Abstract
RNA molecules that bind tightly and specifically to a Rex fusion protein have been isolated from a conformationally constrained pool of random sequence RNAs. The anti-Rex aptamers effectively mimic several features of the wild-type Rex-binding element (XBE). The highest-affinity aptamers effectively compete with the wild-type XBE for binding to the RNA-binding domain of Rex, an arginine-rich motif (ARM), but do not bind to the functionally analogous Rev protein or its ARM. However, characteristic sequence and structural motifs found in some of the anti-Rex aptamers may provide insights into how the Rex protein can interact with other viral RNAs, such as the Rev-responsive element. The anti-Rex aptamers can functionally substitute for the XBE in vivo, a result which supports a previously proposed model for mRNA transport in which the viral genome serves as a platform for assembling a nucleoprotein complex that can co-opt the cellular transport apparatus. Overall, these studies suggest that anti-Rex aptamers may serve as RNA decoys of the Rex protein.
Human T-cell leukemia virus type 1 (HTLV-1) is etiologically associated with adult T-cell leukemia, a malignancy of CD4+ cells (26, 60), and with tropical spastic paraparesis, a degenerative neuropathy (15, 39). Estimates of the numbers of infected individuals range from 10 to 20 million worldwide (38). HTLV-1 replication is largely regulated by the virus-encoded proteins Tax and Rex. Tax is a nuclear trans-activator that forms complexes with cellular transcription factors and stimulates transcription from promoters within the viral long terminal repeat (6, 36, 61). Rex functions posttranscriptionally, enhancing the cytoplasmic appearance of incompletely spliced mRNAs encoding HTLV-1 structural proteins and suppressing the appearance of completely spliced mRNAs encoding regulatory proteins such as Tax and Rex (22, 24, 25, 30). In effect, Rex modulates the switch between the early phase of the life cycle, in which viral regulatory proteins are produced, and the late phase, in which viral particles are assembled (reviewed by Cullen [11]). Because of its central role in regulating viral replication and the progression of infection, Rex is an excellent target for the development of antiviral compounds.
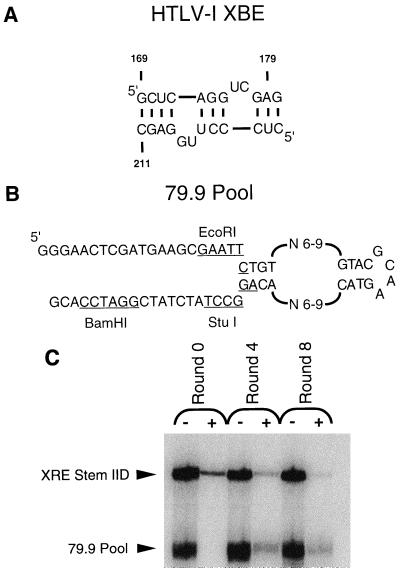
The appearance of singly spliced or unspliced messages in the cytoplasm is mediated in part by the trans-acting Rex protein (25, 30) and the cis-acting Rex-responsive element (XRE) located in the 3′ untranslated region (2, 24, 44). It is likely that Rex also interacts with a portion of the 5′ untranslated region and that RNA transport is dependent on the formation of a ternary complex between Rex and the two distinct Rex-binding sites (3, 14). Mutational analysis has indicated that an arginine-rich motif (ARM) at the amino terminus of Rex contributes to RNA binding (8, 20, 23). The sequences and structures that contribute to the function of the 3′ XRE (referred to as simply the XRE) have been studied by a variety of methods, including site-directed mutation, in vitro selection, chemical protection, nuclease protection and modification interference (5, 9, 21, 24, 50). Overall, these experiments have mapped the primary Rex-binding element (XBE) to stem IID. A minimal version of the Rex-binding element that can still specifically bind the Rex protein contains only 24 residues and folds into a stem-bulge structure (see Fig. 1A) (9).
FIG. 1.
