promoter are indicated. Upstream regulatory sequences consisting of the tandemly repeated 69-bp enhancer and 93-bp DEN region are also shown. The sequence of the NF-κB site within DEN is indicated. Numbering is relative to the start site of transcription, which is marked by the horizontal arrow.
Viral supernatants were harvested from cell cultures, centrifuged to remove cells, and frozen at −70°C for assays of virus titers and injection into mice. Virus titers were determined by infecting M. dunni cells at various dilutions and performing focal immunofluorescence assays on infected cells as described above (46).
Inoculation of mice.
Neonatal (1- to 4-day old) AKR/J mice were injected intraperitoneally with 50 μl of inoculum containing 1 × 106 to 2.5 × 106 infectious units of virus. Control mice were inoculated with tissue culture medium. Necropsy was performed on moribund mice. Animals with the diagnosis of thymic lymphoma showed a massive enlargement of the thymus with fusion of thymic lobes. In most mice, there was also enlargement of liver, spleen, and peripheral lymph nodes.
Isolation of thymic lymphocytes and infectious center assays.
Single-cell suspensions of thymic lymphocytes were prepared by pressing thymus tissues through a wire mesh into RPMI containing 2% inactivated fetal calf serum. Cells were washed once with RPMI and resuspended in the same medium. Between 1 × 102 and 5 × 103 thymic lymphocytes in the presence of 2% Polybrene were plated onto M. dunni fibroblasts, which had been seeded onto 60-mm-diameter culture dishes the day before. After overnight incubation, the medium was changed and cultures were incubated until the cell layer was confluent whereupon immunofluorescent focus assays were performed.
Cell staining.
A total of 106 thymic lymphocytes in 100 μl of MAb 514 supernatant were incubated on ice for 30 min. After incubation, cells were centrifuged at 1,200 × g for 2 min. in an Eppendorf microcentrifuge and washed twice with phosphate-buffered saline (PBS) containing 1% bovine serum albumin and 0.01% sodium azide. Cells were resuspended in 100 μl of FITC-conjugated goat anti-mouse IgG at a 1:100 dilution. Following incubation and washing, cells were resuspended in 100-μl samples of PBS containing hamster anti-CD3 antibody conjugated to phycoerythrin (PE), rat anti-CD4 antibody conjugated to Cy-Chrome, and rat anti-CD8 antibody conjugated to biotin (PharMingen), and NeutraLite avidin conjugated to Cascade blue (Molecular Probes) at dilutions previously determined by titration assays. After a final incubation on ice and wash, cells were resuspended in 0.7 ml of 0.5% paraformaldehyde and analyzed by flow cytometry.
Flow cytometry analysis.
Flow cytometry was performed on a FACS Vantage flow cytometer equipped with an HP 9000 computer running the LYSYS II software (Becton Dickinson Immunocytometry Systems [BDIS], San Jose, Calif.). FITC, PE, and Cy-Chrome were excited with 40 mW of 488-nm-wavelength light from an ILT 5500A argon ion laser (Ion Laser Technology, Salt Lake City, Utah). The argon laser also produced forward and side scatter signals. Cascade blue (Molecular Probes, Eugene, Oreg.) was excited with 50 mW of all lines of ultraviolet light (wavelength, 351 to 365 nm) from an Innova 90-5 argon ion laser (Coherent, Santa Clara, Calif.) spatially separated from the ILT laser. Cy-Chrome fluorescence was collected within the range of wavelengths of 665 to 695 nm. PE fluorescence was collected within the range of wavelengths of 562 to 588 nm, FITC fluorescence was collected within the range of wavelengths of 515 to 545 nm, and CB fluorescence was collected within the range of wavelengths of 420 to 460 nm. The flow cytometer was aligned before each analysis with SPHERO Rainbow Calibration Particles (Spherotech, Libertyville, Ill.), quality controlled with CaliBRITE 3 beads (BDIS), and calibrated with Quantum Size Standards (Flow Cytometry Standards Corp., San Juan, P.R.). Electronic compensation for spectral overlap of the fluorochromes was performed with single-color control samples prepared with the test samples. All data presented were based on analysis of 2 × 104 cells with Paint-a-Gate software (BDIS). Analysis gates were set on isotype controls.
RESULTS
The NF-κB site in the LTR contributes to MCF13 viral pathogenicity.
The 93-bp region between the enhancer and promoter (DEN) of the MCF13 LTR (Fig. 1) is an important determinant of the ability of the MCF13 MLV to induce thymic lymphoma. We showed previously that a deletion of this region significantly reduced the incidence of MCF13-induced lymphomagenesis and increased the latency of disease (46). In addition, we demonstrated that the DEN region controls the LTR-dependent induction of transcription in activated T cells (8) and that the NF-κB site in DEN (Fig. 1) is mainly responsible for this activity (50). To determine whether the NF-κB site also contributes to viral pathogenicity, we generated an MCF13 mutant virus (ΔNF-κB) in which the NF-κB site was replaced with nucleotides corresponding to the BamHI recognition sequence by site-directed mutagenesis (50). The ΔNF-κB mutant virus or comparable amounts of WT MCF13 virus or a mutant virus containing a deletion of the entire DEN region (ΔDEN) was inoculated into newborn AKR/J mice, which were monitored for disease development.
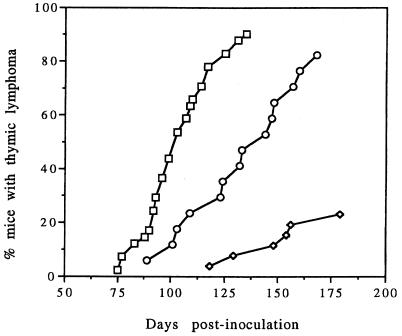
We observed that the ΔNF-κB virus induced tumors only of thymic origin, similar to the WT and ΔDEN viruses. Leukemic animals had a massive enlargement of the thymus with fusion of thymic lobes. In addition, moribund animals had enlarged liver, spleen, and peripheral lymph nodes consistent with the typical features of spontaneous and MCF13-accelerated lymphoma of AKR mice (34, 46). We detected a significant increase in the latency of lymphoma development for the ΔNF-κB virus from that of WT virus (Fig. 2 and Table 1). The mean latency for the ΔNF-κB virus was 131 days compared with 102 days for WT virus (Table 1). Analysis of these data by Student’s t test supported a statistically significant difference between these latency periods (P < 0.01). The ΔNF-κB mean latency, however, was not so great as for the ΔDEN virus, which had a mean latency of 147 days. For the ΔNF-κB virus, we observed only a slight decrease in the incidence of disease from that with WT virus. This was in contrast to the ΔDEN virus for which there was a marked decrease in disease incidence. The values for the mean latency and disease incidence for the WT and ΔDEN viruses that we obtained in this study were similar to those we observed previously (46). Our data for the ΔNF-κB virus indicated that the NF-κB protein-binding site in the DEN region of the MCF13 LTR could account for some, but not all, of the effect of DEN on viral pathogenicity.
