Abstract
The alphavirus Sindbis virus (SV) has a wide host range and infects many types of cultured cells in vitro. The outcome of infection is dependent on the strain of virus used for infection and the properties of the cells infected. To identify cellular determinants of susceptibility to SV infection we mutagenized Chinese hamster ovary (CHO) cells by retroviral insertion with a vector containing the neomycin resistance gene that allowed selection for integration into transcriptionally active genes. Cells were then selected for survival after infection with SV. The most resistant cell line (CHO-18.4m) exhibited delayed virus replication and virus-induced cell death, had a single retroviral insertion, and was defective in SV binding to the cell surface. Further analysis revealed that CHO-18.4m cells were deficient in the expression of the sulfated glycosaminoglycans heparan sulfate and chondroitin sulfate. This further confirms the importance of heparan sulfate as an attachment molecule for SV in vitro and demonstrates the usefulness of this technique for identifying cellular genes that are important for virus replication.
Sindbis virus (SV) is an enveloped, message-sense, single-strand RNA virus that causes rash and arthritis in humans (30) and encephalomyelitis in mice (18). In mice the primary target cells for SV infection are neurons in the brain and spinal cord, and the outcome of central nervous system infection is linked to the efficiency of virus replication and the induction of apoptosis in these cells (19, 24). Newborn mice are very susceptible to fatal SV-induced encephalomyelitis, while in older mice infection is less likely to be fatal. This is linked to the susceptibility of immature neurons to SV-induced apoptosis, while mature neurons often survive infection (23, 24). However, the cellular and host factors responsible for these differences in outcome are largely unknown.
SV has two surface glycoproteins, E1 and E2, which form heterodimers that are trimerized into spikes on the virion surface (36). E2 is an important determinant of virulence and plays a role in the initial binding of the virus to the cell surface. SV, like all alphaviruses, has a wide host range, and the natural cycle of infection requires replication in both vertebrate and invertebrate hosts. Host factors are required not only for virus binding and entry, but also for the replication of plus- and minus-strand RNA, the transcription and translation of mRNA, and the synthesis and processing of the glycoproteins. Thus, there are many steps in the virus life cycle where differences in the availability or specificity of host cell factors could affect virus replication. In addition, the susceptibility of a cell to the induction of apoptosis by SV infection is dependent on the expression of an array of apoptotic and antiapoptotic factors that are still incompletely defined (11).
To begin to identify cellular factors that influence the outcome of SV infection we have used a retroviral gene trap strategy that incorporates the use of a selectable marker to enrich for cells with insertional mutations (6, 42) to generate mutant Chinese hamster ovary (CHO) cells resistant to SV. CHO cells were chosen for this study because they are hypodiploid and have substantial functional hemizygosity at many different loci (14, 15), and mutants have been isolated from CHO cells at a frequency representing genes with a single allele (35), including cells with resistance to SV infection (27, 28). Therefore, CHO cells appear to be a cell line appropriate for genetic mutant isolation by using retroviruses as insertional mutagens. The gene trap approach has been successfully employed for mutagenesis in vitro and in vivo, and the retrovirus can serve as a useful tag for the identification of the disrupted gene (9, 16, 21).
In this study a number of mutant CHO cell clones partially resistant to SV infection were generated. One of these cell lines, CHO-18.4m, had a single retroviral insertion and was highly resistant to SV-induced cell death. This cell line bound SV inefficiently and exhibited delayed virus replication. Because cell surface heparan sulfate is an important attachment molecule for SV (5, 22) we analyzed CHO-18.4m cells for the synthesis of sulfated glycosaminoglycans. The cells were deficient in heparan sulfate, further confirming the importance of heparan sulfate as an attachment molecule for SV in vitro. This technique may be useful for identifying other cellular genes important for virus replication.
MATERIALS AND METHODS
Cell lines.
CHO cells expressing the Moloney murine leukemia virus (Mo-MuLV) ecotropic receptor (CHO-22) were obtained from H. Earl Ruley, Vanderbilt University (1, 16). This cell line was derived from CHO-K1 cells and was used as the host cell for retrovirus infection. CHO-22 and BHK cells were grown in Dulbecco’s modified Eagle medium (DMEM) (GIBCO, Grand Island, N.Y.) supplemented with 10% fetal bovine serum (FBS) (GIBCO).
Viruses.
Three strains of SV were used. AR339 was obtained from the American Type Culture Collection (Manassas, Va.). NSV, a neuroadapted strain of SV, was derived by serial passage of SV AR339 in mouse brain (12). HRSP, a heat-resistant small-plaque strain of AR339, was derived from serial passages of the HR strain in chicken embryo fibroblasts (4). These viruses were plaque purified, stocks were grown, and titers were determined by plaque formation under agar in BHK-21 cells.