Selection of anti-Rex aptamers. (A) Deletion mapping (2), mutational analysis (21), modification interference studies (9), and in vitro selection experiments (5) have all localized the essential Rex-binding element (XBE) to a subdomain of stem IID spanning nucleotides 169 to 180 and 200 to 211 of the RRE. The primary Rex-binding site can be further localized to nucleotides 174 to 180 and 200 to 204 (9). (B) The 79.9 pool contains a randomized internal loop flanked by constant sequence stems and primer-binding sites. Each strand of the internal loop also varies between 6 and 9 residues. The randomized region is large enough to span the core and flanking regions of the XBE and hence should be large enough to interact with the Rex protein. (C) Selected RNAs can compete with the XBE for binding to Rex. Labeled RNA pools from round 0 (unselected), round 4, and round 8 were incubated with labeled stem IID of the XRE in the presence of Rex. After filtration, elution, and precipitation, the labeled RNAs were separated from one another. The positions of the wild-type XRE stem IID and the RNA pools are indicated. The amounts of radiolabeled RNA present in each band were quantitated, and the relative amount of RNA captured by Rex (+) was compared with the relative amount of RNA originally present in each sample prior to filtration (−). The binding affinities of the pools relative to the XBE (binding ratios) were calculated as described in Materials and Methods and were 0.39 for round 0, 2.9 for round 4, and 8.5 for round 8.
Since Rex interacts with a defined RNA substrate, it may prove possible to treat or prevent HTLV-1 infections by using small RNA molecules to decoy Rex and inhibit its function. For example, the human immunodeficiency virus type 1 (HIV-1) TAR element and the HIV-1 Rev-binding element (RBE) are small defined RNA molecules that interact with the HIV-1 Tat and Rev proteins, respectively. When these RNA molecules are expressed in tissue culture cells, they decoy Tat and Rev and inhibit HIV-1 infection (33, 46).
In vitro selection has previously proven to be a valuable tool for identifying novel inhibitors of protein function (12, 18, 32). In particular, we and others have selected RNA molecules (aptamers) from random sequence pools that bind with high affinity and specificity to the Rev protein and disrupt interactions between Rev and the RBE (4, 16, 53). When expressed in cells, the anti-Rev aptamers can suppress Rev function and inhibit viral replication (19).
We have now used an in vitro selection approach to identify high-affinity RNA aptamers that bind to the Rex protein and, more specifically, to the Rex ARM. In addition to potentially serving as inhibitors of Rex function and viral replication, these anti-Rex aptamers should prove useful in more completely defining how RNA molecules mediate Rex responsiveness. For example, it is currently unclear whether the sequence and architecture of the XRE play a role in Rex responsiveness beyond merely providing a binding site for Rex. To address this question, we cloned the anti-Rex aptamers into the XRE in place of the XBE and assayed the resultant constructs for Rex responsiveness. Similarly, it is known that the Rex protein binds to a portion of the Rev-responsive element (RRE) and can functionally substitute for Rev (8, 43). By comparing the sequences and structures of the anti-Rex aptamers with the RRE we can begin to address whether the observed cross-recognition is fortuitous or is the result of a previously unknown overlap in the binding specificities of Rex and Rev.
MATERIALS AND METHODS
Materials.
Oligonucleotide primers were synthesized on a model 391 Applied Biosystems DNA synthesizer and purified by denaturing polyacrylamide gel electrophoresis. All restriction enzymes were from Promega (Madison, Wis.); Taq polymerase (AmpliTaq) was from Perkin-Elmer (Norwalk, Conn.). The GST-Rex fusion protein used in these studies has previously been described and includes the first 79 amino acids of the Rex protein followed by a stop codon at position 80 (5).
Plasmid pCRII was purchased from Invitrogen (Carlsbad, Calif.). Dulbecco’s modified Eagle medium (DMEM) and fetal bovine serum were purchased from Gibco/BRL (Gaithersburg, Md.). The acetyl coenzyme A used in chloramphenicol acetyltransferase (CAT) assays was purchased from Pharmacia (Piscataway, N.J.); [14C]chloramphenicol (CAT assay grade) was obtained from New England Nuclear (Boston, Mass.).
Random sequence RNA pools.
An RNA pool (79.9) that contained two random sequence tracts of 6 to 9 residues and that was designed to form a stem-internal loop-stem structure was used as a starting point for in vitro selection experiments (see Fig. 1B). This pool was previously described by Giver et al. (16). In short, a DNA oligonucleotide corresponding in sequence to the RNA pool was chemically synthesized and amplified by PCR. One of the primers used for amplification contained a T7 RNA polymerase promoter sequence. Approximately 0.5 μg of the amplified template was added to an Ampliscribe transcription reaction (Epicentre Technologies, Madison, Wis.). Approximately 30 μg of the initial RNA pool was obtained following purification on a 10% denaturing polyacrylamide gel.
