Abstract
The 5′ nontranslated RNA (5′NTR) of a genotype 1b hepatitis C virus (HCV-N) directs cap-independent translation of the HCV-N polyprotein with about twofold less efficiency than the 5′NTR of a genotype 1a virus under physiologic conditions (Hutchinson strain, or HCV-H) (M. Honda et al., Virology 222:31–42, 1996). Here, we show by mutational analysis that substitution of the AG dinucleotide sequence at nucleotides (nt) 34 and 35 of HCV-N with GA (present in HCV-H) restores the translational activity to that of the HCV-H 5′NTR both in vitro and in vivo. These nucleotides are located upstream of the minimal essential internal ribosome entry site (IRES), as a 6-nt deletion spanning nt 32 to 37 also increased the translational activity of the HCV-N 5′NTR to that of HCV-H. Thus, the upstream AG dinucleotide sequence has an inhibitory effect on IRES-directed translation. Surprisingly, however, this inhibitory effect was observed only when the translated, downstream RNA sequence contained nt 408 to 929 of HCV (capsid-coding RNA). Further analysis of RNA transcripts containing frameshift mutations demonstrated that the nucleotide sequence of the transcript, and not the amino acid sequence of the expressed capsid protein, determines this difference in translation efficiency. The difference between the translational activities of the HCV-N and HCV-H transcripts was increased when translation was carried out in reticulocyte lysates containing high K+ concentrations, with a sevenfold difference evident at 130 to 150 mM K+. These results suggest that there is an RNA-RNA interaction involving 5′NTR and capsid-coding sequences flanking the IRES and that this is responsible for the reduced IRES activity of the genotype 1b virus, HCV-N.
Hepatitis C virus (HCV) is a positive-stranded, enveloped RNA virus which is classified within the Hepacivirus genus of the family Flaviviridae (3). This virus establishes persistent infection in most infected humans, leading to the development of chronic hepatitis, cirrhosis, and hepatocellular carcinoma (1, 14). There is extensive genetic heterogeneity among different HCV strains, with at least six major genotypes and a series of related subtypes recognized thus far (5, 24). Among these, genotype 1 is predominant worldwide and comprised of two major subtypes, genotypes 1a and 1b (5). Although some clinical studies have found no differences in the clinical expression of liver disease related to the genotype of the infecting virus (15), others have suggested that genotype 1b infections may be more resistant to interferon therapy (17, 30) and may confer greater risk for development of hepatocellular carcinoma than infection with non-genotype 1b strains including genotype 1a viruses (23).
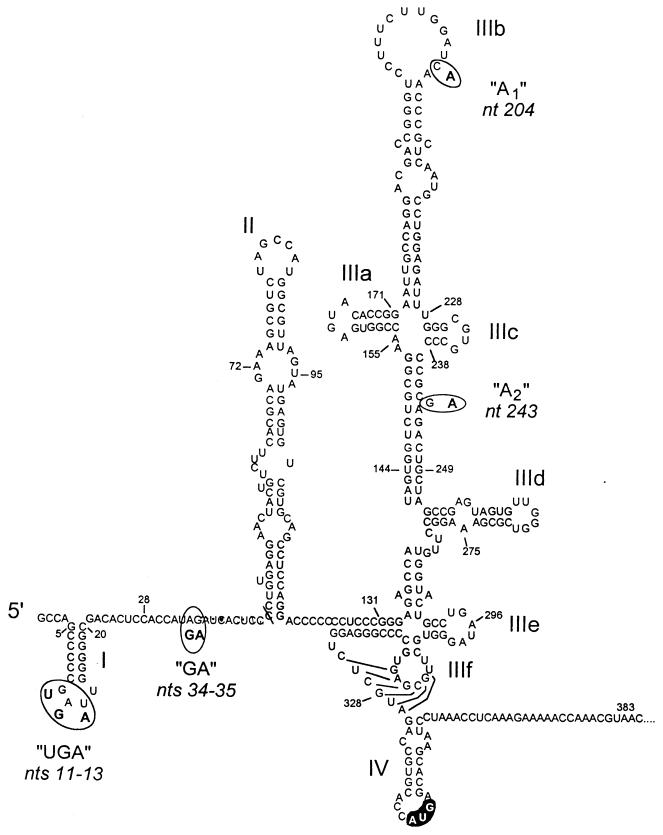
Despite the considerable genetic diversity that exists among different HCV strains, the 5′ nontranslated RNA (5′NTR) sequences of these viruses are relatively invariant. In large part, this reflects the presence of conserved, highly ordered RNA structures within the viral internal ribosome entry site (IRES) which are required for the internal entry of ribosomes on the viral RNA and subsequent cap-independent initiation of translation of the viral polyprotein (8, 12, 19–21, 27, 29). The RNA segment which comprises the IRES occupies most of the 5′NTR, as the 5′ limit of the IRES has been mapped to between nucleotides (nt) 29 and 46 (11, 12). While the 3′ boundary of the IRES is less certain, several recent studies indicate that the activity of the HCV IRES is dependent on sequence located immediately downstream of the initiator AUG (12, 16, 19). The initiator AUG appears to be located within the single-stranded segment of a stem-loop (stem-loop IV) which is formed in part by the capsid protein-coding sequence at the extreme 5′ end of the large open reading frame of HCV (10) (Fig. 1). Mutations which enhanced the stability of this putative stem-loop significantly reduced the efficiency of internal initiation of translation, suggesting that the stem-loop may play a role in controlling translation of the viral polyprotein (10). Larger RNA structures that are upstream of stem-loop IV are absolutely essential for translational activity (12, 21, 28), but the most 5′ stem-loop within the 5′NTR (stem-loop I, nt 5 to 20) appears to have an opposing action, as its deletion enhances the activity of the downstream IRES both in vitro and in vivo (12, 21).
FIG. 1.
Proposed secondary and tertiary RNA structures within the 5′NTR and the immediately downstream segment of the long open reading frame of the genotype 1b virus HCV-N (10, 11). Major structural domains are labeled I, II, IIIa, IIIb, etc.; the initiator AUG codon in stem-loop IV is highlighted. The circled nucleotides indicate differences between the sequences of HCV-N and the genotype 1a HCV-H virus, which are clustered at four loci: UGA, GA, A1, and A2.
We noted previously that the 5′NTR of a genotype 1a virus (Hutchinson strain, or HCV-H) directed translation with greater efficiency than that of a genotype 1b virus (HCV-N) when placed in the context of nearly genome-length viral RNA (12). We found the IRES of the genotype 1a virus to be about twofold more active than the IRES of the genotype 1b virus, both in a cell-free translation system and in transfected Huh-T7 cells in vivo (12). Although it is not certain that a twofold difference in translational efficiency would have a significant impact on either virus replication or disease expression, we considered it likely that the identification of the molecular basis for this difference in activity could contribute to a better understanding of the HCV IRES and the structure of the 5′NTR. This would be important, because the IRES is increasingly recognized as a potential target for antiviral drug development. The sequences of the HCV-H and HCV-N 5′NTRs differ at only seven base positions, located at four separate sites (Fig. 1). Significantly, none of the involved nucleotides are involved in base pairing within recognized secondary or tertiary RNA structures (2, 10).
MATERIALS AND METHODS
Plasmids.
Plasmids pMN2-1G and PMN2/H have been described previously (12). Plasmid pN-CΔE1 contains cDNA representing nt 1 to 1357 (5′NTR, capsid-coding, and 5′ E1-coding sequences) of the HCV-N virus (9) (genotype 1b) genome under the transcriptional control of the T7 promoter. It was constructed by subcloning the XmnI-BamHI fragment of pMN2-1G into pBluescript IISK (Stratagene). pH-CΔE1 is a similar plasmid which contains the 5′NTR of the HCV-H genotype (1a) fused to the 5′ open reading frame of HCV-N. It was constructed by subcloning the XhoI-NheI fragment (containing the T7 promoter sequence and nt 1 to 249 of the 5′NTR) from pMN2/H into pN-CΔE1. pN-CLuc contains the 5′NTR sequences of HCV-N and the first 66 nt of the HCV open reading frame fused in frame to the firefly luciferase sequence, under control of the T7 promoter. It was constructed by inserting a PCR-amplified fragment of pGEM-Luc (Promega) into the AatII and BamHI sites of pN-CΔE1. pH-CLuc was constructed subsequently by inserting the luciferase sequence from pN-CLuc into the AatII and BamHI sites of pH-CΔE1. pN-CAT and pH-CAT contain the 5′NTR sequences of HCV-N and HCV-H, respectively, with 8 nt of the open reading frame fused in frame to the bacterial chloramphenicol acetyltransferase (CAT) sequence under T7 promoter control. These were constructed by inserting the small NheI-BamHI fragment of pWT-CAT (21) into pN-CΔE1 or pH-CΔE1.
