Abstract
The mouse genome contains two related interferon-regulated genes, Mx1 and Mx2. Whereas Mx1 codes for the nuclear 72-kDa protein that interferes with influenza virus replication after interferon treatment, the Mx2 gene is nonfunctional in all laboratory mouse strains examined, since its open reading frame (ORF) is interrupted by an insertional mutation and a subsequent frameshift mutation. In the present study, we demonstrate that Mx2 mRNA of cells from feral mouse strains NJL (Mus musculus musculus) and SPR (Mus spretus) differs from that of the laboratory mouse strains tested. The Mx2 mRNA of the feral strains contains a single long ORF consisting of 656 amino acids. We further show that Mx2 protein in the feral strains is expressed upon interferon treatment and localizes to the cytoplasm much like the rat Mx2 protein, which inhibits vesicular stomatitis virus replication. Furthermore, transfected 3T3 cell lines of laboratory mouse origin expressing Mx2 from feral strains acquire slight resistance to vesicular stomatitis virus.
Mammals have small families of interferon (IFN)-responsive genes that code for structurally related nuclear and cytoplasmic proteins collectively referred to as Mx proteins (2, 18). The Mx1 and Mx2 genes in the mouse show a high degree of sequence similarity (20) and they are both mapped to the same position on chromosome 16. The nuclear Mx1 protein selectively blocks influenza virus multiplication (6). Cells from some laboratory strains synthesize Mx2 mRNA, but their open reading frames (ORFs) are interrupted by an insertion, resulting in a translation frameshift. Alignment of the nonfunctional mouse and functional rat Mx2 sequences has identified a supernumerary cytosine (C) residue at position 1366 of Mx2 mRNA (20). As the Mx2 gene in commonly used strains of laboratory mice is nonfunctional, the antiviral potential or other function of the Mx2 protein is unknown. Swiss mouse 3T3 cells expressing the Mx2 cDNA repaired by site-directed mutagenesis showed slight resistance to the vesicular stomatitis virus (VSV) multiplication (24). In this study, we found that the Mx2 gene from feral strains was functional and could confer resistance to VSV infection.
MATERIALS AND METHODS
Mice.
Two laboratory inbred strains, BALB/c and C57BL/6J, were purchased from Charles River Japan (Yokohama, Japan), and laboratory inbred strain SL/NiA was provided by H. Hiai, Kyoto University, Japan (1). The feral strains NJL (Mus musculus musculus) and SPR (Mus spretus), maintained as inbred lines in our laboratory, were originally captured in Denmark and Spain, respectively.
Cell culture.
Fibroblast cultures were established from 16-day-gestation embryos of BALB/c, NJL, and SPR origin (5). The cells were seeded in 75-cm2 tissue culture flasks (Falcon Labware; Becton Dickinson, Oxnard, Calif.) at a density of 5 × 107 cells/flask in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Gibco-BRL, Gaithersburg, Md.). The medium was changed after one day, and the cells were passaged every third day.
VSV strains.
Recombinant VSV carrying the green fluorescent protein (GFP) gene instead of the G protein gene (VSVΔG*-G) was kindly provided by M. A. Whitt (University of Tennessee, Memphis, Tenn.). Infectivity of VSVΔG*-G on each 3T3 cell line was determined by counting the number of GFP-expressing cells in 10 to 20 microscopic fields (23).
Generation of full-length cDNA.
5′ and 3′ rapid amplification of cDNA ends (RACE) was used with reverse transcription (RT)-PCR to amplify mRNAs (3 μg) extracted from cultured macrophages taken from each mouse strain, using a Marathon cDNA amplification kit (Clontech, Palo Alto, Calif.) to generate double-stranded cDNA. The double-stranded cDNA was ligated with the Marathon cDNA adaptor and purified on a chromaspin-TE1000 column (Clontech) in a total volume of 100 μl. 5′- and 3′-RACE were performed by using the double-stranded cDNA as a template with the Mx2 gene-specific primers GSP1 (5′-TTCTCGTCCACGATACTGCTTT-3′) and GSP2 (5′-AAGTGAGGAGCTGCAGAAGTAC-3′) and the adaptor primers AP1 (5′-CCATCCTAATACGACTCACTATAGGGC-3′) and AP2 (5′-ACTCACTATAGGGCTCGAGCGGC-3′). Additional gene-specific primers (5′GSP [5′-TTCTGTGGAAGGATAGAGGATACC-3′], 5RA seqF [5′-AAGCCGGGAGGGGTGAGTATTG-3′], 5RA seqR [5′-TCCGCACCACATCCACGACCTT-3′], 3′GSP [5′-TCTGAGTGCTAGCACTTATCAGAGC-3′], 3′NES [5′-CAGCAAGTAGATGGGTAGAGGA-3′], and 3′ESEQ [5′-ATCCATGGCTGAGATCTTCC-3′]) were generated based on the sequences of progressively amplified 5′- and 3′-RACE products.
