Abstract
Papovaviruses utilize predominantly cellular DNA replication proteins to replicate their own viral genomes. To appropriate the cellular DNA replication machinery, simian virus 40 (SV40) large T antigen (Tag) binds to three different cellular replication proteins, the DNA polymerase α-primase complex, the replication protein A (RPA) complex, and topoisomerase I. The functionally similar papillomavirus E1 protein has also been shown to bind to the DNA polymerase α-primase complex. Enzyme-linked immunoassay-based protein interaction assays and protein affinity pull-down assays were used to show that the papillomavirus E1 protein also binds to the cellular RPA complex in vitro. Furthermore, SV40 Tag was able to compete with bovine papillomavirus type 1 E1 for binding to RPA. Each of the three RPA subunits was individually overexpressed in Escherichia coli as a soluble fusion protein. These fusion proteins were used to show that the E1-RPA and Tag-RPA interactions are primarily mediated through the 70-kDa subunit of RPA. These results suggest that different viruses have evolved similar mechanisms for taking control of the cellular DNA replication machinery.
During infection, the papovaviruses, small DNA viruses with double-stranded circular chromosomes, employ much of the cellular DNA replication machinery to replicate their own genomes. For a number of years the replication of the simian virus 40 (SV40) genome has been studied as a general model for eukaryotic DNA replication (for a review, see references 31, 32, 47, and 72). The only viral protein required for SV40 DNA replication is the SV40 large T antigen (Tag), the viral DNA replication initiator protein that allows the virus to avoid the host cell regulatory mechanisms (for a review, see reference 13).
SV40 DNA replication is initiated by the binding of Tag to the SV40 origin (for a review, see reference 5). In concert with replication protein A (RPA) and torsional release provided by a topoisomerase, Tag promotes extensive origin unwinding. DNA polymerase α-primase then binds to the template and synthesizes a nascent RNA-DNA chain at the origin (49, 60, 70). The interactions between SV40 Tag, RPA, and DNA polymerase α-primase are essential for forming the initiation complex. Both RPA and Tag bind to DNA polymerase α-primase (15–17, 21, 46, 50, 66, 67, 73) and stimulate DNA polymerase activity (6, 10, 46, 51, 55, 60, 71, 73). Tag also stimulates the primase activity of DNA polymerase α-primase (11, 43, 49, 51). RPA and Tag also interact with each other, and this interaction is important for initiation of SV40 DNA replication (16, 46, 51, 60, 73). The interactions among these three proteins are highly specific. Even though RPA and DNA polymerase α-primase from different species are highly homologous, they show different properties in their interactions with Tag and each other. Thus, RPA and DNA polymerase α-primase from different species demonstrate differing capacities to support SV40 DNA replication (summarized in references 46, 52, 60, and 67).
The RPA heterotrimeric complex consists of three subunits, the 70-kDa subunit (RPA70), the 32-kDa subunit (RPA32), and the 14-kDa subunit (RPA14) (20, 74). Many of the functions of RPA have been found to be associated with RPA70. RPA70 is the major single-stranded DNA (ssDNA)-binding subunit (1, 22–24, 33, 36, 58). RPA70 is also believed to be involved in RPA’s interactions with many other proteins, including DNA polymerase α, p53, VP16, and RPA4 (an apparent homolog of RPA32), among others (6, 16, 18, 23, 24, 27, 35, 36, 41, 42, 78). The other two subunits, RPA32 and RPA14, are also essential for RPA function, since antibodies against RPA32 inhibit SV40 DNA replication in vitro (34) and homologs of all three RPA subunits have been shown to be essential in Saccharomyces cerevisiae (7, 30). RPA32 and RPA14 can form a subcomplex (6, 28, 68), which is believed either to assist in the proper folding of RPA70 or to help in assembly of the RPA heterotrimer. Very little is known about the exact roles of these two smaller subunits. Recent evidence indicates that RPA32 may have a secondary ssDNA-binding domain (2, 58). RPA32 is phosphorylated in a cell cycle-dependent manner, which seems not to be essential for RPA function (14, 29, 39, 42). RPA32 has also been shown to interact with the recombination and repair protein RAD52, the XPA repair protein, and a DNA glycosylase (40, 54, 56). While there has been great interest in further elucidation of the functions of these three subunits, their insolubility when expressed individually has made these questions refractory to biochemical analyses (28, 68).
In attempting to identify the RPA subunit that binds to Tag, Dornreiter et al. (16) did not detect an interaction between Tag and RPA70 by using a Southwestern protein interaction blotting procedure. Lee and Kim (39) have reported that a deletion mutant of RPA32 which forms a RPA heterotrimer with RPA14 and RPA70 is unable to support either the interaction of this RPA complex with Tag or SV40 DNA replication in vitro. Conversely, Braun et al. (6) used the same approach to demonstrate that a similar deletion of RPA32 in a heterotrimeric context interacts productively with Tag. Further, they showed that certain deletions of RPA70, when assembled into a heterotrimer with RPA32 and RPA14, do not interact with SV40 Tag and do not support SV40 DNA replication in vitro (6). A major limitation of both of these studies is that the Tag interaction with RPA was evaluated by abrogation of Tag binding to the RPA heterotrimer. Ideally, one would like to demonstrate in a positive fashion the existence of an RPA subunit or domain that specifically interacts with SV40 Tag.
Like SV40, papillomaviruses use predominantly host cell enzymes to replicate their circular double-stranded DNA genomes (9, 37). The viral protein E1 shares a number of biochemical properties with Tag, such as virus origin binding and DNA helicase activity, and is sufficient to support papillomavirus (PV) DNA replication both in vitro with host cell extracts and in vivo (in some PV strains) (3, 25, 59, 62, 64, 75, 76). Like Tag, E1 also binds to DNA polymerase α-primase (4, 12, 57). Due to E1’s ability to support PV DNA replication, we anticipated that E1, like SV40 Tag, would have to bind to RPA. However, Bonne-Andrea et al. (4) were unable to detect an interaction between E1 and RPA. Another viral protein, E2, a transcriptional activator required for DNA replication of most strains of PV, has been shown to bind to both E1 (61, 63, 75, 77) and RPA (41, 44). Hence, E1 might interact with RPA indirectly via E2. Alternatively, a direct interaction between E1 and RPA complex may be relatively weak and unable to be detected with the experimental methods used by Bonne-Andrea et al. (4). However, since E2 is not required for BPV1 DNA replication in vitro, this indicated to us that BPV1 E1 was likely to bind to the RPA heterotrimer either directly or indirectly via other host factors.
We report here the detection of a direct interaction between E1 and RPA in vitro with both an enzyme-linked immunosorbent assay (ELISA)-based protein-protein interaction assay and a glutathione S-transferase (GST) “pull-down” technique. We show here that the interactions between E1-RPA and Tag-RPA partially inhibit one another, suggesting some shared aspect of their binding domains on RPA. Furthermore, the three subunits of RPA were expressed as fusion proteins and used to show that the interactions between Tag and RPA and E1 and RPA appear to be mediated through RPA70. We have also shown that the RPA70 fusion protein, but not the RPA32 or RPA14 fusion protein, readily inhibits SV40 DNA replication in vitro.
MATERIALS AND METHODS
Materials, proteins, and plasmids.
Restriction enzymes, the large (Klenow) fragment of Escherichia coli DNA polymerase I, T4 polynucleotide kinase, T4 DNA ligase, and rabbit anti-MBP (maltose binding protein) antibody were obtained from New England Biolabs or GIBCO Life Technologies. Calf alkaline phosphatase, horse serum, and calf serum were obtained from GIBCO Life Technologies. Anti-goat immunoglobulin G (IgG) antibody linked to horseradish peroxidase was obtained from Southern Biotechnology Associates. 33′-55′-tetramethylbenzidine was obtained from Sigma. Oligonucleotides were synthesized by Cruachem. Anti-rabbit IgG antibody linked to horseradish peroxidase, E. coli SSB, and [α-32P]dATP were obtained from Amersham-U.S. Biochemicals. Q-Sepharose, SP-Sepharose, glutathione-Sepharose, goat anti-GST antibody, ribonucleotides, and deoxyribonucleotides were purchased from Pharmacia. QIAquick gel extraction kits were purchased from Qiagen Inc. Amylose chromatography resin and pMAL-c2 were obtained from New England Biolabs.