The bicistronic construct pCAT-N(A2)-CLuc contains a T7 transcriptional unit in which the CAT sequence is fused at its 3′ end to the 5′NTR of HCV-N (with a change of G to A at nt 243) and 66 nt of the HCV open reading frame fused in frame with the luciferase sequence. It was constructed by inserting the NheI-XmnI fragment of pN-CLuc into pKCUS2 (gift from B. Clarke, Glaxo-Wellcome). pCAT-N-CLuc and pCAT-H-CLuc are similar constructs containing the 5′NTR sequences of HCV-N and HCV-H, respectively. pCAT-N-CΔE1 is another bicistronic plasmid in which the naturally fused HCV capsid and 5′ E1-coding sequences represent the second cistron. It was constructed by inserting the NheI-BamHI fragment of pN-CΔE1 into pCAT-N-CLuc. The nearly genome-length bicistronic constructs pCAT/N, pCAT/H, pCAT/N(UGA), pCAT/N(GA), pCAT/N(A1), and pCAT/N(A2) were constructed by ligating XmnI-BamHI fragments excised from mutated subgenomic bicistronic plasmids with the large BamHI-XmnI fragment of pMN2-1G (HCV nt 1358 to 9454). pN-CΔE1(H) and pH-CΔE1(H) are monocistronic constructs that contain the 5′NTR of HCV-N and HCV-H, respectively, fused naturally to the HCV-H open reading frame as the second cistron. These were created by inserting the NheI-BamHI fragment of pRC/CMV/HCV-H (gift from H. Lerat) into pN-CΔE1 and pH-CΔE1, respectively.
pCAT-N-CΔE1(Fs) and pCAT-H-CΔE1(Fs) contain frameshift mutations in the capsid-coding region. These were constructed by amplifying the capsid region with the 5′ primer, 5′-ATAGAATTCGACGTCAGTTCCCGGGCGG-3′, which lacks A-409 and the 3′ primer, 5′-TATAGATCTCCTAGGGGGGCCGCCGACGAGCGGA-3′, which inserts a cytosine at nt 769, thus restoring the original reading frame and maintaining its patency. The amplified PCR fragment was inserted into EcoRI and BglII sites of pSP73 (Promega). The resultant plasmid was digested with AatII and AvrII, and the small fragment was cloned into pCAT-N-CΔE1 and pCAT-H-CΔE1 to make pCAT-N-CΔE1(Fs) and pCAT-H-CΔE1(Fs), respectively.
Site-directed mutagenesis of these plasmids (see Results) was carried out by using a standard PCR-based strategy, with Pfu (Pyrococcus furiosus) DNA polymerase. Reactions were for 35 cycles of 95°C for 1 min, 42°C for 1.5 min, and 75°C for 3 min. The sequences of the PCR-manipulated regions and the presence of expected mutations were confirmed by DNA sequencing. Plasmid DNAs were purified on Qiagen-tip 500 columns (Qiagen, Chatsworth, Calif.).
Cells.
Huh-T7 cells (22), which are derived from Huh-7 human hepatocellular carcinoma cells, are stably transformed and constitutively express bacteriophage T7 RNA polymerase. Cell cultures were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin, streptomycin, and 400 μg (active compound) of geneticin (Gibco/BRL) per ml.
Antibodies.
Murine monoclonal antibody to the HCV capsid protein, 27D5G5, was generously provided by Michel Jolivet, BioMerieux. Rabbit polyclonal antibody against CAT was purchased from 5′→3′, Inc. (Boulder, Colo.).
In vitro transcription and translation reactions.
Plasmids were linearized by digestion with BamHI or MluI (pN-CΔE1, pN-CLuc, pN-CAT, pCAT-N-CΔE1, pCAT-N-CLuc, and related mutants) or XhoI (pCAT/N and related mutants). RNA was transcribed by T7 RNA polymerase, using Riboprobe System II (Promega) or Megascript (Ambion) reagents as recommended by the manufacturer. Transcription products were treated with RQ1 DNase (Promega) for 15 min at 37°C, extracted with phenol-chloroform, precipitated with ethanol–7.5 M ammonium acetate, and examined by agarose gel electrophoresis. The concentration of RNA was estimated by spectrophotometry. In some experiments, RNA was separated from nonincorporated nucleotides by chromatography through a Sephadex G-25 column and quantified by gel analysis. In vitro translation was carried out in micrococcal nuclease-treated rabbit reticulocyte lysate, with or without canine pancreatic microsomal membranes (Promega). The effects of varied KCl concentration were determined by using the Flexilysate system (Promega). Unless otherwise specified, translation reactions (25 μl) were programmed with 0.5 to 2.0 μg of RNA and incubated at 30°C for 1 h. Total translation products were separated by 12 or 13% sodium dodecyl sulfate (SDS)-polyacrylamide—6 M urea gel electrophoresis (PAGE) followed by autoradiography. Translation products were quantified by PhosphorImager (Molecular Dynamics, Inc.) analysis.
Stability of HCV-H and HCV-N RNAs in reticulocyte lysate.
Synthetic RNA transcripts of pN-CΔE1 and pH-CΔE1 were trace labeled with 32P (2.4 × 105 cpm/μg of RNA) and used to program reticulocyte lysate for in vitro translation as described above (1 μg of RNA per 25 μl of reaction mixture); 3-μl aliquots of the reaction mixture were sampled at 0, 20, 40, and 60 min and analyzed by electrophoresis in a 2% agarose–0.1% SDS gel. The gel was dried and then subjected to autoradiography and PhosphorImager analysis.
Vaccinia virus-T7 hybrid expression of bicistronic RNA transcripts in transfected Huh-T7 cells.
Huh-T7 cells were seeded into four-well tissue culture chamber slides 24 h prior to transfection. Cells (90% confluent) were infected with recombinant vaccinia virus vTF7-3 (expressing T7 RNA polymerase) (7) in 100 μl of Opti-MEM (Gibco/BRL) at a multiplicity of infection of 10. Following a 1-h incubation at 37°C, the virus inoculum was removed and replaced with a mixture containing 1 μg of bicistronic plasmid DNA [pCAT/N, pCAT/H, pCAT/N(UGA), pCAT/N(GA), pCAT/N(A1), or pCAT/N(A2)] and 3 μl of Lipofectin (Gibco/BRL) in 40 μl of Opti-MEM at 37°C, followed 15 min later by an additional 200 μl of Opti-MEM (12). Cells were subsequently incubated at 37°C in a 5% CO2 atmosphere for 24 h.
Fluorescence image cytometry.
Transfected Huh-T7 cells were washed with phosphate-buffered saline fixed in acetone-methanol (1:1), and stained with rabbit antibody to CAT (1:50 final dilution; 5′→3′, Inc.) and murine monoclonal antibody to the HCV capsid protein (27D5G5; 1:12,000 dilution; generous gift from Michel Jolivet). After a further wash, the cells were incubated with a mixture of fluorescein isothiocyanate (FITC)-conjugated swine antibody to rabbit immunoglobulin (DAKO) and tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat antibody to mouse immunoglobulin (Organon-Teknika-Cappel, Durham, N.C.), each diluted 1:40. The stained cells were washed extensively in phosphate-buffered saline and examined at a magnification of ×40 with a Microphot FXA epifluorescence microscope (Nikon), using filter sets for TRITC or FITC fluorescence. Fluorescence images were recorded with a charge-coupled device camera (Optronics Engineering) and NIH Image acquisition software. Duplicate FITC- and TRITC-labeled images from 30 dually stained cells from each transfected culture were transferred to a DEC 5000/200 workstation (Digital Equipment Corporation), and the boundaries of positive cells were automatically extracted by using IGLOO (Image Graphics Library Object-Oriented) (4) and newly developed algorithms allowing automatic generation of cell masks (13). The contribution of nonspecific labeling to the image was eliminated by subtracting the average grey scale intensity of the background image from the integrated intensity. The ratio of the integrated intensities of the TRITC- and FITC-labeled images was subsequently calculated for each cell.