FIG. 2.
Thymic lymphoma incidence for WT, ΔNF-κB, and ΔDEN MCF13 viruses. Neonatal AKR/J mice were inoculated intraperitoneally with 1 × 106 to 2.5 × 106 infectious units of each virus. The time course of appearance of thymic lymphoma is shown. Symbols: □, WT; ○, ΔNF-κB; ◊, ΔDEN.
TABLE 1.
Effects of LTR deletions upon lymphomagenicity of MCF13 MLV
| Virus | No. of mice inoculated | Disease incidence (%)a | Avg latency ± SEM (days)b |
|---|---|---|---|
| WT | 29 | 93.1 | 102 ± 4.1 |
| ΔNF-κB | 17 | 82.4 | 131 ± 6.5c |
| ΔDEN | 26 | 23.1 | 147 ± 8.8c |
Percentage of inoculated animals that developed thymic lymphoma.
Calculated from animals with thymic lymphoma that were sacrificed or found dead.
Significantly different from the average latency of WT virus as determined by the Student’s t test (P < 0.01).
Differences in the ability of WT and mutant viruses to replicate in the thymus.
The ability of a MLV to replicate efficiently in the thymus is essential for its ability to induce thymic tumors (10, 22, 35, 36). To determine whether differences in thymus infectivity of the viruses could account for their different pathogenic properties, we assessed the percentage of thymic lymphocytes which were infected with MCF13 MLV at various times after inoculation of virus into neonatal mice. To accomplish this, we detected the expression of MCF13 envelope glycoprotein (gp70) on the surface of these cells by an indirect immunofluorescence assay using a primary monoclonal antibody (MAb 514), which specifically recognizes MCF-type glycoproteins (reference 13 and our unpublished data), and a secondary FITC-labeled goat anti-mouse IgG. Stained cells were analyzed by flow cytometry to measure the percentage of cells which expressed the MCF13 gp70 (gp70+ cells). Because we were particularly interested in virus replication during the preleukemic period, we examined thymuses removed from mice 3 to 8 weeks postinoculation (p.i.). We chose this time period because 3 weeks p.i. was the earliest time when we could consistently detect MCF13 gp70 expression on lymphocytes infected with WT virus, and at 10 weeks p.i., mice with frank leukemia began to appear (Fig. 2). This time period coincided with the preleukemic period described by O’Donnell et al. (37).
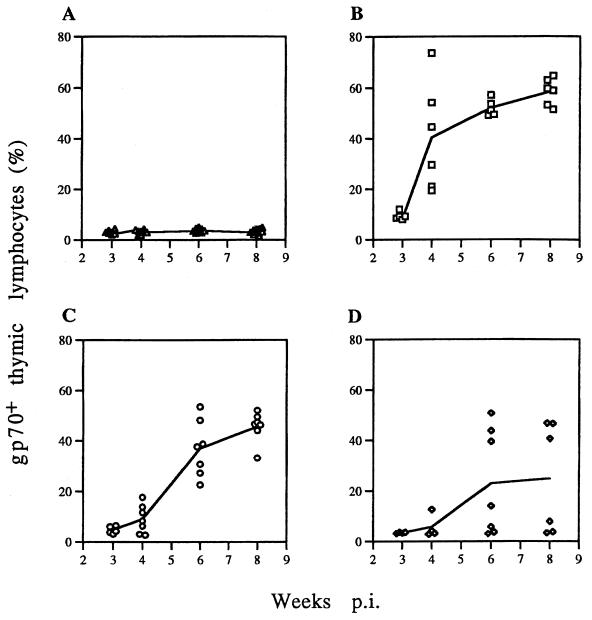
Figure 3 shows the percentage of thymic lymphocytes analyzed by flow cytometry, which were positive for MCF13 gp70, for each animal that was examined. The mean values of the percentages for mice examined at each time point were connected with a solid line. We observed an exponential increase for WT virus-infected cells, with the greatest rise occurring between 3 and 4 weeks p.i. (Fig. 3B). At 8 weeks p.i., the mean value for WT-infected cells corresponded to nearly 60% of thymic lymphocytes. The plot for thymic cells infected with the ΔNF-κB or ΔDEN virus showed that gp70 expression lagged behind the WT-infected cells, with the greatest increase occurring between 4 and 6 weeks (Fig. 3C and D). Furthermore, the mean values for ΔNF-κB and ΔDEN virus at 8 weeks were 46 and 25%, respectively, both significantly less than WT. The plot of gp70+ cells infected by ΔDEN virus showed two strikingly different populations of thymic cells at 6 and 8 weeks (Fig. 3D). Roughly half of the mice had thymic cells with gp70 expression that was greater than the mean (ΔDEN-high), and the other half had gp70 values close to the background level of glycoprotein expression present in control cells (∼4% [Fig. 3A]) (ΔDEN-low).
FIG. 3.
Thymic lymphocytes expressing MCF13 envelope gp70. Lymphocytes were isolated from thymuses removed from mice after virus inoculation at 3, 4, 6, and 8 weeks. A total of 106 lymphocytes were stained with the primary MAb 514, followed with the secondary FITC-conjugated goat anti-mouse IgG. A total of 2 × 104 lymphocytes were analyzed by flow cytometry, and the percentage of gp70+ lymphocytes was calculated with the Paint-a-Gate software (BDIS). Percentages of gp70+ cells for individual mice are shown. Mean values are connected by a solid line. Control (A), WT (B), ΔNF-κB (C), and ΔDEN MCF13 (D) virus were used.