U3Neo Mo-MuLV, used to infect CHO-22 cells, was harvested from the supernatant fluid of Ψ85 virus producer cells (6). This virus has the neomycin resistance gene inserted into the U3 region. Expression of the neomycin resistance gene is dependent on insertion into transcriptionally active cellular promoters (42). For each experiment, fresh producer cell supernatant fluid, passed through a 0.2-μm-pore-size filter, was used as a virus stock. The Mo-MuLV titer was estimated from the number of G418-resistant colonies arising after infection of CHO-22 cells with serial dilutions of virus stocks.
Mo-MuLV infection and selection for provirus-containing, SV-resistant CHO clones.
CHO-22 cells (106) were seeded on 100-mm dishes and the next day were incubated for 2 h with fresh U3Neo Mo-MuLV virus stock at a multiplicity of infection (MOI) of 0.01 in 0.5 ml of a medium containing 8 μg of Polybrene (16) per ml. The medium was removed, and 10 ml of the complete medium was added. One to 2 days later, selection was begun in a medium containing G418 (1 mg/ml) (Mediatech, Inc., Herndon, Va.). Seven to 10 days after the initiation of G418 selection, the second phase of selection was begun by infecting surviving cells with HRSP at an MOI of 5. Five to 7 days later, SV-resistant colonies were picked from the plates by using cloning rings and trypsinization. Individual plates contained 0 to 10 SV-resistant colonies and, to avoid sibling clones, only one colony was generally picked per dish. Cell clones were further expanded in 24-well plates. Some of these clones later died.
Molecular identification of the retroviral integration.
Genomic DNA from mutant CHO-18.4m cells was digested with HindIII, separated by agarose gel electrophoresis, and transferred to a prehybridized Hybond N+ charged nylon membrane (Amersham, Arlington Heights, Ill.). Hybridization was done with an [α-32P]dCTP-labeled PstI-NcoI 387-bp fragment (from pOP13CAT; Stratagene, La Jolla, Calif.) encompassing the 5′ long terminal repeat of Mo-MuLV. The membrane was incubated overnight at 65°C, washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (pH 7.0)–0.1% (wt/vol) sodium dodecyl sulfate (SDS) at room temperature and then in 1× SSC–0.1% SDS at 65°C for 20 min. For autoradiography, film was exposed for at least 2 days at −70°C.
Radiolabeling and purification of virions.
BHK-21 cells were inoculated with SV at an MOI of 1 in DMEM (2% FBS), and 1 h later the virus inoculum was removed and the cells were fed with methionine- and cysteine-free MEM containing 10 μCi of [35S]methionine-cysteine (Trans-label; ICN, Costa Mesa, Calif.) per ml. When more than a 90% cytopathic effect was evident, the supernatant fluid was harvested and clarified. Virus was precipitated in 10% (wt/vol) polyethylene glycol 8000 in 0.5 M NaCl for 2 to 3 h at 4°C and centrifuged at 10,000 × g for 1 h. Pelleted virus was resuspended in NET buffer (10 mM Tris, 3 mM EDTA, 150 mM NaCl [pH 7.4]) and banded in a continuous gradient of 15 to 40% potassium tartrate in phosphate-buffered saline (pH 7.4) at 132,000 × g for 3 h. Banded virus was dialyzed against 0.05 M Tris-Cl (pH 7.4) at 4°C and stored in aliquots at −70°C.
Virus binding assays.
These assays were performed as previously described (39). Briefly, radiolabeled virus (4,000 cpm per well) diluted in 0.4 ml of a cold binding medium (RPMI 1640 without NaHCO3, 0.2% bovine serum albumin, 20 mM HEPES) was added to confluent cells at 4°C. The supernatant and wash fluids were collected from triplicate wells to determine the level of unbound virus. Cells were dissolved in 1% SDS to determine the level of bound virus. The radioactivity in each fraction was counted.
Radioactive amino acid labeling for cellular protein synthesis.
Confluent cells in 25-cm2 culture flasks were infected with SV at an MOI of 15. At various times after infection, 2 ml of a medium containing [35S]methionine-cysteine (50 μCi/ml) was added for 1 h. Cells were then washed with phosphate-buffered saline and lysed with radioimmunoprecipitation assay buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 10 μM EDTA, 0.1% SDS, 1% Triton X-100, 1% deoxycholate). Equal amounts of cell lysates were analyzed on an SDS–15% polyacrylamide gel.
Infectious-center assay.
CHO cells (106) were infected with SV at an MOI of 1 (titers determined in BHK-21 cells) for 2 h at 37°C. Cells were then washed six times with a cold medium to remove virus that had not been internalized and trypsinized. Suspended cells were washed three times with the medium, diluted, and placed onto a confluent monolayer of BHK-21 cells in six-well plates. After incubation for 1 h at 37°C, 0.6% agar in MEM containing 2% FBS was added. Plaques were counted 48 h after incubation.