Sequencing and characterization of Mx2 cDNAs.
After RACE-PCR, amplified full-length cDNA fragments were cloned into pGEM-T Easy vector (Promega, Madison, Wis.). The nucleotide sequences were determined by using an ABI Prism dRhodamine Terminator cycle sequencing kit (Perkin-Elmer, Norwalk, Conn.) with an ABI Prism 310 genetic analyzer (Perkin-Elmer). All of the sequences were compared with that of the Mx2 mRNA reported earlier for BALB/c mice (20), as analyzed by the GENETYX-MAC 7.2.0 program.
Construction of Mx2 expression vector.
After RACE-PCR, the Mx2 cDNA fragments were cloned in the proper orientation into the EcoRI site of plasmid pCI-neo (Promega), which contains the human cytomegalovirus immediate-early enhancer/promoter region and the neomycin phosphotransferase gene.
Transfection of 3T3 cells with Mx2 gene expression plasmid.
3T3 cells (BALB/c embryonic fibroblasts), purchased from Riken Cell Bank (Tsukuba, Japan), were grown in DMEM containing 10% (vol/vol) fetal bovine serum. After transfection with Lipofectin (Gibco-BRL), accomplished essentially as recommended by the manufacturer, clones were selected in medium containing 500 μg of G418 per ml (8, 16).
Mx2 RT-PCR.
Total 3T3 cell RNA (3 μg) was reverse transcribed by using 200 U of SUPERSCRIPT II RT (Gibco-BRL) in a total volume of 20 μl. The oligonucleotide primers for the Mx2 gene were 5′-AGTGAGGAGCTGCAGAAGTACG (bp 1158 to 1179) and 5′-ACTTGGTAGTTCTGTGGAGGTT (bp 1609 to 1588) for the RT-PCR. As an internal control, β-actin mRNA was used in quantitative RT-PCR experiments as described previously (7). The reaction mixture for RT-PCR contained 20 mM Tris-HCl (pH 8.0), 100 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 0.2 mM deoxynucleoside triphosphate (dNTP), 1 μM each primer, and 200 U of SUPERSCRIPT II. PCR was performed in a DNA Thermal Cycler (Perkin-Elmer) with Taq polymerase in 1.5 mM MgCl2, 0.2 μM each primer, and 20 μM each dNTP, as recommended by the supplier. The cycling profile was comprised of an initial denaturating step for 5 min at 94°C followed by 35 cycles at 94°C for 1 min, 60°C for 2 min, 72°C for 1 to 3 min, and a final extension at 72°C for 5 min.
Northern blot analysis.
Total RNA was isolated from embryonic fibroblast cells by using ISOGEN (Nippon Gene, Tokyo, Japan) as instructed by the manufacturer. RNA samples were electrophoresed on formaldehyde gels, transferred, and hybridized as described previously (17). Northern blots were performed by hybridization with 2,161 bp of the Mx2 cDNA (primers 5′GSP [5′-TTCTGTGGAAGGATAGAGGATACC-3′] and 3′GSP [5′-TCTGAGTGCTAGCACTTATCAGAGC-3′]) labelled with [32P]dCTP.
Immunofluorescence staining.
