Abstract
COVID-19 induces acute and persistent neurological symptoms in mild and severe cases. Proposed concomitant mechanisms include direct viral infection and strain, coagulopathy, hypoxia, and neuroinflammation. However, underlying molecular alterations associated with multiple neurological outcomes in both mild and severe cases are majorly unexplored. To illuminate possible mechanisms leading to COVID-19 neurological disease, we retrospectively investigated in detail a cohort of 35 COVID-19 mild and severe hospitalized patients presenting neurological alterations subject to clinically indicated cerebrospinal fluid (CSF) sampling. Clinical and neurological investigation, brain imaging, viral sequencing, and cerebrospinal CSF analyses were carried out. We found that COVID-19 patients presented heterogeneous neurological symptoms dissociated from lung burden. Nasal swab viral sequencing revealed a dominant strain at the time of the study, and we could not detect traces of SARS-CoV-2's spike protein in patients' CSF by multiple reaction monitoring analysis. Patients presented ubiquitous systemic hyper-inflammation and broad alterations in CSF proteomics related to inflammation, innate immunity, and hemostasis, irrespective of COVID-19 severity or neuroimaging alterations. Elevated CSF interleukin-6 (IL6) correlated with disease severity (sex-, age-, and comorbidity-adjusted mean Severe 24.5 pg/ml, 95% confidence interval (CI) 9.62–62.23 vs. Mild 3.91 pg/mL CI 1.5–10.3 patients, p = 0.019). CSF tumor necrosis factor-alpha (TNFα) and IL6 levels were higher in patients presenting pronounced neuroimaging alterations compared to those who did not (sex-, age-, and comorbidity-adjusted mean TNFα Pronounced 3.4, CI 2.4–4.4 vs. Non-Pronounced 2.0, CI 1.4–2.5, p = 0.022; IL6 Pronounced 33.11, CI 8.89–123.31 vs Non-Pronounced 6.22, CI 2.9–13.34, p = 0.046). Collectively, our findings put neuroinflammation as a possible driver of COVID-19 acute neurological disease in mild and severe cases.
Keywords: Coronavirus, Neuroinflammation, COVID-19, Neuro-infectious diseases, Inflammation
Highlights
-
•
Neurological symptoms are dissociated from pulmonary involvement in COVID-19.
-
•
Despite disease heterogeneity, COVID-19 patients shared a single altered CSF proteomic profile.
-
•
Deregulated CSF proteomic pathways related to the innate immune system and hemostasis.
-
•
Mild and severe COVID-19 patients with neurological symptoms display systemic and central nervous system hyper-inflammation.
-
•
Disease severity is associated with increases in CSF IL6.
-
•
Neuroimaging abnormalities are connected to elevated CSF IL6 and TNFα levels.
1. Introduction
COVID-19 impacts multiple organs and tissues beyond the respiratory system, and the association between COVID-19 and acute and persistent neurological disease is increasingly recognized. Neurological manifestations of COVID-19 are frequent and include mild symptoms, such as headaches and anosmia, to severe acute and sub-acute complications, including encephalopathy, encephalitis, coma, stroke, and Guillain-Barré syndrome (Helms et al., 2020a, 2020b; Lechien et al., 2020; Mao et al., 2020; Romero-Sánchez et al., 2020; Chou et al., 2021). COVID-19 survivors, even young patients or those with mild to moderate disease, might experience lingering neurological and psychiatric symptoms, including headache, pain, anxiety, and cognitive disturbance; and increased risk of dementia, psychiatric disorders, and epilepsy (Mazza et al., 2020, 2021; Woo et al., 2020; Taquet et al., 2021a, 2021b, 2022). However, we do not know how COVID-19 induces neurological disturbance.
Viral infections may induce transient or irreversible neural damage directly, by viral infection-mediated tissue damage, or indirectly. Infection-induced inflammation may trigger diffuse brain alterations and symptoms by mechanisms that include aberrant cytokine upregulation, neuro-immune cellular interactions, and blood-brain barrier function (Klein et al., 2017; Lyra e Silva et al., 2022). COVID-19 induces a hyper-inflammatory stage and hypercytokinemia (Webb et al., 2020; Zeng et al., 2020). Acute and sub-acute COVID-19 neurological disturbance is associated with neuroimaging alterations suggestive of inflammation and vascular damage, including hemorrhagic and ischemic stroke (Helms et al., 2020a; Poyiadji et al., 2020; Freitas et al., 2021). Cerebrospinal fluid (CSF) findings may include oligoclonal bands and elevated levels of proteins, glucose, pro-inflammatory cytokines, and IgG, however, white blood cell counts vary and SARS-CoV-2 genetic material detection is rare (Espíndola et al., 2020, 2021; Helms et al., 2020a; Freitas et al., 2021; Etter et al., 2022; Siahaan et al., 2022). Interleukin-6 (IL6) and Tumor Necrosis Factor-alpha (TNFα) are central innate immunity mediators upregulated in COVID-19 patients’ blood and CSF that have an already described importance for viral infection-induced neurological disturbance (Burton and Johnson, 2012; Figueiredo et al., 2019; Zeng et al., 2020; Espíndola et al., 2021; Santa Cruz et al., 2021; Guasp et al., 2022). Common neuropathological findings in COVID-19 deceased patients include signs of hypoxia, glial activation, immune infiltrates, and vascular damage (Lee et al., 2020; Solomon et al., 2020; Thakur et al., 2021; Yang et al., 2021; Lee MH et al., 2022). Viral material is found in COVID-19 post-mortem brain tissue, although inconsistently (Matschke et al., 2020; Thakur et al., 2021; Yang et al., 2021; Crunfli et al., 2022). Overall, current available clinical and pre-clinical evidence supports concomitant mechanisms, including neuroinflammation, coagulopathy, and hypoxia, at the core of COVID-19-induced neuropathogenesis, particularly in severe cases (Solomon, 2021; Lyra e Silva et al., 2022).
However, available clinical data that connects COVID-19 disease severity, neuroinflammation, and neuroimaging alterations is limited. Most available evidence for COVID-19-induced neuropathological mechanisms comes from severe or deceased COVID-19 cases, which may not reflect mechanisms in mild cases. Additionally, the multiple possible neurological outcomes following COVID-19 infection could imply distinct underlying molecular alterations, a matter that needs to be further explored. Understanding the acute disease in detail is important to tackling disease mechanisms that may be associated with persistent symptoms in survivors. Furthermore, identifying altered central nervous system (CNS) molecules in COVID-19 neurological disease may lead to new therapeutic targets and blood-based biomarkers to monitor patients quickly during interventional therapies.
We hypothesized that COVID-19 severity and associated pulmonary burden and systemic inflammation, as well as aspects of viral strain and possible viral protein neuroinvasion, would lead to neuroinflammation and, thus, neurological disease. In this study, we conducted a detailed clinical, molecular, and neuroimaging examination in COVID-19 patients with neurological symptoms. We explored relationships between COVID-19 severity, systemic and CNS hyper-inflammation, CSF proteome, viral material, neuroimaging findings, and neurological outcomes. Given the critical roles of IL6 and TNFα in innate inflammatory pathways, their associations with COVID-19 severity, and already described importance for viral infection-induced neurological disturbance (reviewed by Lyra e Silva et al., 2022), we measured these two cytokines’ levels in patients CSF. As main findings, we showed that COVID-19 patients presented heterogeneous neurological features associated with a single altered CSF proteome pattern, despite disease severity, pulmonary burden, or the presence of pronounced neuroimaging alterations. CSF TNFα is ubiquitously elevated in mild and severe patients, but CSF IL6 is preferentially elevated in severe cases. Finally, both cytokines were higher in cases with pronounced neuroimaging alterations. Our results put neuroinflammation as a possible main driver of COVID-19 neurological disease in mild and severe cases.
2. Methods
2.1. COVID-19 cohort
We retrospectively analyzed data from COVID-19-confirmed hospitalized patients from D’Or São Luiz Network Hospitals in Rio de Janeiro (Brazil) and subject to clinically indicated CSF analysis from April to November 2020. Suspected COVID-19 was defined based on symptoms clinically compatible with COVID-19 (World Health Organization, 2020a). Confirmation of COVID-19 was based on detecting SARS-CoV-2's E, RDRP, or N genes RNA by reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) assays (Allpex, 2019 n-CoV assay #RP10252W) on the nasopharyngeal or nasal swabs or by blood detection of anti-SARS-CoV-2 IgG/IgM antibodies. The National Commission for Research Ethics (CONEP) from the Brazilian Ministry of Health and the Committee for Research Ethics of D’Or Institute of Research and Education (IDOR) approved the study protocol and all amendments, CAAE #29496920.8.0000.5262; CAAE #41576620.7.0000.5249.
2.1.1. Clinical data analysis
An independent data board reviewed clinical, laboratory, and imaging data with access to unblinded data. Clinical data were extracted from the patients’ medical records, including medical history and comorbidities inquired by the medical team to patients and relatives at hospital admission anamnesis, clinical characteristics at hospital admission, in-hospital symptoms, complications, and medication used, according to an approved clinical research form (CRF) (International Severe Acute Respiratory and Emerging Infection Consortium World Health Organization, 2020a) and clinical characterization protocol (CCP) (International Severe Acute Respiratory and Emerging Infection Consortium World Health Organization, 2020b) created by members of the International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC) in collaboration with the World Health Organization. Multimorbidity was defined by the presence of two or more chronic illnesses. Detailed laboratory results were also collected, and qualitative and quantitative estimations of lung disease caused by COVID-19 were determined by computerized tomography (CT) scan examination.
COVID-19 severity classification followed the “Ordinal Scale for Clinical Improvement” proposed by a special World Health Organization (WHO) committee (World Health Organization, 2020b). Mild disease included hospitalized patients who did not receive oxygen therapy or received oxygen by masks or nasal cannula. Severe disease included hospitalized patients who required at least one of the following treatments: oxygen by non-invasive ventilation; high-flow oxygen; intubation; and mechanical ventilation with or without additional organ support.
An experienced neurologist reviewed patients’ clinical data and defined a major neurological complaint. Encephalitis was defined as presenting altered mental status (altered level of consciousness, lethargy, or personality change) for at least 24h and two or more of the following criteria: a) seizures not attributable to a pre-existing condition; b) new-onset focal neurologic finding; c) elevated CSF white blood cell count (above 5 cells/mm3); d) acute neuroimaging finding consistent with encephalitis; e) abnormal electroencephalography consistent with encephalitis, or f) fever (above 38 °C) within 3-days of symptom onset (Venkatesan et al., 2013).
2.1.2. Neuroimaging protocols and analysis
All neuroimaging data were acquired during hospitalization. Brain CT and/or MRI images were analyzed by two experienced neuroradiologists blinded to patient clinical data, according to a pre-established structured report. Patients with normal brain scans or findings not associated with COVID-19 were classified as negative findings. Patients solely presenting intracranial hypertension signs were assigned to a specific group defined as moderate neuroimaging findings. Patients with other discoveries related to COVID-19 encephalitis, ischemic lesions, stroke, or demyelinating lesions (associated or not to intracranial hypertension) were read as pronounced neuroimaging changes and assigned to another group. CT scans (with or without contrast administration) were acquired in multidetector scanners with volumetric data acquisition. MRI acquisitions were performed on 1.5 T equipment, following standardized multiplanar protocols for brain, spine, and/or vessel studies.
2.2. Systemic inflammatory analysis
2.2.1. COVID-19 hyperinflammatory syndrome score (cHIS)
Blood test data most proximal to CSF sampling were retrieved to compute cHIS as previously described (Webb et al., 2020). The score was calculated using reported fever at admission or hospitalization and blood indicators of cytokinaemia (C-reactive protein (CRP) above 15 mg/dL or triglyceride concentration above 150 mg/dL or blood IL6 concentration above 15 pg/mL); hematological dysfunction (neutrophil to lymphocyte ratio (NLR) above 10, less than 110 billion/L platelets, or hemoglobin bellow 9.2 g/dL); coagulopathy (D-dimer concentration above 1.5 μg/mL); hepatic injury (lactate dehydrogenase concentration above 400 U/L or aspartate aminotransferase above 100 U/L); macrophage activation (ferritin concentration above 700 μg/L).