Competitor RNAs.
tRNA (Boehringer Mannheim, Indianapolis, Ind.) was used as a nonspecific competitor for RNA binding to Rex. Stem IID of the XRE was used as a specific competitor and was transcribed from XhoI-cut pGEM-XRE. Plasmid pGEM-XRE contains residues 143 through 223 of the XRE (numbering as in Bogerd et al. [9]); some polylinker sequences were also present in the resultant transcript.
In vitro selection of anti-Rex aptamers.
Anti-Rex aptamers were isolated from the 79.9 pool by iterative selection for binding followed by amplification of bound species. In each round of selection, tRNA and Rex protein (see Table 1 for amounts) were incubated for 10 min at ambient temperature in 30 μl of 1× binding buffer (50 mM Tris-HCl [pH 8.0], 50 mM KCl). The RNA pool in 20 μl of 1× binding buffer was heated to 90°C for 2 min, was cooled to ambient temperature over 10 min to equilibrate conformers, and in some rounds was passed over HAWP 25 modified cellulose filters (Millipore, New Bedford, Mass.) to remove filter-binding sequences. The solutions containing the Rex protein and the RNA pool were mixed; in some rounds a specific competitor RNA, the XRE, was also thermally equilibrated and added to the mixture (see Table 1). The final reaction mix (60 μl) was incubated at ambient temperature for 60 min. The amounts and final concentrations of tRNA, Rex, pool RNA, and XRE were varied during the course of selection as detailed in Table 1. RNA-protein complexes were separated from free RNA by vacuum filtration (5 mm Hg) over HAWP 25 modified cellulose filters (Millipore). Following application of the RNA-protein mixtures, filters were washed twice with 500 μl of 1× binding buffer. The final two rounds of selection included a dilution step following the binding reaction; the dilution step would have favored the retention of RNA-protein complexes with low off-rates. RNAs that were coretained with Rex were eluted from the filters with 400 μl of 2× PK buffer (0.2 M Tris-Cl [pH 7.6], 2.5 mM EDTA, 0.3 M NaCl, 2% SDS) for 30 min at 75°C. The eluate was extracted with phenol-chloroform and precipitated with ethanol, and the pellet was resuspended in 25 μl of water.
TABLE 1.
79.9 selection conditions
| Round | tRNA concn (μM) | 79.9 pool concn (μM) | Rex (pmol) | Stem IID concn (μM) | Prefiltration | Dilution |
|---|---|---|---|---|---|---|
| 1 | 1.3 | 1.3 | 1.8 | 3× | ||
| 2 | 6.6 | |||||
| 3 | 0.9 | |||||
| 4 | 2.2 | |||||
| 5 | 0.9 | 3× | ||||
| 6 | 3.4 | |||||
| 7 | 4.2 | Yesa | ||||
| 8 | 13 | 8.6 | Yesb |
Diluted to a final volume of 1 ml in 1× binding buffer. This reaction was allowed to equilibrate an additional 10 min and then filtered.
Diluted to 800 μl in 1× binding buffer, and then an additional 200 μl of XRE-containing solution was added for a final concentration of 6.9 μM. This reaction was allowed to equilibrate an additional 10 min and then filtered.
RNA for subsequent rounds of selection was synthesized by a combination of reverse transcription, PCR amplification, and in vitro transcription. A portion of the extracted RNA (10 μl) was reverse transcribed in a reaction mix (20 μl) that contained 40 mM KCl, 50 mM Tris-Cl (pH 8.0), 6 mM MgCl2, 0.4 mM deoxynucleoside triphosphates (dNTPs), 5 mM dithiothreitol (DTT), 2.5 μM primer 20.86 (5′ CGTGGATCCGATAGATAGGC 3′), and 5 U of avian myeloblastosis virus reverse transcriptase (RT) (Seikagaku, St. Petersburg, Fla.). dNTPs, DTT, and RT were added following an initial annealing step (3 min at 75°C; 5 min at ambient temperature). The reaction mix was then incubated at 42°C for 45 min. For PCR amplification, 15 μl of this reaction mix was diluted in 85 μl of PCR mix (10 mM Tris-Cl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2, 5% acetamide, 0.05% NP-40, 200 μM dNTPs, 2.5 U of Taq polymerase, 0.5 μM primer 37.17 [5′ GGTAATACGACTCACTATAGGGAACTCGATGAAGCGA 3′], and 0.5 μM primer 20.86). The reaction mix was thermally cycled (94°C, 45 s; 45°C, 1 min; 72°C, 2 min) until a product band of the correct size was observed on an agarose gel. Control reactions without template were carried out in parallel; no amplification products were detected in control reactions. PCR template (5 μl [50% of a precipitated PCR mix resuspended in Tris-EDTA, pH 8.0]) was added to an Ampliscribe transcription reaction, and RNA transcripts were isolated from a denaturing polyacrylamide gel.