Cultured cells from peritoneal macrophages prepared as described previously (11) were treated for 18 h with 1,000 IU of mouse alpha/beta IFN (IFN-α/β) (Sigma Chemical Co., St. Louis, Mo.)/ml. Macrophages were then fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS) at pH 7.3 for 30 min. The cells were permeabilized with 0.5% Triton X-100 in PBS, washed, and exposed to the monoclonal antibody 2C12, which cross-reacts with Mx proteins from many species (22, 25). The cells were then washed, exposed to tetramethyl rhodamine isothiocyanate-conjugated rabbit anti-mouse immunoglobulin G (Dako Co., Glostrup, Denmark), and mounted in 0.1 M Tris-HCl (pH 8.8) and glycerol. Finally, the cells were viewed and photographed with a BH2-RFCA microscope (Olympus Optical Co., Tokyo, Japan) equipped for epifluorescence.
Statistical analysis.
Data were expressed as means ± standard errors of the means. Statistical significance was evaluated by using Fisher’s protected least significant difference test. P values less than 0.05 were considered statistically significant.
RESULTS
Characterization of cDNAs encoding the Mx2 gene in the feral mouse strains.
Mx2 mRNAs of laboratory mouse strains BALB/c and CBA/J have ORFs interrupted by an insertional mutation (20). The Mx2 cDNA sequences from two feral strains, NJL and SPR, were compared with that from the BALB/c strain. It was indicated previously that the insertion of cytosine at position 1366 (C-1366) of BALB/c Mx2 mRNA resulted in two overlapping ORFs encoding nonfunctional proteins. As shown in Fig. 1, C-1366 was also present in Mx2 mRNA of two feral strains, but the adenine (A) at position 1367 was deleted. Therefore, the Mx2 mRNA of feral strains is composed of a single long ORF, and the putative Mx2 protein that is a product of this ORF consists of 656 amino acids (Fig. 2).
FIG. 1.
The nucleotide and amino acid sequences of Mx2 mRNA in cells from strains NJL and SPR of feral mice.
FIG. 2.
Comparison of the nucleotide and amino acid sequences of Mx2 proteins of BALB/c and wild-type strains NJL and SPR. The inserted A residue at position 1367 is found in Mx2 mRNA of BALB/c where the two long ORFs overlap. The Mx2 mRNA of the wild-type strains contained a single long ORF, as the A was excluded.
Mx2 mRNA is highly expressed in IFN-α/β-activated wild-type embryonic cells.
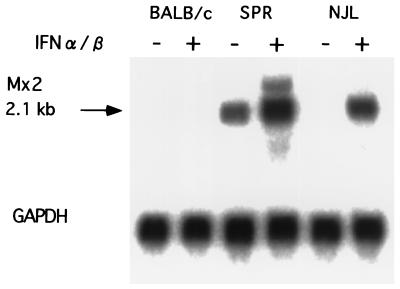
To determine the expression pattern of Mx2, total RNAs derived from embryonic fibroblast cells of BALB/c, NJL, and SPR strains were isolated and analyzed by Northern blots. We treated the cells with 500 IU of mouse IFN-α/β per ml for 18 h and found that cells from both feral strains yielded higher concentrations of 2.1-kb mRNAs than the BALB/c cells when total RNA was hybridized with an Mx2 probe (Fig. 3). No detectable 2.1-kb mRNA was found in BALB/c cells treated with IFN-α/β or in nontreated cells. NJL cells lacked a detectable level of 2.1-kb Mx2 mRNA in the case of cells not treated with IFN-α/β, whereas SPR Mx2 mRNA was detected even in such cells.
FIG. 3.
Northern blot analysis of Mx2 expression. Each lane contained 20 μg of total RNA isolated from embryonic cells. Embryonic cells were treated with 500 IU of IFN-α/β per ml for 18 h or were left untreated. The Mx2 cDNA (top) was used as a probe and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA (bottom) was used as a control for the probe.
Cytoplasmic localization of Mx2 expressed in two wild-type strains.