Assessment of cell surface GAGs.
Cells were treated with 6 mIU of heparinase I (EC 4.2.2.7) (Sigma Chemical Co., St. Louis, Mo.) per ml for 1 h at room temperature as previously described (5), and the effect on virus binding was assessed. The amount of cell surface heparan sulfate and chondroitin sulfate was assayed quantitatively by labeling cells with 35SO42− and precipitating glycosaminoglycans (GAGs) with cetylpyridinium chloride (CPC) (7). Sixty-millimeter-diameter dishes containing 106 cells were labeled for 24 h in 5 ml of sulfate-free DMEM containing 10% dialyzed FBS, 100 U of penicillin per ml, and 50 μCi of 35SO42− (New England Nuclear, Beverly, Mass.). Cells were lysed with 0.1 N NaOH, exhaustively digested with pronase, and precipitated with 1.25% CPC according to the protocol of J. D. Esko (7). Chondroitin sulfate A at a concentration of 4 mg/ml was included as a carrier. GAGs were treated with 200 mIU of chondroitinase ABC (Sigma) or mock treated and then reprecipitated with CPC containing a carrier GAG, and radioactivity was counted by liquid scintillation. Counts were normalized to the protein concentration of the original cell lysate, as determined by the Bradford assay (Bio-Rad, Hercules, Calif.) with bovine serum albumin as a standard.
RESULTS
Generation of SV-resistant CHO mutant cells by retroviral insertional mutagenesis.
CHO-22 cells were infected with U3Neo Mo-MuLV and selected first with G418 and then with the HRSP strain of SV. Although the U3Neo vector is inserted randomly, only those insertions which juxtapose the neomycin resistance gene and a cellular promoter will result in G418 resistance, since the U3 promoter is disrupted in this virus (3, 20, 31, 34, 42).
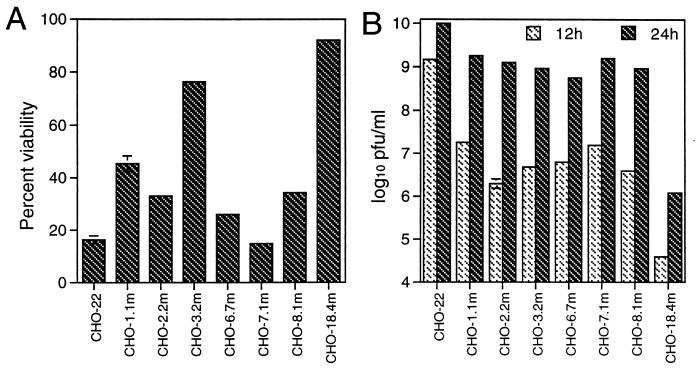
In principle, because a mammalian cell expresses only 10,000 to 20,000 genes, a library of 2 × 105 Neor cell clones should be enough to disrupt all readily targeted genes. We screened more than 2 × 106 Neor cell clones by HRSP infection. Approximately 1 in 1,000 G418-resistant colonies survived infection with HRSP. Approximately two-thirds of these colonies died thereafter, usually associated with productive virus replication. Finally, a total of 76 resistant clones were obtained. These mutant clones were then tested to confirm their resistance to HRSP infection. Different levels of resistance, as judged by viability (Fig. 1A) and virus production (Fig. 1B), were evident. Among these clones, CHO-18.4m was the most resistant to HRSP infection and was selected for further studies.
FIG. 1.
Viabilities and virus production levels of different representative CHO cell clones after HRSP infection. Parent CHO-22 cells and seven clones of mutant CHO cells were infected with HRSP at a BHK MOI of 5. (A) At 36 h after infection, cell viabilities were assessed by trypan blue exclusion. (B) Virus production at 12 and 24 h after infection was determined by plaque assay on BHK-21 cells. Error bars indicate standard deviations.
Characterization of the resistance of CHO-18.4m cells to SV infection.
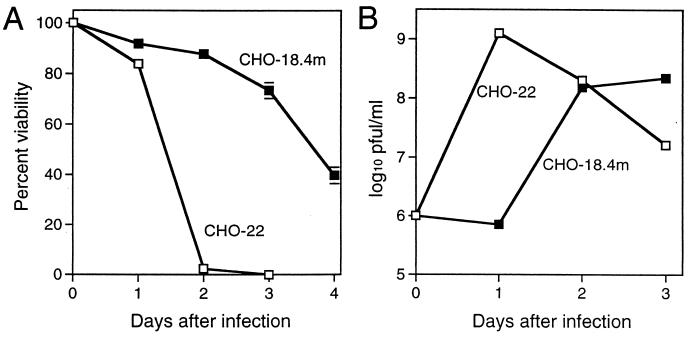
CHO-18.4m cells showed no visible plaques under agar after infection by HRSP while the expected small plaques (about 1 to 2 mm in diameter) were obtained in the parent CHO-22 cells (Table 1). There were no significant differences in plaque size after infection with the more neurovirulent AR339 and NSV strains of SV (12, 25), but CHO-18.4m cells were less susceptible to infection since the number of plaques formed was 5- to 10-fold lower than the number formed in CHO-22 cells (Table 1). The CHO-18.4m cells survived longer after infection (Fig. 2A), and the growth of HRSP in CHO-18.4m cells was delayed by 24 h compared to growth in CHO-22 cells (Fig. 2B).