RESULTS
Relative efficiency of translation directed by genotype 1a (HCV-H) and genotype 1b (HCV-N) 5′NTRs in vitro.
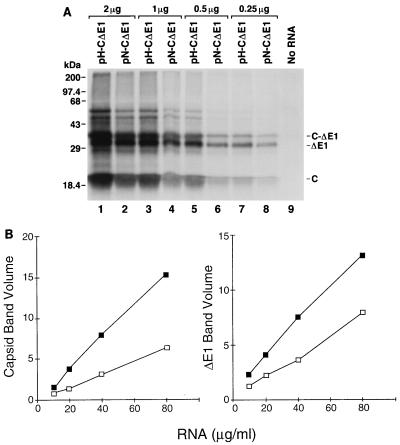
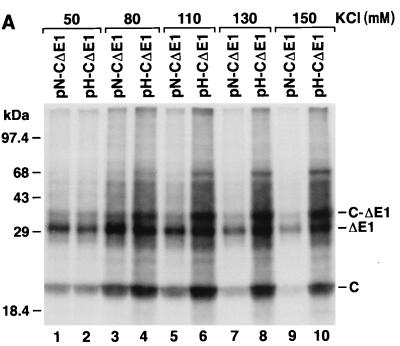
To better quantify the difference in translational activities of the IRESs of HCV-N and HCV-H, we examined the abilities of RNAs transcribed in vitro from plasmids pN-CΔE1 and pH-CΔE1 to direct translation of HCV proteins in rabbit reticulocyte lysates. These transcripts represented the 5′NTR sequences of HCV-N and HCV-H, respectively, fused naturally to nt 342 to 1357 of the open reading frame of HCV-N. The in vitro translation reactions (25 μl each) were programmed with 0.25 to 2 μg of RNA (10 to 80 μg/ml) and supplemented with microsomal membranes to allow signalase cleavage at the capsid-E1 junction. The major products of translation included the 21-kDa capsid protein, a 30-kDa truncated, glycosylated E1 protein (ΔE1), and a 34-kDa unprocessed precursor protein (C-ΔE1) (Fig. 2A). Lesser quantities of two higher-molecular-mass proteins (46 and 54 kDa) were also observed in some experiments. Although no efforts were made to specifically identify these proteins, they likely represent aggregates of the major products. At each RNA concentration, greater quantities of all of these proteins were produced by the HCV-H transcripts. PhosphorImager (Molecular Dynamics) analysis indicated that the quantities of the capsid and ΔE1 proteins increased in a linear fashion over the range of the RNA concentrations used to program translation (Fig. 2B). On average, pH-CΔE1 transcripts produced 2.4-fold more capsid protein and 1.8-fold more ΔE1 compared with pN-CΔE1 transcripts.
FIG. 2.
Translation of RNA transcripts containing genotype 1a HCV-H or genotype 1b HCV-N 5′NTR sequences. (A) SDS-PAGE of products of translation from rabbit reticulocyte lysates programmed with 2 (lanes 1 and 2), 1 (lanes 3 and 4), 0.5 (lanes 5 and 6), 0.25 (lanes 6 and 8), or 0 (lane 9) μg of RNA derived from pH-CΔE1 (containing the 5′NTR of HCV-H; lanes 1, 3, 5, and 7) or pN-CΔE1 (containing the 5′NTR of HCV-N; lanes 2, 4, 6, and 8) per 25-μl reaction. (B) Quantitative analysis (relative PhosphorImager band volume) of HCV capsid (left) and ΔE1 (right) proteins produced in reticulocyte lysates programmed with increasing concentrations of RNA transcribed from pH-CDE1 (■) or pN-CΔE1 (□).
To exclude the possibility that the difference in the amounts of capsid and ΔE1 proteins produced by the pN-CΔE1 and pH-CΔE1 transcripts could be due to differences in stability of these RNAs, we programmed reticulocyte lysate with trace-labeled RNA transcripts. Reactions were sampled at 20, 40, and 60 min, and the integrity of the RNA was determined by gel electrophoresis. Approximately 60% of each RNA template was degraded during the first 20 min of the reaction (data not shown), but there were no differences in the rates of degradation of the pN-CΔE1 and pH-CΔE1 RNAs. These results were confirmed by RNase protection assays using an RNA probe targeted to the capsid-coding region, which also showed no differences in the rate of disappearance of the protected RNA fragment following addition of pN-CΔE1 and pH-CΔE1 transcripts to reticulocyte lysate (data not shown). Taken together, these results confirm that the 5′NTR of HCV-H is about twice as active as that of HCV-N in directing translation of the HCV polyprotein in rabbit reticulocyte lysates.
Genetic basis of the difference in translational activities of HCV-H and HCV-N.
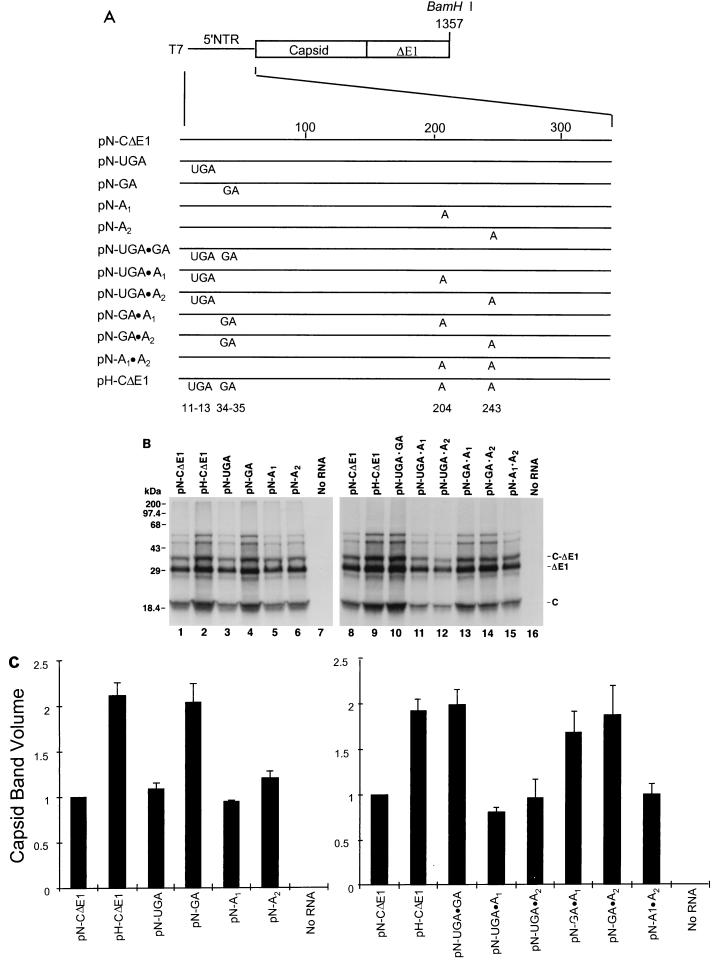
As indicated above, the 5′NTR sequences of the these two viruses differ at seven nucleotide positions (Fig. 1). To determine which of these differences in the nucleotide sequences of these RNAs were responsible for the variation in translational activity, we constructed a series of chimeric 5′NTR constructs in which the four nucleotide substitution groups present in the HCV-H sequence (Fig. 1) were systematically introduced into the background of the HCV-N sequence in pN-CΔE1 (Fig. 3A). Plasmids pN-UGA, pN-GA, pN-A1, and pN-A2 contain the HCV-H substitutions at nt 11 to 13 (GAU→UGA), 34 and 35 (AG→GA), 204 (C→A), and 243 (G→A), respectively. RNAs transcribed from these plasmids in vitro were used to program reticulocyte lysates for translation at a concentration of 40 μg/ml (Fig. 3B). The amounts of capsid and ΔE1 proteins produced from pN-GA transcripts (Fig. 3B, lane 4) were nearly equivalent to those produced by pH-CΔE1 transcripts (lane 2), while pN-UGA, pN-A1, and pN-A2 transcripts (lanes 3, 5, and 6) had translational activities approximating that of RNA transcribed from pN-CΔE1 (lane 1). These differences were reproducible in three separate experiments and were confirmed by PhosphorImager analysis (Fig. 3C).