2.2.2. Blood cytokine evaluation
We retrieved 16 serum samples from COVID-19 patients collected during hospitalization (median 0 days, interquartile range 0–5.5, maximum 44 days apart CSF collection date), stored in polypropylene tubes, and frozen at −80 °C until molecular analysis. Control serum came from the above specified five healthy donors (negative RT-qPCR for SARS-CoV-2). Before assays, samples were thawed and kept on ice. Cytokines were measured using the Human Cytokine/Chemokine Magnetic Bead Panel kit (Millipore, #HCYTOMAG-60K) following manufacturers’ instructions. Results were read at a MagPix Luminex xMAP instrument and analyzed with XPonent software. Data is represented as a fold change computed by subtracting the control mean value (X) from COVID-19 individual values (Y) and dividing by the control mean, [(Y-X)/X]. Raw data (pg/mL) was used for statistical analysis. Multiple comparisons are corrected by FDR, Q = 5%. IL3 was only detected in one COVID-19 sample and was further excluded from the analysis.
2.3. CSF collection and analysis
Irrespective of patients' grouping, after lumbar puncture, CSF was immediately processed for routine laboratory analysis consisting of cell counts, total protein, glucose, lactate, microbiological analysis, and the opening pressure estimation. For the COVID-19 group, 14 CSF samples were also investigated for the presence of SARS-CoV-2 RNA and other neuropathogens using the Biomanguinhos (E + P1) RT-qPCR kit (FIOCRUZ, Brazil), XGEN Master COVID-19 (Mobius Brazil), XGEN Viral Meningitis Panel (Mobius, Brazil) or FilmArray Meningitis/Encephalitis Panel (bioMérieux, Brazil). Oligoclonal bands IgG investigation results were available for four COVID-19 patients (HYDRASYS FOCUSING, Sebia, France). For all patients from all groups, the remaining cell-free CSF supernatants were stored in polypropylene tubes and immediately frozen at −80 °C until used for molecular analysis described next.
2.3.1. NPH pre-pandemic uninfected controls
CSF samples from ten donors diagnosed with NPH and subject to lumbar puncture to drain fluid excess in 2019 were used as uninfected control.
2.3.2. CSF IL6 and TNFα quantification
Before assays, CSF samples were thawed and kept on ice. TNFα concentration was measured using Cayman TNFα (human) ELISA kit (#589201). Calibrators were diluted in a blocking solution (5% bovine serum albumin, 0.05% Tween-20 in phosphate-buffered saline). Samples were tested undiluted. IL6 levels were measured using a Human IL6 Quantikine ELISA kit (R&D Systems, #D6050). Before analysis, samples were diluted (1:2) in a solution provided by the kit (RD6F). Samples, quality controls, and calibrators were run in duplicates for all targets, following manufacturers’ indications. Standard curves were calculated using a 4-parameter logistic regression model. Low undetermined levels were expressed as 0 pg/mL.
2.3.3. LC-MS/MS shotgun proteomics analysis
COVID-19 and control CSF samples first received a protease inhibitor cocktail (Halt Protease Inhibitor Cocktail, Thermo Scientific, #78430). The CSF samples’ protein amount was quantified using the BCA and diluted 1:1 v/v in buffer (Tris-HCL 100 mM and 2M Urea). Aiming to obtain a higher quality of buffer exchange and protein digestion, we performed the FASP protocol for tryptic digestion (Distler et al., 2016) in 20 μg of protein, briefly described in washing steps to buffer exchange in a microcolumn tip (10 kDa MW cut off), and tryptic digestion performed in the column. Samples were reduced, alkylated, and later digested using trypsin. Digested peptides from each sample were resuspended in 0.1% FA. The separation of tryptic peptides was performed on an ACQUITY MClass System (Waters Corporation). 1 μg of each digested sample was loaded onto a Symmetry C18 5 μm, 180 μm × 20 mm precolumn (Waters Corp.) used as a trapping column and subsequently separated by a 120-min reversed-phase gradient at 300 nL/min (linear-gradient, 3–55% ACN over 90 min) using an HSS T3 C18 1.8 μm, 75 μm × 150 mm nanoscale and LC column (Waters Corp.) maintained at 30 °C. The gradient elution Water-Formic Acid (99.9/0.1, v/v) was used as eluent A and Acetonitrile Formic Acid (99.9/0.1, v/v) as B. The Separated peptides were analyzed by a High Definition Synapt G2-Si Mass spectrometer directly coupled to the chromatographic system. Differential protein expression was evaluated with a data-independent acquisition (DIA) of shotgun proteomics analysis by Expression configuration mode (Mse). The mass spectrometer operated in “Expression Mode”, switching between low (4 eV) and high (25–60 eV) collision energies on the gas cell, using a 1.0s scan time per function over 50–2000 m/z. All spectra have been acquired in Ion Mobility Mode by applying a 1.000 m/s wave velocity for the ion separation and a 175 m/s transfer wave velocity. The processing of low and elevated energy added to the data of the reference lock mass ([Glu1]-Fibrinopeptide B Standard, Waters Corp.) provides a time-aligned inventory of accurate mass retention time components for both the low and elevated-energy (EMRT, exact mass retention time). Each sample was run in three technical replicates.
Continuum LC-MS data from three replicate experiments for each sample was processed for qualitative and quantitative analysis using the software Progenesis (Waters Corp.). The qualitative identification of proteins was obtained by searching the Homo sapiens database (UniProt KB/Swiss-Prot Protein reviewed). Search parameters were set as automatic tolerance for precursor ions and product ions, a minimum of one fragment ions matched per peptide, a minimum of three fragment ions matched per protein, a minimum of one unique peptide matched per protein, 2 missed cleavage, carbamidomethylation of cysteines as fixed modification and oxidation of methionines as variable modifications, FDR of the identification algorithm <1%.
Label-free quantitative analysis was obtained using the relative abundance intensity integrated with Progenesis software, using all peptides identified for normalization. The expression analysis was performed considering technical replicates available for each experimental condition, following the hypothesis that each group is an independent variable. It was considered only proteins present in two out of three technical replicates, and a statistical cut-off of ANOVA >0.05 was adopted. The dataset containing statistically significant deregulated proteins was used for Protein Interaction and pathway-enrichment analysis performed in Cytoscape, and in silico analyses were performed in an R environment. For Heatmap, volcano plot and PCA analysis, it was used a homemade Python language open source software OmicScope (Reis-de-Oliveira et al., 2023). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD033979 and 10.6019/PXD033979.
2.3.4. Multiple reaction monitoring (MRM) for SARS-CoV-2's spike protein
Approximately 500 fmols of digested peptides from a recombinant spike protein produced by Cell Culture Engineering Lab (COPPER/UFRJ)(Alvim et al., 2020) were spiked in tryptic peptides from an E. Coli total protein extract and further loaded onto a Symmetry C18 5 μm, 180 μm × 20 mm precolumn (Waters Corp.) used as trapping column and subsequently separated by a 90 min reversed-phase gradient at 300 nL/min (linear-gradient, 3–55% ACN over 90 min) using an HSS T3 C18 1.8 μm, 75 μm × 150 mm nanoscale and LC column (Waters Corp.) at 40 °C. For the gradient elution, Water-Formic Acid (99.9/0.1, v/v) was used as eluent A and Acetonitrile Formic Acid (99.9/0.1, v/v) as B. The Separated peptides were analyzed by a High Definition Synapt G2-Si Mass spectrometer directly coupled to the chromatographic system.
The generated raw file was imported into the Skyline software. The Spike protein was chosen as a FASTA file as a reference for the theoretical tryptic peptides, isolating the mass of the peptides of interest (parent ions) used for the MS/MS selection (fragment ions) and their retention time to create the MRM method. Four peptides’ transitions (543.2774++; 570.3035++; 609.7987++, and 679.8386++) were chosen, and their respective fragments of the recombinant spike protein considering only peptides with at least three fragment ions and 8–25 amino acids. The Skyline software created the collision energy to be applied for each peptide. The MRM method for searching for the Spike protein was applied to all CSF samples.
2.4. SARS-CoV-2's genome sequencing, assembly, and phylogeny
Five SARS-CoV-2 nasopharyngeal swabs positive samples (collected during hospitalization) with viral genes N1 or N2 amplification Ct < 30 were sequenced. Sequencing libraries were prepared using the QIAseq FX DNA Library Prep kit (QIAGEN, Germany), and reactions were sequenced on the Illumina MiSeq platform (Illumina, USA) with a v3 (600 cycles) cartridge. A custom pipeline for data quality control and consensus genome assembly was used (Moreira et al., 2021). The viral genomes were classified into Pango lineages using the Pangolin tool v2.4.2. The assembly and classification are available in Suppl. Table 6. A dataset (n = 58) containing only lineages identified in Rio de Janeiro (Brazil) between April and September 2020 was created using genomes publicly available on the GISAID EpiCoV database to corroborate the classification (See Suppl. Table 6). The dataset was aligned with minimap18, and a maximum likelihood phylogeny was inferred with IQ-tree v2.0.319 under the GTR + F + I + G4 model (Li, 2018).
2.5. Statistical analysis
Statistical analyses were performed using Prism 9 Software, v 9.0.2 or Matlab R2019b (Mathworks, USA), or IBM SPSS Statistics version 29.0.2.0 (20). Statistical analysis for proteomic and MRM analyses are described in the above sessions. Categorical variables are expressed as frequency and proportion (No, %) and analyzed using Fisher's exact or Chi-squared tests, followed by post-hoc Chi-Squared pairwise comparison. Continuous variables were checked for normal distribution using the D'Agostino & Pearson and Shapiro-Wilk normality tests. As indicated, normally distributed data were expressed as the mean and standard error of the mean (SEM) or standard deviation (SD). Variances were compared using the F test. Groups were compared using a t-test, Welch's t-test (correction for different variances). To identify possible confounders in analyses, we considered previously reported associations between sex, aging, multimorbidity, cardiovascular, diabetes, neurological, and neuropsychiatric comorbidities with the risk for severe COVID-19 and chronic systemic and CNS inflammation (De Felice and Ferreira, 2014; Friedman et al., 2019; Grande et al., 2020; Guan et al., 2020; Williamson et al., 2020; Zeng et al., 2020; Evans et al., 2021; Jun et al., 2021; Lau et al., 2021; Tahira et al., 2021; Taquet et al., 2021c; ten Caten, 2021; De Felice et al., 2022). Thus, group comparisons for cHIS and CSF neuroinflammatory biomarkers (IL6 and TNFα) utilized ANCOVA using sex, age, diabetes, and the numbers of cardiovascular, neurological/psychiatric, and all other comorbidities as covariates. Heteroskedasticity was checked using the White and F tests. The equality of variances was checked using Levene's test. Covariates correlation was checked using Pearson's correlation tests and no high correlation (r > 0.8) was found. Residuals' normal distribution was checked using the Kolmogorov-Smirnov and Shapiro-Wilk tests. Because residuals for ANCOVA analyses using CSF IL6 levels as a dependent variable did not follow a normal distribution, statistical analyses were performed after log transformation (LogIL6). Due to the small sample size, CSF IL6 and TNFα comparisons between neurological diagnostic groups was performed using Kruskal-Wallis followed by Dunn's test. Non-parametric data were expressed as the median and interquartile range (IQR) and analyzed using the Mann-Whitney test. Multiple comparisons were corrected by the FDR method when indicated. The level of statistical significance was set at 5%. Missing or unavailable data were not included in the statistical analysis.