The modified MBP expression vector encoding a thrombin cleavage site and a larger polylinker (pMAL-cT) was kindly provided by M. Xue and W. Ruyechan. The T7 pET expression vectors for RPA and the RPA subunits (p3a-RPA14/32, p11d-RPA70, and p11d-tRPA) were kindly provided by L. Henricksen and M. Wold (28). Rabbit anti-RPA and monoclonal antibodies against the two larger subunits of RPA were described previously (14).
SV40 Tag was expressed in Sf9 cells infected with a recombinant baculovirus expression vector and purified from lysates by immunoaffinity chromatography (38, 53, 65, 69). The GST-E1 fusion protein and E1 were purified as previously described (45, 61). Briefly, GST-E1 was purified by affinity chromatography with glutathione-Sepharose, and E1 was cleaved from the GST domain and further purified by conventional anion-exchange chromatography. The RPA heterotrimeric complex was overproduced in E. coli with the T7 expression system (p11d-tRPA) (28) and purified to near homogeneity as described previously (8).
Construction of RPA subunit fusion protein expression vectors.
The cDNA sequences encoding the individual subunits of RPA were subcloned into pMAL-c2 or pMAL-cT (a derivative of pMAL-c2 with a linker encoding a thrombin cleavage site). Both expression vectors generate fusion proteins containing the MBP domain as the N-terminal portion. To prepare pMAL-RPA14, the coding region of RPA14 was cleaved from p3a-RPA14/32 with XbaI and BamHI and subcloned into pBS SKII(−). The RPA14 fragment was digested from this construct with NcoI, treated with DNA polymerase I Klenow fragment and deoxynucleotides, and then digested with HindIII. After agarose gel purification, the RPA14 fragment was ligated into the pMAL-cT vector (which had previously been digested with BamHI, treated with DNA polymerase I Klenow fragment and deoxynucleotides, digested with HindIII, and isolated via agarose gel purification [with the QIAquick gel extraction kit]).
To prepare the RPA32 expression vector, pMAL-RPA32, p3a-RPA14/32 was digested with NdeI, treated with DNA polymerase I Klenow fragment and deoxynucleotides, and digested with HindIII, and the RPA32 coding fragment was purified from an agarose gel (with the QIAquick gel extraction kit). The RPA32 fragment was then ligated into the pMAL-c2 vector (which had been digested with XbaI, treated with DNA polymerase I Klenow fragment and deoxynucleotides, digested with HindIII, and isolated from an agarose gel).
To prepare pMAL-RPA70, two oligonucleotides, 5′-GATCTGAATTCCTGGTACCGCGTGGTTC-3′ and 5′-CATGGAACCACGCGGTACCAGGAATTCA-3′, were synthesized, annealed to one another, treated with T4 polynucleotide kinase, and inserted into p11d-RPA70 (cleaved with NcoI and BglII) upstream of the coding region of RPA70. The RPA70 coding region was cleaved from this new construct with EcoRI and ligated into pMAL-c2 which had been dephosphorylated following EcoRI digestion. This step generated two constructs with inserts of opposite orientation. Transformants were screened by restriction digestion with KpnI to identify clones with the insert in the proper orientation.
All RPA subunit fusion expression plasmids obtained were analyzed for induction of the predicted fusion protein and were subjected to DNA sequencing and immunoblot analysis to confirm the identities of the fusion proteins generated.
Expression and purification of RPA subunit fusion proteins.
pMAL-RPA32 and pMAL-RPA14 were transformed into BL21 (DE3) pLysS, and pMAL-RPA70 was transformed into DH5α. Cells were grown to an optical density at 600 nm of 0.6 and induced with IPTG (isopropyl-β-d-thiogalactopyranoside) at 0.4 mM. The cells were collected 4 h after the addition of IPTG. The cell pellets were frozen in liquid nitrogen, thawed, and resuspended in lysis buffer (50 mM Tris [pH 7.4], 1 mM EDTA, 0.1% [vol/vol] Nonidet P-40, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). BL21 (DE3) pLysS cells were autolysed after resuspension and sonicated to shear the DNA. DH5α cells were lysed by one passage through a Parr cell disruption bomb at 2,000 lb/in2. All lysates were subjected to centrifugation (170,000 × g for 30 min at 4°C), and the supernatants were each applied to an amylose column (2.5 by 2 cm). The columns were subsequently washed with 12 bed volumes of column buffer (20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM Na2EDTA), and the fusion proteins were eluted with the same buffer containing 10 mM maltose. The eluted proteins were detected with the Bio-Rad protein assay reagent, and fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% [wt/vol] acrylamide) and Coomassie blue staining.
ELISAs.
ELISAs were carried out in 96-well vinyl plates at room temperature. To prepare the immobilized substrate (also referred to as the solid phase, or target protein), wells were coated for 60 min with purified protein (as indicated in the figure legends) in 50 μl of TBS (25 mM Tris-HCl [pH 7.4], 150 mM NaCl). The wells were then washed with TBST (TBS with 0.1% [vol/vol] either NP-40 or Triton X-100) and blocked with 5% (wt/vol) dry milk and 2% (vol/vol) serum (either calf or horse) in TBST for 45 min to overnight. After being washed three times with TBST, various amounts of the challenging protein (as indicated in the figure legends) were added to the wells in 50 μl of TBST supplemented with 1 mM MgCl2, 1 mM CaCl2, and 40 U of micrococcal nuclease/ml and incubated for 30 min. After being washed three times with TBST, the plates were incubated with the appropriate anti-challenging protein primary antibody (as indicated in the figure legends) diluted in TBST with 0.5% (wt/vol) dry milk and 1% (vol/vol) calf or horse serum for 60 min at room temperature. The plates were then washed three times with TBST and incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (1:5,000 in TBST with 0.5% [wt/vol] dry milk and 1% [vol/vol] calf or horse serum) for 60 min. After being washed seven times with TBST and once with TBS, the plate wells were incubated with 50 μl of visualization buffer (110 mM sodium acetate [pH 5.5]) containing the chromogenic substrate 33′-55′-tetramethylbenzidine (1 mg/ml) and hydrogen peroxide (0.0075% [vol/vol]). After 10 min the reaction was stopped by the addition of 50 μl of 2 M sulfuric acid. The assays were quantified spectrophotometrically by absorbance at 450 nm. Each assay was performed at least five times. Each figure depicts data from a representative experiment.
In the competition binding assays the target protein was attached to the ELISA plate wells and the wells were then blocked as described above. A constant amount of the challenging protein was mixed with various amounts of competitor protein, and the mixtures were preincubated for 30 min at room temperature. These mixtures were then incubated with the immobilized protein in the wells of the ELISA plates. Binding of the challenging protein was evaluated as described above. Each assay was performed at least five times, and the data shown is from one representative experiment.
Protein affinity pull-down assay.
Using the modified procedures of Weisshart et al. (73), protein affinity pull-down assays were performed to examine the interaction between RPA and BPV1 E1. GST and GST-E1 were bacterially expressed and bound to glutathione-Sepharose as described previously (73). These were used in the construction of 0.1-ml columns that were each equilibrated in 10 column volumes of binding buffer (30 mM HEPES-KOH [pH 7.9], 50 mM KCl, 7 mM MgCl2, 0.25 mM EDTA, 0.05% [vol/vol] NP-40). Purified RPA, 0.01 mg, was diluted to 0.1 mg/ml in binding buffer, applied to each column, and recycled twice. The columns were each washed with 10 to 20 ml of wash buffer (30 mM HEPES-KOH [pH 7.9], 100 mM NaCl, 7 mM MgCl2). The volume displaced by the last 0.3 ml of wash buffer was collected for the wash analysis. Bound proteins were eluted with 0.3 ml of elution buffer (30 mM HEPES-KOH [pH 7.9], 1% [wt/vol] SDS, 300 mM β-mercaptoethanol).
Samples were separated on 15% (wt/vol) SDS-PAGE gels and electrophoretically transferred to nitrocellulose membranes. These membranes were blocked with 5% (wt/vol) dry milk and 1% (vol/vol) calf serum diluted in TBST (with NP-40). The nitrocellulose membranes were incubated with monoclonal mouse antisera against RPA70 and RPA32 (1:100 dilution of cell culture supernatant in 0.5% [wt/vol] dry milk, 0.2% [vol/vol] calf serum, and TBST) and peroxidase-conjugated goat anti-mouse antibody (Pierce) (80 ng/ml diluted in TBST). RPA subunits were visualized by chemiluminescence detection (Pierce) in accordance with the manufacturer’s instructions.