TABLE 1.
Plaque formation in CHO-18.4m and CHO-22 cells after infection with three different strains of SV
| SV strain | Virulence in mice | Cell line | No. of plaques | Plaque sizea |
|---|---|---|---|---|
| HRSP | +/− | CHO-22 | 5 × 107 | Small |
| CHO-18.4m | 0 | |||
| AR339 | ++ | CHO-22 | 2 × 108 | Large |
| CHO-18.4m | 3 × 107 | Large | ||
| NSV | ++++ | CHO-22 | 3 × 108 | Large |
| CHO-18.4m | 5 × 107 | Large |
HRSP plaques in CHO-18.4m cells under agar were too small to be seen 3 days after infection.
FIG. 2.
Characteristics of CHO-18.4m and CHO-22 cells after HRSP infection. Cells were infected with HRSP at a BHK MOI of 5. Viability (A) was assessed by trypan blue exclusion, and virus production (B) was assessed by plaque assay in BHK-21 cells. Error bars indicate standard deviations.
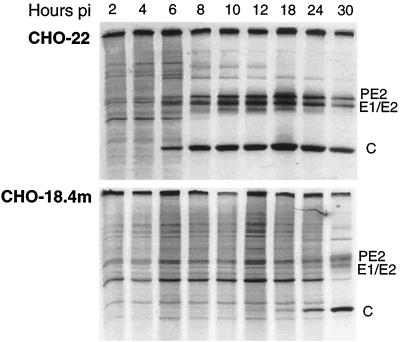
To determine whether the production of infectious virus was inhibited at an early or late step in replication, the production of HRSP viral structural proteins (PE2, E1 or E2, and C) was assessed (Fig. 3). Viral proteins were detectable 6 h after infection in CHO-22 cells but not until 18 h after infection in CHO-18.4m cells. Cellular protein synthesis was eventually shut off in both cell lines, but this was also delayed in CHO-18.4m cells.
FIG. 3.
Protein synthesis in CHO-18.4m and CHO-22 cells after HRSP infection. The synthesis of viral proteins and the shutoff of host protein synthesis are delayed in CHO-18.4m cells compared to CHO-22 cells. Cells were infected with HRSP at a BHK MOI of 15 and pulse labeled with [35S]methionine for 1 h at the indicated times postinfection (pi).
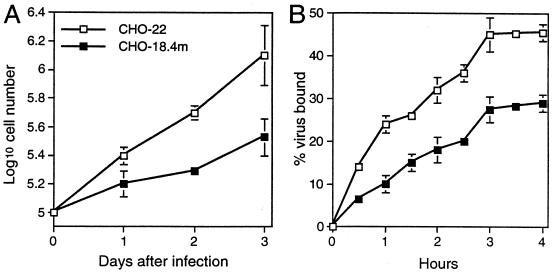
To characterize further the resistance of CHO-18.4m cells to SV infection, virus binding was analyzed. CHO-18.4m cells grew more slowly than CHO-22 cells (Fig. 4A). Therefore, the cell numbers were adjusted to be equal when the binding assays were performed. The binding of 35S-labeled HRSP virus to both cell lines reached a plateau at 3 h, but the binding to CHO-18.4m cells was less efficient than the binding to CHO-22 cells (Fig. 4B).
FIG. 4.
Comparison of cell proliferation and HRSP binding to CHO-18.4m and CHO-22 cells. (A) Proliferation of cells cultured at 37°C in DMEM supplemented with 10% FBS and nonessential amino acids. (B) Binding of 35S-labeled HRSP. Error bars indicate standard deviations.
Infectious-center assay of HRSP-infected CHO-18.4m and CHO-22 cells.
We further analyzed the comparative efficiencies of SV infection of CHO-18.4m and CHO-22 cells by an infectious-center assay, which assesses productive infection of individual cells. After 2 h of incubation with HRSP at an MOI (as determined in BHK cells) of 1, 106 cells from each CHO cell clone were analyzed for their ability to initiate infection when overlaid on BHK cells. Only 1.2 × 102 CHO-18.4m cells (0.012%) were scored as infectious, whereas 2 × 104 CHO-22 cells (2%) were scored as infectious. The ratio of infectious CHO-22 cells to infectious CHO-18.4m cells was therefore approximately 100:1, a difference that is larger than that found in virus binding.