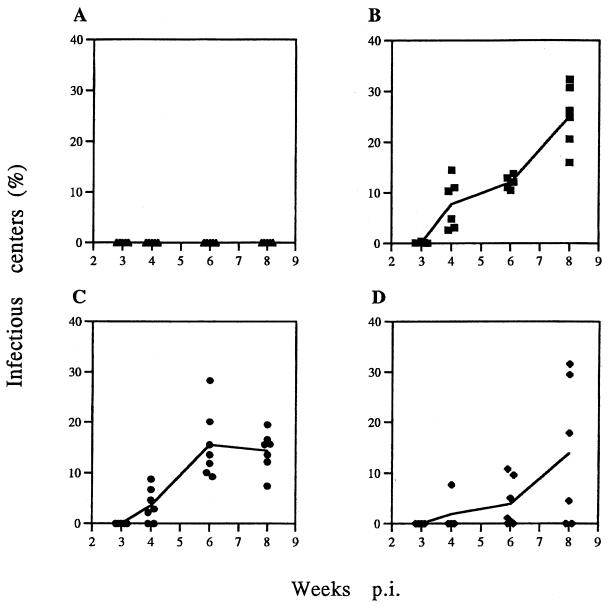
As an additional measure of virus replication in the thymus, we performed infectious center assays with the same thymuses that were used to detect MCF13 gp70 expression. We again performed a time course study from 3 to 8 weeks p.i. The mean values of thymic lymphocytes which produced infectious WT virus increased from 0.12% at 3 weeks to 25% at 8 weeks (Fig. 4B). Infectious ΔNF-κB and ΔDEN mutant viruses were not detectable until 4 weeks p.i. (Fig. 4C and D). At 8 weeks, the production of infectious virus for both mutants was only 14% of thymic lymphocytes. Thus, compared with WT virus, both mutant viruses replicated more slowly and to lower levels. Similar to our analysis of gp70 expression of mice inoculated with ΔDEN, we detected two different populations of thymuses producing either higher or lower levels of infectious virus compared with the mean values at 6 and 8 weeks p.i. Animals in these two populations corresponded to the same animals which segregated into the two populations expressing either high or low levels of gp70 on their thymic cells.
FIG. 4.
Infectious center assays of thymic lymphocytes. Lymphocytes were isolated from thymuses removed from mice after virus inoculation at 3, 4, 6, and 8 weeks. A total of 1 × 102 to 5 × 103 lymphocytes were plated onto M. dunni fibroblasts. The cell cultures were allowed to grow to confluency. Cells were stained with MAb 514 and FITC-conjugated goat anti-mouse IgG. Infectious center assay data are represented as percentages of the numbers of plated thymic lymphocytes. Each point corresponds to an individual mouse. Mean values are connected by a solid line. Control (A), WT (B), ΔNF-κB (C), and ΔDEN MCF13 (D) virus were used.
Because we had detected two different populations of ΔDEN virus, we wished to determine whether reversions or secondary mutations in the LTR could account for their different abilities to replicate in the thymus. To accomplish this, we examined the LTR sequences of two isolates from each population by DNA sequencing. Infectious virus was rescued from thymic lymphocytes with either high or low levels of gp70 expression and infectious centers. LTR sequences of the ΔDEN proviruses were amplified by PCR from cellular DNA, which was isolated from M. dunni cells infected with the rescued virus. The DNA sequence of the PCR product indicated that there were no changes in the LTRs of ΔDEN viruses from either the high- or low-level gp70-expressing population (data not shown). However, mutations elsewhere in the ΔDEN viral genome could be responsible for our observations and would require further analysis to detect.
Identification of thymic lymphocyte subpopulations in which virus replication occurs.
Because we observed that replication of the ΔNF-κB and ΔDEN viruses in thymic lymphocytes was reduced from that of WT virus, we asked whether these different MCF13 viruses replicated in different subpopulations of cells. Using multiparametric flow cytometry, we identified the phenotype of thymic lymphocytes which were infected with WT or mutant viruses from 3 to 8 weeks p.i. For this analysis, we first selected thymic lymphocytes which expressed MCF13 gp70 with the Paint-a-Gate software. Subsequently, we calculated the percentage of the selected gp70+ cells represented by each subpopulation of thymic lymphocytes (Table 2). Thymic subsets were identified by staining with fluorescence-labeled antibodies to the CD3, CD4, and CD8 cell surface antigens, which distinguish five major subpopulations of thymic T cells. As shown in Table 2, these five subpopulations were the immature CD3− CD4− CD8−, CD3− CD4+ CD8+, and CD3+ CD4+ CD8+ cells and the more mature CD3+ CD4+ CD8− and CD3+ CD4− CD8+ cells (14, 39, 41). Additional thymic cell populations, which have been identified by others, i.e., CD3− CD4+ CD8−, CD3− CD4− CD8+, and CD3+ CD4− CD8− (14, 23, 24, 41), were too low in number to be reproducibly detected and, hence, were not included in our analysis. For the analysis of the gp70+ cells infected with ΔDEN virus, we included only the ΔDEN-high mice (Fig. 3D and 4D).
TABLE 2.