For each experiment, the levels of GST and GST-E1 bound to the beads were evaluated to ensure that the control beads (GST) had equal or greater levels of protein bound than the test beads (GST-E1). For every experiment, more GST was bound to the matrix than GST-E1.
SV40 in vitro DNA replication assays.
SV40 in vitro DNA replication assays were carried out with 293 cell cytosolic extracts (S100). For each set of in vitro assays, a master reaction mixture was prepared and divided into aliquots for each assay. Standard 10-μl reaction mixtures contained 40 mM creatine phosphate (sodium salt); 20 mM Tris buffer (pH 7.5 at 25 mM and room temperature); 7 mM MgCl2; 0.5 mM dithiothreitol; 4 mM ATP; 200 μM (each) CTP, GTP, and UTP; 100 μM (each) dCTP, dGTP, dTTP; 25 μM dATP with 0.005 mCi of [α-32P]dATP; 30 ng of supercoiled DNA template (pSV011); 0.024 U of creatine phosphokinase; 525 ng of SV40 Tag; 4.6 μl (40 to 50 μg) of human 293 cell S100 extract; and various amounts of recombinant MBP-RPA subunit, MBP, or E. coli SSB. Replication reaction mixtures were assembled on ice and incubated at 37°C for 60 min. The reactions were stopped by placing them on ice. Squares of DE81 paper were dotted with 2 μl of each reaction mixture and washed four times with 0.5 M Na2HPO4 for 3 min each. The squares were then rinsed, first with water and then with ethanol (95% [vol/vol]). The squares were then dried and subjected to scintillation counting. The remainder of each reaction mixture was treated with SDS-EDTA-proteinase K (1% [wt/vol], 10 mM, and 50 μg/ml, respectively) for 20 min at 37°C. The mixtures were extracted with phenol-chloroform and ethanol precipitated. The products were analyzed by electrophoresis on 0.8% (wt/vol) agarose gels in 0.5× TBE. The gels were dried, and the radioactive products were analyzed with a Bio-Rad molecular imaging PhosphorImager system.
RESULTS
Interaction of RPA with BPV1 E1 protein.
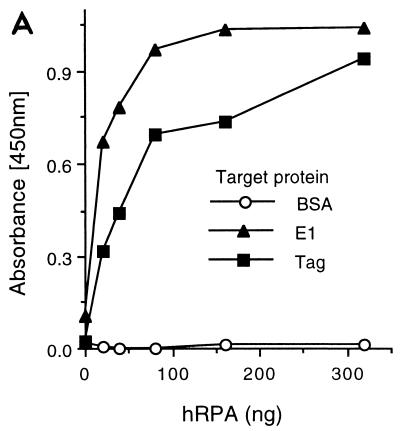
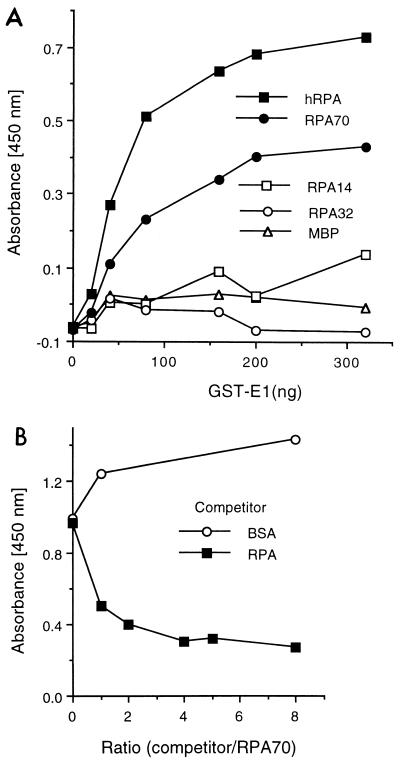
It has previously been reported that SV40 Tag interacts with RPA (6, 16, 46, 73). Due to the similarities in function between papillomavirus E1 proteins and SV40 Tag, we wished to determine whether BPV1 E1 and RPA also interact. To address this question, we first used the ELISA-based protein-protein interaction assay to evaluate binding between RPA and E1. Purified E1 or Tag was immobilized in the wells of ELISA plates and, following blocking of the wells, incubated with increasing concentrations of RPA. Binding of RPA to the negative control, bovine serum albumin (BSA), remained at background levels, whereas binding to E1 increased as a function of RPA concentration (Fig. 1A). The level of interaction between RPA and E1 was comparable to that observed between RPA and SV40 Tag (Fig. 1A) and similar to those previously reported between RPA and Tag (16, 46, 73).
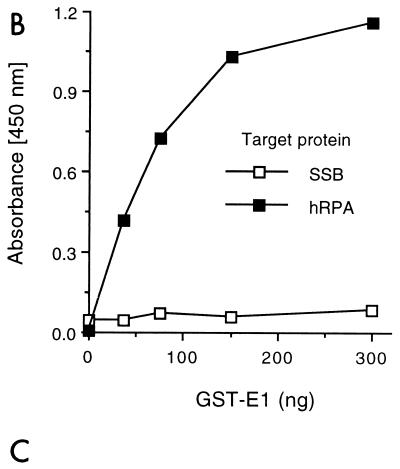
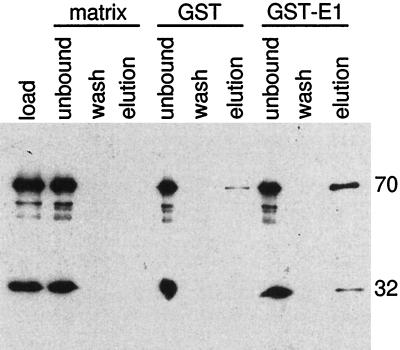
FIG. 1.
BPV1 E1 interacts with RPA. (A) RPA binding to immobilized E1. Five hundred nanograms of either Tag, E1, or BSA was immobilized in wells of a 96-well ELISA plate and challenged with increasing amounts of bacterially produced hRPA for 30 min as described in Materials and Methods. Interactions were detected with rabbit polyclonal antibody against hRPA and horseradish peroxidase-conjugated anti-rabbit antibody. The wells were incubated with a chromogenic substrate, and absorbance was measured at 450 nm as described in Materials and Methods. (B) GST-E1 binding to immobilized RPA. Two hundred nanograms of hRPA, E. coli SSB, or BSA was bound to ELISA wells and challenged with increasing amounts of GST-E1. Bound GST-E1 was detected with goat anti-GST antibody and horseradish peroxidase-coupled anti-goat antibody. Binding was evaluated as described in Materials and Methods. Background (GST-E1 binding to BSA) for each GST-E1 concentration used was subtracted from the experimental results prior to presentation. (C) Immunoblot of GST-E1 affinity pull-down assay. Ten micrograms of hRPA was applied to 0.1-ml columns of matrix alone (glutathione Sepharose) (matrix), GST-bound matrix (GST), or GST-E1-bound matrix (GST-E1). After washing and elution of the microcolumns, the various samples were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with monoclonal antibodies against the two large subunits of RPA (70 and 32 kDa, as indicated on the right) as described in Materials and Methods. Lanes: load, 0.2% of the total hRPA applied to each column; unbound, 0.2% of the unbound hRPA; wash, 0.5% of the last 0.3 ml of column wash; elution, 0.5% of the elution.
The interaction ELISA was also performed in the reverse orientation. Purified RPA was used as the target protein (solid phase), and GST-E1 was applied as the challenging protein (Fig. 1B). E. coli SSB, an ssDNA-binding protein which does not support BPV1 DNA replication (48), was used as a negative control. The binding of GST-E1 to RPA increased with increasing GST-E1 concentrations. In contrast, there was no detectable interaction between GST-E1 and E. coli SSB.