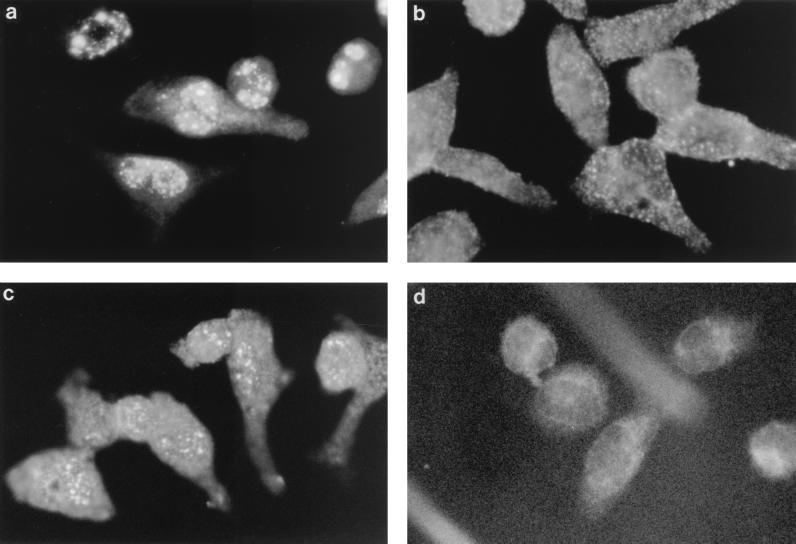
The Mx1+ laboratory mouse strains A2G and SL/NiA are resistant to influenza virus, and Mx1 protein is localized in the nucleus (4, 6, 10). We have reported that the feral strains NJL and SPR also are positive for Mx1 protein (12). Rat Mx2 cDNA is closely related to mouse Mx2 cDNA and codes for a cytoplasmic protein that inhibits VSV but not influenza virus replication (14). We therefore explored whether the Mx2 protein of the feral mouse strains synthesized in response to IFN is localized in the cytoplasm. Peritoneal macrophages were collected from each mouse strain and were treated for 18 h with 1,000 IU of IFN-α/β per ml. Proteins reacting with antibody 2C12, which cross-reacts with Mx1 and Mx2 proteins, accumulated both in the nuclei and in the cytoplasm of the macrophages from feral strains NJL and SPR (Fig. 4). Additionally, protein reacting with antibody 2C12 in the cytoplasm of the feral strains gave a punctate staining pattern. In contrast, no staining was observed in macrophages from BALB/c and C57BL/6 mice. These results suggested that the positive expression and localization of Mx2 protein in the feral strains were identical to those of the rat.
FIG. 4.
Detection of Mx2 protein in IFN-treated peritoneal macrophages from feral mouse strains by immunofluorescence with the anti-Mx monoclonal antibody 2C12 as described previously (3). The cells shown are from the influenza virus-resistant strain SL/NiA (a), wild-type strain SPR (b), wild-type strain NJL (c), and susceptible strain C57BL/6 (d). All cells were treated with 500 IU of IFN-α/β per ml for 18 h.
Permanently transfected cell lines constitutively express Mx2 proteins.
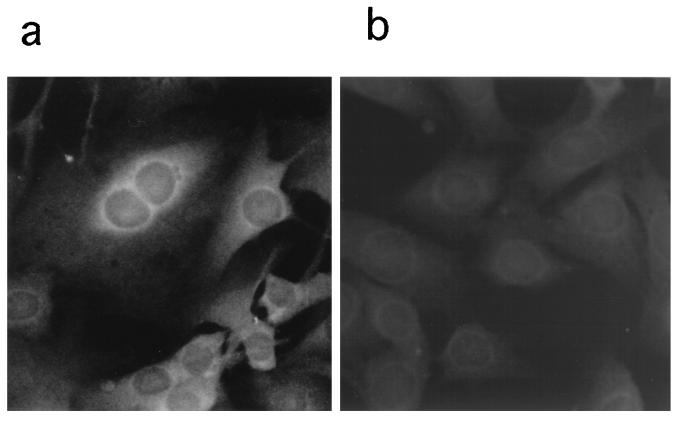
The Mx2 cDNA constructs with plasmid Mx2-pCI-neo were transfected into 3T3 cell lines. The 3T3 cells are derived from the BALB/c mouse strain lacking functional Mx genes and therefore are unable to synthesize endogenous Mx proteins (11, 19). Individual clones of stably transfected cells containing the Mx2 cDNA constructed from feral strains NJL and SPR were tested for Mx2 protein expression by immunofluorescence staining with specific antibody 2C12. In each clone, 80 to 90% of cells expressed Mx2 protein (Fig. 5). The cytoplasm of the transfected 3T3 cells accumulating Mx2 protein from feral strains gave a punctate staining pattern, whereas the cytoplasm of cells transfected with control pCI-neo showed no staining. Constitutive expression of these Mx2 proteins did not significantly affect the growth rates of the transfected 3T3 cell lines.