FIG. 3.
Genetic basis for the difference in translational activities of the HCV-N and HCV-H 5′NTRs. (A) Organization of T7 transcriptional units within chimeric constructs containing the unique nucleotide sequences of HCV-H placed individually and in combination within the background of the HCV-N 5′NTR and immediately downstream segment of the open reading frame encoding C-ΔE1 (pN-CΔE1). Protein-coding regions are shown as rectangles; noncoding RNA is shown as a solid line. Map positions of the base substitutions are shown at the bottom. (B) SDS-PAGE of products of translation from reticulocyte lysates programmed with RNAs transcribed from the plasmids depicted in panel A. The two panels represent results from separate experiments. The gel positions of the capsid (C), ΔE1, and nonprocessed C-ΔE1 protein products are shown at the right. (C) PhosphorImager analysis of capsid protein quantities produced in translation experiments such as those shown in panel B. The measured band volumes were normalized to that of reactions programmed with RNA from pN-CΔE1 (=1.0). The error bars represent the standard error calculated from the results of three separate experiments.
We also studied chimeric transcripts which contained combinations of two nucleotide substitution groups: pN-UGA · GA, pN-UGA · A1, pN-UGA · A2, pN-GA · A1, pN-GA · A2, and pN-A1 · A2 (Fig. 3A). RNA transcripts derived from pN-UGA · GA, pN-GA · A1, and pN-GA · A2 (Fig. 3B, lanes 10, 13, and 14), which all contain the GA substitution at nt 34 and 35, were translated with an efficiency equivalent to that for pH-CΔE1 (Fig. 3B, lane 9). In contrast, translation of the remaining chimeric transcripts, which contain AG at this locus (pN-UGA · A1, pN-UGA · A2, and pN-A1 · A2; lanes 11, 12, and 15), did not exceed that of pN-CΔE1 RNA (lane 8). These findings were reproducible and confirmed by PhosphorImager analysis (Fig. 3C). They strongly support the conclusion that the AG→GA substitution at nt 34 and 35 is responsible for the increased translational activity of the HCV-H 5′NTR.
Base substitutions at nt 34 and 35 influence the efficiency of translation only on transcripts containing the complete capsid-coding region.
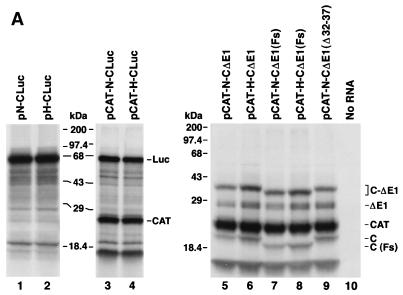
In an effort to develop a sensitive reporter assay to test the effects of these nucleotide substitutions on IRES activity in vivo (see below), we constructed plasmids (pN-CLuc and pH-CLuc) in which sequence encoding the reporter protein, firefly luciferase, was fused in frame at nt 408 of the HCV sequence in pN-CΔE1 and pH-CΔE1, thus replacing the sequence downstream of nt 66 of the HCV open reading frame. We also constructed dicistronic variants of these plasmids (pCAT-N-CLuc and pCAT-H-CLuc), in which sequence encoding bacterial CAT was placed upstream of the HCV 5′NTR. Synthetic RNA transcripts derived from these plasmids were used to program reticulocyte lysates for translation. Surprisingly, we found that the type of 5′NTR sequence (HCV-H or HCV-N) had no influence on the translational efficiency of either the monocistronic (Fig. 4A, lane 1 versus lane 2) or dicistronic (lane 3 versus lane 4) transcripts. The absence of a significant difference in the translational activities of the monocistronic transcripts (pN-CLuc and pH-CLuc) was confirmed by direct assay of the translation products for luciferase activity (data not shown), while the absence of a difference between the dicistronic transcripts (pCAT-N-CLuc and pCAT-H-CLuc) was confirmed by PhosphorImager analysis of the relative quantities of luciferase and CAT produced in each reaction (Fig. 4B). Similarly, we found no differences in the translational activities of dicistronic RNA transcripts which contained the HCV-H base substitutions within the background of the HCV-N 5′NTR [pCAT-N(UGA)-CLuc, pCAT-N(GA)-CLuc, pCAT-N(A1)-CLuc, and pCAT-N(A2)-CLuc] (data not shown).
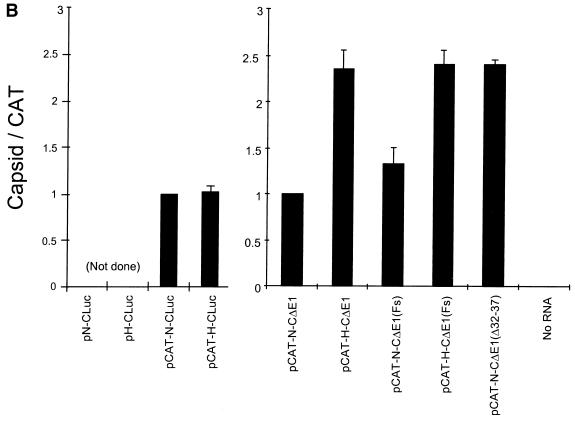
FIG. 4.
(A) SDS-PAGE of products of translation from reticulocyte lysates programmed with monocistronic (lanes 1 and 2) or dicistronic (lanes 3 and 4) RNAs in which the HCV capsid sequence beyond nt 66 of the open reading frame was replaced with luciferase sequence. The major luciferase product appears as a band of ∼67 kDa, while CAT migrates with an apparent molecular mass of ∼24 kDa. Lanes 5 to 10 show a further analysis of the requirements for downstream HCV coding sequence. In pCAT-N-CΔE1(Fs) and pCAT-H-CΔE1(Fs) transcripts, the HCV capsid coding sequences contain −1 and +1 frameshift mutations at nt 409 and 769, respectively, resulting in the expression of the C(Fs) protein which contains nonsense sequence replacing much of the natural capsid sequence. Nucleotides 32 to 37 are deleted from the HCV-N 5′NTR sequence within pCAT-N-CΔE1(Δ32-37). The gel positions of the capsid (C), ΔE1, nonprocessed C-ΔE1, luciferase (Luc), and CAT protein products are shown at the right. The most rapidly migrating band observed in lanes 3 to 9 is an unidentified product of the dicistronic RNAs that appears to be derived from the upstream CAT reading frame by an uncertain initiation event. The quantity of this product was closely tied to the quantity of CAT produced in these reactions, and it was not considered in calculating the quantitative estimates of IRES activity shown in Fig. 4B. (B) PhosphorImager analysis of products of translation reactions programmed with dicistronic RNAs in panel A. For each pair of reactions programmed with RNAs containing the HCV-N or HCV-H 5′NTRs, the ratio of the band volumes of the capsid protein and CAT were normalized to the ratio obtained with RNA containing the HCV-N 5′NTR (pCAT-N-CLuc or pCAT-N-CΔE1 = 1.0). Error bars represent the standard error calculated from replicate experiments.
To determine whether the absence of a translational difference could be due specifically to the presence of the luciferase sequence, we assessed the translational activity of monocistronic transcripts derived from pN-CAT and pH-CAT (21). These constructs contain the CAT reporter protein sequence fused in frame to HCV capsid-coding sequence but preserve only 8 nt of the original HCV open reading frame. As with the luciferase reporter transcripts, there were no differences in the amount of CAT produced from these RNAs in rabbit reticulocyte lysates (data not shown).