PCR products (1 μg) from the fourth and eighth rounds of selection were ligated into a TA cloning vector (Invitrogen) according to the protocol provided. Sequence data were generated from recombinant clones by standard dideoxy sequencing protocols.
Binding assays.
Pool RNAs were assayed for the ability to be coretained with the Rex protein on modified cellulose filters. Unselected and selected RNA pools were internally labeled by including 1 μl of [α-32P]UTP (3,000 Ci/mmol) (Dupont NEN) in an Ampliscribe transcription reaction. RNA transcripts were gel purified and resuspended in distilled water. As in the selection procedure, tRNA and Rex fusion protein were mixed and incubated at ambient temperature for 10 min. Pool RNA was thermally equilibrated at 75°C for 2 min followed by cooling to ambient temperature over 10 min. The solutions containing the Rex protein and the pool RNA were mixed in 1× binding buffer (0.96 μM final concentration of pool RNA, 9.6 μM tRNA, and 20 nM Rex fusion protein in a 40-μl final volume). The binding reaction was incubated at ambient temperature for 1 h, the mixture was filtered, and the filters were washed twice with 500 μl of 1× binding buffer. The amount of radioactive RNA that was coretained with protein was quantitated with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). The amount of RNA that bound to the filter alone in the absence of protein (generally less than 1% of applied counts) was independently determined and subtracted from the protein-dependent signal.
Competition assays.
In competition assays, pool RNAs (20 μl) were thermally equilibrated (90°C for 2 min, followed by cooling to ambient temperature over 10 min) and combined with stem IID of the XRE (20 μl), an excess of tRNA (20 μl), and 15 nM Rex protein in 1× binding buffer (0.59 μM pool RNAs, 0.64 μM stem IID, and 6.6 μM tRNA in a 60-μl final volume). The reaction was incubated, filtered, and eluted as in the selection experiments. After precipitation, samples were resuspended in 4 μl of stop dye (7 M urea, 1× TBE, 0.1% bromophenol blue) and electrophoretically separated on a denaturing 10% polyacrylamide gel. The amount of radioactivity in each band was determined with a PhosphorImager (Molecular Dynamics). Binding ratios were calculated by the formula {[(counts filtered)pool − background]/[(counts unfiltered)pool − background]}/{[(counts filtered)stem IID − background]/[(counts unfiltered)stem IID − background]}. The binding ratio represent an estimate of the aggregate affinity of pool RNAs for Rex relative to wild-type stem IID.
Clones were assayed in a similar fashion. RNA transcripts were generated from PCR-amplified templates and purified on a 10% denaturing polyacrylamide gel. Clones and stem IID of the XRE were thermally equilibrated (90°C for 2 min, followed by cooling to ambient temperature over 10 min) in 1× binding buffer in separate tubes. Rex fusion protein and tRNA were also mixed together and allowed to equilibrate for 10 min. An excess of anti-Rex aptamer and stem IID were then added to Rex and tRNA (0.45 μM stem IID, 0.64 μM clone, 0.6 μM tRNA, and 15 nM Rex fusion protein in a 60-μl final volume), and the final mixture was allowed to equilibrate at ambient temperature for 1 h. The reaction mixture was filtered over nitrocellulose, and binding ratios were calculated as described above. In some instances the aptamer all but eliminated binding of stem IID to Rex, and additional stem IID (0.9 μM final concentration) was added to the reaction mix to increase its signal during quantitation. The additional stem IID did not have a significant effect on the final binding ratios.
RNA titration experiments.