Subpopulations of selected gp70+ thymic cells
| Wk p.i. | Virus | gp70+ thymic subpopulation (%)a
|
||||
|---|---|---|---|---|---|---|
| CD3− CD4− CD8− | CD3− CD4+ CD8+ | CD3+ CD4+ CD8+ | CD3+ CD4− CD8+ | CD3+ CD4+ CD8− | ||
| 3 | WT | 1.3 ± 0.8 | 9.6 ± 2.3 | 80.0 ± 3.7 | 1.9 ± 1.8 | 4.5 ± 2.2 |
| ΔNF-κB | 1.2 ± 0.4 | 6.6 ± 1.8 | 77.9 ± 1.8 | 2.4 ± 0.9 | 8.5 ± 1.8 | |
| ΔDEN | 1.1 ± 0.4 | 4.7 ± 1.4 | 82.4 ± 1.1 | 2.1 ± 0.4 | 6.9 ± 0.6 | |
| 4 | WT | 0.2 ± 0.1 | 11.1 ± 4.4 | 82.3 ± 3.0 | 0.6 ± 0.1 | 4.5 ± 1.8 |
| ΔNF-κB | 0.9 ± 0.5 | 12.1 ± 4.9 | 77.9 ± 5.3 | 1.1 ± 0.7 | 5.5 ± 1.9 | |
| ΔDEN | 1.3 ± 0.7 | 8.9 ± 4.8 | 76.0 ± 2.4 | 2.7 ± 1.7 | 8.1 ± 3.6 | |
| 6 | WT | 0.6 ± 0.2 | 18.2 ± 4.1 | 71.0 ± 4.1 | 2.0 ± 0.4 | 5.5 ± 1.0 |
| ΔNF-κB | 0.4 ± 0.2 | 13.5 ± 3.9 | 78.6 ± 3.4 | 0.8 ± 0.3 | 5.0 ± 1.1 | |
| ΔDEN | 0.7 ± 0.6 | 8.7 ± 5.1 | 83.6 ± 4.8 | 1.4 ± 0.6 | 4.1 ± 1.7 | |
| 8 | WT | 1.1 ± 0.3 | 15.5 ± 2.3 | 71.9 ± 3.5 | 1.6 ± 0.5 | 6.7 ± 1.1 |
| ΔNF-κB | 0.5 ± 0.1 | 15.6 ± 4.3 | 75.4 ± 3.0 | 0.9 ± 0.3 | 5.8 ± 2.1 | |
| ΔDEN | 1.2 ± 1.1 | 17.9 ± 9.8 | 71.5 ± 9.1 | 1.9 ± 1.0 | 4.5 ± 1.3 | |
Values are means ± standard deviations of the percentages of selected gp70+ thymic lymphocytes represented by the T-cell subpopulations. Thymuses from four to seven mice, which were inoculated with medium or WT, ΔNF-κB, or ΔDEN MCF13 virus, were individually analyzed for each time point.
As shown in Table 2, we observed that at all time points for WT and mutant viruses, the gp70+ cells were comprised predominantly of CD3+ CD4+ CD8+ and CD3− CD4+ CD8+ cells. For all of the viruses, the CD3+ CD4+ CD8+ cells represented a large majority of the gp70+ cells (71 to 84%) and the CD3− CD4+ CD8+ cells constituted the next largest subpopulation (5 to 18%). The only subpopulation in which the percentage of ΔDEN-infected gp70+ cells was significantly lower than WT-infected cells from 3 to 6 weeks p.i. was CD3− CD4+ CD8+. For the other subpopulations, there were no consistently observed significant differences between WT and either mutant virus. These data also showed that the percentages of subpopulations of gp70+ cells were similar to those of uninfected thymic cells from control mice with the exception of the most immature CD3− CD4− CD8− subpopulation (compare Tables 2 and 3). We detected significantly lower percentages of gp70+ CD3− CD4− CD8− cells than for uninfected cells at all times. These decreases ranged from 3- to 27-fold, depending on the virus and time point, with the greatest differences observed at 4 weeks p.i. for WT virus and 6 weeks p.i. for both mutant viruses.
TABLE 3.
Subpopulations of control thymic cells
| Wk p.i. | Control thymic subpopulation (%)a
|
||||
|---|---|---|---|---|---|
| CD3− CD4− CD8− | CD3− CD4+ CD8+ | CD3+ CD4+ CD8+ | CD3+ CD4− CD8+ | CD3+ CD4+ CD8− | |
| 3 | 3.3 ± 0.9 | 14.2 ± 1.8 | 72.4 ± 1.9 | 1.1 ± 0.4 | 6.5 ± 1.2 |
| 4 | 4.1 ± 0.9 | 14.9 ± 2.8 | 70.2 ± 4.1 | 1.1 ± 0.4 | 7.1 ± 1.5 |
| 6 | 6.2 ± 1.4 | 16.0 ± 2.6 | 65.3 ± 4.3 | 1.5 ± 0.4 | 7.8 ± 0.5 |
| 8 | 6.1 ± 2.3 | 14.4 ± 3.0 | 68.7 ± 3.6 | 1.2 ± 0.4 | 6.8 ± 1.5 |
Values are means ± standard deviations of the percentages of uninfected thymic lymphocytes represented by the T-cell subpopulations. Thymuses from four to eight mice, which were inoculated with tissue culture medium, were individually analyzed for each time point.
Increase in percentage of virus-infected cells in thymic subpopulations.
Our analysis of subpopulations of gp70+ cells as discussed above did not reveal whether there were any changes over time in the number of virus-infected cells in each subpopulation. Any differences in the replication rates of WT and mutant viruses could identify thymic cell types involved in viral transformation. Determining whether this type of change occurred required that we first select each subpopulation of thymic cells and then calculate the percentage of gp70+ cells for each subpopulation, i.e., the reverse of what we did to characterize the virus-infected cells as described above.
Table 4 shows that for WT virus as well as both mutant MCF13 viruses, increases of virus-infected cells occurred in all of the subpopulations from 3 to 8 weeks p.i. The greatest change occurred in the CD3− CD4+ CD8+ subpopulation, where there was a ninefold increase for WT virus. The change in this subpopulation for the ΔNF-κB and ΔDEN viruses was even greater, with increases of 43- and 84-fold, respectively. The second largest change occurred in the CD3+ CD4+ CD8+ subpopulation. For these cells, the change for WT virus was 7-fold, and for ΔNF-κB and ΔDEN-high, it was 15- and 17-fold, respectively. By 8 weeks p.i., the values of gp70+ cells of the CD3− CD4+ CD8+ and CD3+ CD4+ CD8+ subpopulations reached between 35 and 59% for the three viruses. The CD3+ CD4− CD8+ and CD3+ CD4+ CD8− subpopulations had lower gp70+ percentage changes from 3 to 8 weeks p.i., and the percentages of virus-infected cells for these two subpopulations at 8 weeks were also lower than those for the CD3− CD4+ CD8+ and CD3+ CD4+ CD8+ subpopulations.