In contrast to these results, dicistronic transcripts in which the truncated HCV open reading frame (CΔE1) represented the downstream cistron (pCAT-N-CΔE1 and pCAT-H-CΔE1) retained the increased translational activity observed with the HCV-H 5′NTR in earlier analyses of monocistronic transcripts (Fig. 4A, lane 5 versus lane 6; Fig. 4B). The greater translational activity of the HCV-H 5′NTR was evident whether these reaction mixes were supplemented or not supplemented with microsomal membranes (data not shown). Although the base substitutions at nt 34 and 35 of the HCV-H 5′NTR are responsible for its greater translational activity (Fig. 3), these results indicate that the difference in translational activity that these substitutions confer is evident only when the transcript contains natural HCV sequences downstream of nt 408.
To further define the downstream sequence required for the enhanced translational activity of the HCV-H 5′NTR, we compared nearly genome-length dicistronic RNA transcripts containing the HCV-H or HCV-N 5′NTR fused naturally to HCV-N sequence extending to nt 9454 of the HCV genome (plasmids pCAT/N and pCAT/H) (12). While these RNAs produced equivalent amounts of CAT in reticulocyte lysates, PhosphorImager analyses indicated that 1.8-fold more capsid protein was produced from pCAT/H than from pCAT/N (data not shown). Thus, the inclusion of HCV sequence beyond the truncated CΔE1 segment did not influence the relative translational activities of the HCV-H and HCV-N 5′NTRs. We next examined the translational activities of dicistronic transcripts which contained only the capsid-coding sequence within the downstream cistron. These RNAs were prepared by runoff transcription of MluI-digested pCAT-N-CΔE1 and pCAT-H-CΔE1 plasmid DNAs; they terminated at nt 929 of the HCV genome. We again noted significantly greater production of capsid protein by transcripts containing the HCV-H 5′NTR (data not shown). Thus, we conclude that the capsid-coding sequence between nt 408 and 929 is required for expression of the difference between the translational activities of the HCV-N and HCV-H 5′NTRs.
Expression of the capsid protein is not required for the enhanced translational activity of HCV-H.
Experiments with chimeric polioviruses in which the native picornavirus IRES was replaced by the IRES of HCV suggested that the HCV capsid protein might play a role in facilitating the internal initiation of translation (16). If this hypothesis is correct, the fact that we observed differences in the translational activities of the HCV-H and HCV-N 5′NTRs only when they were fused to the native HCV capsid sequence could be explained by differences in the interaction of these 5′NTRs with the capsid protein. Alternatively, the requirement for capsid sequence could reflect an interaction of this RNA with sequences within the 5′NTR. To distinguish between these two possibilities, we introduced frameshift mutations into the capsid coding sequence of pCAT-N-CΔE1 and pCAT-H-CΔE1. The resulting plasmids, pCAT-N-CΔE1(Fs) and pCAT-H-CΔE1(Fs), each contain two mutations: a −1 frameshift at nt 409 and a +1 frameshift at nt 769, the latter of which restores the original reading frame and maintains its patency to the end of the truncated E1 sequence. Thus, although transcripts produced from these plasmids differ by only 2 nt from the RNAs transcribed from pCAT-N-CΔE1 and pCAT-H-CΔE1, they encode a markedly altered capsid protein, C(Fs), which consists of nonsense sequence between residues 23 and 143, or for approximately 63% of its sequence.
As shown in Fig. 4A, both C(Fs) and unprocessed C(Fs)ΔE1 migrated more rapidly than the native capsid and unprocessed CΔE1 proteins in SDS-PAGE (compare lanes 7 and 8 with lanes 5 and 6). However, significantly greater quantities of these nonsense proteins were expressed from pCAT-H-CΔE1(Fs) compared with pCAT-N-CΔE1(Fs) transcripts (compare lanes 7 and 8). By PhosphorImager analysis, there was approximately 1.8-fold more C(Fs) produced from transcripts containing the HCV-H 5′NTR (Fig. 4B). Since previous experiments had demonstrated that the difference in translational activity required the presence of sequence between nt 408 and 929, these data strongly suggest that the difference in IRES activity is dependent on the nucleotide sequence of the RNA and not the amino acid sequence of the protein which it encodes.
The AG dinucleotide sequence at nt 34 and 35 of HCV-N has an inhibitory effect on translation.
The 5′ limit of the minimal essential HCV IRES sequence has not been precisely mapped. However, several studies suggest that it is located downstream of the dinucleotide sequence at nt 34 and 35, which we found to be primarily responsible for the difference in the translational activities of HCV-H and HCV-N transcripts (Fig. 3) (20). Since the nature of the sequence at nt 34 and 35 was found to have such an important effect on the translational activity of HCV transcripts, it was of interest to further investigate whether sequence in this region is required for the internal initiation of translation on these RNAs. Thus, we created a 6-nt deletion mutation (nt 32 to 37) in pCAT-N-CΔE1, resulting in plasmid pCAT-N-CΔE1(Δ32-37). Interestingly, dicistronic transcripts derived from this plasmid retained robust IRES activity. The amount of capsid protein produced from the pCAT-N-CΔE1(Δ32-37) transcripts (Fig. 4A, lane 9) was approximately 2.4-fold higher than the amount produced by transcripts from pCAT-N-CΔE1 (lane 5) and approximately equivalent to the amount produced by transcripts containing the HCV-H 5′NTR (lane 6). These results indicate that the AG dinucleotide sequence at nt 34 and 35 is inhibitory to translation directed by the IRES in pCAT-N-CΔE1. They were confirmed by PhosphorImager analysis in replicate experiments (Fig. 4B). Thus, the sequence at nt 34 and 35 is located upstream of the 5′ end of the minimal IRES, even though the presence of an AG dinucleotide at this position has an inhibitory effect on the activity of the IRES within the genotype 1b virus. Since we have recently shown that a substitution of the sequence at nt 45 and 46 significantly reduces IRES activity (11), these results map the 5′ border of the IRES to the segment between nt 38 and 46, or at the 5′ end of stem-loop II (Fig. 1).
The dinucleotide sequence at nt 34 and 35 determines the efficiency of internal initiation of translation in vivo.
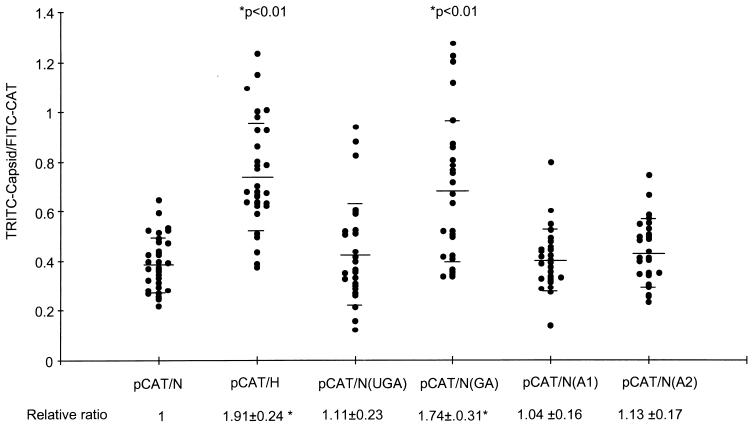
Because the studies described above were carried out in vitro with a cell-free translation system, it was important to compare the translational activities of the HCV-H and HCV-N 5′NTRs in vivo. For this purpose, we used the hybrid virus vTF7-3 (7) to direct RNA transcription in Huh-T7 cells (22) transfected with the nearly genome-length, bicistronic plasmids pCAT/H and pCAT/N. CAT and capsid protein expression were measured by quantitative, single-cell, double-label, indirect immunofluorescence cytometry. This method allows the ratio of digitally recorded, integrated intensities of the TRITC-labeled capsid image and the FITC-labeled CAT image to be calculated for individual transfected cells (see Materials and Methods for details). As the level of CAT reflects the abundance of the bicistronic RNA transcripts within each cell, the ratio of capsid protein to CAT should be proportional to the strength of the IRES in each transcript.