To estimate the dissociation constants of aptamer-Rex complexes, the amount of RNA retained on filters was determined as a function of RNA concentration. Transfer RNA (1 μg) was combined with 18 nM Rex and varying concentrations of thermally equilibrated (75°C for 3 min; ambient temperature for 10 min) aptamers in 1× binding buffer (50-μl final volume). The reaction mixture was incubated for 1 h at ambient temperature and filtered, and the amount of RNA retained was quantitated with a PhosphorImager. To determine the specific activity of an RNA sample, 5 μl of a 300 nM solution was spotted onto a modified cellulose filter and separately quantitated on a PhosphorImager. The number of moles of RNA bound by protein was estimated based on the number of counts retained on modified cellulose filters and the specific activity of the RNA. Moles of RNA retained was plotted against the concentration of RNA in each reaction mix. A simple bimolecular interaction between Rex and anti-Rex aptamers was consistent with the binding data, and dissociation constants were estimated by a least squares fit of the data (Kaleidograph; Abelbeck Software, Reading, Pa.).
Mobility shift assays.
Interactions between aptamers and ARM proteins were monitored with a mobility shift assay. Following thermal equilibration in 1× binding buffer (75°C for 2 min; ambient temperature for 10 min), 10 ng of 32P-end-labeled aptamer RNA was mixed with 2 μg of tRNA and varying amounts of either HTLV-1 Rex fusion protein or HIV-1 Rev protein in 10 μl of 1× binding buffer. This reaction mixture was allowed to equilibrate for 1.5 h at ambient temperature. The reaction was gently mixed with 2 μl of 6× nondenaturing dye (0.25% bromophenol blue, 40% glycerin in double-distilled water) immediately prior to gel electrophoresis. RNA-protein complexes were resolved from free RNA on an 8% native polyacrylamide gel (19:1 bis-acrylamide, 1× TBE; 5 W). Electrophoresis was carried out at ambient temperature until the bromophenol blue had run 12 cm into the gel. Bands were visualized with a PhosphorImager.
Mobility shift experiments were also carried out with a minimal version of the 8-5 aptamer (min-apt) and peptides corresponding to the Rex and Rev ARMs. The minimal aptamer was transcribed from a hemiduplex generated from the following DNA oligonucleotides: T7 primer, 5′ GAAATTAATACGACTCACTATAG 3′; template, 5′ GGGCGTACCGTCGTACTTGCGTACCGGCGCCCTATAGTGAGTCGTATTAATT 3′. The sequence of the template spans the conserved core of aptamer 8-5 and is similar to the sequence of clone 39b. The following peptides were used for mobility shift experiments and represent the ARMs of Rex and Rev, respectively: Rex ARM, MPKTRRRPRRSQRKRP-am (57); Rev ARM, suc-TRQARRNRRRRWRERQRAAAAR-am (48), where “suc” indicates succinylation of the amino terminus and “am” indicates amidation of the carboxy terminus. These peptides were synthesized by Amgen (Boulder, Colo.), and their purity was verified by HPLC and mass spectrometry. The end-labeled minimal aptamer and various amounts of Rex ARM peptide or Rev ARM peptide were incubated together in 10 μl of modified binding buffer (50 mM Tris-HCl, 50 mM KCl, 1 mM MgCl2, 5% glycerol, 5 ng of RNA) at 4°C for 1 h. Aptamer-peptide complexes were separated from free aptamer on a 10% native polyacrylamide gel (59:1 bis-acrylamide, 0.5× TBE; 350 V) at 4°C.
Construction of XRE chimeras.
Plasmids in which aptamer sequences were inserted into the XRE in place of the XBE were constructed. Plasmid pGEM REM 8 was used as the backbone for the construction of aptamer-XRE chimeras. This plasmid is an XRE deletion mutant and fails to support Rex responsiveness in vivo (2). The plasmid contains a unique BglII site in place of stem IID of the XRE, as well as an XbaI site at each end of the XRE (24). Aptamer inserts 8-5 and 9A were prepared by PCR amplification of aptamer templates cloned into pCRII. The primers used for amplification introduce BglII sites at the 5′ and 3′ ends of the insert and have the following sequences: forward 8-5 primer, 5′ GGGAGATCTCGAGATAGGCCGGCGC 3′; reverse 8-5 primer, 5′ GGGAGATCTGTAGGCGACGGTA 3′; forward 9A primer, 5′ GGGAGATCTCGAGCCTGTCCGGTGA 3′; reverse 9A primer, 5′ GGGAGATCTGTCGACGGGTACG 3′. Underlined sequences correspond to residues originally found in the aptamer.