TABLE 4.
gp70+ cells in each subpopulation
| Wk p.i. | Virus | % of gp70+ cells in subpopulationa
|
||||
|---|---|---|---|---|---|---|
| CD3− CD4− CD8− | CD3− CD4+ CD8+ | CD3+ CD4+ CD8+ | CD3+ CD4− CD8+ | CD3+ CD4+ CD8− | ||
| 3 | WT | 2.3 ± 0.8 | 5.9 ± 1.6 | 8.7 ± 1.9 | 13.6 ± 3.5 | 4.5 ± 1.1 |
| ΔNF-κB | 1.1 ± 0.9 | 0.8 ± 0.2 | 2.9 ± 0.3 | 5.5 ± 3.1 | 2.7 ± 0.9 | |
| ΔDEN | 0.8 ± 0.3 | 0.6 ± 0.2 | 2.7 ± 0.3 | 3.4 ± 1.3 | 2.4 ± 0.3 | |
| 4 | WT | 1.3 ± 0.6 | 28.3 ± 13.4 | 34.6 ± 16.4 | 18.5 ± 10.4 | 17.5 ± 10.1 |
| ΔNF-κB | 1.1 ± 0.3 | 5.6 ± 3.3 | 8.4 ± 5.3 | 6.5 ± 1.6 | 4.7 ± 1.7 | |
| ΔDENb | 1.2 ± 0.0 | 6.7 ± 0.0 | 10.1 ± 0.0 | 3.2 ± 0.0 | 3.2 ± 0.0 | |
| 6 | WT | 3.3 ± 1.9 | 43.3 ± 2.8 | 51.4 ± 1.8 | 23.9 ± 2.4 | 24.6 ± 3.6 |
| ΔNF-κB | 1.4 ± 0.8 | 23.1 ± 10.9 | 29.9 ± 13.3 | 10.8 ± 3.7 | 12.0 ± 5.5 | |
| ΔDENb | 1.3 ± 0.8 | 33.0 ± 16.7 | 43.2 ± 20.4 | 17.4 ± 7.4 | 13.3 ± 5.4 | |
| 8 | WT | 5.2 ± 2.5 | 50.8 ± 4.4 | 59.0 ± 5.7 | 23.0 ± 4.5 | 27.7 ± 5.4 |
| ΔNF-κB | 1.8 ± 0.8 | 34.5 ± 6.4 | 43.9 ± 5.1 | 13.7 ± 3.9 | 15.2 ± 3.0 | |
| ΔDENb | 1.7 ± 1.0 | 50.2 ± 9.6 | 44.9 ± 4.8 | 15.7 ± 2.7 | 16.5 ± 2.1 | |
Values are means ± standard deviations of the percentages of gp70+ cells in the T-cell subpopulations. Each subpopulation was selected by the Paint-a-Gate software, and the percentage of gp70+ cells of each subpopulation was calculated. Thymuses from one to seven mice, which were inoculated with medium or WT, ΔNF-κB, or ΔDEN MCF13 virus, were individually analyzed for each time point.
Thymuses from only ΔDEN-high mice were analyzed.
The largest difference between WT and mutant viruses was detectable for the CD3− CD4+ CD8+ cells at early times (i.e., 3 and 4 weeks p.i.). At 3 weeks p.i., we observed 7- and 10-fold differences between WT- and ΔNF-κB- or ΔDEN-infected cells, respectively. At 8 weeks p.i., the percentages of gp70+ cells of all subpopulations were approximately the same for WT and mutant viruses except for the CD3− CD4− CD8− subset, where percentages of WT-infected cells remained greater than cells infected by either mutant virus. Thus, WT and mutant virus infection occurred to the greatest extent in the CD3− CD4+ CD8+ and CD3+ CD4+ CD8+ cells.
Characterization of thymic tumors produced by WT and mutant MCF13 MLVs.
As described above, both mutant viruses produced thymic lymphoma similar to the WT MCF13. We identified the phenotype of these tumor cells to determine whether there were any differences that were dependent upon the inoculated virus. Table 5 shows that the tumor phenotypes for all three viruses were similar to each other. Furthermore, we observed that the majority of tumors had a heterogeneous phenotype and were oligoclonal. The thymic cell type which was present in all tumors was CD3+ CD4+ CD8+, and in the majority of tumors induced by all three viruses, this subpopulation was the predominant cell type. These observations were consistent with the results of our analysis of thymic subpopulations during the preleukemic period, which showed that WT and both mutant viruses infected the same subpopulations of cells and that the subpopulation with the largest percentage of virus-infected cells was CD3+ CD4+ CD8+. Although all thymic subpopulations were detectable in these tumors, the CD3− CD4− CD8− cells usually constituted an insignificant percentage of tumor cells, which reflected our detection of very low levels of virus infection of these cells during the preleukemic period.
TABLE 5.
Phenotypes of thymic lymphomasa
| Virus | Lymphoma | % of tumor cells in thymic subpopulation
|
||||
|---|---|---|---|---|---|---|
| CD3− CD4− CD8− | CD3− CD4+ CD8+ | CD3+ CD4+ CD8+ | CD3+ CD4− CD8+ | CD3+ CD4+ CD8− | ||
| WT | 1 | 0.5 | 19.1 | 61.3 | 2.6 | 14.3 |
| 2 | 0.1 | 10.0 | 83.1 | 4.9 | 1.3 | |
| 3 | 0.1 | 11.0 | 85.1 | 2.1 | 0.8 | |
| 4 | 0.1 | 0.6 | 7.8 | 90.7 | 0.0 | |
| 5 | 0.2 | 3.1 | 92.4 | 3.0 | 0.5 | |
| 6 | 0.1 | 9.8 | 74.8 | 11.4 | 0.7 | |
| 7 | 0.3 | 0.6 | 97.3 | 0.3 | 1.1 | |
| 8 | 0.0 | 0.4 | 93.6 | 0.4 | 5.3 | |
| 9 | 0.8 | 0.6 | 68.7 | 9.4 | 13.7 | |
| 10 | 0.4 | 40.6 | 51.7 | 1.9 | 2.4 | |
| 11 | 0.4 | 16.4 | 70.2 | 1.3 | 9.0 | |
| ΔNF-κB | 1 | 0.1 | 2.5 | 33.3 | 59.6 | 0.3 |
| 2 | 0.4 | 3.7 | 90.8 | 3.7 | 0.4 | |
| 3 | 0.1 | 2.4 | 91.4 | 5.0 | 0.5 | |
| 4 | 0.5 | 2.8 | 79.4 | 1.4 | 13.8 | |
| ΔDEN | 1 | 0.4 | 5.5 | 78.1 | 0.9 | 13.4 |
| 2 | 1.5 | 30.9 | 60.7 | 0.5 | 3.8 | |
| 3 | 0.1 | 1.5 | 84.1 | 0.1 | 14.0 | |
| 4 | 7.6 | 4.6 | 35.7 | 40.7 | 2.2 | |
| 5 | 0.2 | 0.3 | 15.1 | 0.5 | 80.7 | |
T-cell phenotype of cells isolated from individual thymic lymphomas induced by WT, ΔNF-κB, or ΔDEN MCF13 virus.