3. Results
3.1. COVID-19 patients presented heterogeneous neurological features
Thirty-five patients from 11 hospitals were sequentially included in this study. Patients' inclusion and exclusion criteria are described in Suppl. Table 1. They were classified according to disease severity considering clinical data (World Health Organization, 2020b). Patients' inclusion schema, main clinical presentation/disease severity, and summary of the available data are presented in Suppl. Fig. 1. For statistical comparisons, deceased patients (n = 5) joined the Severe group. Patients’ ages ranged from 26 to 87 years old (mean 55.6), with 12 females (34.3%) (Table 1). Only one patient did not require an intensive care unit (ICU) hospitalization. Most patients presented at least one comorbidity (29/35) and 23 (65.7%) presented multimorbidity as defined by the presence of two or more chronic diseases (Table 1). Individualized clinical data are shown in Suppl. Table 2. As expected, mild disease patients were younger, presented fewer comorbidities, including cardiovascular diseases and diabetes, and had shorter hospitalization, including fewer days in an ICU (Table 1). Male patients showed a 5.5-fold increased risk for severe disease. Severe patients presented more dyspnea at hospital admission than mild patients. Also, bacterial pneumonia, hypertension, sepsis, and acute kidney failure were associated with disease severity. Severe patients reported less myalgia at admission than mild ones.
Table 1.
Patients' clinical profile and non-neurological symptoms and complications. Categorical variables are expressed as the number of positive cases (No) and percentage (%). Normal distributed continuous variables are expressed as mean and standard deviation (SD). Non-parametric continuous variables are expressed as the median and interquartile range (IQR). *p < 0.05, **p < 0.01, ****p < 0.0001, Fisher's exact test, t-test or Mann-Whitney test. &Comparisons made with ANCOVA using sex, age, diabetes, and numbers of neurological, cardiovascular, and other comorbidities as covariates, F(1,27) = 3.2. (.) Non-applicable. (ACE) angiotensin-converting enzyme, and (ARBs) angiotensin receptors blockers. Multimorbidity is defined by the presence of two or more chronic illnesses.
| All patients | Severe | Mild | Odds ratio (p-value) | Pronounced neuroimaging alterations | Negative or moderate neuroimaging alterations | Odds ratio (p-value) | |
|---|---|---|---|---|---|---|---|
| All participants, n (%) | 35 | 13 (37.1) severe 5 (14.3) dead | 17 (48.6) | . | 10 (28.6) | 12 (34.3) moderate 13 (37.1) negative |
·· |
| Age [years], mean (SD) | 55.6 (17.0) | 61.8 (15.9) | 49.1 (15.9) | (0.024*) | 57.1 (18.7) | 55.0 (16.6) | (0.751) |
| Sex, male (%) | 23 (65.7) | 15 (83.3) | 8 (47.1) | 5.5 (0.035*) | 17 (68.0) | 6 (60.0) | 0.71 (0.706) |
| Clinical profile, No (%) | |||||||
| Number of comorbidities, median (IQR) | 2.00 (1.0–4.0) | 3.00 (2.0–4.0) | 2.0 (0.0–3.0) | (0.045*) | 3.0 (2.0–4.0) | 2.0 (1.0–3.0) | (0.212) |
| Multimorbidity, No (%) | 23 (65.7) | 14 (77.8%) | 9 (52.9%) | 3.1 (0.164) | 9 (90.0) | 14 (56.0) | 7.1 (0.113) |
| Number of cardiovascular diseases, median (IQR) | 0.0 (0.0–1.0) | 1.0 (0.0–2.25) | 0.0 (0.0–0.5) | (0.025*) | 0.5 (0.0–1.25) | 0.0 (0.0–1.0) | (0.872) |
| Hypertension, No (%) | 14 (40.0) | 10 (55.6) | 4 (23.5) | 4.1 (0.086) | 4 (40.0) | 10 (40.0) | 1.0 (1.00) |
| Coronary heart disease, No (%) | 5 (14.3) | 4 (22.2) | 1 (5.9) | 4.6 (0.338) | 1 (10.0) | 4 (16.0) | 0.58 (1.00) |
| Heart failure, No (%) | 2 (5.7) | 1 (5.6) | 1 (5.9) | 0.9 (1.00) | 0 (0.0) | 2 (8.0) | 0.0 (1.00) |
| Congenital heart disease, No (%) | 2 (5.7) | 2 (11.1) | 0 (0) | ∞ (0.486) | 1 (10.0) | 1 (4.0) | 2.67 (1.00) |
| Acute myocardial infarction, No (%) | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Number of metabolic disorders, median (IQR) | 0.0 (0.0–1.0) | 0.0 (0.0–1.0) | 0.0 (0.0–1.0) | (0.546) | 0.5 (0.0–2.0) | 0.0 (0.0–1.0) | (0.339) |
| Obesity, No (%) | 5 (14.3) | 1 (5.6) | 4 (23.5) | 0.2 (0.177) | 1 (10.0) | 4 (16.0) | 0.58 (1.00) |
| Dyslipidemia, No (%) | 3 (8.6) | 1 (5.6) | 2 (11.8) | 0.4 (0.603) | 1 (10.0) | 2 (8.0) | 1.28 (1.00) |
| Diabetes, No (%) | 7 (20.0) | 7 (38.9) | 0 (0.0) | ∞ (0.008**) | 4 (40.0) | 3 (12.0) | 4.89 (0.155) |
| Hypothyroidism, No (%) | 4 (11.4) | 2 (11.1) | 2 (11.8) | 0.9 (1.00) | 2 (20.0) | 2 (8.0) | 2.875 (0.561) |
| Hematologic disease, No (%) | 1 (2.9) | 0 (0) | 1 (5.9) | 0.0 (0.486) | 0 (0.0) | 1 (4.0) | 0.0 (0.706) |
| Malignant neoplasm, No (%) | 4 (11.4) | 3 (16.7) | 1 (5.9) | 3.2 (0.603) | 1 (10.0) | 3 (12.0) | 0.815 (1.00) |
| Chronic pulmonary disease, No (%) | 3 (8.6) | 1 (5.6) | 2 (11.8) | 0.4 (0.603) | 2 (20.0) | 1 (4.0) | 6.0 (0.190) |
| Smoking, No (%) | 2 (5.7) | 0 (0.0) | 2 (11.8) | 0.0 (0.229) | 0 (0.0) | 2 (8.0) | 0.0 (1.00) |
| Chronic renal disease, No (%) | 4 (11.4) | 3 (16.7) | 1 (5.9) | 3.2 (0.603) | 2 (20.0) | 2 (8.0) | 2.875 (0.561) |
| AIDS/HIV, No (%) | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Hepatitis C, No (%) | 1 (2.9) | 0 (0) | 1 (5.9) | 0.0 (0.486) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Other comorbidities, No (%) | 14 (40.0) | 8 (44.4) | 6 (35.3) | 1.5 (0.733) | 3 (30.0) | 11 (44.0) | 0.545 (0.704) |
| Drugs in use at hospital admission, No (%) | |||||||
| Immunosuppressants | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| ACE inhibitors | 3 (8.6) | 3 (16.7) | 0 (0) | ∞ (0.229) | 2 (20.0) | 1 (4.0) | 6.0 (0.190) |
| ARBs | 3 (8.6) | 2 (11.1) | 1 (5.9) | 2.0 (1.00) | 0 (0.0) | 3 (12.) | 0.0 (0.542) |
| Anticoagulants | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 1 (10.0) | 0 (0.0) | ∞ (0.286) |
| Corticosteroids | 2 (5.7) | 1 (5.6) | 1 (5.9) | 0.09 (1.00) | 1 (10.0) | 1 (4.0) | 2.67 (1.0) |
| Clinical signs and symptoms at hospital admission, No (%) | |||||||
| Fever, >37.5 °C | 22 (62.9) | 11 (61.1) | 11 (64.7) | 0.9 (1.00) | 4 (40.0) | 18 (72.0) | 0.26 (0.123) |
| Cough | 20 (57.1) | 10 (55.6) | 10 (58.8) | 0.9 (1.00) | 3 (30.0) | 17 (68.0) | 0.20 (0.062) |
| Sore throat | 4 (11.4) | 2 (11.1) | 2 (11.8) | 0.9 (1.00) | 0 (0.0) | 4 (16.0) | 0.0 (0.303) |
| Rhinorrhea | 2 (5.7) | 0 (0) | 2 (11.8) | 0.0 (0.229) | 0 (0.0) | 2 (8.0) | 0.0 (1.0) |
| Dyspnea | 17 (48.6) | 12 (66.7) | 5 (29.4) | 4.8 (0.044*) | 3 (30.0) | 14 (56.0) | 0.34 (0.264) |
| Rash | 1 (2.9) | 0 (0) | 1 (5.9) | 0.0 (0.486) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Hypertension | 2 (5.7) | 1 (5.6) | 1 (5.9) | 0.9 (1.00) | 0 (0.0) | 2 (8.0) | 0.0 (1.0) |
| Myalgia | 11 (31.4) | 1 (5.6) | 10 (58.8) | 0.04 (0.001**) | 0 (0.0) | 11 (44%) | 0.0 (0.015*) |
| Arthralgia | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Diarrhoea | 4 (11.4) | 2 (11.1) | 2 (11.8) | 0.9 (1.00) | 2 (20.0) | 2 (8.0) | 2.875 (0.561) |
| Hospitalization | |||||||
| Duration [days], median (IQR) | 17 (7–53) | 46.5 (28.25–72.5) | 7.0 (3–11.5) | (<0.001****) | 33.5 (17–58) | 12.0 (6.5–37) | (0.212) |
| ICU stay [days], median (IQR) | 11 (5–36) | 36.0 (23–64) | 6.0 (2.5–10.75) | (<0.001***) | 36.0 (10–60) | 9.0 (4–27.75) | (0.086) |
| Main in-hospital non-neurological symptoms and complications, No (%) | |||||||
| Fever | 3 (8.6) | 2 (11.1) | 1 (5.9) | 2.0 (1.00) | 0 (0.0) | 3 (12.) | 0.0 (0.542) |
| Sinusitis | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Bacterial pneumonia | 6 (17.1) | 6 (33.3) | 0 (0) | ∞ (0.019*) | 2 (20.0) | 4 (16.0) | 1.31 (1.00) |
| Acute severe respiratory distress syndrome | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Tracheobronchitis | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 1 (10.0) | 0 (0.0) | ∞ (0.286) |
| Dyspnea | 3 (8.6) | 2 (11.1) | 1 (5.9) | 2.0 (1.00) | 0 (0.0) | 3 (12.) | 0.0 (0.542) |
| Pulmonary fibrosis | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Pulmonary thromboembolism | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Hemoptysis | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 1 (10.0) | 0 (0.0) | ∞ (0.286) |