The pGEM REM 8 backbone was linearized with BglII and ligated to the 8-5 or 9A aptamer DNAs that were also cleaved with BglII. Recombinant clones were isolated, and the XRE chimeras were excised from the pGEM background by restriction with XbaI. The XbaI restriction fragments were filled in by T4 DNA polymerase to form blunt ends, and the blunt restriction fragments were ligated into a unique, blunted ClaI site in plasmid pCMV138 (previously described by Luo et al. [35]). Plasmid pCMV138 is identical to pDM138 (29) except that it contains the cytomegalovirus (CMV) promoter region in place of the simian virus 40 promoter. The sequences of the inserts in the final recombinant clones were verified by standard dideoxy sequencing methods.
Transfection experiments.
CV-1 African green monkey kidney cells were grown in 1× DMEM containing 11% heat-inactivated fetal calf serum, 35 mM Na2CO3, and 25 mM HEPES (pH 7.1). Twenty-four hours prior to transfection, cells were seeded at 75% confluency in 60-mm culture dishes. Transient transfection assays for Rex function were performed by a calcium phosphate-mediated DNA transfection protocol previously described by Lin and Green (34) and included the following modifications. Each transfection mixture contained 1 μg of a given CMV promoter-driven wild-type XRE or XRE chimera reporter plasmid, 0.75 μg of the pcREX expression plasmid, and 2 μg of a β-galactosidase expression plasmid (pRSV-β-gal; Promega) as an internal control for both transfection efficiency and gene expression. The total DNA concentration was adjusted to 10 μg with a plasmid containing the CMV promoter (but no downstream insert) or with pUC119 DNA. Transfected cells were harvested 48 h posttransfection. CAT levels were determined with a CAT assay previously described by Hope et al. (28); β-galactosidase levels were determined using a β-galactosidase assay previously described by Lin and Green (34).
RESULTS
In vitro selection of anti-Rex aptamers.
The minimal Rex-binding element (XBE) has previously been shown by deletion mapping (2), mutational analysis (21), modification interference studies (9), and in vitro selection experiments (5) to form a relatively short stem-bulge RNA secondary structure in which the identities of as few as 10 residues may be critical for Rex-binding function (Fig. 1A). The compactness of the XBE is similar to the compactness observed for other RNA molecules that bind ARMs (16, 53, 55, 58, 59).
In order to identify high-affinity, functional mimics of the Rex-binding element, a similarly compact pool (79.9) was employed as a starting point for in vitro selection experiments (Fig. 1B). The 79.9 pool contained two random sequence tracts of 12 to 18 residues pinioned between constant sequence stems. Since the 79.9 pool could have contained only around 1011 possible species and since 1013 molecules were introduced into the first round of selection, all possible sequences and stem-like structures were likely included within the pool (16). In consequence, all possible Rex-binding elements that were similar in complexity to the wild-type XBE should have been present.
Complexes between Rex and members of the RNA pool were isolated by coimmobilization on nitrocellulose filters. The captured RNAs were eluted and amplified by reverse transcription, PCR, and in vitro transcription. In order to select the highest-affinity binding species, a selection scheme that became progressively more stringent at each round was employed (Table 1). tRNA was included in the binding reaction as a nonspecific competitor, and its concentration was increased through the course of selection. Stem IID of the XRE (containing the XBE) was introduced as a specific competitor in the third round, and its concentration was also progressively increased. The concentration of the Rex fusion protein target was decreased either by adding less target to the binding reaction or by diluting the binding reaction prior to capturing Rex-RNA complexes by filtration.
The selected population was assayed for the ability to bind Rex after four and eight rounds of selection and amplification. In the presence of nonspecific (tRNA) and specific (XBE) competitors, only 1.3% of the native RNA population could bind to Rex, while 7% of the population from the fourth round of selection could bind Rex, as could 13% from the eighth round of selection. The RNA populations from rounds 0, 4, and 8 were also assayed in direct competition with the wild-type binding element (Fig. 1C). Labeled pool and XBE RNAs were mixed with limiting amounts of Rex. The binding reactions were allowed to come to equilibrium over the course of an hour and filtered over nitrocellulose, and bound nucleic acids were eluted and precipitated. Since RNA was in excess over the target protein, different species should be retained in proportion to their relative affinities for Rex. The binding ratios for pool to wild-type RNAs were determined following electrophoretic separation and quantitation with a PhosphorImager. While unselected RNAs had a binding ratio of 0.39, the RNA population from round 4 had a binding ratio of 2.9 and the population from round 8 had a binding ratio of 8.5 (Fig. 1C); in other words, by the eighth cycle the selected RNA population as a whole could bind Rex almost ninefold better than the wild-type XBE. The increase in binding ratio from round 4 to round 8 suggested that the pool had been winnowed until only the best binding species were retained.