DISCUSSION
Our previous studies have shown that besides the direct repeats, which comprise the enhancer element, in the LTR of the MCF13 MLV, the region between the enhancer and promoter (DEN) is another important determinant of viral pathogenicity (46). In this report we have demonstrated that an important function of the DEN region is its ability to regulate virus replication in the thymus during the preleukemic period. Previous reports by others have shown that enhancer sequences, which affect pathogenicity, do not play a role in viral replication in this target tissue (12, 16, 19) and, thus, must contribute to tumorigenesis in another manner. We have observed that DEN is the sole region in the LTR which regulates transcription in activated T cells in vitro (8). This activity of the DEN region also may be important for activation of proviral transcription in the target cells for MCF replication in the thymus. We observed that the ΔNF-κB protein-binding site in DEN contributed to the regulation of viral replication in the thymus and, thus, to MCF13 pathogenicity. However, a mutant virus lacking the NF-κB site was less attenuated in pathogenicity than one missing the entire DEN region. This result suggested that additional protein-binding sites, which we previously identified in DEN (50), also contribute to MCF virus pathogenicity. In addition, it must be kept in mind that the deletion of the DEN region alters the spacing between the enhancer and promoter in the LTR, which also could contribute to the greater attenuation of the ΔDEN mutant. Although the mutant viruses differed from WT virus in their ability to replicate in the thymus, they replicated in M. dunni fibroblasts as efficiently as WT virus (data not shown).
It has been shown for several other retroviruses, such as human immunodeficiency virus (32), human T-cell leukemia virus type 1 (1, 29, 40), simian immunodeficiency virus (48), bovine leukemia virus (6), and avian leukosis virus (5, 11), that NF-κB plays a regulatory role in viral pathogenicity and/or transcriptional regulation. NF-κB is a multigene family of transcriptional regulators, which control a wide range of genes involved in immune and inflammatory responses (2, 3). Most of the cortical cells in the thymus, which include the CD3− CD4− CD8−, CD3− CD4+ CD8+, and CD3+ CD4+ CD8+ subpopulations, contain p50 homodimers as the predominant constitutive form of NF-κB (31, 47). p50 homodimers have been shown to function as a transcriptional repressor (21, 38). At the same time, these same subpopulations contain relatively high levels of inducible forms of NF-κB, mainly corresponding to p50/RelA and p50/c-Rel (47), which are transcriptional activators (2). Induction of these NF-κB heterodimers has been shown to occur during positive selection of a minor population of CD3+ CD4+ CD8+ cells (7, 31). Our data indicate that NF-κB contributes to the regulation of MCF13 infection of thymic subpopulations, which contain inducible forms of transcriptionally active NF-κB. Whether virus infection of these cells results in the induction of NF-κB, which further enhances infectivity in a type of autoregulatory fashion, is an intriguing question regarding the mechanism of MCF tumorigenesis which we are currently addressing.
Our data showed that WT virus and both mutant viruses infected the same subpopulations of thymic cells. However, the rate at and degree to which these cells were infected by the mutant viruses were lower than for WT virus. The subpopulation in which we saw the greatest difference in early viral infection between WT virus and both mutant viruses was CD3− CD4+ CD8+ (Table 4), which suggested that this cell type may be a major target for MCF13 transformation. This subpopulation is thought to provide the precursors to the CD3+ CD4+ CD8+ cells, another thymic cell type in which virus infection occurred at high levels. Previous observations by others of the high levels of MCF MLV infection of CD3+ CD4+ CD8+ cells (9, 37) was puzzling because the vast majority of these cells do not normally proliferate (14, 39, 41), a sine qua non for most retrovirus replication. Studies have shown that most of the CD3+ CD4+ CD8+ cells are destined to undergo apoptosis (14, 39). Our detection of MCF13 infection of the precursors of these nonproliferating cells suggests that virus replication may occur in the CD3− CD4+ CD8+ precursor subpopulation, which continues to differentiate into the CD3+ CD4+ CD8+ cells, after which virus replication ceases. This hypothesis would explain the discrepancy between the lower percentage of thymic cells which produce infectious virus and the higher percentage of thymic cells which express viral glycoprotein, which we and others have seen (22, 33). To test this idea, it would be necessary to isolate these two subpopulations and examine virus replication in each.
In addition, the most immature CD3− CD4− CD8− cells may be another important target for virus infection, as this is the only subpopulation in which the percentage of WT virus-infected cells for the most part remained greater than both mutant viruses throughout the preleukemic period. Because MLVs require cell division for their replication, it has been proposed that some immature blast cell in the thymus is the target for virus infection and transformation (9, 37). In this study, we have identified these immature cells to be the CD3− CD4− CD8− and CD3− CD4+ CD8+ subpopulations, both of which are considered early thymic precursors and contain large fractions of proliferating cells (14, 41). Thus, it is possible that viral glycoprotein expression detectable in more mature subsets is a result of infection of these early precursor cells, which then continue to differentiate.
Because WT virus and both mutant viruses infected the same thymic subsets, it was not surprising that these viruses induced thymic tumors with similar phenotypes. The phenotypic heterogeneity of these tumors also has been observed for T-cell lymphomas induced by the Moloney MLV in rats as well as those induced by the SL3-3 virus in AKR mice (18, 27). Although the majority of SL3-3 lymphomas contained CD4+ CD8+ cells, only a third of the Moloney MLV-induced tumors contained cells with this phenotype. This difference in the predominance of this cell type in rat and AKR tumors may be a result of differences in both the genetic background of the host and LTR sequences of the oncogenic MLV.
Our data indicate that the different rates of replication in the thymus are a major determinant of the difference in pathogenicity between the WT virus and the ΔNF-κB and ΔDEN mutant viruses. These results support the idea that transcriptional regulatory sequences in the DEN region of the LTR contribute to the control of virus replication in thymic lymphocytes. However, DEN may be required for additional steps in tumorigenicity, such as the regulation of transcription of cellular oncogenes, since we have observed that DEN sequences control transcription in activated T cells (8). We have also observed that NF-κB activation of transcription of the viral genome in thymic target cells is an important determinant of virus replication during the early stages of tumorigenesis.