To further confirm the binding of BPV1 E1 to RPA, protein affinity pull-down assays were performed. This method was recently used to analyze the interaction between RPA and various truncations of SV40 Tag (73). Purified RPA was applied to glutathione, GST, and GST-E1 matrices. After extensive washing, the bound proteins were eluted. The load, wash, and eluted proteins were analyzed for the presence of RPA by immunoblotting. As observed in a representative experiment (Fig. 1C), RPA was consistently detected in elutions from the GST-E1 columns. In about half of the experiments performed, trace amounts of RPA could be detected in elutions from the GST columns (as shown in Fig. 1C), although in each of these instances, the amount of RPA that bound to the GST matrix was always far less than the amount that bound to the GST-E1 matrix. This suggests that there is a weak, nonspecific interaction between RPA and GST. No RPA was ever detected in the elutions from the glutathione-Sepharose columns. These results indicate that RPA also binds to E1 immobilized on an agarose matrix.
Tag inhibits RPA-E1 binding.
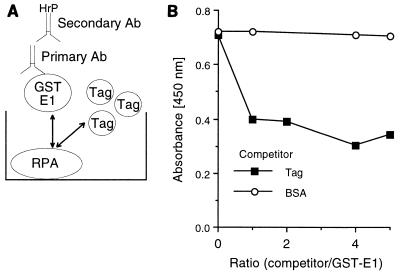
Since both SV40 Tag and BPV1 E1 protein bind to RPA, and they are the initiator proteins for viral DNA replication, we suspected that Tag and E1 may bind to the same or overlapping regions of RPA. To address this question, we carried out competition ELISAs. Purified RPA was immobilized in the ELISA wells. The E1 fusion protein, GST-E1, was used as the challenging protein. A constant amount of GST-E1 was mixed with increasing concentrations of competitor Tag prior to incubation with the immobilized RPA (Fig. 2A). The plates were then incubated with a primary goat anti-GST antibody and the appropriate enzyme-linked secondary antibody (in this case, anti-goat IgG linked to horseradish peroxidase) and evaluated as described in Materials and Methods. Binding of GST-E1 to RPA was efficiently inhibited by an equal amount of Tag. BSA, the negative control, had no effect on E1-RPA binding even at a fivefold molar excess (Fig. 2B). The converse experiment was also performed (using E1 as a competitor for the binding of RPA to immobilized Tag) and demonstrated that E1 can inhibit the Tag-RPA interaction (data not shown).
FIG. 2.
Tag competes with E1 for binding to RPA. (A) Graphic representation of the molecular interactions in the competition ELISA. GST-E1 (the challenge protein) binds to hRPA (shown immobilized to the ELISA well as the target protein). If the binding of SV40 Tag to RPA is competitive with the E1-RPA interaction, then increasing levels of Tag will compete with GST-E1 binding, resulting in a decreased amount of bound GST-E1. After binding, GST-E1 is recognized by goat anti-GST IgG, and the goat IgG is visualized with the secondary anti-goat IgG horseradish peroxidase (HrP)-linked antibody (Ab). (B) RPA (100 ng) was immobilized on ELISA plate wells. The challenging protein (300 ng of GST-E1) was first mixed with increasing amounts of competitor and then incubated with the immobilized RPA. The competitors were Tag and BSA. The indicated ratios are based on protein mass. Bound GST-E1 was detected with goat anti-GST antibody and HrP-coupled secondary antibody as described in Materials and Methods.
Expression and purification of RPA subunit proteins.
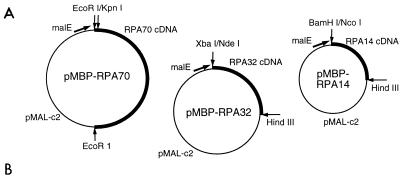
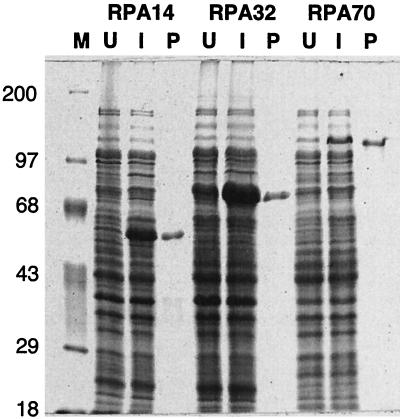
Since the interactions between RPA and the viral initiator proteins play such an important role in viral DNA replication, we wished to determine which of the three RPA subunits interacts with Tag and E1. However, when the RPA subunits are expressed individually, they either are insoluble, are difficult to purify, or aggregate with other proteins (28, 68). Therefore, the cDNA sequences encoding the individual RPA subunits were subcloned into expression vectors which generate fusions of the cloned protein with the C terminus of a bacterial MBP domain (Fig. 3A). The three resulting RPA subunit-MBP fusion proteins were expressed at high levels with a significant proportion of the protein in the soluble fraction. In cells containing the RPA32 or RPA14 expression plasmids, the fusion proteins are the predominant proteins in the cells, while cells containing the RPA70 expression plasmid produce much less fusion protein (Fig. 3B).
FIG. 3.
Construction of fusion vectors and expression and purification of RPA subunit fusion proteins (MBP-RPA14, MBP-RPA32, and MBP-RPA70) in E. coli. (A) Expression vectors were constructed with pMAL-c2 and cDNAs encoding the three RPA subunits as described in Materials and Methods. The indicated restriction sites were used in the cloning steps. Each vector encodes a fusion protein consisting of the MBP domain (encoded by malE sequences) fused to the indicated RPA subunit. The fusion proteins were induced and purified as described in Materials and Methods. (B) Purified fusion proteins (P), whole-cell lysates from uninduced cells (U) and induced cells (I), and molecular mass markers (M; in kilodaltons) were separated by SDS-PAGE and visualized by Coomassie blue staining as described in Materials and Methods.
The expressed MBP-RPA subunit fusion proteins were subjected to amylose affinity chromatography. All three fusion proteins were purified to near homogeneity as evaluated by Coomassie blue staining of SDS polyacrylamide gels (Fig. 3B). The sizes of the three fusion proteins were as expected. DNA sequencing of the expression plasmids and immunoblotting of both bacterial extracts and purified protein preparations confirmed the identity of each of the RPA subunit fusion proteins.
RPA subunit binding to BPV1 E1 protein.
The purified RPA subunit fusion proteins were used in protein binding ELISAs to identify the subunit responsible for RPA binding to E1. ELISA wells were coated with RPA subunits or the RPA complex, blocked, and challenged with GST-E1. As shown in Fig. 4A, E1 bound to the RPA complex. MBP-RPA70 was the only subunit with appreciable binding to E1. Neither MBP-RPA32 nor MBP-RPA14 demonstrated a measurable interaction with E1. GST alone did not show detectable binding to RPA or any of the RPA subunits (data not shown). The complementary assay, in which either GST-E1 or E1 was immobilized in ELISA wells and challenged with the individual subunit proteins, confirmed these results in that the only RPA subunit fusion protein to show binding to E1 was MBP-RPA70 (data not shown). Protein affinity pull-down assays consistently demonstrated a higher affinity of GST-E1 with MBP-RPA70 than with the other subunits (data not shown). These results suggest that RPA70 is the subunit that is primarily responsible for the E1-RPA interaction.
FIG. 4.
E1 interactions with the recombinant RPA subunit fusion proteins. (A) MBP, RPA complex, or one of the RPA subunit fusion proteins (MBP-RPA14, MBP-RPA32, MBP-RPA70) was bound to wells of ELISA plates (500 ng/well). Each series of wells was challenged with increasing amounts of GST-E1. The plates were then incubated with anti-GST antibody, and binding was evaluated as described in the legend to Fig. 2. (B) RPA competes with RPA70 for binding to E1. GST-E1 (150 ng/well) was immobilized to wells of an ELISA plate as the target protein. The challenging protein, MBP-RPA70 (200 ng/well), was first mixed with increasing amounts of either the competitor, hRPA (RPA), or the control protein, BSA. The mixtures were then incubated with the immobilized GST-E1. Bound RPA70 was detected by incubation with rabbit anti-MBP antibody, followed by horseradish peroxidase-coupled anti-rabbit antibody and a chromogenic substrate as described in Materials and Methods.