In these double-label experiments, approximately 30 to 40% of the cells stained positively for both CAT and capsid proteins (not shown) after transfection with the bicistronic plasmids. Thirty of these cells were sampled from each transfected culture and subjected to quantitative analysis (Fig. 5). Although there was considerable cell-to-cell variation in the capsid/CAT (TRITC/FITC) ratio, on average this ratio was approximately 1.9-fold higher in cells transfected with pCAT/H than in those transfected with pCAT/N (P < 0.01). Thus, these results are remarkably concordant with the in vitro data and confirm that the 5′NTR of HCV-H has greater intrinsic activity than the 5′NTR of HCV-N in vivo. We next analyzed cells transfected with variants of pCAT/N which contained the individual groups of base substitutions present in the 5′NTR of HCV-H: pCAT/N(UGA), pCAT/N(GA), pCAT/N(A1), and pCAT/N(A2) (Fig. 1). As pCAT/N(GA) was the only one of these plasmids to express the capsid protein at levels equivalent to that of pCAT/H (1.74-fold-higher than pCAT/N; P < 0.01), these results confirmed that the dinucleotide sequence at nt 34 and 35 of HCV-H is responsible for its increased translational activity in vivo.
FIG. 5.
Quantitative, single-cell, indirect immunofluorescence cytometry for HCV capsid and CAT proteins, following infection of Huh-T7 cells with vTF7-3 and transfection with plasmid DNAs containing nearly genome-length HCV sequences with the 5′NTRs of HCV-N or HCV-H (N, pCAT/N; H, pCAT/H) (see text) or pCAT/N derivatives containing the indicated nucleotide substitutions present in the HCV-H 5′NTR (Fig. 1). Each point represents the ratio of capsid (TRITC label) to CAT (FITC label) content for a single cell. Horizontal bars depict means ± standard deviations (values shown below each data set). P values indicate significant differences from values obtained in cells transfected with pCAT-N-CΔE1.
The AG dinucleotide sequence and downstream coding region influence the optimal KCl concentrations for IRES-directed translation in vitro.
The results described above are consistent with an interaction between RNA segments positioned upstream and downstream of the IRES that is detrimental to translation of the HCV-N sequence (see Discussion). If this interaction were due to base pairing between the flanking RNA segments, it should be enhanced at higher ionic strengths, leading to a greater reduction in the translational activity of the IRES. To test this hypothesis, we assessed the translational activity of pN-CΔE1 and pH-CΔE1 transcripts in the cell-free system at KCl concentrations ranging from 50 to 150 mM. These experiments demonstrated a striking difference in the translational activities of the HCV-H and HCV-N 5′NTRs at higher concentrations of KCl (Fig. 6A). pN-CΔE1 transcripts were optimally translated at about 80 mM KCl, with significantly reduced translation at higher KCl concentrations. In contrast, the translation of pH-C-ΔE1 transcripts was remarkably preserved at higher KCl concentrations, resulting in a much increased difference in the relative translational activities of the two transcripts. At 150 mM KCl, translation of pH-CΔE1 was approximately sevenfold that of pN-CΔE1 (Fig. 6A; compare lanes 9 and 10).
FIG. 6.
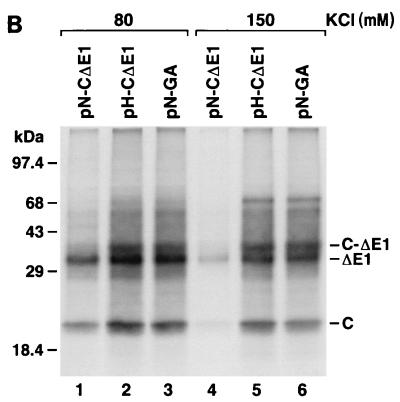
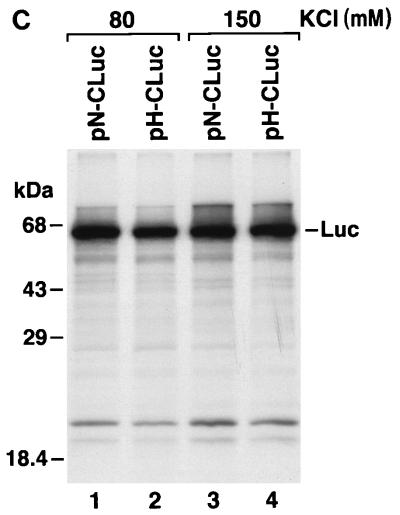
(A) SDS-PAGE of products of translation of pN-CΔE1 (lanes 1, 3, 5, 7, and 9) and pH-CΔE1 (lanes 2, 4, 6, 8, and 10) transcripts in rabbit reticulocyte lysates containing increasing concentrations of KCl. (B) Translation of pN-CΔE1 (lanes 1 and 4), pH-CΔE1 (lanes 2 and 5), and pN-GA (Fig. 3A) (lanes 3 and 6) transcripts in rabbit reticulocyte lysates containing 80 mM (lanes 1 to 3) and 150 mM (lanes 4 to 6) KCl. (C) Translation of pN-CLuc (lanes 1 and 3) and pH-CLuc (lanes 2 and 4) transcripts in rabbit reticulocyte lysates containing 80 mM (lanes 1 and 2) and 150 mM (lanes 3 and 4) KCl. Luc, luciferase.
To determine whether the differing responses to increased concentrations of KCl were due to the sequence differences that we had found to be responsible for the increased translational activity of HCV-H, we assessed the translational activity of pN-GA transcripts at 80 and 150 mM KCl. Although pN-GA differs from pN-CΔE1 only by the GA-for-AG substitution at nt 34 and 35, pN-GA transcripts behaved similarly to pH-CΔE1 transcripts, with relatively efficient translation even at 150 mM KCl (Fig. 6B; compare lanes 4 to 6). Thus, the marked sensitivity of the HCV-N IRES to increasing concentrations of KCl is related to the AG dinucleotide sequence at nt 34 and 35. Similar experiments demonstrated that replacement of the HCV-N open reading frame (downstream of nt 409) with the luciferase-coding sequence in pN-CLuc also eliminated the substantial suppression of HCV-N translation at higher KCl concentrations (compare lanes 1 and 3 in Fig. 6C with lanes 1 and 4 in Fig. 6B). Thus, as in the experiments described above, the reduced translation of the HCV-N transcripts at high concentrations of KCl is determined by a combination of sequences within RNA segments flanking the IRES. These data are thus supportive of the hypothesis that there is an interaction between these flanking RNA segments that is detrimental to IRES-directed translation in HCV-N.