ACKNOWLEDGMENTS
We thank Yihua Zhang for technical assistance and Eric Van Buren for help in performing the flow cytometry experiments and the many discussions regarding analysis of the data. We also thank Stephen Lerman, director of the Core Flow Cytometry Facility of the Karmanos Cancer Institute and the Department of Immunology and Microbiology at Wayne State University, for helpful suggestions on flow cytometry work. We appreciate the critical reading of this manuscript by A. Galy and Y.-C. Kong. We are grateful to Leonard Evans for his continuous generosity in providing us with the MCF-specific monoclonal antibody.
This work was supported by Public Health Service grant CA44166 from the National Institutes of Health to F.K.Y.
REFERENCES
- 1.Arima N, Molitor J A, Smith M R, Kim J H, Daitoku Y, Greene W C. Human T-cell leukemia virus type I Tax induces expression of the Rel-related family of κB enhancer-binding proteins: evidence for a pretranslational component of regulation. J Virol. 1991;65:6892–6899. doi: 10.1128/jvi.65.12.6892-6899.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baeuerle P A, Henkel T. Function and activation of NF-κB in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 3.Barnes P J, Karin M. Nuclear factor-κB—a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 4.Boral S L, Okenquist A, Lenz J. Identification of the SL3-3 virus enhancer core as a T-lymphoma cell-specific element. J Virol. 1989;63:76–84. doi: 10.1128/jvi.63.1.76-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowers W J, Baglia L A, Ruddell A. Regulation of avian leukosis virus long terminal repeat-enhanced transcription by C/EBP-Rel interactions. J Virol. 1996;70:3051–3059. doi: 10.1128/jvi.70.5.3051-3059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks P A, Nyborg J K, Cockerell G L. Identification of an NF-κB binding site in the bovine leukemia virus promoter. J Virol. 1995;69:6005–6009. doi: 10.1128/jvi.69.10.6005-6009.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen D, Rothenberg E V. Molecular basis for developmental changes in interleukin-2 gene inducibility. Mol Cell Biol. 1993;13:228–237. doi: 10.1128/mcb.13.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Yoshimura F K. Identification of a region of a murine leukemia virus long terminal repeat with novel transcriptional regulatory activities. J Virol. 1994;68:3308–3316. doi: 10.1128/jvi.68.5.3308-3316.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cloyd M W. Characterization of target cells for MCF viruses in AKR mice. Cell. 1983;32:217–225. doi: 10.1016/0092-8674(83)90512-3. [DOI] [PubMed] [Google Scholar]
- 10.Cloyd M W, Hartley J W, Rowe W P. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J Exp Med. 1980;151:542–552. doi: 10.1084/jem.151.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curristin S M, Bird K J, Tubbs R J, Ruddell A. VBP and RelA regulate avian leukosis virus long terminal repeat-enhanced transcription in B cells. J Virol. 1997;71:5972–5981. doi: 10.1128/jvi.71.8.5972-5981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis B R, Chandy K G, Brightman B K, Gupta S, Fan H. Effects of nonleukemogenic and wild-type Moloney murine leukemia virus on lymphoid cells in vivo: identification of a preleukemic shift in thymocyte subpopulations. J Virol. 1986;60:423–430. doi: 10.1128/jvi.60.2.423-430.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans L H, Morrison R P, Malik F G, Portis J, Britt W J. A neutralizable epitope common to the envelope glycoproteins of ecotropic, polytropic, xenotropic, and amphotropic murine leukemia viruses. J Virol. 1990;64:6176–6183. doi: 10.1128/jvi.64.12.6176-6183.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fowlkes B J, Pardoll D M. Molecular and cellular events of T cell development. Adv Immunol. 1989;44:207–264. doi: 10.1016/s0065-2776(08)60643-4. [DOI] [PubMed] [Google Scholar]
- 15.Golemis E, Li Y, Fredrickson T N, Hartley J W, Hopkins N. Distinct segments within the enhancer region collaborate to specify the type of leukemia induced by nondefective Friend and Moloney viruses. J Virol. 1989;63:328–337. doi: 10.1128/jvi.63.1.328-337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallberg B, Schmidt J, Luz A, Pederson F S, Grundstrom T. SL3-3 enhancer factor 1 transcriptional activators are required for tumor formation by SL3-3 murine leukemia virus. J Virol. 1991;65:4177–4181. doi: 10.1128/jvi.65.8.4177-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanecak R, Pattengale P K, Fan H. Deletion of a GC-rich region flanking the enhancer element within the long terminal repeat sequence alters the disease specificity of Moloney murine leukemia virus. J Virol. 1991;65:5357–5363. doi: 10.1128/jvi.65.10.5357-5363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hays E F, Bristol G C, McDougall S, Klotz J L, Kronenberg M. Development of lymphoma in the thymus of AKR mice treated with the lymphomagenic virus SL3-3. Cancer Res. 1989;49:4225–4230. [PubMed] [Google Scholar]
- 19.Holland C A, Thomas C Y, Chattopadhyay S K, Koehne C, O’Donnell P V. Influence of enhancer sequences on thymotropism and leukemogenicity of mink cell focus-forming viruses. J Virol. 1989;63:1284–1292. doi: 10.1128/jvi.63.3.1284-1292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishimoto A, Takimoto M, Adachi A, Kakuyama M, Kato S, Kakimi K, Fukuoka K, Ogiu T, Matsuyama M. Sequences responsible for erythroid and lymphoid leukemia in the long terminal repeats of Friend mink cell focus-forming and Moloney murine leukemia virus. J Virol. 1987;61:1861–1866. doi: 10.1128/jvi.61.6.1861-1866.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang S M, Tran A C, Grilli M, Lenardo M J. NF-kappa B subunit regulation in nontransformed CD4+ T lymphocytes. Science. 1992;256:1452–1456. doi: 10.1126/science.1604322. [DOI] [PubMed] [Google Scholar]
- 22.Kawashima K, Ikeda H, Hartley J W, Stockert E, Rowe W P, Old L J. Changes in expression of murine leukemia virus antigens and production of xenotropic virus in the late preleukemic period in AKR mice. Proc Natl Acad Sci USA. 1976;73:4680–4684. doi: 10.1073/pnas.73.12.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraft D L, Weissman I L, Waller E K. Differentiation of CD3−4−8− human fetal thymocytes in vivo: characterization of a CD3−4+8− intermediate. J Exp Med. 1993;178:265–277. doi: 10.1084/jem.178.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kydd R, Lundberg K, Vremec D, Harris A, Shortman K. Intermediate steps in thymic positive selection. J Immunol. 1995;155:3806–3814. [PubMed] [Google Scholar]