Analysis of retroviral insertion in CHO-18.4m cells.
To determine the number of retroviral integrations into the cellular genomes of CHO-18.4m cells, DNA isolated from CHO-18.4m and CHO-22 cells was digested with HindIII and subjected to Southern blot analysis with the neomycin resistance gene probe (Fig. 5). A band of 7.2 kb was seen in CHO-18.4m DNA but not in CHO-22 DNA. Therefore, only one copy of the retroviral vector was inserted into CHO-18.4m cells, consistent with the disruption of a single gene.
FIG. 5.

Retrovirus integration in CHO-18.4m cells. Southern blot analysis of the neomycin resistance gene in CHO-18.4m and CHO-22 cells. Genomic DNAs were digested with HindIII, separated on a 0.6% agarose gel, transferred to a membrane, and hybridized with a 32P-labeled probe for the neomycin resistance gene.
Analysis of the presence of GAGs on the cell surface.
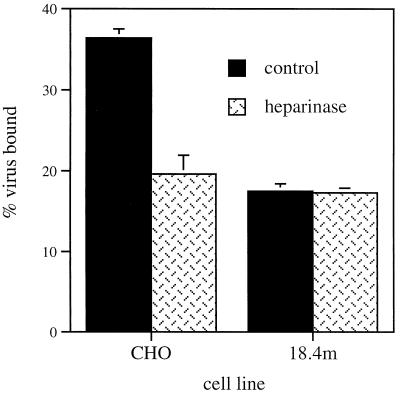
Since heparan sulfate was recently identified as an important initial cell surface binding element for SV (5, 22) and CHO-18.4m cells had a defect in virus binding, cells were analyzed to determine whether CHO-18.4m cells might be deficient in GAGs (Fig. 6). The pretreatment of cells with heparinase significantly decreased the binding of SV to CHO-22 cells but did not affect SV binding to CHO-18.4m cells, indicating that CHO-18.4m cells lack heparan sulfate.
FIG. 6.
Effect on virus binding of treating cells with heparinase. CHO-22 and CHO-18.4m cells were treated with heparinase, or were mock treated, and assayed for the binding of 35S-labeled HRSP after 1 h at 4°C. Error bars indicate standard deviations.
In order to quantitate the relative amounts of cell surface sulfated GAGs on CHO-18.4m cells, cells were cultured in the presence of 35SO4 for 24 h. Three separate 60-mm-diameter dishes were assayed for each cell line. GAGs were isolated, and the amount of 35S incorporated was quantitated. GAGs from CHO-18.4m cells contained only 3% as much label as those from CHO-22 cells (57.3 ± 16.2 and 1,792.8 ± 726.3 cpm per μg of protein, respectively), indicating either a severe reduction in the sulfation of GAGs or a reduction in the synthesis of heparan sulfate and chondroitin sulfate. The small amount of 35S-labeled GAGs present on CHO-18.4m cells was completely sensitive to digestion with chondroitinase ABC, indicating a total lack of cell surface heparan sulfate (after digestion with chondroitinase ABC, −3.27 ± 0.54 and 1,167.3 ± 413.4 cpm per μg of protein for CHO-18.4m and CHO-22 cells, respectively).
DISCUSSION
SV infects most vertebrate cell lines and causes apoptosis. Cells have different susceptibilities to SV infection and to SV-induced apoptosis depending on cell type and maturity both in vivo and in vitro. Cellular factors are therefore important in influencing the process of virus infection. For example, BHK cells are killed faster and produce a higher titer of virus than CHO cells. A mosquito cell line, C6/36, is not killed by AR339; however, this virus induces cell death in most vertebrate cell lines. The retroviral gene trap is a useful tool to generate cell mutants expressing altered phenotypes and to pinpoint the genomic loci associated with these mutant phenotypes. We have shown that cells resistant to SV infection can be developed by retroviral insertional mutagenesis. The mutant cells isolated were shown to lack heparan sulfate, a cell surface GAG that is responsible for the initial binding of SV to the surfaces of cells in culture (5, 22).
Using the retroviral gene trap approach to selecting cells resistant to SV infection produced 76 mutant CHO cell clones. However, none of these clones was completely resistant to HRSP, indicating that SV replication involves many cellular genes and that those that are essential for virus replication may also be essential for cell survival. Partial resistance to SV infection has also been a characteristic of CHO cells selected after chemical mutagenesis (27, 28). We assume that the viral selection of mutant cells was successful, despite the fact that resistance to HRSP-induced apoptosis was not complete, because the efficiency of infection of the mutant cells was markedly reduced. During the initial selection there were on average fewer than 10 cell colonies surviving in a 100-mm-diameter culture plate. Any virus produced became diluted in the overlying medium. The cellular mutations selected led to inefficient infection, which is most apparent when the MOI is low. This may be the reason that many of the cell clones died after they were propagated and thus were exposed to more concentrated virus.