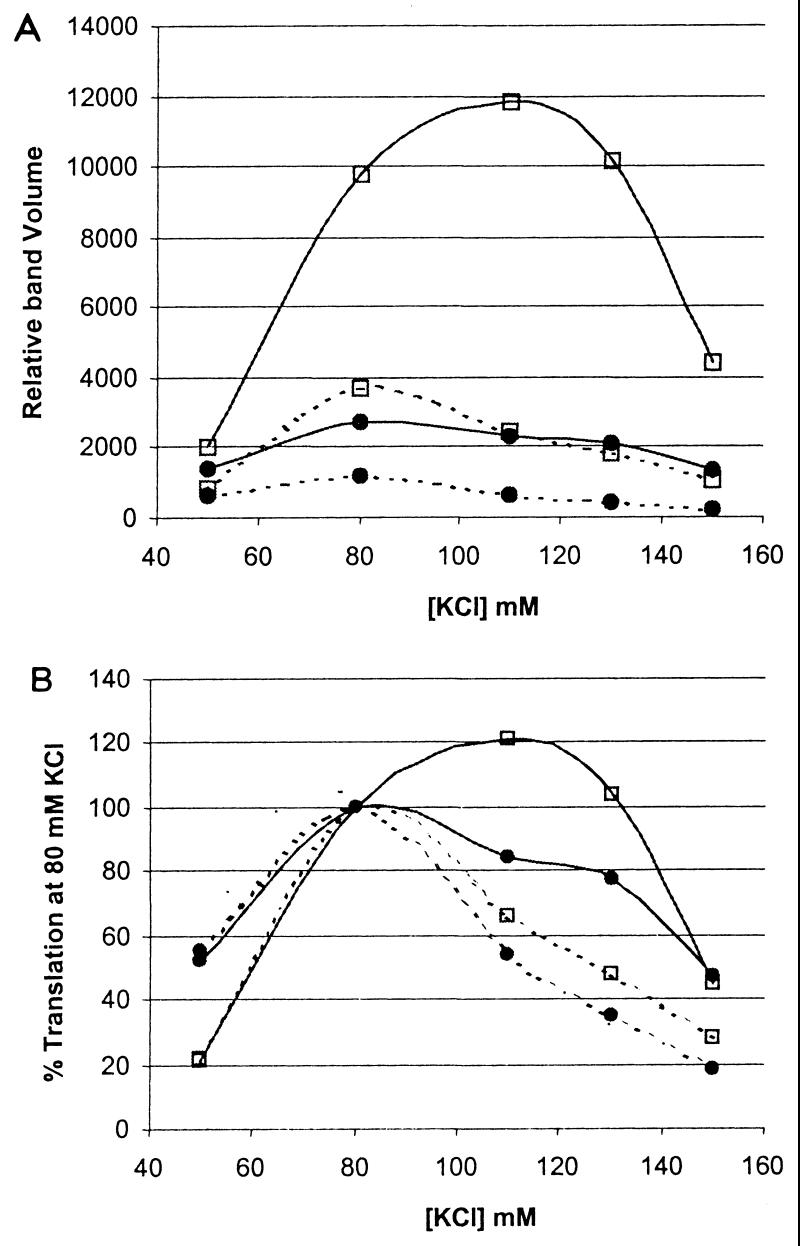
Finally, we determined whether a similar interaction might exist between the HCV-H 5′NTR and HCV-H polyprotein-coding sequence. Monocistronic transcripts derived from pN-CΔE1 and pH-CΔE1(H), or pN-CΔE1(H) and pH-CΔE1, contain homologous (H-H and N-N for simplicity) or heterologous (N-H and H-N) combinations of the 5′NTRs and polyprotein-coding sequences of HCV-H and HCV-N, respectively. These were used to program reticulocyte lysates under a range of KCl concentrations. At 80 mM KCl, the H-N chimeric sequence translated with about 2.5-fold-greater activity than the N-N sequence (Fig. 7A), consistent with earlier observations. Although the H-N chimera demonstrated an optimal KCl concentration of about 80 mM, it remained reasonably well translated at 130 or even 150 mM KCl, relative to translation of the N-N chimera. Strikingly, H-H transcripts were the most active translationally, with an optimal KCl concentration of 110 mM KCl, and with translation at 130 mM that was equal to or exceeded that at 80 mM KCl (Fig. 7B). These results were reproducible in separate experiments. The results shown in Fig. 7 indicate that either the 5′NTR or the open reading frame of HCV-N contributes to a lessening of translational activity when fused to HCV-H sequence to form a chimera. In this context, the suppressive effect of the 5′NTR is greater than that of the polyprotein-coding region.
FIG. 7.
Translation of monocistronic transcripts containing each possible combination of the HCV-H and HCV-N 5′NTRs and downstream coding sequences in rabbit reticulocyte lysates supplemented with increasing concentrations of KCl. Products of translation were separated by SDS-PAGE and quantified by PhosphorImager analysis. Relative band volumes were calculated based on the number of methionine residues in each of the two expressed polyproteins. pN-CΔE1 (···•···) and pH-CΔE1(H) (···▫···), or pN-CΔE1(H) (—•—) and pH-CΔE1 (—▫—), contain homologous or chimeric combinations of the 5′NTRs and polyprotein coding sequences of HCV-H and HCV-N, respectively. (A) Quantity of translation product produced at each KCl concentration. (B) Translation efficiency of RNA transcripts relative to translation at 80 mM KCl.
DISCUSSION
The results described in this report have important implications for the structure of HCV RNA, although their relevance to potential differences in the pathogenesis of infections with genotypes 1a and 1b is much less certain, as discussed below. First of all, it is important to note that the seven base substitutions which distinguish the 5′NTRs of the HCV-H and HCV-N viruses are representative of differences in the sequences of other genotype 1a and 1b viruses. We compared the complete 5′NTR sequences of 25 different genotype 1a and 1b viruses from GenBank (many other 5′NTR sequences listed in GenBank lack the extreme 5′ end). The most frequent differences between the 1a and 1b strains were those which distinguish the HCV-H and HCV-N strains, at nt 11 to 13 (UGA versus GAU in 1a and 1b, respectively), nt 34 and 35 (GA versus AG), nt 204 (A versus C), and nt 243 (A versus G). Importantly, all 17 genotype 1b sequences contained the AG dinucleotide sequence at nt 34 and 35, while this sequence was present in only 1 of 8 genotype 1a strains. Thus, the differences that we have found in the translational activities of the HCV-H and HCV-N 5′NTRs likely typify differences between most genotype 1a and 1b viruses.
Although the reduced translational activity of the genotype 1b 5′NTR was shown to be due to an inhibitory effect of the AG dinucleotide sequence at nt 34 and 35 (Fig. 3 and 4), this difference was expressed only when transcripts contained the complete HCV capsid coding sequence (nt 1 to 929). Previous studies have suggested that sequence downstream of the 5′NTR, within the most 5′ 30 nt of the open reading frame, may influence the activity of the HCV IRES (10, 12, 19). However, the region identified as influencing translational activity in this study lies much further downstream. We found no difference in the translational activities of transcripts containing the 1a and 1b 5′NTRs and the first 66 nt of the HCV open reading frame fused to the luciferase-coding sequence (Fig. 4). Thus, the downstream nucleotide sequence required for expression of the difference between genotypes 1a and 1b in translational activities resides between nt 408 and 929 of the HCV genome. The primary nucleotide sequences of the genotype 1a and 1b viruses that we studied differ at approximately 12% of base positions within this segment. A further striking observation was that deletion of nt 32 to 37, including the AG dinucleotide sequence, from the HCV-N 5′NTR did not impair but actually enhanced translation from dicistronic transcripts. Thus, both the upstream and downstream determinants of the greater translational activity of the genotype 1a 5′NTR are located outside the minimal essential IRES sequence.
The fact that RNA sequences within the coding region as well as upstream of the IRES act cooperatively to influence the activity of the IRES suggests that these upstream and downstream RNA segments physically interact with each other in a fashion that may influence the secondary or tertiary structure of RNA within the IRES, or otherwise alters its availability to interact with the 40S ribosome subunit (18). Alternatively, it is possible that an interaction between upstream sequence at nt 34 and 35 and sequence within the capsid-coding region is inhibitory to passage of ribosomes on the coding sequence, thus leading to premature termination of translation or a slower rate of translation. We did not observe any HCV-specific products of translation with a molecular mass less than that of the capsid protein among the products of the in vitro translation reactions, but very small polypeptide products would not be detected in the 11 to 12% polyacrylamide gels used in these studies.
The interaction between the upstream and downstream RNA sequences that we have identified could involve base pair formation, multiple RNA-protein contacts, or both. Smith and Simmonds (26) recently have proposed the existence of conserved secondary RNA structures within the 5′ 167 nt of the HCV open reading frame. This region overlaps with the segment of the coding sequence which we identified as influencing genotype 1b translational activity. It is tempting to speculate that these newly recognized structures form higher-ordered interactions with the extreme 5′ end of the viral RNA and that these might function as cis-active signals in RNA replication. The data shown in Fig. 7 indicate that the HCV-H sequence possesses the ability to interact with either the 5′NTR or downstream coding region of the HCV-N sequence. Despite this, the HCV-H 5′NTR is capable of robust translation of its open reading frame, even at high KCl concentrations (Fig. 7). Thus, if this interaction occurs within the isolated HCV-H sequence, it appears to be far less stable than in the HCV-N sequence.