- 25.Laimins L A, Gruss P, Pozzatti R, Khoury G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984;49:183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lander M R, Chattopadhyay S K. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ecotropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J Virol. 1984;52:695–698. doi: 10.1128/jvi.52.2.695-698.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazo P A, Klein-Szanto A J P, Tsichlis P N. T-cell lymphoma lines derived from rat thymomas induced by Moloney murine leukemia virus: phenotypic diversity and its implications. J Virol. 1990;64:3948–3959. doi: 10.1128/jvi.64.8.3948-3959.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Golemis E, Hartley J W, Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987;61:693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindholm P F, Marriott S J, Gitlin S D, Bohan C A, Brady J N. Induction of nuclear NF-κB DNA binding activity after exposure of lymphoid cells to soluble tax protein. New Biol. 1990;2:1034–1043. [PubMed] [Google Scholar]
- 30.LoSardo J, Boral A L, Lenz J. Relative importance of elements within the SL3-3 virus enhancer for T-cell specificity. J Virol. 1990;64:1756–1763. doi: 10.1128/jvi.64.4.1756-1763.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore N C, Girdlestone J, Anderson G, Owen J J, Jenkinson E J. Stimulation of thymocytes before and after positive selection results in the induction of different NF-kappa B/Rel protein complexes. J Immunol. 1995;155:4653–4660. [PubMed] [Google Scholar]
- 32.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 33.Nowinski R C, Doyle T. Cellular changes in the thymuses of preleukemic AKR mice: correlation with changes in the expression of murine leukemia viruses. Cell. 1977;12:341–353. doi: 10.1016/0092-8674(77)90110-6. [DOI] [PubMed] [Google Scholar]
- 34.Nowinski R C, Hays E F. Oncogenicity of AKR endogenous leukemia viruses. J Virol. 1978;27:13–18. doi: 10.1128/jvi.27.1.13-18.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Donnell P V, Stockert E, Obata Y, Old L J. Leukemogenic properties of AKR dualtropic (MCF) viruses: amplification of murine leukemia virus-related antigens on thymocytes and acceleration of leukemia development in AKR mice. Virology. 1981;112:548–563. doi: 10.1016/0042-6822(81)90301-9. [DOI] [PubMed] [Google Scholar]
- 36.O’Donnell P V, Nowinski R C, Stockert E. Amplified expression of murine leukemia virus (MuLV)-coded antigens on thymocytes and leukemia cells of AKR mice after infection by dualtropic (MCF) MuLV. Virology. 1982;119:450–464. doi: 10.1016/0042-6822(82)90104-0. [DOI] [PubMed] [Google Scholar]
- 37.O’Donnell P V, Woller R, Chu A. Stages in development of mink cell focus-inducing (MCF) virus-accelerated leukemia in AKR mice. J Exp Med. 1984;160:914–934. doi: 10.1084/jem.160.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plaksin D, Baeuerle P A, Eisenbach L. KBF1 (p50 NF-kappa B homodimer) acts as a repressor of H-2Kb gene expression in metastatic tumor cells. J Exp Med. 1993;177:1651–1662. doi: 10.1084/jem.177.6.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothenberg E V. The development of functionally responsive T cells. Adv Immunol. 1992;58:85–214. doi: 10.1016/s0065-2776(08)60487-3. [DOI] [PubMed] [Google Scholar]
- 40.Ruben S, Proteat H, Tan T, Kawakami K, Roeder R, Haseltine W, Rosen C. Cellular transcription factors and regulation of IL-2 receptor gene expression by HTLV-I tax gene product. Science. 1988;241:89–91. doi: 10.1126/science.2838905. [DOI] [PubMed] [Google Scholar]
- 41.Scollay R. T-cell subset relationships in thymocyte development. Curr Opin Immunol. 1991;3:204–209. doi: 10.1016/0952-7915(91)90051-2. [DOI] [PubMed] [Google Scholar]
- 42.Sitbon M, Nichio J, Wehrly H, Lodmell D, Chesebro B. Use of a focal immunofluorescence assay on live cells for quantitation of retroviruses: distinction of host range classes in virus mixture and virological cloning of dual-tropic murine leukemia viruses. Virology. 1985;141:110–118. doi: 10.1016/0042-6822(85)90187-4. [DOI] [PubMed] [Google Scholar]
- 43.Speck N A, Renjifo B, Golemis E, Fredrickson T N, Hartley J W, Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990;4:233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- 44.Speck N A, Renjifo B, Hopkins N. Point mutations in the Moloney murine leukemia virus enhancer identify a lymphoid-specific viral core motif and 1,3-phorbol myristate acetate-inducible element. J Virol. 1990;64:543–550. doi: 10.1128/jvi.64.2.543-550.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thornell A, Hallberg B, Grundstrom T. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol Cell Biol. 1988;8:1625–1637. doi: 10.1128/mcb.8.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tupper J C, Chen H, Hays E F, Bristol G C, Yoshimura F K. Contributions to transcriptional activity and to viral leukemogenicity made by sequences within and downstream of the MCF13 murine leukemia virus enhancer. J Virol. 1992;66:7080–7088. doi: 10.1128/jvi.66.12.7080-7088.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weih F, Carrasco D, Bravo R. Constitutive and inducible Rel/NF-κB activities in mouse thymus and spleen. Oncogene. 1994;9:3289–3297. [PubMed] [Google Scholar]
- 48.Winandy S, Renjifo B, Li Y, Hopkins N. Nuclear factors that bind two regions important to transcriptional activity of the simian immunodeficiency virus long terminal repeat. J Virol. 1992;66:5216–5223. doi: 10.1128/jvi.66.9.5216-5223.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshimura F K. Identification of a DNA fragment from a molecularly cloned mink cell focus-inducing murine leukemia virus specific for xenotropic virus-related sequences. J Virol. 1982;43:348–351. doi: 10.1128/jvi.43.1.348-351.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshimura F K, Cankovic M, Smeltz R, Ibrahim S. Identification of nucleotide sequences that regulate transcription of the MCF13 murine leukemia virus long terminal repeat in activated T cells. J Virol. 1997;71:2572–2576. doi: 10.1128/jvi.71.3.2572-2576.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]