To confirm that the binding of E1 to MBP-RPA70 represents E1 binding to the RPA complex, we carried out competition binding assays with the RPA complex and MBP-RPA70. Increasing amounts of competitor protein (either RPA complex or BSA) were mixed with a fixed amount of the challenging protein (MBP-RPA70) before the mixtures were incubated with the immobilized target protein, GST-E1. MBP-RPA70 binding to GST-E1 was evaluated as described in Materials and Methods. Binding of MBP-RPA70 to GST-E1 was significantly inhibited by an equal amount of RPA complex but was not inhibited by BSA (Fig. 4B). Taken together these results indicate that the interaction between E1 and RPA70 is an apt reflection of the interaction between E1 and the RPA heterotrimer.
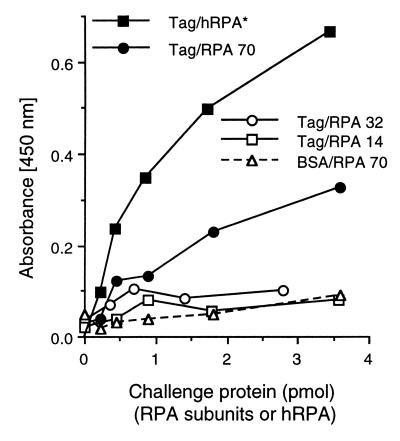
RPA subunit binding to Tag.
Heterotrimeric RPA complexes formed with an RPA70 subunit with amino acids 170 to 327 deleted have been reported not to bind to SV40 Tag (6). However, there has been no report of a positive interaction between either the RPA70 subunit or this putative binding region and SV40 Tag. Since RPA70 deletions lose the ability to bind to Tag, and since Tag competes with E1 for binding to the RPA complex and the E1-RPA complex interaction appears to be mediated by RPA70, we examined whether the Tag-RPA interaction is also mediated by RPA70. Tag was immobilized in ELISA plate wells and challenged with each of the three RPA subunit fusion proteins. Interactions were detected with an anti-MBP antibody. Only RPA70 showed a significant interaction with Tag; RPA32 and RPA14 did not show any detectable binding to Tag (Fig. 5). Competition assays similar to those shown in Fig. 4 demonstrated that RPA70 and the RPA complex compete with one another for binding to Tag (data not shown).
FIG. 5.
Tag interactions with the recombinant RPA subunit fusion proteins. Five hundred nanograms of BSA (dashed line) or Tag (solid lines) was bound to ELISA plate wells. The wells were then incubated with increasing levels of RPA (solid squares) or the RPA subunit fusion proteins (MBP-RPA14, open squares; MBP-RPA32, open circles; MBP-RPA70, solid circles and open triangles). Bound RPA or RPA subunits were detected with either rabbit anti-RPA (∗) or rabbit anti-MBP antibody. Levels of binding were evaluated as described in Materials and Methods. Note that the different primary antibody used to evaluate the binding of the RPA complex prevents meaningful quantitative comparison of RPA binding with RPA subunit binding.
Inhibition of SV40 in vitro DNA replication by RPA70.
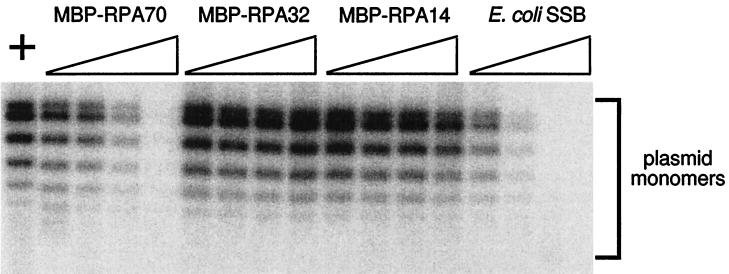
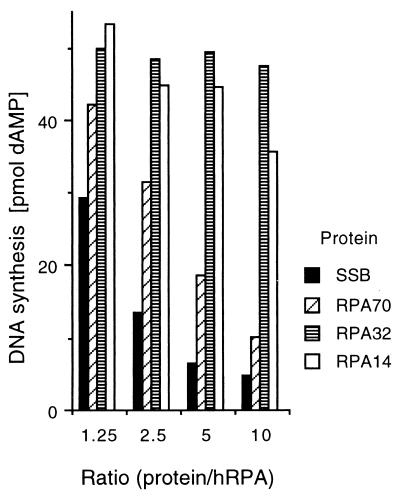
The RPA-Tag interaction has been shown to be critical for SV40 DNA replication in vitro (46, 73). Since MBP-RPA70 competes with RPA for binding to Tag, it was anticipated that high levels of exogenously added MBP-RPA70 would inhibit or squelch SV40 in vitro DNA replication by sequestering Tag from binding to the RPA complex in human cell extracts. To test this hypothesis, various proteins, including the purified recombinant RPA subunits, were titrated into in vitro SV40 DNA replication reaction mixtures. Of the three subunits of RPA, only MBP-RPA70 demonstrated a significant dose-dependent inhibitory effect. MBP-RPA32 did not have any inhibitory effect on SV40 in vitro DNA replication, whereas MBP-RPA14 showed only a slight inhibition and only at very great molar excess. (Note that the experiment shown in Fig. 6 was performed based on protein mass. The lower molecular mass of the MBP-RPA14 fusion protein, compared to those of the MBP-RPA32 and MBP-RPA70 fusion proteins, resulted in the addition of much higher molar levels of MBP-RPA14 than of the other subunits.) E. coli SSB also inhibited SV40 replication in vitro. The negative control, the MBP domain alone, did not show measurable inhibition of SV40 DNA replication (data not shown). Experiments utilizing a partially reconstituted SV40 DNA replication system dependent upon RPA showed that MBP-RPA70 is incapable of supporting SV40 DNA replication in vitro in the absence of the RPA heterotrimer (26).
FIG. 6.
Inhibition of SV40 DNA replication in vitro by the recombinant RPA subunits. Recombinant MBP-RPA14, MBP-RPA32, MBP-RPA70, or E. coli SSB was added to in vitro SV40 DNA replication reactions. The reaction mixtures were assembled and incubated at 37°C for 60 min as described in Materials and Methods. (Top) The products were separated on a 0.8% (wt/vol) agarose gel, and the gel was dried and subjected to phosphorimage analysis. Lane +, control reaction containing 0.3 mg of MBP/ml; each series of four lanes shows products from reactions with increasing levels of MBP-RPA70, MBP-RPA32, MBP-RPA14, and E. coli SSB, as indicated. The wedges indicate the increasing amounts of protein added to the reactions at final concentrations of 0.038, 0.075, 0.15, and 0.3 mg/ml. (Bottom) Histogram of the amount of DNA synthesis in each in vitro DNA replication assay. The values on the x axis indicate the approximate ratios of the added protein to the endogenous hRPA in the human cell extract. (The level of hRPA in each reaction due to the cell extract is estimated to be ∼0.03 mg/ml.) Total DNA synthesis was measured as described in Materials and Methods and is expressed as picomoles of dAMP incorporated in 60 min in a 0.05-ml reaction mixture. The control reaction (with no added protein) produced 55 pmol of dAMP synthesis, while the MBP (0.3 mg/ml) control resulted in 48 pmol of incorporated dAMP.
DISCUSSION
Specific interactions between SV40 Tag, RPA, and DNA polymerase α-primase are well documented, and there is a body of evidence suggesting that the physical associations among the three DNA replication proteins are important for viral DNA replication (10, 19, 43, 46, 55, 60, 67, 71). The functional similarities between BPV1 E1 and SV40 Tag suggested to us that E1 might interact with RPA and polymerase α-primase. We have previously detected in vitro interactions between E1 and polymerase α-primase (44), consistent with reports from other laboratories (4, 12, 57). In this study an ELISA-based protein interaction assay and a protein affinity pull-down assay were employed to demonstrate that BPV1 E1 binds to RPA. This interaction is seen regardless of which protein is immobilized as the solid phase on the ELISA plate. Therefore, like SV40 Tag, BPV1 E1 binds to both of the cellular replication complexes, RPA and DNA polymerase α-primase.
Since both viral initiator proteins have convergently evolved to bind to the same cellular replication proteins, and since the cellular replication proteins must retain their functions and protein-protein interactions during DNA replication, it is not unlikely that these viral proteins bind to the same or similar domains of the cellular replication proteins. Indeed, both Tag and E1 have been shown to bind to the two largest subunits of DNA polymerase α-primase (11, 12, 15, 17, 67). If this were the case for the interactions of Tag and E1 with RPA, one would predict that the presence of one of these viral proteins would negatively affect the level of binding between the other viral protein and RPA. This was shown to be true (Fig. 2). Furthermore, both viral proteins bound to only one of the three RPA subunits, RPA70 (Fig. 4 and 5). Another laboratory has also demonstrated that SV40 Tag binds to the 70-kDa subunit of RPA (20a). These results support the hypothesis that these viral proteins, E1 and Tag, bind to the cellular RPA complex via the same or similar domains on RPA.