The CHO-18.4m cell line was the most resistant to HRSP infection and showed a deficiency in virus binding. Southern blot analysis showed that only one copy of the retroviral vector was inserted into cellular DNA. The cellular gene disrupted was therefore likely to be involved in the expression of a molecule that participates in virus binding. We chose the gene trap approach to mutagenesis to enhance our ability to identify cellular genes important for SV replication. However, we were able to sequence only 9 bp of the flanking sequence, which was insufficient to identify the cellular gene disrupted (data not shown), and therefore we turned to the candidate gene approach based on our previous studies of molecules important for SV binding to CHO cells (5) to identify the cellular defect.
Several attempts have been made to identify a SV receptor(s). The first studies were reported by Smith and Tignor (33), who showed that the receptor for SV on neuroblastoma cells is a protein that binds neurovirulent SV better than avirulent SV. Particular molecules that have been suggested include a 90-kDa surface protein on human lymphoblastoid cells (26), the high-affinity laminin receptor (67 kDa) on BHK-21 cells (43), and a 74-kDa glycoprotein on N18 mouse neuroblastoma cells (41). Most recently, some of this confusion may have been resolved by the identification of heparan sulfate as important for the initial attachment of SV to cells in tissue culture (5, 22). It is possible that some of these putative receptors are heparan sulfate-bearing proteoglycans.
Mutant cell lines which escape killing by other viruses have been reported, and a variety of mechanisms for resistance have been identified. Mutant cell lines resistant to herpes simplex virus killing have mutations in surface proteoglycan synthesis, rendering them resistant to virus attachment and entry (2, 13). In other cases, defects of postentry processing and progeny virus maturation have been identified (17, 40, 44). Previously described CHO cells resistant to SV infection show defects in endosomal acidification required for virus entry (28), in furin required for processing PE2 to E2 (44), and in virus binding (27). Our CHO-18.4m cells were deficient in cell surface heparan sulfate and appear to be resistant to SV infection by a mechanism similar to that found for herpes simplex virus (32). We assume that, as for herpes simplex virus (29, 37), there are additional surface molecules that mediate SV entry into cells but that these surface molecules are less effective for initial virus binding and thus, when expressed alone, initiate infection inefficiently.
Cellular receptors have a critical role in virus-cell interaction and the spread of infection. The receptors for some viruses participate both in initial virus binding and also in virus entry. For instance, domains 1 and 2 of intercellular adhesion molecule 1, the receptor for most rhinoviruses, bind the virus, and domains 3 and 4 mediate penetration and uncoating (10). However, for many viruses penetration is mediated through molecules other than the original attachment molecule. For instance, CD4 is the initial binding protein for HIV, but entry is dependent on subsequent binding to a member of the G-protein coupled chemokine receptor family (8).
Strains of SV differ in virulence, and this is often correlated with the efficiency of binding to neural cells (38). AR339 causes fatal encephalitis in young mice, while NSV, one of the most virulent strains of SV, kills both young and adult mice. HRSP, which forms small plaques in cell monolayers, is the least virulent strain and does not reproducibly kill newborn mice (25). HRSP, AR339, and NSV showed a similar order of virulence in both CHO-18.4m and CHO-22 cells. The same virus inoculum produced different numbers of plaques in different cell lines, indicating differences in the efficiency of infection. For example, an HRSP inoculum that formed 108 plaques in BHK-21 cells formed only 106 to 107 plaques in CHO-22 cells. Successful infection is therefore determined by both the virus infectivity and the cell susceptibility. Further studies will be required to identify the specific gene disrupted and to identify the secondary receptor used efficiently by NSV to infect CHO-18.4m cells lacking heparan sulfate.
ACKNOWLEDGMENTS
We thank Jeffrey Esko for helpful discussions.
This work was supported by Public Health Service grant NS18596 (D.E.G.), a postdoctoral fellowship from the National Multiple Sclerosis Society (A.P.B.), and a predoctoral scholarship from the Taiwan National Defense Medical Center (J.-T.J.).