FIG. 5.
Detection of Mx2 protein by immunofluorescence with the anti-Mx monoclonal antibody 2C12 in stably transfected 3T3 cells expressing the Mx2 gene from feral strains NJL and SPR after IFN treatment. The transfected mouse 3T3 cells express the Mx2 gene from feral strains (a) and control 3T3-pCI-neo (b). Both cell lines were treated with 500 IU of IFN-α/β per ml for 18 h.
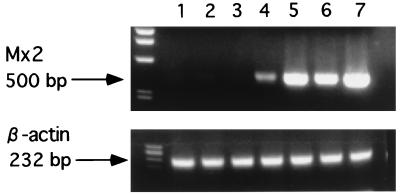
The relative concentrations of Mx2 mRNAs in transfected cells with the Mx2 cDNA construct from feral strains (clones N2, N4, S1, and S2) were much higher than those in the control cells (Fig. 6).
FIG. 6.
Total RNA from transfected 3T3 cells was reverse transcribed and PCR amplified by using Mx2 primers (top) and β-actin primers (bottom). Lanes: 1, 3T3 cells; 2 and 3, control 3T3-pCI-neo; 4 and 5, 3T3 cells expressing Mx2 from SPR (S1 and S2); 6 and 7, 3T3 cells expressing Mx2 from NJL (N2 and N4). An aliquot of each PCR product was electrophoresed on a 1.2% agarose gel and visualized with ethidium bromide.
VSV replication is inhibited in cells expressing Mx2 protein from feral mouse strains.
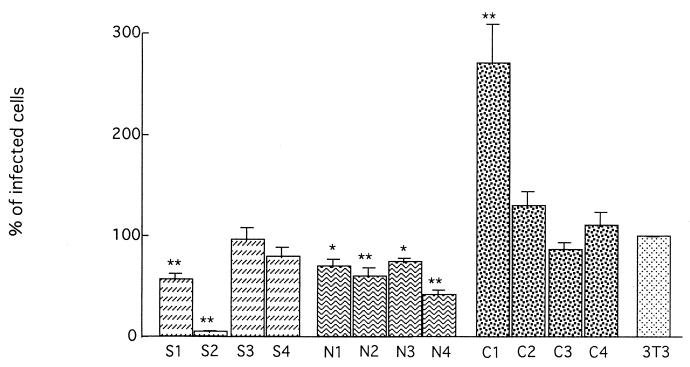
Since Mx proteins have been shown to exert an antiviral effect against influenza virus and VSV infection in vitro (6, 8, 21), the infectivities of VSVΔG*-G were examined on each 3T3 cell clone expressing mouse Mx2 protein from feral strains. Of the cell lines expressing the cytoplasmic Mx2 protein of feral strains NJL and SPR, N2, N4, S1, and S2 showed resistance to viral infection, as they gave rise to significantly lower numbers of infected cells than those of control 3T3 cells (Fig. 7).
FIG. 7.
The infectivity of VSVΔG*-G in Mx2 cDNA-transfected cell lines. S1 to S4, transfected 3T3 cells expressing Mx2 of SPR; N1 to N4, transfected 3T3 cells expressing Mx2 of NJL; C1 to C4, 3T3 cells transfected with pCI-neo. The infectivity on 3T3 cells is expressed as 100%. Shown are mean values ± standard errors of the means (n = 10). Significance levels at P < 0.05 (∗) and P < 0.01 (∗∗) compared with 3T3 cells are indicated.
DISCUSSION
In all laboratory strains tested so far, the mouse Mx2 gene is nonfunctional (20). No Mx2 gene was detected in IFN-α/β-treated cells from strain A2G, which is one of the strains expressing the functional Mx1 gene. Cells from BALB/c and other laboratory mouse strains tested synthesize low levels of Mx2 mRNA under physiological conditions, but its ORF is interrupted by a nucleotide insertion with subsequent frameshift (20).