| Pleural effusion | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 1 (10.0) | 0 (0.0) | ∞ (0.286) |
| Hypotension | 2 (5.7) | 2 (11.1) | 0 (0) | ∞ (0.486) | 1 (10.0) | 1 (4.0) | 2.67 (1.0) |
| Hypertension | 5 (14.3) | 5 (27.8) | 0 (0) | ∞ (0.045*) | 3 (30.0) | 2 (8.0) | 4.93 (0.128) |
| Tachycardia | 3 (8.6) | 3 (16.7) | 0 (0) | ∞ (0.229) | 1 (10.0) | 2 (8.0) | 1.28 (1.00) |
| Ventricular arrhythmia | 3 (8.6) | 3 (16.7) | 0 (0) | ∞ (0.229) | 3 (30.0) | 0 (0.0) | ∞ (0.018*) |
| Atrial fibrillation | 2 (5.7) | 2 (11.1) | 0 (0) | ∞ (0.486) | 1 (10.0) | 1 (4.0) | 2.67 (1.0) |
| Mitral valve regurgitation | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 1 (10.0) | 0 (0.0) | ∞ (0.286) |
| Cardiac arrest | 2 (5.7) | 2 (11.1) | 0 (0) | ∞ (0.486) | 1 (10.0) | 1 (4.0) | 2.67 (1.0) |
| Cardiogenic shock | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 1 (10.0) | 0 (0.0) | ∞ (0.286) |
| Bleeding/Haemorrhage | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Venous thrombosis | 4 (11.4) | 3 (16.7) | 1 (5.9) | 3.2 (0.603) | 1 (10.0) | 3 (12.) | 0.815 (1.00) |
| Disseminated intravascular coagulation | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Hemodynamic instability | 3 (8.6) | 3 (16.7) | 0 (0) | ∞ (0.229) | 2 (20.0) | 1 (4.0) | 6.0 (0.190) |
| Severe anaemia | 3 (8.6) | 3 (16.7) | 0 (0) | ∞ (0.229) | 1 (10.0) | 2 (8.0) | 1.28 (1.00) |
| Blood transfusion | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Bacteremia | 4 (11.4) | 3 (16.7) | 1 (5.9) | 3.2 (0.603) | 1 (10.0) | 3 (12.0) | 0.815 (1.00) |
| Sepsis/septic shock | 9 (25.7) | 8 (44.4) | 1 (5.9) | 12.8 (0.018*) | 4 (40.0) | 5 (20.0) | 2.67 (0.393) |
| Hepatitis | 1 (2.0) | 0 (0) | 1 (5.9) | 0.0 (0.486) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Hepatic dysfunction | 2 (5.7) | 1 (5.6) | 1 (5.9) | 0.9 (1.00) | 1 (10.0) | 1 (4.0) | 2.67 (1.0) |
| Acute kidney failure | 6 (17.1) | 6 (33.3) | 0 (0) | ∞ (0.019*) | 2 (20.0) | 4 (16.0) | 1.31 (1.00) |
| Urinary retention | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 1 (10.0) | 0 (0.0) | ∞ (0.286) |
| Pyelonephritis/pyeloureteritis | 2 (5.7) | 2 (11.1) | 0 (0) | ∞ (0.486) | 1 (10.0) | 1 (4.0) | 2.67 (1.00) |
| Hyperglycemia | 3 (8.6) | 3 (16.7) | 0 (0) | ∞ (0.229) | 1 (10.0) | 2 (8.0) | 1.28 (1.00) |
| Metabolic acidosis | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Acute diarrhoea | 2 (5.7) | 2 (11.1) | 0 (0) | ∞ (0.486) | 0 (0.0) | 2 (8.0) | 0.0 (1.00) |
| Acute colonic pseudo-obstruction | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 1 (10.0) | 0 (0.0) | ∞ (0.286) |
| Gastrointestinal bleeding | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 1 (10.0) | 0 (0.0) | ∞ (0.286) |
| Rhabdomyolysis | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 1 (10.0) | 0 (0.0) | ∞ (0.286) |
| Facial hyperemia/oedema | 2 (5.7) | 1 (5.6) | 1 (5.9) | 0.9 (1.00) | 0 (0.0) | 2 (8.0) | 0.0 (1.00) |
| Pharmacodermia | 2 (5.7) | 1 (5.6) | 1 (5.9) | 0.9 (1.00) | 0 (0.0) | 2 (8.0) | 0.0 (1.00) |
| Drug-induced hypersensitivity syndrome | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 0 (0.0) | 1 (4.0) | 0.0 (1.00) |
| Drugs during hospitalization, number of patients in use (%) | |||||||
| Immunoglobulins | 2 (5.7) | 0 (0) | 2 (11.8) | 0.0 (0.229) | 0 (0.0) | 2 (8.0) | 0.0 (1.00) |
| Corticosteroids | 18 (51.4) | 11 (61.1) | 7 (41.2) | 2.2 (0.318) | 6 (60.0) | 12 (48.0) | 1.625 (0.711) |
| Antibiotics | 28 (80.0) | 11 (61.1) | 12 (70.6) | 3.3 (0.228) | 8 (80.0) | 20 (80.0) | 1.00 (1.00) |
| Anticoagulants | 24 (68.6) | 13 (72.2) | 11 (64.7) | 1.4 (0.725) | 7 (70.0) | 17 (68.0) | 1.10 (1.00) |
| Antivirals | 10 (28.6) | 8 (44.4) | 2 (11.8) | 6.00 (0.060) | 4 (40.0) | 6 (24.0) | 2.1 (0.421) |
| Antipsychotics | 2 (5.7) | 1 (5.6) | 1 (5.9) | 0.9 (1.00) | 0 (0.0) | 2 (8.0) | 0.0 (1.00) |
| Antidepressants | 1 (2.9) | 1 (5.6) | 0 (0) | ∞ (1.00) | 1 (10.0) | 0 (0.0) | ∞ (0.286) |
| COVID-19-associated Hyperinflammatory Syndrome (cHIS), adjusted means (No ≥ 2, %) | 2.6 (23, 65.7) | 3.2 (15, 88.9) | 2.0 (8, 47.06) | 5.6 (0.035* and 0.085&) | 2.7 (7, 70.0) | 2.6 (16, 64.0) | 1.3 (1.00 and 0.88&) |
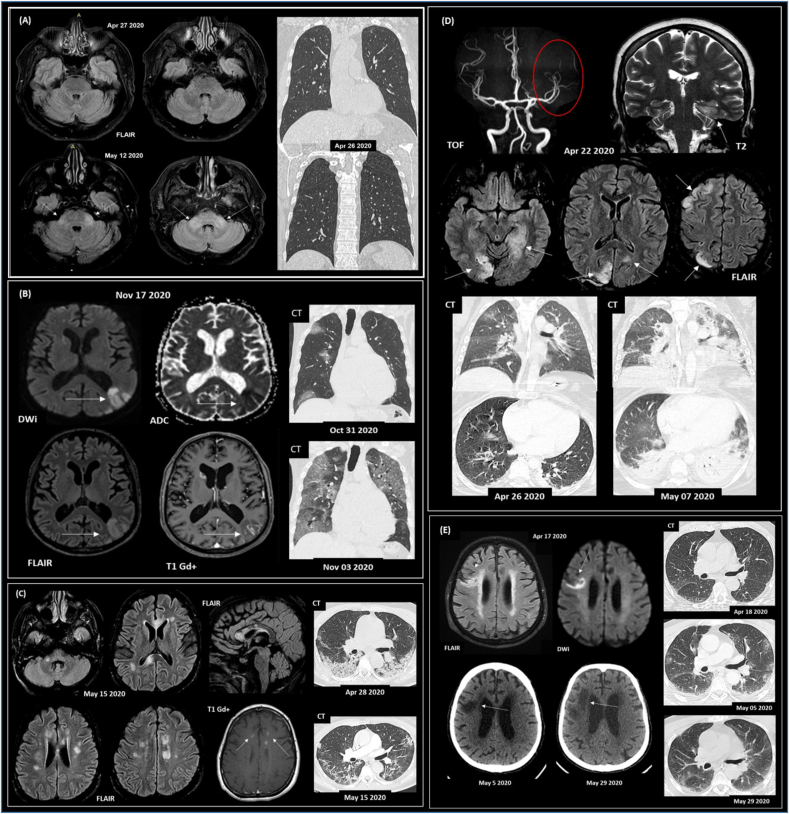
All patients had at least one brain CT and/or MRI exam (performed at median one day, IQR 0–4 days apart from lumbar puncture). Twelve (34.3%) patients only presented signs of intracranial hypertension (moderate neuroimaging finding). Ten (28.6%) patients showed focal or diffuse CT or MRI changes associated with COVID-19 and were assigned as a pronounced group. Representative brain and chest imaging from pronounced cases are shown in Fig. 1, and a radiological description for each patient is provided (Suppl. Table 2). The main radiological findings were related to demyelinating lesions, encephalitis, and stroke.
Fig. 1.
Representative paired neuroimaging and chest CT findings from COVID-19 patients presenting neurological symptoms. (A) Subject #16 (65 y/o; male) with encephalitis and a positive result for SARS-CoV-2 RT-qPCR in the CSF screening. Initial symptoms of fever and vomiting, evolving with drowsiness, ataxia, acute diplopia and convergent strabismus. No signs of viral pneumonia were detected on the CT scan. Case previously reported (Freitas et al., 2021). (B) Subject #25 (83 y/o; male; deceased) with acute CNS ischemic lesions (vasculitis); viral pneumonia, fever, respiratory distress, and rhabdomyolysis. Coronal chest CT images (lung window) show multiple areas of round ground-glass opacities, mainly on the right lung, with increasing extent after three days. (C) Subject #14 (50 y/o; female) presented acute demyelinating brain lesions and COVID-19 pneumonia. Axial CT images show areas of consolidation distributed predominantly at peripheral and posterior areas of the lower lobes and improvement in lung infiltrates after 17 days. (D) Subject #27 (29 y/o; female; deceased) with a previous diagnosis of Systemic Lupus Erythematosus, presenting CNS vasculitis; refractory epileptic status; and viral pneumonia. Evolution to progressive global systemic failure. Chest CT shows mild round ground-glass opacity on the middle lobe and upper right lung. Eleven days later, abnormalities increased with diffuse consolidation, mainly in the lower lobes. (E) Subject #23 (78 y/o; male; deceased) presented a subacute ischemic stroke with an altered level of consciousness, seizures, and coma; viral pneumonia. Axial CT images show no lung parenchymal findings consistent with viral pneumonia that progresses to mild patchy ground-glass opacity in the lungs' periphery 17 days after. Lung abnormalities are almost entirely resolved after 41 days. Subjects (B–E) with positive RT-qPCR for SARS-CoV-2 in the nasopharyngeal swab. Abbreviations: FLAIR, DWi, ADC, T1, T1 Gd+, T2 refer to MRI sequences, and Gd + represents venous contrast. Time of flight (TOF) arterial angiographic MRI sequence.
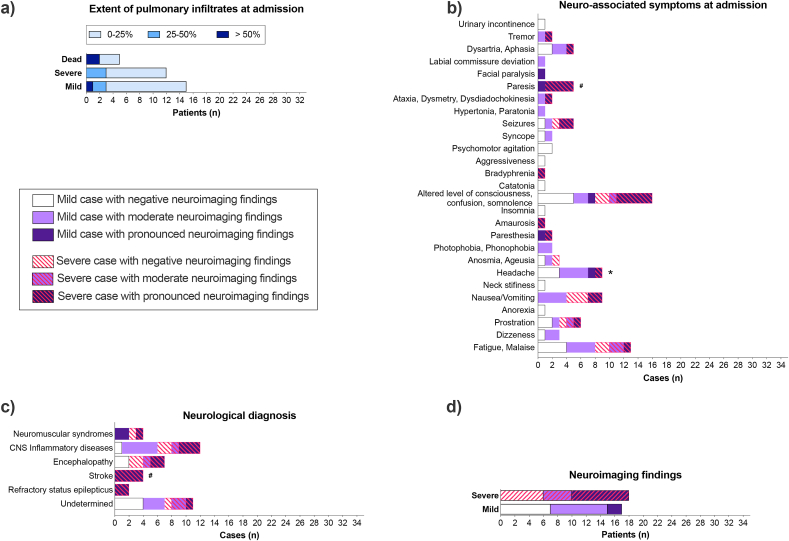
While most patients did not present extensive pulmonary infiltrates in CT scans at hospital admission (75% of patients with less than 25% of pulmonary infiltrates' extent, only 9.4% of patients with more than 50%, Fig. 2a), 85.7% (n = 30) of the patients presented neurological symptoms early at this point (Fig. 2b). Headache was most prevalent in mild diseased patients at admission and hospitalization (Fig. 2b; Suppl. Fig. 2d). A neurological diagnosis was determined in 68.6 % of patients (Fig. 2c). Seven patients (20%) presented neurological or psychiatric comorbidity (See Suppl. Fig. 2a). Severe patients showed a tendency (Odds ratio 6.0, p = 0.0599, Fisher's exact test) to have a higher prevalence of pronounced neuroimaging changes than mild ones (Fig. 2d). Among reported symptoms, pronounced neuroimaging abnormalities were associated with paresis (Fig. 2b). Stroke events (Fig. 2d) and ventricular arrythmia (Table 1) were associated with pronounced neuroimaging findings. Further, patients with neurological/neuropsychiatric comorbidities were at risk of developing pronounced neuroimaging alterations associated with COVID-19 (Suppl. Fig. 2a).