As indicated above, the 5′NTRs of the genotype 1a and 1b viruses that we studied are representative of the HCV genotypes which comprise the majority of virus strains identified within either the United States or Japan and which have been suggested (although not proven) to cause infections with subtly different natural histories (17, 23, 25, 30). Could a minimal difference in the translational activities of these viruses at physiologic ionic conditions, such as that characterized here, lead to significant differences in replication capacity and possibly pathogenicity? Unfortunately, it is not possible to address this question in the absence of cell culture systems which are permissive for HCV infection and which mimic the replicative environment of the hepatocyte in vivo. Nonetheless, the fact that the AG dinucleotide sequence had similar effects on HCV translation both in rabbit reticulocyte lysates and in transfected Huh-T7 cells suggests that its apparent inhibition of IRES function is not cell type specific. The effect we observed is thus likely to be present during infection in the liver.
It is also important to note that minimal differences in translational activity can be compounded into substantial differences in replication capacity, if translation is a rate-limiting step in virus replication. Other work in our laboratory has shown that mutations within the 5′NTR of hepatitis A virus, which confer only a 4- to 6-fold increase in the internal initiation of translation by this picornavirus in monkey kidney cells, result in a marked increase in the size of replication foci and a 10-fold increase in virus yields in these cells (6, 22). Thus, while there is no direct evidence that the twofold greater translational activity of the genotype 1a 5′NTR has a significant impact on viral replication or pathogenesis in vivo, this possibility cannot be excluded at present.
ACKNOWLEDGMENTS
This work was supported in part by grants RO1-AI32599 and U19-AI40035 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Alter M J, Margolis H S, Krawczynski K, Judson F N, Mares A, Alexander W J, Hu P Y, Miller J K, Gerber M A, Sampliner R E, Meeks E L, Beach M J. The natural history of community-acquired hepatitis C in the United States. N Engl J Med. 1992;327:1899–1905. doi: 10.1056/NEJM199212313272702. [DOI] [PubMed] [Google Scholar]
- 2.Brown E A, Zhang H, Ping L-H, Lemon S M. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo Q-L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 4.Coggins J M. Image and graphics library, object-oriented (IGLOO) manual. Chapel Hill, N.C: Department of Computer Science, University of North Carolina; 1992. [Google Scholar]
- 5.Davidson F, Simmonds P, Ferguson J C, Jarvis L M, Dow B C, Follett E A C, Seed C R G, Krusius T, Lin C, Medgyesi G A, Kiyokawa H, Olim G, Duraisamy G, Cuypers T, Saeed A A, Teo D, Conradie J, Kew M C, Lin M, Nuchaprayoon C, Ndimbie O K, Yap P L. Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5′ non-coding region. J Gen Virol. 1995;76:1197–1204. doi: 10.1099/0022-1317-76-5-1197. [DOI] [PubMed] [Google Scholar]
- 6.Day S P, Murphy P, Brown E A, Lemon S M. Mutations within the 5′ nontranslated region of hepatitis A virus RNA which enhance replication in BS-C-1 cells. J Virol. 1992;66:6533–6540. doi: 10.1128/jvi.66.11.6533-6540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukushi S, Katayama K, Kurihara C, Ishiyama N, Hoshino F B, Ando T, Oya A. Complete 5′ noncoding region is necessary for the efficient internal initiation of hepatitis C virus RNA. Biochem Biophys Res Commun. 1994;199:425–432. doi: 10.1006/bbrc.1994.1246. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi N, Higashi H, Kaminaka K, Sugimoto H, Esumi M, Komatsu K, Hayashi K, Sugitani M, Suzuki K, Tadao O, Mizuno K, Shikata T. Molecular cloning and heterogeneity of the human hepatitis C virus (HCV) genome. J Hepatol. 1993;17:S94–S107. doi: 10.1016/s0168-8278(05)80432-5. [DOI] [PubMed] [Google Scholar]
- 10.Honda M, Brown E A, Lemon S M. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 11.Honda M, Beard M R, Ping L-H, Lemon S M. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honda M, Ping L-H, Rijnbrand R C A, Amphlett E, Clarke B, Rowlands D, Lemon S M. Structural requirements for initiation of translation by internal ribosomal entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 13.Kim D, Charlton J D, Coggins J M, Mohler J L. Semi-automated nuclear shape analysis of prostatic carcinoma and benign prostatic hyperplasia. Anal Quant Cytol Histol. 1994;16:400–415. [PubMed] [Google Scholar]
- 14.Kiyosawa K, Sodeyama T, Tanaka E, Gibo Y, Yoshizawa K, Nakano Y, Furata S, Akahane Y, Nishioka K, Purcell R H, Alter H J. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology. 1990;12:671–675. doi: 10.1002/hep.1840120409. [DOI] [PubMed] [Google Scholar]
- 15.Lau J Y, Davis G L, Prescott L E, Maertens G, Lindsay K L, Qian K, Mizokami M, Simmonds P Hepatitis Interventional Therapy Group. Distribution of hepatitis C virus genotypes determined by line probe assay in patients with chronic hepatitis C seen at tertiary referral centers in the United States. Ann Intern Med. 1996;124:868–876. doi: 10.7326/0003-4819-124-10-199605150-00002. [DOI] [PubMed] [Google Scholar]
- 16.Lu H H, Wimmer E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc Natl Acad Sci USA. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nousbaum J-B, Pol S, Nalpas B, Landais P, Berthelot P, Bréchot C Collaborative Study Group. Hepatitis C virus type 1b (II) infection in France and Italy. Ann Intern Med. 1995;122:161–168. doi: 10.7326/0003-4819-122-3-199502010-00001. [DOI] [PubMed] [Google Scholar]
- 18.Pestova T V, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynolds J E, Kaminiski A, Kettinen H J, Carroll A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynolds J E, Kaminski A, Carroll A R, Clarke B E, Rowlands D J, Jackson R J. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA. 1996;2:867–878. [PMC free article] [PubMed] [Google Scholar]
- 21.Rijnbrand R, Bredenbeek P, van der Straaten T, Whetter L, Inchauspe G, Lemon S, Spaan W. Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 22.Schultz D E, Honda M, Whetter L E, McKnight K L, Lemon S M. Mutations within the 5′ nontranslated RNA of cell culture-adapted hepatitis A virus which enhance cap-independent translation in cultured African green monkey kidney cells. J Virol. 1996;70:1041–1049. doi: 10.1128/jvi.70.2.1041-1049.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silini E, Bottelli R, Asti M, Bruno S, Candusso M E, Brambilla S, Bono F, Iamoni G, Tinelli C, Mondelli M U, Ideo G. Hepatitis C virus genotypes and risk of hepatocellular carcinoma in cirrhosis: a case-control study. Gastroenterology. 1996;111:199–205. doi: 10.1053/gast.1996.v111.pm8698200. [DOI] [PubMed] [Google Scholar]
- 24.Simmonds P, Holmes E C, Cha T-A, Chan S-W, McOmish F, Irvine B, Yap P L, Kolberg J, Urdea M S. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 25.Smith D B, Davidson F, Yap P L, Brown H, Kolberg J, Detmer J, Urdea M, Simmonds P, Mellor J, Neville J, Prescott L, Dow B C, Follett E A C, Skuldamrongpanich T, Tanprasert S, Nuchaprayoon C, Lin C K, Kew M C, Crookes R, Conradie J D, Lin M, Seed C, De Olim G A B, Martins I A. Levels of hepatitis C virus in blood donors infected with different viral genotypes. J Infect Dis. 1996;173:727–730. doi: 10.1093/infdis/173.3.727. [DOI] [PubMed] [Google Scholar]
- 26.Smith D B, Simmonds P. Characteristics of nucleotide substitutions in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J Mol Evol. 1997;45:238–246. doi: 10.1007/pl00006226. [DOI] [PubMed] [Google Scholar]
- 27.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C, Le S, Ali N, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zein N N, Persing D H. Hepatitis C genotypes: current trends and future implications. Mayo Clin Proc. 1996;71:458–462. doi: 10.4065/71.5.458. [DOI] [PubMed] [Google Scholar]