Our sequence analysis showed that the putative coding region of the Mx2 mRNA in the feral strains NJL and SPR differed from that of laboratory strains because of the absence of A at position 1367. Hence, the Mx2 mRNA of these two feral strains contains a single long ORF and should be functional. It was indeed demonstrated by Northern blotting that IFN-treated embryonic cells from feral strains strongly expressed the Mx2 gene. In contrast, expression of the BALB/c Mx2 gene was not detected even after treatment with IFN-α/β. Immunostaining experiments indicated that Mx2 protein was localized in the cytoplasm, and a punctate staining pattern was observed. Furthermore, by RT-PCR using an Mx2-specific primer, it was confirmed that transfected 3T3 cell lines from Mx2 cDNA from feral strains expressed a high level of Mx2 mRNA. Therefore, we demonstrated a functional Mx2 gene in the mouse, although it remains to be determined how the Mx2 protein interferes with virus infection.
Rat cells synthesize three related proteins, Mx1, Mx2, and Mx3, upon IFN treatment (14). Rat Mx1 protein is predominantly localized in the nucleus and protects cells against influenza virus but only weakly against VSV. Rat Mx2 protein is cytoplasmic and protects against VSV. Rat Mx3 is also cytoplasmic, but it lacks activity. Though rat Mx2 differs from rat Mx3 in only eight amino acids (Leu-36, Ala-190, Lys-232, Ser-518, Ser-561, Lys-564, Arg-588, and His-630), mouse Mx2 matches rat Mx2 at four of these amino acids (Ala-190, Ser-518, Ser-561, and Arg-588) and rat Mx3 at two of them (Val-36 and Glu-232) (20). Johannes et al. (13) demonstrated direct evidence of the presence of anti-VSV determinants in the C terminus of rat Mx2, especially in the region between Arg-588 and His-630 (15, 24). However, the change of His in rat Mx2 protein to Gln in the mouse protein at position 630 might be a very drastic alteration, suggesting that this His residue plays a key role in rat Mx2 protein. On the other hand, the cells transfected with mouse Mx2 cDNA repaired by the exclusion of a C residue at position 1366 showed a high degree of resistance to VSV, when the mouse 3-hydroxy-3-methylglutaryl coenzyme A reductase gene promoter was used (24). In the present study, the degree of resistance of Mx2-transfected cells was limited. This may be due to (i) the His-to-Gln change at position 630 or (ii) the difference of the promoter activity of expression vector and/or cell line used. It was recently found that VSV is less Mx sensitive than members of the Bunyaviridae family such as La Crosse virus, Hanta virus, and Rift Valley fever virus (9). It will be necessary to examine the antiviral potential of functional Mx2-expressing cell lines against bunyavirus infections.
REFERENCES
- 1.Abujiang P, Yamada Y, Haller O, Kobayashi H, Kamoto T, Lu L M, Ogawa M, Ishimoto A, Katoh H, Kanehira K, Ikegami S, Fukumoto M, Hiai H. The origin of SL family mice. Lab Anim Sci. 1996;46:410. [PubMed] [Google Scholar]
- 2.Aebi M, Fäh J, Hurt N, Samuel C E, Thomis D, Bazzigher L, Pavlovic J, Haller O, Staeheli P. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol Cell Biol. 1988;9:5062–5072. doi: 10.1128/mcb.9.11.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnheiter H, Skuntz S, Noteborn M, Chang S, Meier E. Transgenic mice with intracellular immunity to influenza virus. Cell. 1990;62:51–61. doi: 10.1016/0092-8674(90)90239-b. [DOI] [PubMed] [Google Scholar]
- 4.Arnheiter H, Haller O. Antiviral state against influenza virus neutralized by microinjection of antibodies to interferon-induced Mx proteins. EMBO J. 1988;7:1315–1320. doi: 10.1002/j.1460-2075.1988.tb02946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnheiter H, Staeheli P. Expression of interferon dependent resistance to influenza virus in mouse embryo cells. Arch Virol. 1983;76:127–137. doi: 10.1007/BF01311696. [DOI] [PubMed] [Google Scholar]