The competition of challenge-target binding by soluble competitor proteins exhibited an unusual plateau effect (Fig. 2B and 4B). While the addition of equal amounts of competitor resulted in the expected 50% inhibition of binding to the target protein, one would predict that the signal of bound challenge protein would continue to decay as more competitor was added. Little additional inhibition was seen. A similar result was seen when RPA was immobilized and GST-E1 was incubated with the immobilized RPA and increasing levels of RPA in solution (26). These results seem to indicate that at high protein concentrations, binding to immobilized target protein is favored over binding to soluble protein. This effect precludes quantitative comparison of binding to immobilized and soluble proteins. However, the clear differences seen at low competitor/target ratios, and between competitors previously shown to bind to the target protein and the control competitor, argue that these competitions are qualitatively informative.
There exist two conflicting reports in the literature: one suggests that it is RPA32 that interacts with SV40 Tag, while the other indicates that RPA70 interacts with Tag and RPA32 does not (6, 39). Both studies utilized partial RPA subunit deletion mutants that nonetheless assemble into heterotrimeric RPA complexes. This approach was used in these studies because the individual RPA subunits are predominantly insoluble when the subunits are expressed individually. However, by nature this approach is susceptible to experimental artifact in that a “positive” result is defined by abrogation of binding. A loss of binding by a truncation mutant could be due to a secondary effect, such as altered protein conformation at a second site. Our goal was to demonstrate which RPA subunit specifically interacts with SV40 Tag and the BPV1 E1 protein by using a positive binding assay. The vectors described in this report that direct the expression of soluble RPA subunit fusion proteins have allowed us to address this question. We have demonstrated that it is the RPA70 subunit that is primarily responsible for the binding of Tag and E1 to the RPA heterotrimer. We hope that these vectors that direct the expression of predominantly soluble RPA subunit fusion proteins will be useful in addressing other questions pertaining to RPA function.
While our findings indicate that RPA70 mediates the interactions between RPA and the viral initiator proteins, we cannot totally exclude roles for RPA32 and RPA14 in these interactions. It is possible that RPA32 and/or RPA14 may allosterically modulate the structure of RPA70 and thereby affect RPA’s interaction with E1. This could account for the apparently lower binding levels of E1 to MBP-RPA70 than to the RPA heterotrimeric complex (Fig. 4A). Alternatively, RPA32 and RPA14 may have no effect on the E1-RPA70 interaction, and the apparently lower binding may be an experimental artifact caused by expressing RPA70 as a recombinant fusion protein. Although we have detected no such interactions, our experiments also do not exclude the possibility of direct interactions between E1 and RPA14 or RPA32.
Bonne-Andrea et al. (4) were unable to detect an interaction between E1 and RPA by either E1 protein affinity chromatography or protein interaction ELISAs (using E1 as a target protein to adsorb RPA from human cell extracts). These results apparently conflict with our own. The different results are likely attributable to the differing techniques used in the two studies. In the study by Bonne-Andrea et al., crude cell extracts were used as the source of RPA. The ELISA-based assay and the protein affinity pull-down assay in our study utilize purified proteins during critical protein interaction steps. No one has reported detecting the Tag-RPA interaction by coprecipitation from crude extracts. Therefore, it would not be surprising if the E1-RPA interaction was not detectable by this method. The interaction between these viral initiator proteins and RPA is relatively weak compared to their interactions with other cellular replication proteins (65a). However, it must be recognized that in vivo this interaction likely does not occur alone but rather in concert with other protein-protein interactions (such as Tag or E1 and polymerase α-primase, and polymerase α-primase and RPA), resulting in a much higher overall affinity.
Of the three recombinant RPA subunits, only MBP-RPA70 inhibited SV40 in vitro DNA replication reactions to a significant degree. MBP-RPA14 showed a very slight inhibition, but only at very high molar levels, while no inhibition of SV40 DNA replication was detected upon addition of MBP-RPA32 (Fig. 6). Therefore, it is possible that interactions between MBP-RPA70 and Tag prevented Tag from binding to the native RPA complex in these reactions, thus inhibiting SV40 DNA replication. Polymerase α-primase has also been shown to interact with RPA via RPA70. Therefore, it is also possible that MBP-RPA70 is abrogating this interaction. Further, it has been well documented that RPA70 is the major ssDNA-binding subunit of the RPA complex (1, 22–24, 33, 36, 58), and ssDNA-binding proteins, such as E. coli SSB and yeast RPA, can inhibit SV40 in vitro DNA replication in the presence of human RPA (Fig. 6) (44). It is therefore unclear whether the inhibitory effect of MBP-RPA70 on SV40 DNA replication in vitro is caused by squelching of the Tag-RPA interaction, by squelching of the RPA-polymerase α interaction, or by MBP-RPA70 competing with RPA for ssDNA binding. Further experiments designed to identify the minimal Tag-binding domain of RPA70 will help to address this question.
It has been shown that the RPA32 and RPA14 homologs are essential in S. cerevisiae, and mutants exhibit phenotypes consistent with a block in DNA replication (7, 30). Furthermore, antibodies against RPA32 subunits have been shown to inhibit SV40 DNA replication in vitro (33). Therefore, it was surprising that high levels of MBP-RPA14 or MBP-RPA32 did not inhibit SV40 DNA replication in vitro. One possible explanation is that the MBP domain is precluding these proteins from interacting with their natural partners, so that the fusion proteins do not compete with the RPA complex for essential interactions. Alternatively, the DNA replication functions of RPA14 and RPA32 may be merely to stabilize the RPA70 subunit, as previously suggested (74). While RPA14 and RPA32 may have additional functions (in DNA repair or recombination, for example), their requirement to form the heterotrimer so that RPA70 can support DNA replication may complicate the analysis of any other roles the smaller subunits may play. The ability of antibodies against RPA32 to inhibit SV40 DNA replication may be due to steric limitations at a very crowded DNA replication fork. It is still unclear whether RPA32 and RPA14 play a direct, vital role in DNA replication.
We propose that the polyomaviruses and the papillomaviruses have evolved similar mechanisms to appropriate the cellular DNA replication machinery. Both types of viruses have evolved a protein that recognizes the viral origin, acts as a DNA helicase, and binds to the two cellular DNA replication complexes, RPA and DNA polymerase α-primase. We propose that as these two cellular complexes are recruited to the viral DNA replication fork, they in turn recruit the other required cellular DNA replication proteins.
ACKNOWLEDGMENTS
This work was supported in part by a U.S. Public Health Service Research Grant (GM56406) and an American Cancer Society research grant (GMC 87550) and in part by funds provided by the State of New York and the SUNY Buffalo School of Medicine and Biomedical Sciences.
We thank L. A. Henricksen and M. Wold for the human RPA (hRPA) expression vectors and M. Xue and W. Ruyechan for the pMALcT plasmid. N. Luke assisted in the subcloning of the MBP-RPA70 expression vector. We also thank J. Newman for technical assistance, K. Fien, J.-S. Liu, S.-R. Kuo, and members of the L. Read laboratory and the Center for Microbial Pathogenesis for helpful scientific discussions, and K. Fien and E. Niles for comments on drafts of the manuscript.