Fig. 2.
COVID-19 induces neurological disturbances despite the low extent of pulmonary infiltrates. (A) Pulmonary infiltrates extent in CT scans at hospital admission (n = 33). (B-D) Patients were grouped as presenting negative (white bars), moderate (light purple) or pronounced (dark purple) neuroimaging (CT/MRI) findings related to COVID-19 hospitalization (n = 35). Dashed lines represent severe disease or death. CNS inflammatory diseases include meningitis, encephalitis, meningoencephalitis, encephalomyelitis, myelitis, CNS vasculitis, and acute disseminated encephalomyelitis (ADEM). Neuromuscular syndromes include Guillain-Barré Syndrome (GBS), cranial neuropathy, peripheral facial paralysis, and myopathy. See Suppl. Table 2 for individualized data. *p < 0.017, disease severity as an outcome, Fisher's exact test; #p < 0.017, neuroimaging positivity as an outcome, Chi-Square test, followed by pairwise comparison with Bonferroni correction. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Patients had a CSF laboratory investigation (performed at 14 days [median], interquartile range [IQR] 8–25 days apart from the COVID-19 initial symptoms; median 3 days, IQR 0.5–16 days after hospital admission). Clinical indication for lumbar punction procedure and routine CSF laboratory findings did not differ between COVID-19 severe and mild patients (Suppl. Figs. 2b and c). Only three patients presented CSF white blood cell (WBC) counts above five per mm3 (ranging from 9 to 65 cells/mm3). Increased CSF opening pressures were observed in 13 patients (ranging from 21 to 35 cmH2O), protein levels were high in twelve patients (ranging from 41 to 100 mg/dL), and glucose was increased in seven patients (from 81 to 144 mg/dL), see Suppl. Table 3. The microbiological screening was negative in all samples. One patient (#16) had SARS-CoV-2 mRNA and oligoclonal bands detected in the CSF. This patient was diagnosed with meningoencephalitis, and his case is also described elsewhere (Freitas et al., 2021).
We sequenced the SARS-CoV-2 genome found on the nasopharyngeal swab of four patients. Phylogeny analysis revealed the presence of the viral lineage B.1.33 (Suppl. Fig. 3, Suppl. Tables 4 and 5). To investigate the presence of SARS-CoV-2 spike protein in the CNS of COVID-19 patients presenting neurological symptoms, we performed a multiple reaction monitoring (MRM) analysis in a representative subset of COVID-19 patients’ CSF (n = 16, eight patients with mild disease, six with severe illness, and two deceased; four patients with pronounced, seven with moderate, and five with no neuroimaging alterations). First, we used tryptic peptides from a recombinant spike protein to construct the MRM method and validate the ion transitions (MS1 and MS2 level) and the retention time of the desired peptides (Suppl. Fig. 4a). Once the MRM method was established, we applied the technique to COVID-19 and 10 pre-pandemic uninfected control CSF samples (samples from normal pressure hydrocephalus (NPH) patients collected in 2019, before COVID-19 was detected in Brazil). As a result, we found no evidence of spike protein in the tested samples (Suppl. Fig. 4b). Comparisons between COVID-19 and NPH CSF samples' white blood cell count and protein, glucose, and lactate levels are presented in Suppl. Table 3.
3.2. Systemic and CNS hyper-inflammation are associated with COVID-19 neurological symptoms and neuroimaging alterations
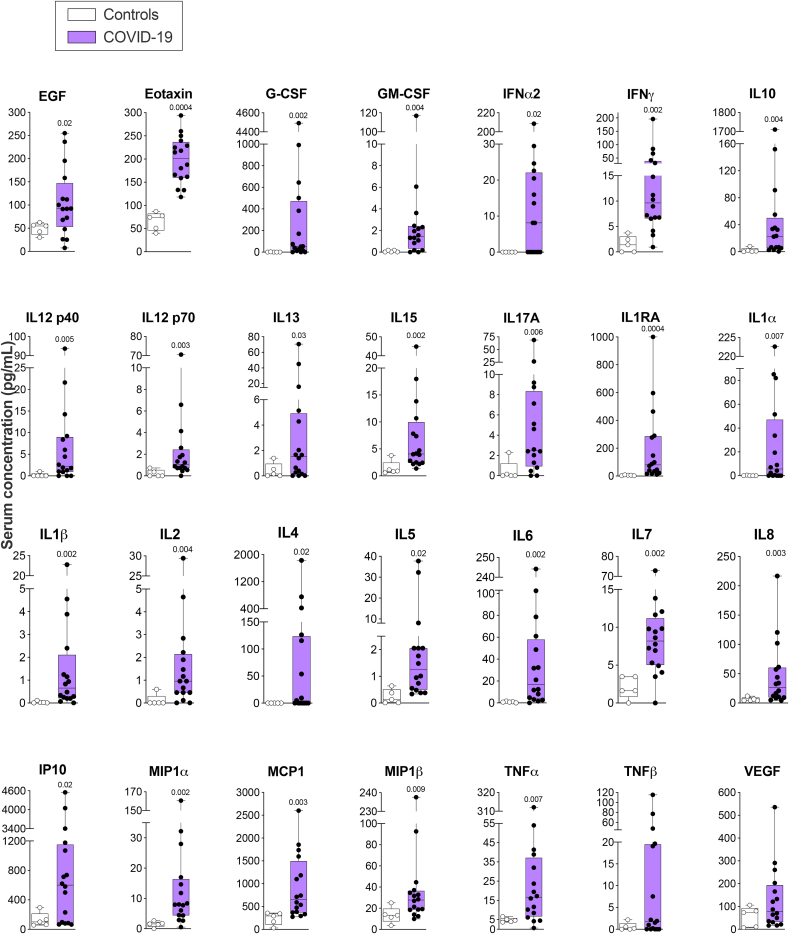
Clinical and blood laboratory data analysis revealed signs of COVID-19 hyper-inflammation (cHIS ≥2) in 23 (66%) patients proximal to CSF. The frequency of patients presenting hyper-inflammation (cHIS ≥2) was higher in the severe group (Table 1). After controlling for age, sex, and comorbidities, we found a tendency for a relationship between higher cHIS score and COVID-19 severity (Table 1). We also measured a panel of 28 cytokines from 16 patients (collected at 0 [median], IQR 0–5.5, days from lumbar puncture) and five healthy donors' serum. Patients’ cytokine analysis was restricted to sample availability. COVID-19 neurological patients displayed ubiquitous increases in circulating cytokine levels compared to controls (Fig. 3).
Fig. 3.
COVID-19 patients with neurological symptoms present modified systemic inflammatory biomarkers. Data are shown as the median, interquartile range (box), and min-max points (whiskers). Symbols indicate individual patients (n = 5 control, 16 COVID-19). Mann-Whitney test corrected for False Discovery Rate (FDR) using the two-stage step-up (Benjamini, Krieger, and Yekutieli) method. Discoveries are signalized by the above q-values (FDR-adjusted p-values).
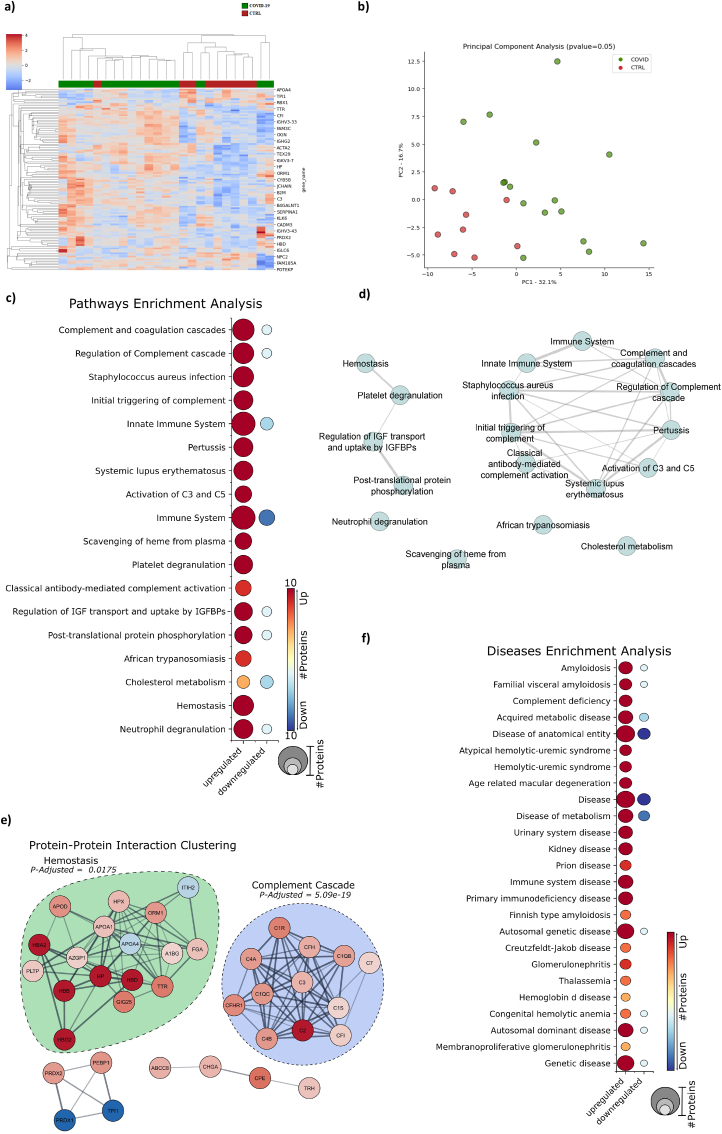
Turning to the central nervous system, we performed large-scale mass-spectrometry-based shotgun proteomics in a subset of COVID-19 patients’ CSF compared to uninfected controls. Due to restrictions on sample availability, the same subgroup analyzed by MRM was used for proteomics analyses. As a result, we found that COVID-19 induced an altered CSF proteomic pattern compared to NPH, presenting 116 statistically significantly deregulated proteins. Hierarchical clustering analysis, considering the protein expression levels of each patient for similarity, showed a tendency to group COVID-19 patients (Fig. 4a). However, it was not sufficient to completely distinguish them from the control group. PCA analysis was also performed to confirm the trend among COVID-19 patients, making it possible to distinguish the groups (Fig. 4b and Suppl. Fig. 5b). Nonetheless, two samples still overlapped (Fig. 4a and b).
Fig. 4.
COVID-19 patients with neurological symptoms display cerebrospinal fluid (CSF)-altered proteomic patterns related to inflammation, innate immunity, and hemostasis. (A) Hierarchical clustering analysis from uninfected normal pressure hydrocephalus (NPH) controls (CTRL, n = 10) and COVID-19 (n = 16) CSF using the measured relative abundance of the proteins. (B) Enrichment analysis using the statistically significant COVID-19 dysregulated proteins searched against a public databank to obtain up and downregulated proteins matched with biological pathways. (C) Disease enrichment analysis of COVID-19 statistically significant dysregulated proteins. (D) The enriched pathways' interaction evidences the connection between the pathways modulated by COVID-19. (E) Protein-Protein interaction clustering analysis of the COVID-19 statistically significant dysregulated proteins.
Pathway enrichment analysis showed that COVID-19 modulated proteins related to the complement and coagulation cascade, immune response, and metabolic processes (Fig. 4c). Altered proteins clustered in two main pathway groups: innate immune system/complement and hemostasis/clotting cascade deregulation (Fig. 4d). Protein-protein clustering interaction analysis further highlighted that part of the dysregulated proteins clustered to homeostasis and complement cascade response (Fig. 4e). Alteration of proteins significantly overlapped with amyloidosis, inflammatory response, metabolic diseases, and hemolytic diseases (Fig. 4f). Gene ontology (GO) enrichment analysis supported dysregulations at vesicle transport and secretion, oxygen and cholesterol molecular functioning, as well as innate and adaptive immune responses (Suppl. Fig. 5c). We have confronted our dataset of the COVID-19 patients’ CSF dysregulated proteins against a databank with tissue protein markers showing a high enrichment with protein markers of CSF and CNS tissue (Suppl. Fig. 5d), increasing the reliability of our data. Finally, we found only a few proteins significantly deregulated when comparing CSF proteomic data from mild and severe cases or cases presenting negative, moderate, or pronounced neuroimaging findings. These sub-groups were not distinguished within the COVID-19 patients.