- 6.Dreiding P, Staeheli P, Haller O. Interferon-induced protein Mx accumulates in nuclei of mouse cells expressing resistance to influenza viruses. Virology. 1985;140:192–196. doi: 10.1016/0042-6822(85)90460-x. [DOI] [PubMed] [Google Scholar]
- 7.Farina S F, Girard L J, Vanin E F, Nienhius A W, Bodine D M. Dysregulated expression of GATA-1 following retrovirus-mediated gene transfer into murine hematopoietic stem cells increases erythropoiesis. Blood. 1995;86:4124–4133. [PubMed] [Google Scholar]
- 8.Fortunati E, Bout A, Zanta M A, Valerio D, Scarpa M. In vitro and in vivo gene transfer to pulmonary cells mediated by cationic liposomes. Biochim Biophys Acta. 1996;1306:55–62. doi: 10.1016/0167-4781(95)00217-0. [DOI] [PubMed] [Google Scholar]
- 9.Haller O, Frese M, Kochs G. Mx proteins: mediators of innate resistance to RNA viruses. Rev Sci Tech O I E (Off Int Epizoot) 1998;17:220–230. doi: 10.20506/rst.17.1.1084. [DOI] [PubMed] [Google Scholar]
- 10.Haller O, Acklin M, Staeheli P. Influenza virus resistance of wild mice: wild-type and mutant Mx alleles occur at comparable frequencies. J Interferon Res. 1987;7:647–656. doi: 10.1089/jir.1987.7.647. [DOI] [PubMed] [Google Scholar]
- 11.Haller O, Arnheiter H, Gresser I, Lindenmann J. Genetically determined, interferon-dependent resistance to influenza virus in mice. J Exp Med. 1979;149:601–612. doi: 10.1084/jem.149.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin H K, Yamashita T, Ochiai K, Haller O, Watanabe T. Characterization and expression of the Mx1 gene in wild mouse species. Biochem Genet. 1998;36:311–322. doi: 10.1023/a:1018741312058. [DOI] [PubMed] [Google Scholar]
- 13.Johannes L, Arnheiter H, Meier E. Virus specificity of rat Mx2 protein depends on subcellular localization. J Interferon Res. 1991;11:S50. [Google Scholar]
- 14.Meier E, Kunz G, Haller O, Arnheiter H. Activity of rat Mx proteins against a rhabdovirus. J Virol. 1990;64:6263–6269. doi: 10.1128/jvi.64.12.6263-6269.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noteborn M, Arnheiter H, Richter-Mann L, Browning H, Weissmann C. Transport of the murine Mx protein into the nucleus is dependent on a basic carboxyl-terminal sequence. J Interferon Res. 1987;7:657–669. doi: 10.1089/jir.1987.7.657. [DOI] [PubMed] [Google Scholar]
- 16.Pavlovic J, Zurcher T, Haller O, Staeheli P. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human Mx A protein. J Virol. 1990;64:3370–3375. doi: 10.1128/jvi.64.7.3370-3375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Staeheli P. Interferon-induced proteins and the antiviral state. Adv Virus Res. 1990;38:147–200. doi: 10.1016/s0065-3527(08)60862-3. [DOI] [PubMed] [Google Scholar]
- 19.Staeheli P, Grob R, Meier E, Sutcliffe J G, Haller O. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol Cell Biol. 1988;8:4518–4523. doi: 10.1128/mcb.8.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staeheli P, Sutcliffe G. Identification of a second interferon-regulated murine Mx gene. Mol Cell Biol. 1988;8:4524–4528. doi: 10.1128/mcb.8.10.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell. 1986;44:147–158. doi: 10.1016/0092-8674(86)90493-9. [DOI] [PubMed] [Google Scholar]
- 22.Staeheli P, Dreiding P, Haller O, Lindenmann J. Polyclonal and monoclonal antibodies to the interferon-inducible protein Mx of influenza virus-resistant mice. J Biol Chem. 1985;260:1821–1825. [PubMed] [Google Scholar]
- 23.Takada A, Robison C, Goto H, Sanchez A, Murti K G, Whitt M A, Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zurcher T, Pavlovic J, Staeheli P. Mouse Mx2 protein inhibits vesicular stomatitis virus but not influenza virus. Virology. 1992;187:796–800. doi: 10.1016/0042-6822(92)90481-4. [DOI] [PubMed] [Google Scholar]
- 25.Zurcher T, Pavlovic J, Staeheli P. Nuclear localization of mouse Mx1 protein is necessary for inhibition of influenza virus. J Virol. 1992;66:5059–5066. doi: 10.1128/jvi.66.8.5059-5066.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]