REFERENCES
- 1.Bochkarev A, Pfuetzner R A, Edwards A M, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 2.Bochkareva E, Frappier L, Edwards A M, Bochkarev A. The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J Biol Chem. 1998;273:3932–3936. doi: 10.1074/jbc.273.7.3932. [DOI] [PubMed] [Google Scholar]
- 3.Bonne-Andrea C, Santucci S, Clertant P. Bovine papillomavirus E1 protein can, by itself, efficiently drive multiple rounds of DNA synthesis in vitro. J Virol. 1995;69:3201–3205. doi: 10.1128/jvi.69.5.3201-3205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonne-Andrea C, Santucci S, Clertant P, Tillier F. Bovine papillomavirus E1 protein binds specifically DNA polymerase alpha but not replication protein A. J Virol. 1995;69:2341–2350. doi: 10.1128/jvi.69.4.2341-2350.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borowiec J A, Dean F B, Bullock P A, Hurwitz J. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell. 1990;60:181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- 6.Braun K A, Lao Y, He Z, Ingles C J, Wold M S. Role of protein-protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase alpha by multiple mechanisms. Biochemistry. 1997;36:8443–8454. doi: 10.1021/bi970473r. [DOI] [PubMed] [Google Scholar]
- 7.Brill S J, Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 8.Brill S J, Stillman B. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature. 1989;342:92–95. doi: 10.1038/342092a0. [DOI] [PubMed] [Google Scholar]
- 9.Chow L T, Broker T R. Papillomavirus DNA replication. Intervirology. 1994;37:150–158. doi: 10.1159/000150373. [DOI] [PubMed] [Google Scholar]
- 10.Collins K L, Kelly T J. Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase alpha-primase. Mol Cell Biol. 1991;11:2108–2115. doi: 10.1128/mcb.11.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins K L, Russo A A, Tseng B Y, Kelly T J. The role of the 70 kDa subunit of human DNA polymerase alpha in DNA replication. EMBO J. 1993;12:4555–4566. doi: 10.1002/j.1460-2075.1993.tb06144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conger K L, Liu J S, Kuo S R, Chow L T, Wang T S F. Human papillomavirus DNA replication—interactions between the viral E1 protein and two subunits of human DNA polymerase alpha/primase. J Biol Chem. 1999;274:2696–2705. doi: 10.1074/jbc.274.5.2696. [DOI] [PubMed] [Google Scholar]
- 13.Depamphilis M J, Bradley M K. Replication of SV40 and polyoma virus chromosomes. In: Salzman N P, Howley P M, editors. The papovaviridae. New York, N.Y: Plenum Press; 1986. pp. 99–246. [Google Scholar]
- 14.Din S, Brill S J, Fairman M P, Stillman B. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes Dev. 1990;4:968–977. doi: 10.1101/gad.4.6.968. [DOI] [PubMed] [Google Scholar]
- 15.Dornreiter I, Copeland W C, Wang T S. Initiation of simian virus 40 DNA replication requires the interaction of a specific domain of human DNA polymerase alpha with large T antigen. Mol Cell Biol. 1993;13:809–820. doi: 10.1128/mcb.13.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dornreiter I, Erdile L F, Gilbert I U, von Winkler D, Kelly T J, Fanning E. Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dornreiter I, Hoss A, Arthur A K, Fanning E. SV40 T antigen binds directly to the large subunit of purified DNA polymerase alpha. EMBO J. 1990;9:3329–3336. doi: 10.1002/j.1460-2075.1990.tb07533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutta A, Ruppert J M, Aster J C, Winchester E. Inhibition of DNA replication factor RPA by p53. Nature. 1993;365:79–82. doi: 10.1038/365079a0. [DOI] [PubMed] [Google Scholar]
- 19.Erdile L F, Collins K L, Russo A, Simancek P, Small D, Umbricht C, Virshup D, Cheng L, Randall S, Weinberg D, et al. Initiation of SV40 DNA replication: mechanism and control. Cold Spring Harbor Symp Quant Biol. 1991;56:303–313. doi: 10.1101/sqb.1991.056.01.037. [DOI] [PubMed] [Google Scholar]
- 20.Fairman M P, Stillman B. Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Fanning, E. Personal communication.
- 21.Gannon J V, Lane D P. p53 and DNA polymerase alpha compete for binding to SV40 T antigen. Nature. 1987;329:456–458. doi: 10.1038/329456a0. [DOI] [PubMed] [Google Scholar]
- 22.Gomes X V, Henricksen L A, Wold M S. Proteolytic mapping of human replication protein A: evidence for multiple structural domains and a conformational change upon interaction with single-stranded DNA. Biochemistry. 1996;35:5586–5595. doi: 10.1021/bi9526995. [DOI] [PubMed] [Google Scholar]
- 23.Gomes X V, Wold M S. Functional domains of the 70-kilodalton subunit of human replication protein A. Biochemistry. 1996;35:10558–10568. doi: 10.1021/bi9607517. [DOI] [PubMed] [Google Scholar]
- 24.Gomes X V, Wold M S. Structural analysis of human replication protein A. Mapping functional domains of the 70-kDa subunit. J Biol Chem. 1995;270:4534–4543. doi: 10.1074/jbc.270.9.4534. [DOI] [PubMed] [Google Scholar]
- 25.Gopalakrishnan V, Khan S A. E1 protein of human papillomavirus type 1a is sufficient for initiation of viral DNA replication. Proc Natl Acad Sci USA. 1994;91:9597–9601. doi: 10.1073/pnas.91.20.9597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han, Y., and T. Melendy. Unpublished data.
- 27.He Z, Brinton B T, Greenblatt J, Hassell J A, Ingles C J. The transactivator proteins VP16 and GAL4 bind replication factor A. Cell. 1993;73:1223–1232. doi: 10.1016/0092-8674(93)90650-f. [DOI] [PubMed] [Google Scholar]
- 28.Henricksen L A, Umbricht C B, Wold M S. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. . (Erratum, 269:16519.) [PubMed] [Google Scholar]
- 29.Henricksen L A, Wold M S. Replication protein A mutants lacking phosphorylation sites for p34cdc2 kinase support DNA replication. J Biol Chem. 1994;269:24203–24208. [PubMed] [Google Scholar]
- 30.Heyer W D, Rao M R, Erdile L F, Kelly T J, Kolodner R D. An essential Saccharomyces cerevisiae single-stranded DNA binding protein is homologous to the large subunit of human RP-A. EMBO J. 1990;9:2321–2329. doi: 10.1002/j.1460-2075.1990.tb07404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurwitz J, Dean F B, Kwong A D, Lee S H. The in vitro replication of DNA containing the SV40 origin. J Biol Chem. 1990;265:18043–18046. [PubMed] [Google Scholar]
- 32.Kelly T J. SV40 DNA replication. J Biol Chem. 1988;263:17889–17892. [PubMed] [Google Scholar]
- 33.Kenny M K, Lee S H, Hurwitz J. Multiple functions of human single-stranded-DNA binding protein in simian virus 40 DNA replication: single-strand stabilization and stimulation of DNA polymerases alpha and delta. Proc Natl Acad Sci USA. 1989;86:9757–9761. doi: 10.1073/pnas.86.24.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kenny M K, Schlegel U, Furneaux H, Hurwitz J. The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J Biol Chem. 1990;265:7693–7700. [PubMed] [Google Scholar]
- 35.Keshav K F, Chen C, Dutta A. Rpa4, a homolog of the 34-kilodalton subunit of the replication protein A complex. Mol Cell Biol. 1995;15:3119–3128. doi: 10.1128/mcb.15.6.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D K, Stigger E, Lee S H. Role of the 70-kDa subunit of human replication protein A (I). Single-stranded DNA binding activity, but not polymerase stimulatory activity, is required for DNA replication. J Biol Chem. 1996;271:15124–15129. doi: 10.1074/jbc.271.25.15124. [DOI] [PubMed] [Google Scholar]
- 37.Lambert P F. Papillomavirus DNA replication. J Virol. 1991;65:3417–3420. doi: 10.1128/jvi.65.7.3417-3420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanford R E. Expression of simian virus 40 T antigen in insect cells using a baculovirus expression vector. Virology. 1988;167:72–81. doi: 10.1016/0042-6822(88)90055-4. [DOI] [PubMed] [Google Scholar]
- 39.Lee S H, Kim D K. The role of the 34-kDa subunit of human replication protein A in simian virus 40 DNA replication in vitro. J Biol Chem. 1995;270:12801–12807. doi: 10.1074/jbc.270.21.12801. [DOI] [PubMed] [Google Scholar]
- 40.Lee S H, Kim D K, Drissi R. Human xeroderma pigmentosum group A protein interacts with human replication protein A and inhibits DNA replication. J Biol Chem. 1995;270:21800–21805. doi: 10.1074/jbc.270.37.21800. [DOI] [PubMed] [Google Scholar]
- 41.Li R, Botchan M. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 42.Lin Y L, Chen C, Keshav K F, Winchester E, Dutta A. Dissection of functional domains of the human DNA replication protein complex replication protein A. J Biol Chem. 1996;271:17190–17198. doi: 10.1074/jbc.271.29.17190. [DOI] [PubMed] [Google Scholar]
- 43.Matsumoto T, Eki T, Hurwitz J. Studies on the initiation and elongation reactions in the simian virus 40 DNA replication system. Proc Natl Acad Sci USA. 1990;87:9712–9716. doi: 10.1073/pnas.87.24.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melendy, T. Unpublished data.