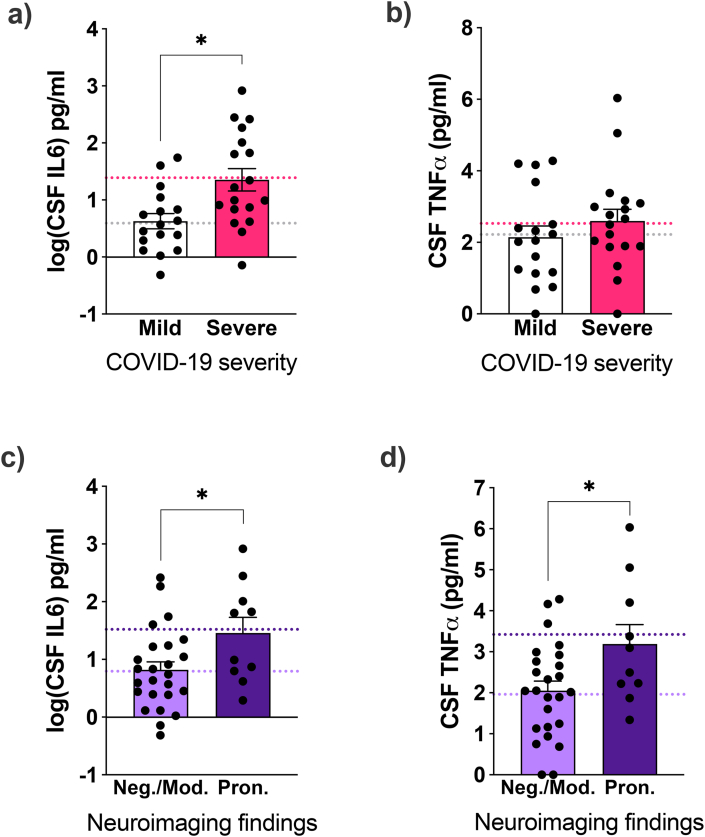
Subsequentially, we measured two key pro-inflammatory cytokine levels (IL6 and TNFα) in COVID-19 patients’ CSF. COVID-19 patients presented higher levels of both cytokines than uninfected controls (Suppl. Fig. 6 a, b). However, we did not detect differences when comparing patients by their neurological diagnosis (Suppl. Fig. 6 c, d). After accounting for the possible confounding effects of age, sex, and comorbidities, CSF IL6 was higher in severe compared to mild cases (Fig. 5a and b), while both CSF IL6 and TNFα were elevated in cases with pronounced neuroimaging findings (Fig. 5c and d).
Fig. 5.
COVID-19 severity and neuroimaging abnormalities are linked to cerebrospinal fluid (CSF) neuroinflammatory biomarkers in COVID-19 patients presenting neurological symptoms. COVID-19 patients were grouped by (a, b) disease severity or by (c, d) presenting negative or moderate (Neg./Mod.) or pronounced (Pron.) neuroimaging alterations. Data are shown as mean and SEM. Symbols indicate individual patients. *p < 0.05, ANCOVA using sex, age, diabetes, and numbers of neurological, cardiovascular, and other comorbidities as covariates. (a) F(1,27) = 6.3, p = 0.019; (b) F(1,27) = 0.25, p = 0.624; (c) F(1,27) = 4.4, p = 0.046; (d) F(1,27) = 5.9, p = 0.022. Dashed lines indicate model-adjusted means.
4. Discussion
Our results collectively link systemic and CNS inflammation to neurological alterations in hospitalized COVID-19 patients. In accordance with previous studies, we observed that COVID-19 induces heterogeneous neurological illnesses and brain structural changes. However, heterogeneous cases showed a homogeneous pattern of CSF proteomic alterations. Despite a possible basal inflammation in NPH uninfected controls (Lolansen et al., 2021) that could underestimate our results, our proteomic analysis revealed broad proteomic changes related to neuroinflammation, innate immunity activation, and response to infections in COVID-19 patients. Innate immunity activation in the CNS results in altered glial and blood-brain barrier functioning, circulating immune cell recruitment, and the release of cytokines (Klein et al., 2017). Those changes alter neurotransmitter release, cerebral metabolism, oxygen consumption, and blood flow, resulting in acute behavioral changes and neurological symptoms (Klein et al., 2017). Additionally, maladaptive immune response may induce synapse and tissue damage (Vasek et al., 2016; Figueiredo et al., 2019; Garber et al., 2019), and chronic neuroinflammation has been associated with psychiatric and neurodegenerative diseases (Frost et al., 2019; De Sousa et al., 2021; De Felice et al., 2022; Lyra e Silva et al., 2022). We found a cluster of proteins related to complement cascade activated in COVID-19 patients' CSF. In the adult CNS, complement activation and synapse tagging trigger synaptic elimination by microglia in animal models of neuro-infectious diseases (Vasek et al., 2016; Figueiredo et al., 2019). Accordingly, research on COVID-19's post-mortem brain samples supports pathologic microglia and complement involvement (Thakur et al., 2021; Yang et al., 2021; Lee MH et al., 2022). Additionally, we found CNS proteomic alterations related to hemostasis that could be behind the frequently observed vascular, hypoxic, and ischemic pathology in severe post-mortem brains (Thakur et al., 2021).
Hyper-inflammation is overt in COVID-19 patients and has been previously linked to COVID-19 severity and mortality (Webb et al., 2020; Zeng et al., 2020). Our cohort included only neurological cases, and we found that most patients, irrespectively of neuroimaging findings and particularly the severe cases, presented evidence of systemic hyper-inflammation and hypercytokinemia at the time of CSF sampling. In accordance with previous studies (Espíndola et al., 2021; Guasp et al., 2022), TNFα and IL6 were found to be high in COVID-19 patients' CSF and blood. Moreover, CSF IL6 was higher in severe cases, and both CSF IL6 and TNFα were upregulated in cases with pronounced neuroimaging alterations. These findings link CNS inflammation to systemic inflammation in COVID-19 patients with neurological symptoms, supporting the hypothesis that systemic inflammation associated with severe disease and circulating cytokines could drive COVID-19 neurological disease (Klein et al., 2017; Lyra e Silva et al., 2022). Our study further contributes to the identification of key biomarkers associated with COVID-19-induced CNS disturbance and neuroimaging alterations that could be incorporated as additional analysis in situations where CSF examination is already clinically required and contribute to therapeutic protocol elaboration. This knowledge could also base the development of blood-based biomarkers, possibly enabling monitoring of patients’ response to therapeutics even before symptoms improve. We have recently established that brain TNFα upregulation triggers microglial-mediated synaptic engulfing in a mouse model of CNS viral infection leading to memory impairment (Figueiredo et al., 2019). In a COVID-19 longitudinal cohort, we showed that acute and persistent blood TNFα is associated with persistent neuroimaging and memory alterations up to one-year post-COVID (Sudo et al., 2024). Infliximab treatment, an anti-TNFα monoclonal antibody, alleviated memory deficits and synapse pathology in infected mice. Both IL6 and TNFα blocking treatments are trialed for COVID-19 treatment (Angriman et al., 2021; World Health Organization, 2021). It would be beneficial to evaluate if these treated patients develop fewer neurological symptoms. It is also important to consider that persistent chronic inflammation is associated with neurodegenerative and neuropsychiatric diseases onset (De Felice et al., 2020; Lyra e Silva et al., 2022). As biomarker research evolves to enable minimally invasive measurements of neuroinflammation in the blood, this could be translated to long-term patient monitoring, which is particularly important for those at risk of developing persistent or long-term negative neurological outcomes.
We hypothesized that viral strain characteristics could be behind the development of important neurological symptoms in COVID-19. However, SARS-CoV-2 sequencing of four neurological cases revealed the B.1.33 lineage, the same lineage most COVID-19 patients in Brazil were infected with during the study's period (Candido et al., 2020). It would be essential to understand how our results replicate in larger cohorts, considering post-vaccination and the onset of other viral strains, such as omicron. We also hypothesized that direct virus infection or the presence of viral proteins could be driving neuropathology in COVID-19 patients (Rhea et al., 2020; Lyra e Silva et al., 2022). One out of fourteen CSF samples tested positive for SARS-CoV-2 mRNA. Indeed, SARS-CoV-2 genetic material is rarely detected in patients' CSF (Espíndola et al., 2020). Additionally, we found no traces of Spike protein in patients' CSF. However, SARS-CoV-2 material has been previously reported in patients' post-mortem samples. Therefore, we cannot exclude the possibility of brain parenchyma infection.
Survivors of critical illness and respiratory distress syndrome frequently experience neurological disturbance and long-lasting cognitive decline (Sasannejad et al., 2019). Accordingly, we found that severe COVID-19 patients presented more pronounced systemic inflammation and neuroimaging alterations. Our study design does not differentiate the neuropathological contributions of infection specifically by SARS-CoV-2 from any-cause hypoxia and critical illness. However, most patients in our cohort did not present extensive pulmonary damage in CT scans at hospital admission while presenting a neurological symptom as the primary clinical presentation. Others also report COVID-19 neurological cases in which neurological complaints appeared before respiratory symptoms (Siahaan et al., 2022).
Finally, our study's strengths include the multicentric design and the collection of in-depth supportive clinical, neurological, neuroimaging, and laboratory data allied to a broad-scoped CSF proteomic and targeted biomarker analysis. However, the retrospective and cross-sectional design, the lack of neurological evaluation for COVID-19 patients before infection or at a follow-up, and the low sample size limit our conclusions. Thus, it will be essential to see how our findings replicate in larger cohorts using blood-born biomarkers. Additionally, future longitudinal cohort studies allied with pre-clinical investigation will be important to provide evidence for the causality of associations herein reported.
Funding
This work was supported by the Brazilian Ministry of Health and the Brazilian National Council for Scientific and Technological Development (CNPq), the Carlos Chagas Filho Research Support Foundation (FAPERJ), the São Paulo Research Foundation (FAPESP), the Rede Corona-ômica BR MCTI/FINEP affiliated to RedeVírus/MCTI, the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (MEC/CAPES) and the D’Or Institute for Research and Education (IDOR). FGQBA and TPP received stipends from CNPq and FAPERJ.
The study's sponsors had no role in the study design, in the collection, analysis, or interpretation of data, in the writing of the manuscript, or in the decision to submit the paper for publication. The corresponding authors had full access to all the data in the study and had the final responsibility for submitting it for publication.
CRediT authorship contribution statement
Fernanda G.Q. Barros-Aragão: Writing – review & editing, Writing – original draft, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Talita P. Pinto: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation. Victor C. Carregari: Writing – review & editing, Validation, Methodology, Investigation, Formal analysis. Nathane B.S. Rezende: Writing – review & editing, Methodology, Investigation, Formal analysis, Data curation. Thaís L. Pinheiro: Writing – review & editing, Investigation, Formal analysis, Data curation. Guilherme Reis-de-Oliveira: Writing – review & editing, Investigation, Formal analysis. Mauro J. Cabral-Castro: Writing – review & editing, Investigation, Formal analysis. Daniel C. Queiroz: Investigation, Formal analysis, Data curation. Paula L.C. Fonseca: Writing – review & editing, Investigation, Formal analysis, Data curation. Alessandro L. Gonçalves: Investigation, Formal analysis. Gabriel R. de Freitas: Investigation, Formal analysis, Data curation, Conceptualization. Felipe K. Sudo: Conceptualization. Paulo Mattos: Conceptualization. Fernando A. Bozza: Conceptualization. Renato S. Aguiar: Writing – review & editing, Methodology, Investigation, Formal analysis, Data curation, Conceptualization, Methodology, Investigation, Formal analysis. Rosana S. Rodrigues: Writing – review & editing, Methodology, Investigation, Formal analysis, Data curation. Carlos O. Brandão: Writing – review & editing, Methodology, Investigation, Conceptualization. Andrea S. Souza: Writing – review & editing, Methodology, Investigation, Formal analysis, Data curation. Fernanda G. De Felice: Writing – review & editing, Methodology, Investigation, Formal analysis, Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Conceptualization. Fernanda Tovar-Moll: Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
none.