Novel, high-affinity RNA ligands for Rex.
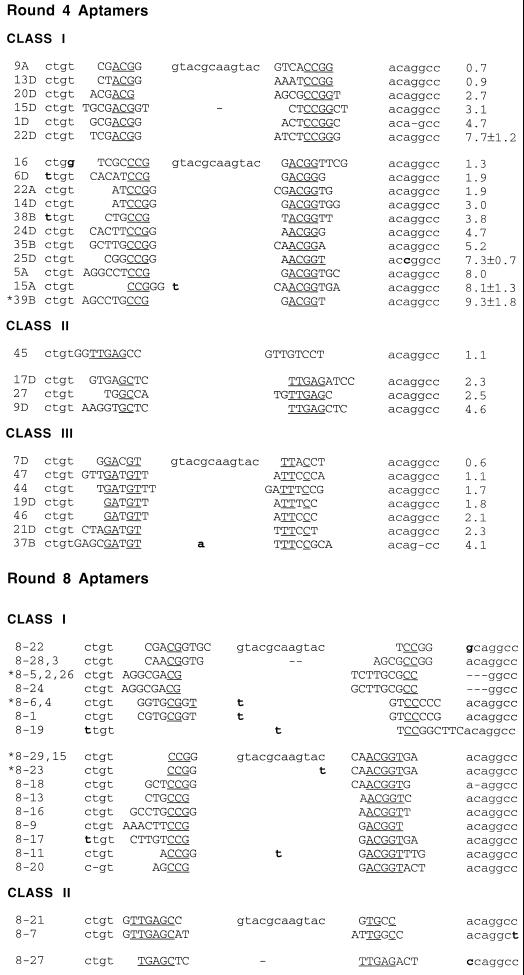
Individuals from the fourth and eighth rounds of selection were cloned and sequenced, and sequences were aligned. The round 4 aptamers could be divided into three families based on their primary sequences (Fig. 2). The relative binding affinities of the round 4 aptamers were determined in competition with stem IID (Fig. 2) and ranged from 0.6-fold of wild-type levels to roughly 9-fold higher than wild-type levels. The highest-affinity aptamers could be found in class I, while aptamers of intermediate affinity were dispersed between all three families.
FIG. 2.
Anti-Rex aptamer sequences. The sequences of aptamers from round 4 and round 8 were obtained and were aligned based on sequence similarities. The aptamers could be separated into three distinct classes. By the eighth round of selection, the class III aptamers had been largely eliminated from the population, leaving the class I and class II aptamers as the predominant species. Sequences derived from the random region are in uppercase letters, completely conserved nucleotides within an aptamer class are underlined, and mutations in constant regions are shown in boldface type. Binding ratios from competition assays are shown to the right of each aptamer from round 4; all binding ratios are averages of at least two trials.
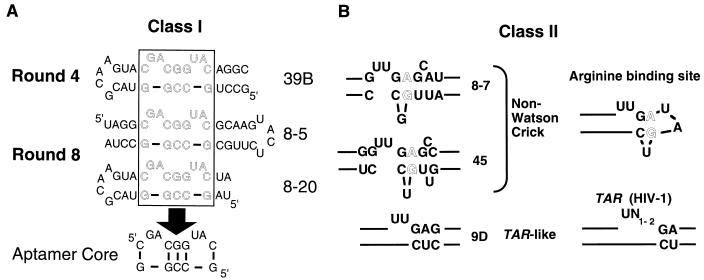
Two of the three families were predicted to form a consensus secondary structure. Because the constant regions of the original pool were constrained by base pairing and only a few additional base pairs could be formed by residues within the random regions, it is likely that the predicted structures are accurate. The most abundant family of aptamers, class I, was predicted to form a stem structure containing a centrally paired 5′ CGG … CCG 3′ motif (Fig. 3A). Interestingly, variants in which the predicted stem could be oriented either toward or away from the constant sequence stem-loop were isolated. A highly conserved adenosine is found 5′ to the centrally paired region in many class I clones (30 of 32), while a uridine is located 3′ to this structure in a majority of the clones (25 of 32). These residues are predicted to participate in bulges that varied in size from 2 to 5 nucleotides. Neither the predicted structure of the consensus class I aptamer nor the predicted structures of individual class I aptamers closely resembled the wild-type XBE, even though it should have been possible to isolate XBE-like molecules from the 79.9 pool. Class II molecules are predicted to contain a 5′ GAG … CUC 3′ pairing flanked by a bulge-loop containing two uridine residues (Fig. 3B). This sequence and structural motif is reminiscent of an arginine-binding pocket that is found in HIV-1 TAR (41) and that has been predicted to exist in the XBE (5). Class III molecules were characterized by a conserved consensus sequence 5′ NGAUGU(1–13) … AUUCCCN 3′ that could not be readily modeled as a stable secondary structure.