REFERENCES
- 1.Albritton L M, Tseng L, Scadden D T, Cunningham J M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 2.Banfield B W, Leduc Y, Esford L, Schubert K, Tufaro F. Sequential isolation of proteoglycan synthesis mutants by using herpes simplex virus as a selective agent: evidence for a proteoglycan-independent virus entry pathway. J Virol. 1995;69:3290–3298. doi: 10.1128/jvi.69.6.3290-3298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barklis E, Mulligan R C, Jaenisch R. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell. 1986;47:391–399. doi: 10.1016/0092-8674(86)90596-9. [DOI] [PubMed] [Google Scholar]
- 4.Burge B, Pfefferkorn E. Isolation and characterization of conditional lethal mutants of Sindbis virus. Virology. 1966;30:204–213. doi: 10.1016/0042-6822(66)90096-1. [DOI] [PubMed] [Google Scholar]
- 5.Byrnes A P, Griffin D E. Binding of Sindbis virus to cell surface heparan sulfate. J Virol. 1998;72:7349–7356. doi: 10.1128/jvi.72.9.7349-7356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang W, Hubbard C, Friedel C, Ruley H E. Enrichment of insertional mutants following retrovirus gene trap selection. Virology. 1993;193:737–747. doi: 10.1006/viro.1993.1182. [DOI] [PubMed] [Google Scholar]
- 7.Esko J D. Current protocols in molecular biology. New York, N.Y: Wiley; 1993. Preparation and analysis of glycoconjugates; pp. 1721–1729. [Google Scholar]
- 8.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–876. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 9.Frankel W, Potter T A, Rosenberg N, Lenz J, Rajan T V. Retroviral insertional mutagenesis of a target allele in a heterozygous murine cell line. Proc Natl Acad Sci USA. 1995;82:6600–6604. doi: 10.1073/pnas.82.19.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greve J M, Forte C P, Marlor C W, Meyer A M, Hoover-Litty H, Wunderlich D, McClelland A. Mechanisms of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM-1. J Virol. 1991;65:6015–6023. doi: 10.1128/jvi.65.11.6015-6023.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin D E, Hardwick J M. Regulators of apoptosis on the road to persistent alphavirus infection. Annu Rev Microbiol. 1997;51:565–592. doi: 10.1146/annurev.micro.51.1.565. [DOI] [PubMed] [Google Scholar]
- 12.Griffin D E, Johnson R T. Role of the immune response in recovery from Sindbis virus encephalitis in mice. J Immunol. 1977;118:1070–1075. [PubMed] [Google Scholar]
- 13.Gruenheid S, Gatzke L, Meadows H, Tufaro F. Herpes simplex virus infection and propagation in a mouse L cell mutant lacking heparan sulfate proteoglycans. J Virol. 1993;67:93–100. doi: 10.1128/jvi.67.1.93-100.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta R S, Chan D Y, Siminovitch L. Evidence for functional hemizygosity at the Emtr locus in CHO cells through segregation analysis. Cell. 1978;14:1007–1013. doi: 10.1016/0092-8674(78)90354-9. [DOI] [PubMed] [Google Scholar]
- 15.Gupta R S, Chan D Y, Siminovitch L. Evidence for variation in the number of functional gene copies at the AmaR locus in Chinese hamster cell lines. J Cell Physiol. 1978;97:461–467. doi: 10.1002/jcp.1040970321. [DOI] [PubMed] [Google Scholar]
- 16.Hubbard S C, Walls L, Ruley H E, Muchmore E A. Generation of Chinese hamster ovary cell glycosylation mutants by retroviral insertional mutagenesis. J Biol Chem. 1994;269:3717–3724. [PubMed] [Google Scholar]
- 17.Inocencio N M, Moehring J M, Moehring T J. A mutant CHO-K1 strain with resistance to Pseudomonas exotoxin A is unable to process the precursor fusion glycoprotein of Newcastle disease virus. J Virol. 1993;67:593–595. doi: 10.1128/jvi.67.1.593-595.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson A C, Moench T R, Griffin D E. The pathogenesis of spinal cord involvement in the encephalomyelitis of mice caused by neuroadapted Sindbis virus infection. Lab Investig. 1987;56:418–423. [PubMed] [Google Scholar]
- 19.Jackson A C, Moench T R, Trapp B D, Griffin D E. Basis of neurovirulence in Sindbis virus encephalomyelitis of mice. Lab Investig. 1988;58:503–509. [PubMed] [Google Scholar]
- 20.Jaenisch R, Jahner D, Nobis P, Simon I, Lohler J, Harbers K, Grotkopp D. Chromosomal position and activation of retroviral genomes inserted into the germ line of mice. Cell. 1981;24:519–529. doi: 10.1016/0092-8674(81)90343-3. [DOI] [PubMed] [Google Scholar]
- 21.King W, Patel M D, Lobel L I, Goff S P, Nguyen-Huu M C. Insertion mutagenesis of embryonal carcinoma cells by retroviruses. Science. 1985;228:554–558. doi: 10.1126/science.3838595. [DOI] [PubMed] [Google Scholar]
- 22.Klimstra W B, Ryman K D, Johnston R E. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol. 1998;72:7357–7366. doi: 10.1128/jvi.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine B, Huang Q, Isaacs J T, Reed J C, Griffin D E, Hardwick J M. Conversion of lytic to persistent alphavirus infection by the bcl-2 cellular oncogene. Nature. 1993;361:739–742. doi: 10.1038/361739a0. [DOI] [PubMed] [Google Scholar]