- 45.Melendy T, Sedman J, Stenlund A. Cellular factors required for papillomavirus DNA replication. J Virol. 1995;69:7857–7867. doi: 10.1128/jvi.69.12.7857-7867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melendy T, Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J Biol Chem. 1993;268:3389–3395. [PubMed] [Google Scholar]
- 47.Melendy T, Stillman B. SV40 DNA replication. In: Eckstein F, Lilly D M J, editors. Nucleic acids and molecular biology. Vol. 6. Berlin, Germany: Springer Verlag; 1992. pp. 129–158. [Google Scholar]
- 48.Muller F, Seo Y S, Hurwitz J. Replication of bovine papillomavirus type 1 origin-containing DNA in crude extracts and with purified proteins. J Biol Chem. 1994;269:17086–17094. [PubMed] [Google Scholar]
- 49.Murakami Y, Eki T, Hurwitz J. Studies on the initiation of simian virus 40 replication in vitro: RNA primer synthesis and its elongation. Proc Natl Acad Sci USA. 1992;89:952–956. doi: 10.1073/pnas.89.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murakami Y, Hurwitz J. DNA polymerase alpha stimulates the ATP-dependent binding of simian virus tumor T antigen to the SV40 origin of replication. J Biol Chem. 1993;268:11018–11027. [PubMed] [Google Scholar]
- 51.Murakami Y, Hurwitz J. Functional interactions between SV40 T antigen and other replication proteins at the replication fork. J Biol Chem. 1993;268:11008–11017. [PubMed] [Google Scholar]
- 52.Murakami Y, Wobbe C R, Weissbach L, Dean F B, Hurwitz J. Role of DNA polymerase alpha and DNA primase in simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1986;83:2869–2873. doi: 10.1073/pnas.83.9.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murphy C I, Weiner B, Bikel I, Piwnica-Worms H, Bradley M K, Livingston D M. Purification and functional properties of simian virus 40 large and small T antigens overproduced in insect cells. J Virol. 1988;62:2951–2959. doi: 10.1128/jvi.62.8.2951-2959.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagelhus T A, Haug T, Singh K K, Keshav K F, Skorpen F, Otterlei M, Bharati S, Lindmo T, Benichou S, Benarous R, Krokan H E. A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J Biol Chem. 1997;272:6561–6566. doi: 10.1074/jbc.272.10.6561. [DOI] [PubMed] [Google Scholar]
- 55.Nasheuer H P, von Winkler D, Schneider C, Dornreiter I, Gilbert I, Fanning E. Purification and functional characterization of bovine RP-A in an in vitro SV40 DNA replication system. Chromosoma. 1992;102:S52–S59. doi: 10.1007/BF02451786. [DOI] [PubMed] [Google Scholar]
- 56.Park M S, Ludwig D L, Stigger E, Lee S H. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J Biol Chem. 1996;271:18996–19000. doi: 10.1074/jbc.271.31.18996. [DOI] [PubMed] [Google Scholar]
- 57.Park P, Copeland W, Yang L, Wang T, Botchan M R, Mohr I J. The cellular DNA polymerase alpha-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc Natl Acad Sci USA. 1994;91:8700–8704. doi: 10.1073/pnas.91.18.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Philipova D, Mullen J R, Maniar H S, Lu J, Gu C, Brill S J. A hierarchy of SSB promoters in replication protein A. Genes Dev. 1996;10:2222–2233. doi: 10.1101/gad.10.17.2222. [DOI] [PubMed] [Google Scholar]
- 59.Santucci S, Bonne-Andrea C, Clertant P. Bovine papillomavirus type 1 E1 ATPase activity does not depend on binding to DNA nor to viral E2 protein. J Gen Virol. 1995;76:1129–1140. doi: 10.1099/0022-1317-76-5-1129. [DOI] [PubMed] [Google Scholar]
- 60.Schneider C, Weisshart K, Guarino L A, Dornreiter I, Fanning E. Species-specific functional interactions of DNA polymerase alpha-primase with simian virus 40 (SV40) T antigen require SV40 origin DNA. Mol Cell Biol. 1994;14:3176–3185. doi: 10.1128/mcb.14.5.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sedman J, Stenlund A. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 1995;14:6218–6228. doi: 10.1002/j.1460-2075.1995.tb00312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sedman J, Stenlund A. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J Virol. 1998;72:6893–6897. doi: 10.1128/jvi.72.8.6893-6897.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seo Y S, Muller F, Lusky M, Gibbs E, Kim H Y, Phillips B, Hurwitz J. Bovine papilloma virus (BPV)-encoded E2 protein enhances binding of E1 protein to the BPV replication origin. Proc Natl Acad Sci USA. 1993;90:2865–2869. doi: 10.1073/pnas.90.7.2865. . (Erratum, 90:5878.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seo Y S, Muller F, Lusky M, Hurwitz J. Bovine papilloma virus (BPV)-encoded E1 protein contains multiple activities required for BPV DNA replication. Proc Natl Acad Sci USA. 1993;90:702–706. doi: 10.1073/pnas.90.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simanis V, Lane D P. An immunoaffinity purification procedure for SV40 large T antigen. Virology. 1985;144:80–100. doi: 10.1016/0042-6822(85)90308-3. [DOI] [PubMed] [Google Scholar]
- 65a.Simmons, D. Personal communication.
- 66.Smale S T, Tjian R. T-antigen DNA polymerase alpha complex implicated in simian virus 40 DNA replication. Mol Cell Biol. 1986;6:4077–4087. doi: 10.1128/mcb.6.11.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stadlbauer F, Voitenleitner C, Bruckner A, Fanning E, Nasheuer H P. Species-specific replication of simian virus 40 DNA in vitro requires the p180 subunit of human DNA polymerase alpha-primase. Mol Cell Biol. 1996;16:94–104. doi: 10.1128/mcb.16.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stigger E, Dean F B, Hurwitz J, Lee S H. Reconstitution of functional human single-stranded DNA-binding protein from individual subunits expressed by recombinant baculoviruses. Proc Natl Acad Sci USA. 1994;91:579–583. doi: 10.1073/pnas.91.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stillman B, Gerard R D, Guggenheimer R A, Gluzman Y. T antigen and template requirements for SV40 DNA replication in vitro. EMBO J. 1985;4:2933–2999. doi: 10.1002/j.1460-2075.1985.tb04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsurimoto T, Melendy T, Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990;346:534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- 71.Tsurimoto T, Stillman B. Multiple replication factors augment DNA synthesis by the two eukaryotic DNA polymerases, alpha and delta. EMBO J. 1989;8:3883–3889. doi: 10.1002/j.1460-2075.1989.tb08567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 73.Weisshart K, Taneja P, Fanning E. The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication. J Virol. 1998;72:9771–9781. doi: 10.1128/jvi.72.12.9771-9781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wold M S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 75.Yang L, Li R, Mohr I J, Clark R, Botchan M R. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature. 1991;353:628–632. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]
- 76.Yang L, Mohr I, Fouts E, Lim D A, Nohaile M, Botchan M. The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc Natl Acad Sci USA. 1993;90:5086–5090. doi: 10.1073/pnas.90.11.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang L, Mohr I, Li R, Nottoli T, Sun S, Botchan M. Transcription factor E2 regulates BPV-1 DNA replication in vitro by direct protein-protein interaction. Cold Spring Harbor Symp Quant Biol. 1991;56:335–346. doi: 10.1101/sqb.1991.056.01.040. [DOI] [PubMed] [Google Scholar]
- 78.Zhang D, Frappier L, Gibbs E, Hurwitz J, O’Donnell M. Human RPA (hSSB) interacts with EBNA1, the latent origin binding protein of Epstein-Barr virus. Nucleic Acids Res. 1998;26:631–637. doi: 10.1093/nar/26.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]