Acknowledgements
We thank Dr Castilho's research group for kindly providing us with the SARS-CoV-2's recombinant spike protein. We would also like to thank BSc Daniele Freitas and BSc Fabiane Ferreira Nascimento for support in clinical data research, Dr. Ana Paula Sampaio and Dr. Ananssa Silva for laboratory managing support, and B.Sc. Jhonata Sousa for technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbih.2024.100805.
Contributor Information
Fernanda G. De Felice, Email: fernanda.defelice@queensu.ca.
Fernanda Tovar-Moll, Email: fernanda.tovarmoll@idor.org.
List of abbreviations
- CCP
clinic characterization protocol
- cHIS
COVID-19 hyperinflammatory syndrome score
- CNS
central nervous system
- CONEP
Brazilian National Commission for Research Ethics
- IDOR
D’Or Institute of Research and Education
- CRF
clinical research form
- CSF
cerebrospinal fluid
- DIA
data-independent acquisition
- GO
Gene ontology
- ICU
intensive care unit
- IH
intracranial hypertension
- IL6
interleukin-6
- IQR
interquartile range
- ISARIC
International Severe Acute Respiratory and Emerging Infection Consortium
- CT
Computerized tomography
- MRI
Magnetic resonance imaging
- NPH
normal pressure hydrocephalus
- RBC
red blood cell
- RT-qPCR
reverse transcriptase-quantitative polymerase chain reaction
- SD
standard deviation
- SEM
standard error of the mean
- SRM
selective reaction monitoring
- WBC
white blood cells
- WHO
World Health Organization
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
We have shared anonymized individualized patient data at Table 1. The mass spectrometry proteomics data deposited at ProteomeXchange Consortium, PRIDE partner repository (PXD033979 10.6019/PXD033979)
References
- Alvim R.G., Lima T.M., Rodrigues D.A.S., Marsili F.F., Bozza V.B.T., Higa L.M., Monteiro F.L., Leitão I.C., Carvalho R.S., Galliez R.M., Castineiras T.M.P.P., Nobrega A., Travassos L.H., Ferreira Jr OC., Tanuri A., Vale A.M., Castilho L.R. An affordable anti-SARS-COV-2 spike protein ELISA test for early detection of IgG seroconversion suited for large-scale surveillance studies in low-income countries. medRxiv. 2020 doi: 10.1101/2020.07.13.20152884. [DOI] [Google Scholar]
- Angriman F., Ferreyo B., Burry L., Fan E. Interleukin-6 receptor blockade in patients with COVID-19: placing clinical trials into context. Lancet Respir. Med. 2021;9:655–664. doi: 10.1016/S2213-2600(21)00139-9. https://pubmed.ncbi.nlm.nih.gov/33930329/ Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton M.D., Johnson R.W. Interleukin-6 trans-signaling in the senescent mouse brain is involved in infection-related deficits in contextual fear conditioning. Brain Behav. Immun. 2012;26:732–738. doi: 10.1016/j.bbi.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candido D.S., et al. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369(1979):1255–1260. doi: 10.1126/science.abd2161. https://pubmed.ncbi.nlm.nih.gov/32703910/ Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S.H.-Y., Beghi E., Helbok R., Moro E., Sampson J., Altamirano V., Mainali S., Bassetti C., Suarez J.I., McNett M., GCS-NeuroCOVID Consortium and ENERGY Consortium Global Incidence of neurological manifestations among patients hospitalized with COVID-19-A report for the GCS-NeuroCOVID Consortium and the ENERGY Consortium. JAMA Neurol. 2021;4 doi: 10.1001/jamanetworkopen.2021.12131. http://www.ncbi.nlm.nih.gov/pubmed/33974053 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crunfli F., et al. Morphological, cellular, and molecular basis of brain infection in COVID-19 patients. Proc Natl Acad Sci U S A. 2022;119 doi: 10.1073/pnas.2200960119. https://www.pnas.org/doi/abs/10.1073/pnas.2200960119 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice F.G., Ferreira S.T. Inflammation, defective insulin signaling, and mitochondrial dysfunction as common molecular denominators connecting type 2 diabetes to Alzheimer disease. Diabetes. 2014;63:2262–2272. doi: 10.2337/db13-1954. https://pubmed.ncbi.nlm.nih.gov/24931033/ Available at: [DOI] [PubMed] [Google Scholar]
- De Felice F.G., Gonçalves R.A., Ferreira S.T. Impaired insulin signalling and allostatic load in Alzheimer disease. Nature Reviews Neuroscience 2022. 2022;23(4 23):215–230. doi: 10.1038/s41583-022-00558-9. https://www.nature.com/articles/s41583-022-00558-9 Available at: [DOI] [PubMed] [Google Scholar]
- De Felice F.G., Tovar-Moll F., Moll J., Munoz D.P., Ferreira S.T. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the central nervous system. Trends Neurosci. 2020 doi: 10.1016/j.tins.2020.04.004. https://linkinghub.elsevier.com/retrieve/pii/S0166223620300916 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa V.L., Araújo S.B., Antonio L.M., Silva-Queiroz M., Colodeti L.C., Soares C., Barros-Aragão F., Mota-Araujo H.P., Alves V.S., Coutinho-Silva R., Eduardo B., Savio L., Ferreira S.T., da Costa R., Clarke J.R., Figueiredo C.P. Innate immune memory mediates increased susceptibility to Alzheimeŕs disease-like pathology in sepsis surviving mice. Brain Behav. Immun. 2021 doi: 10.1016/j.bbi.2021.04.001. https://pubmed.ncbi.nlm.nih.gov/33838250/ Available at: [DOI] [PubMed] [Google Scholar]
- Distler U., Kuharev J., Navarro P., Tenzer S. Label-free quantification in ion mobility–enhanced data-independent acquisition proteomics. Nature Protocols 2016. 2016;11(4 11):795–812. doi: 10.1038/nprot.2016.042. https://www.nature.com/articles/nprot.2016.042 Available at: [DOI] [PubMed] [Google Scholar]
- Espíndola O., Siqueira M., Soares C., Lima M., Leite A., Araujo A., Brandão C., Silva M. Patients with COVID-19 and neurological manifestations show undetectable SARS-CoV-2 RNA levels in the cerebrospinal fluid. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.05.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espíndola O.M., Gomes Y.C.P., Brandão C.O., Torres R.C., Siqueira M., Soares C.N., Lima M.A.S.D., Leite A.C.C.B., Venturotti C.O., Carvalho A.J.C., Torezani G., Araujo A.Q.C., Silva M.T.T. Inflammatory cytokine patterns associated with neurological diseases in COVID-19. Ann. Neurol. 2021 doi: 10.1002/ana.26041. http://www.ncbi.nlm.nih.gov/pubmed/33547819 00 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etter M.M., et al. Severe Neuro-COVID is associated with peripheral immune signatures, autoimmunity and neurodegeneration: a prospective cross-sectional study. Nat. Commun. 2022;13(1 13):1–21. doi: 10.1038/s41467-022-34068-0. https://www.nature.com/articles/s41467-022-34068-0 2022. Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L.E., Taylor J.L., Smith C.J., Pritchard H.A.T., Greenstein A.S., Allan S.M. Cardiovascular comorbidities, inflammation, and cerebral small vessel disease. Cardiovasc. Res. 2021;117:2575–2588. doi: 10.1093/cvr/cvab284. [DOI] [PubMed] [Google Scholar]
- Figueiredo C.P., et al. Zika virus replicates in adult human brain tissue and impairs synapses and memory in mice. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-11866-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas GR de, Figueiredo M.R., Vianna A., Brandão C.O., Torres-Filho H.M., Martins A.F.A., Tovar-Moll F., Barroso P.F. Clinical and radiological features of severe acute respiratory syndrome coronavirus 2 meningo-encephalitis. Eur. J. Neurol. 2021;28:3530–3532. doi: 10.1111/ene.14687. https://onlinelibrary.wiley.com/doi/full/10.1111/ene.14687 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman E.M., Mroczek D.K., Christ S.L. Multimorbidity, inflammation, and disability: a longitudinal mediational analysis. Ther Adv Chronic Dis. 2019;10 doi: 10.1177/2040622318806848. https://journals.sagepub.com/doi/10.1177/2040622318806848 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost P.S., Barros-Aragão F., da Silva R.T., Venancio A., Matias I., Lyra e Silva N.M., Kincheski G.C., Pimentel-Coelho P.M., De Felice F.G., Gomes F.C.A., Ferreira S.T., Figueiredo C.P., Clarke J.R. Neonatal infection leads to increased susceptibility to Aβ oligomer-induced brain inflammation, synapse loss and cognitive impairment in mice. Cell Death Dis. 2019;10:323. doi: 10.1038/s41419-019-1529-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber C., Soung A., Vollmer L.L., Kanmogne M., Last A., Brown J., Klein R.S. T cells promote microglia-mediated synaptic elimination and cognitive dysfunction during recovery from neuropathogenic flaviviruses. Nat. Neurosci. 2019;22:1276–1288. doi: 10.1038/s41593-019-0427-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande G., Marengoni A., Vetrano D.L., Roso-Llorach A., Rizzuto D., Zucchelli A., Qiu C., Fratiglioni L., Calderón-Larrañaga A. Multimorbidity burden and dementia risk in older adults: the role of inflammation and genetics. 2020. https://alz-journals.onlinelibrary.wiley.com/doi/10.1002/alz.12237 Available at: [DOI] [PMC free article] [PubMed]
- Guan W., et al. Comorbidity and its impact on 1590 patients with covid-19 in China: a Nationwide analysis. Eur. Respir. J. 2020 doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasp M., Muñoz-Sánchez G., Martínez-Hernández E., Santana D., Carbayo Á., Naranjo L., Bolós U., Framil M., Saiz A., Balasa M., Ruiz-García R., Sánchez-Valle R., Group T.B.N.-C.S. CSF biomarkers in COVID-19 associated encephalopathy and encephalitis Predict long-term outcome. Front. Immunol. 2022:1600. doi: 10.3389/fimmu.2022.866153. https://www.frontiersin.org/articles/10.3389/fimmu.2022.866153/full 0. Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms J., Kremer S., Merdji H., Clere-Jehl R., Schenck M., Kummerlen C., Collange O., Boulay C., Fafi-Kremer S., Ohana M., Anheim M., Meziani F. Neurologic features in severe SARS-CoV-2 infection. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms J., Kremer S., Merdji H., Schenck M., Severac F., Clere-Jehl R., Studer A., Radosavljevic M., Kummerlen C., Monnier A., Boulay C., Fafi-Kremer S., Castelain V., Ohana M., Anheim M., Schneider F., Meziani F. Delirium and encephalopathy in severe COVID-19: a cohort analysis of ICU patients. Crit. Care. 2020;24:491. doi: 10.1186/s13054-020-03200-1. https://ccforum.biomedcentral.com/articles/10.1186/s13054-020-03200-1 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC), World Health Organization (WHO) Formulário DE REGISTO DE CASO CENTRAL COVID-19. 2020. https://isaric.org/wp-content/uploads/2020/10/ISARIC-COVID-19-CORE-CRF_PT.pdf Available at:
- International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC), World Health Organization (WHO) Protocolo de Caracterização Clínica para Infecções Emergentes Severas do ISARIC/OMS: Coronavírus. 2020. https://isaric.org/document-library/?fwp_media_tax4=protocol Available at:
- Jun T., Nirenberg S., Weinberger T., Sharma N., Pujadas E., Cordon-Cardo C., Kovatch P., Huang K. Analysis of sex-specific risk factors and clinical outcomes in COVID-19. Communications Medicine 2021. 2021;1(1 1):1–8. doi: 10.1038/s43856-021-00006-2. https://www.nature.com/articles/s43856-021-00006-2 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R.S., Garber C., Howard N. Infectious immunity in the central nervous system and brain function. Nat. Immunol. 2017;18:132–141. doi: 10.1038/ni.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau E.S., McNeill J.N., Paniagua S.M., Liu E.E., Wang J.K., Bassett I.V., Selvaggi C.A., Lubitz S.A., Foulkes A.S., Ho J.E. Sex differences in inflammatory markers in patients hospitalized with COVID-19 infection: Insights from the MGH COVID-19 patient registry. PLoS One. 2021;16 doi: 10.1371/journal.pone.0250774. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0250774 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechien J.R., et al. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur. Arch. Oto-Rhino-Laryngol. 2020:1–11. doi: 10.1007/s00405-020-05965-1. http://www.ncbi.nlm.nih.gov/pubmed/32253535 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H., Perl D.P., Nair G., Li W., Maric D., Murray H., Dodd S.J., Koretsky A.P., Watts J.A., Cheung V., Masliah E., Horkayne-Szakaly I., Jones R., Stram M.N., Moncur J., Hefti M., Folkerth R.D., Nath A. Microvascular injury in the brains of patients with covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2033369. http://www.nejm.org/doi/10.1056/NEJMc2033369 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.H., Perl D.P., Steiner J., Pasternack N., Li W., D M., F S., I H.-S., R J., MN S., JT M., M H., RD F., A N. Neurovascular injury with complement activation and inflammation in COVID-19. Brain. 2022 doi: 10.1093/brain/awac151. https://pubmed.ncbi.nlm.nih.gov/35788639/ Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. https://academic.oup.com/bioinformatics/article/34/18/3094/4994778 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolansen S.D., Rostgaard N., Oernbo E.K., Juhler M., Simonsen A.H., MacAulay N. Inflammatory markers in cerebrospinal fluid from patients with hydrocephalus: a systematic Literature review Rizzo R. Dis. Markers. 2021;2021:1–12. doi: 10.1155/2021/8834822. https://www.hindawi.com/journals/dm/2021/8834822/ Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyra e Silva N.M., Barros-Aragão F.G.Q., De Felice F.G., Ferreira S.T. Inflammation at the crossroads of COVID-19, cognitive deficits and depression. Neuropharmacology. 2022;209 doi: 10.1016/j.neuropharm.2022.109023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D., Miao X., Li Y., Hu B. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020 doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschke J., et al. Neuropathology of patients with COVID-19 in Germany: a post-mortem case series. Lancet Neurol. 2020;19:919–929. doi: 10.1016/S1474-4422(20)30308-2. www.thelancet.com/ Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza M.G., De Lorenzo R., Conte C., Poletti S., Vai B., Bollettini I., Melloni E.M.T., Furlan R., Ciceri F., Rovere-Querini P., Benedetti F. Anxiety and depression in COVID-19 survivors: role of inflammatory and clinical predictors. Brain Behav. Immun. 2020;89:594–600. doi: 10.1016/j.bbi.2020.07.037. Available at:/pmc/articles/PMC7390748/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza M.G., Mariagrazia P., De Lorenzo R., Cristiano M., Sara P., Roberto F., Fabio C., Patrizia R.-Q., Francesco B. Persistent psychopathology and neurocognitive impairment in COVID-19 survivors: effect of inflammatory biomarkers at three-month follow-up. Brain Behav. Immun. 2021 doi: 10.1016/j.bbi.2021.02.021. http://www.ncbi.nlm.nih.gov/pubmed/33639239 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira F.R.R., Bonfim D.M., Zauli D.A.G., Silva J.P., Lima A.B., Malta F.S.V., Ferreira A.C.S., Pardini V.C., Magalhães W.C.S., Queiroz D.C., Souza R.M., Geddes V.E.V., Costa W.C., Moreira R.G., Faria N.R., Voloch C.M., Souza R.P., Aguiar R.S. Epidemic spread of SARS-CoV-2 lineage B.1.1.7 in Brazil. Viruses. 2021;13 doi: 10.3390/v13060984. https://www.mdpi.com/1999-4915/13/6/984/htm 984 13:984 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyiadji N., Shahin G., Noujaim D., Stone M., Patel S., Griffith B. COVID-19–associated acute hemorrhagic Necrotizing encephalopathy: CT and MRI features. Radiology. 2020 doi: 10.1148/radiol.2020201187. http://pubs.rsna.org/doi/10.1148/radiol.2020201187 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-de-Oliveira G., Carregari V.C. Martins-de-Souza D (2023) OmicScope. https://zenodo.org/records/8074722 Available at:
- Rhea E.M., Logsdon A.F., Hansen K.M., Williams L.M., Reed M.J., Baumann K.K., Holden S.J., Raber J., Banks W.A., Erickson M.A. The S1 protein of SARS-CoV-2 crosses the blood–brain barrier in mice. Nat. Neurosci. 2020 doi: 10.1038/s41593-020-00771-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Sánchez C.M., et al. Neurologic manifestations in hospitalized patients with COVID-19: the ALBACOVID registry. Neurology. 2020 doi: 10.1212/WNL.0000000000009937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa Cruz A., Mendes-Frias A., Oliveira A.I., Dias L., Matos A.R., Carvalho A., Capela C., Pedrosa J., Castro A.G., Silvestre R. Interleukin-6 is a biomarker for the development of Fatal severe acute respiratory syndrome coronavirus 2 pneumonia. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.613422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasannejad C., Ely E.W., Lahiri S. Long-term cognitive impairment after acute respiratory distress syndrome: a review of clinical impact and pathophysiological mechanisms. Crit. Care. 2019;23:1–12. doi: 10.1186/s13054-019-2626-z. https://ccforum.biomedcentral.com/articles/10.1186/s13054-019-2626-z Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siahaan Y.M.T., Puspitasari V., Pangestu A. COVID-19-Associated encephalopathy: Systematic review of case reports. J. Clin. Neurol. 2022;18 doi: 10.3988/jcn.2022.18.2.194. https://pubmed.ncbi.nlm.nih.gov/35196749/ Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon I., Normandin E., Bhattacharyya S., Mukeji S., Ali A., Adams G., Hornick J., Padera R., Sabeti P. Neuropathological features of covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2019373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon T. Neurological infection with SARS-CoV-2 — the story so far. Nat. Rev. Neurol. 2021;17:65–66. doi: 10.1038/s41582-020-00453-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo F.K., et al. Cognitive, behavioral, neuroimaging and inflammatory biomarkers after hospitalization for covid-19 in Brazil. Brain Behav. Immun. 2024;115:434–447. https://linkinghub.elsevier.com/retrieve/pii/S0889159123003185 Available at: [Google Scholar]
- Tahira A.C., Verjovski‐Almeida S., Ferreira S.T. Alzheimer’s & Dementia; 2021. Dementia Is an Age‐independent Risk Factor for Severity and Death in COVID‐19 Inpatients; pp. 1–14.https://onlinelibrary.wiley.com/doi/10.1002/alz.12352 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taquet M., Dercon Q., Luciano S., Geddes J.R., Husain M., Harrison P.J. Incidence, co-occurrence, and evolution of long-COVID features: a 6-month retrospective cohort study of 273,618 survivors of COVID-19. PLoS Med. 2021;18 doi: 10.1371/journal.pmed.1003773. https://journals.plos.org/plosmedicine/article?id=10.1371/journal.pmed.1003773 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taquet M., Geddes J.R., Husain M., Luciano S., Harrison P.J. 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: a retrospective cohort study using electronic health records. Lancet Psychiatr. 2021;8:416–427. doi: 10.1016/S2215-0366(21)00084-5. http://www.ncbi.nlm.nih.gov/pubmed/33836148 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taquet M., Luciano S., Geddes J.R., Harrison P.J. Bidirectional associations between COVID-19 and psychiatric disorder: retrospective cohort studies of 62 354 COVID-19 cases in the USA. Lancet Psychiatr. 2021;8:130–140. doi: 10.1016/S2215-0366(20)30462-4. www.thelancet.com/psychiatry Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taquet M., Sillett R., Zhu L., Mendel J., Camplisson I., Dercon Q., Harrison P.J. Neurological and psychiatric risk trajectories after SARS-CoV-2 infection: an analysis of 2-year retrospective cohort studies including 1 284 437 patients. Lancet Psychiatr. 2022;9:815–827. doi: 10.1016/S2215-0366(22)00260-7. https://pubmed.ncbi.nlm.nih.gov/35987197/ Available at: March 6, 2023] [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Caten F., et al. In-depth analysis of laboratory parameters reveals the interplay between sex, age, and systemic inflammation in individuals with COVID-19. Int. J. Infect. Dis. 2021;105:579–587. doi: 10.1016/j.ijid.2021.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur K.T., et al. COVID-19 neuropathology at Columbia university irving medical center/New York presbyterian hospital. Brain. 2021 doi: 10.1093/brain/awab148. PMC8083258/?report=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasek M.J., et al. A complement–microglial axis drives synapse loss during virus-induced memory impairment. Nature. 2016;534:538–543. doi: 10.1038/nature18283. http://www.nature.com/doifinder/10.1038/nature18283 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan A., et al. Case definitions, diagnostic algorithms, and priorities in encephalitis: consensus statement of the international encephalitis Consortium. Clin. Infect. Dis. 2013;57:1114–1128. doi: 10.1093/cid/cit458. Available at:/pmc/articles/PMC3783060/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B.J., Peltan I.D., Jensen P., Hoda D., Hunter B., Silver A., Starr N., Buckel W., Grisel N., Hummel E., Snow G., Morris D., Stenehjem E., Srivastava R., Brown S.M. Clinical criteria for COVID-19-associated hyperinflammatory syndrome: a cohort study. Lancet Rheumatol. 2020;2:e754–e763. doi: 10.1016/S2665-9913(20)30343-X. www.thelancet.com/rheumatology Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson E.J., et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo M.S., Malsy J., Pöttgen J., Seddiq Zai S., Ufer F., Hadjilaou A., Schmiedel S., Addo M.M., Gerloff C., Heesen C., Schulze Zur Wiesch J., Friese M.A. Frequent neurocognitive deficits after recovery from mild COVID-19. Brain Commun. 2020;2 doi: 10.1093/braincomms/fcaa205. https://academic.oup.com/braincomms/article/doi/10.1093/braincomms/fcaa205/5998660 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization WHO COVID-19: case Definitions. COVID-19: surveillance, case investigation and epidemiological protocols. 2020. https://www.who.int/publications/i/item/WHO-2019-nCoV-Surveillance_Case_Definition-2020.2 Available at:
- World Health Organization WHO R&D blueprint novel coronavirus COVID-19 therapeutic trial synopsis. 2020. https://www.who.int/blueprint/priority-diseases/key-action/COVID-19_Treatment_Trial_Design_Master_Protocol_synopsis_Final_18022020.pdf Available at:
- Yang A.C., et al. Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature. 2021;595:565–571. doi: 10.1038/s41586-021-03710-0. http://www.ncbi.nlm.nih.gov/pubmed/34153974 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z., Yu H., Chen H., Qi W., Chen L., Chen G., Yan W., Chen T., Ning Q., Han M., Wu D. Longitudinal changes of inflammatory parameters and their correlation with disease severity and outcomes in patients with COVID-19 from Wuhan, China. Critical Care 2020. 2020;24(1 24):1–12. doi: 10.1186/s13054-020-03255-0. https://ccforum.biomedcentral.com/articles/10.1186/s13054-020-03255-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, 2021. Solidarity trial plus: an international randomized trial of additional treatments for COVID-19 in hospitalized patients who are all receiving the local standard of care Available at: ISRCTN. https://www.isrctn.com/ISRCTN18066414. (Accessed 17 August 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have shared anonymized individualized patient data at Table 1. The mass spectrometry proteomics data deposited at ProteomeXchange Consortium, PRIDE partner repository (PXD033979 10.6019/PXD033979)