- 24.Lewis J, Wesselingh S L, Griffin D E, Hardwick J M. Alphavirus-induced apoptosis in mouse brains correlates with neurovirulence. J Virol. 1996;70:1828–1835. doi: 10.1128/jvi.70.3.1828-1835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lustig S, Jackson A C, Hahn C S, Griffin D E, Strauss E G, Strauss J H. The molecular basis of Sindbis virus neurovirulence in mice. J Virol. 1988;62:2329–2336. doi: 10.1128/jvi.62.7.2329-2336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maassen J A, Terhorst C. Identification of a cell-surface protein involved in the binding site of Sindbis virus on human lymphoblastoic cell lines using a heterobifunctional cross-linker. Eur J Biochem. 1981;115:153–158. doi: 10.1111/j.1432-1033.1981.tb06211.x. [DOI] [PubMed] [Google Scholar]
- 27.Mento S J, Siminovitch L. Isolation and preliminary characterization of Sindbis virus-resistant Chinese hamster ovary cells. Virology. 1981;111:320–330. doi: 10.1016/0042-6822(81)90336-6. [DOI] [PubMed] [Google Scholar]
- 28.Moehring J M, Moehring T J. Strains of CHO-K1 cells resistant to Pseudomonas exotoxin A and cross-resistant to diphtheria toxin and viruses. Infect Immun. 1983;41:998–1009. doi: 10.1128/iai.41.3.998-1009.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 30.Niklasson B, Espmark A, LeDuc J W, Gargan T P, Ennis W A, Tesh R B, Main A J., Jr Association of a Sindbis-like virus with Ockelbo disease in Sweden. Am J Trop Med Hyg. 1984;33:1212–1217. doi: 10.4269/ajtmh.1984.33.1212. [DOI] [PubMed] [Google Scholar]
- 31.Reddy S, DeGregori J V, von Melchner H, Ruley H E. Retrovirus promoter-trap vector to induce lacZ gene fusions in mammalian cells. J Virol. 1991;65:1507–1515. doi: 10.1128/jvi.65.3.1507-1515.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shieh M T, WuDunn D, Montgomery R I, Esko J D, Spear P G. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J Cell Biol. 1992;116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith A L, Tignor G H. Host receptors for two strains of Sindbis virus. Arch Virol. 1980;66:11–26. doi: 10.1007/BF01315041. [DOI] [PubMed] [Google Scholar]
- 34.Sorge J, Cutting A E, Erdman V D, Gautsch J W. Integration-specific retrovirus expression in embryonal carcinoma cells. Proc Natl Acad Sci USA. 1984;81:6627–6631. doi: 10.1073/pnas.81.21.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanley P, Chaney W. Control of carbohydrate processing: the lec1A CHO mutation results in partial loss of N-acetylglucosaminyltransferase I activity. Mol Cell Biol. 1985;5:1204–1211. doi: 10.1128/mcb.5.6.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terry-Allison T, Montgomery R I, Whitbeck J C, Xu R, Cohen G H, Eisenberg R J. HveA (herpesvirus entry mediator A), a coreceptor for herpes simplex virus entry, also participates in virus-induced cell fusion. J Virol. 1998;72:5802–5810. doi: 10.1128/jvi.72.7.5802-5810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tucker P C, Griffin D E. The mechanism of altered Sindbis virus neurovirulence associated with a single amino acid change in the E2 glycoprotein. J Virol. 1991;65:1551–1557. doi: 10.1128/jvi.65.3.1551-1557.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tucker P C, Lee S H, Bui N, Martinie D, Griffin D E. Amino acid changes in the Sindbis virus E2 glycoprotein that increase neurovirulence improve entry into neuroblastoma cells. J Virol. 1997;71:6106–6112. doi: 10.1128/jvi.71.8.6106-6112.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tufaro F, Snider M D, McKnight S L. Identification and characterization of a mouse cell mutant defective in the intracellular transport of glycoproteins. J Cell Biol. 1987;105:647–657. doi: 10.1083/jcb.105.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ubol S, Griffin D E. Identification of a putative alphavirus receptor on mouse neural cells. J Virol. 1991;65:6913–6921. doi: 10.1128/jvi.65.12.6913-6921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Melchner H, Ruley H E. Identification of cellular promoters by using a retrovirus promoter trap. J Virol. 1989;63:3227–3233. doi: 10.1128/jvi.63.8.3227-3233.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K-S, Kuhn R J, Strauss E G, Ou S, Strauss J H. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J Virol. 1992;66:4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watson D G, Moehring J M, Moehring T J. A mutant CHO-K1 strain with resistance to Pseudomonas exotoxin A and alphaviruses fails to cleave Sindbis virus gycoprotein PE2. J Virol. 1991;65:2332–2339. doi: 10.1128/jvi.65.5.2332-2339.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]