FIG. 3.
Sequence and structural motifs in anti-Rex aptamers. Aptamers were grouped into families based on comparisons between the sequences and predicted secondary structures of anti-Rex aptamers isolated from round 4 and round 8. (A) Class I aptamers. The similar sequences and stem-bulge structures characteristic of class I aptamers could be found in either orientation relative to the capping stem-loop structure. For example, aptamers 39B and 8-20 are in one orientation, while aptamer 8-5 is in the opposite orientation. A common set of nucleotides (boxed region) are found in the most populous, highest-affinity binding species. (B) Class II aptamers. The Class II aptamers contain a sequence and predicted structural motif that is similar to an arginine-binding motif previously observed in HIV-1 TAR elements and other RNA molecules. For example, aptamer 9D (left) contains a sequence and predicted structural motif similar to HIV-1 TAR (right). Similarly, aptamers 8-7 and 45 (left) contain non-Watson-Crick base pairs similar to arginine-binding motifs previously identified by in vitro selection (right) (49). Lines indicate nucleotides that extend outside of the highlighted domain.
By round 8, only class I and II molecules remained in the population, with class I molecules predominating. In addition, the diversity of the class I molecules observed in round 4 had decreased significantly, indicating that the population had been pared to a more optimal subset of high-affinity structures. Both orientations of the class I motif were still present in the population. The class I aptamers that remained in the population following round 8 contained in general a core sequence and structural motif that was even more tightly constrained than the class I sequence and structural motif observed following round 4 (Fig. 3A). A number of class I aptamers that contained this core motif were further assayed for the ability to bind Rex (data not shown). The highest-affinity aptamer, 8-5, was chosen for further characterization and development as a potential mimic of the XBE.
Characterization of the 8-5 aptamer.
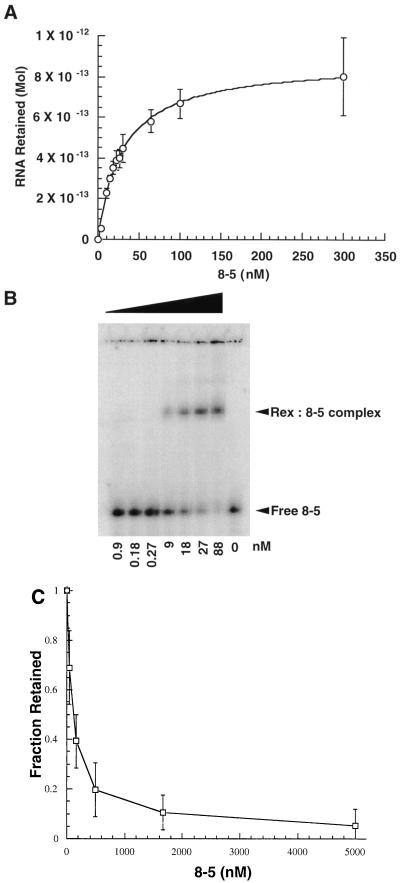
Aptamer 8-5 was assayed for the ability to bind to Rex. A binding curve was generated by incubating increasing amounts of RNA with a constant amount of Rex protein and capturing aptamer-Rex complexes by filtration. The Kapp of the aptamer-Rex complex was estimated to be 30 nM (Fig. 4A). In a complementary experiment, increasing amounts of protein were incubated with a constant amount of RNA and the formation of the aptamer-Rex complex was monitored by a gel mobility shift. The formation of the aptamer-Rex complex was again found to be concentration dependent, and the Kapp was similar (25 nM [Fig. 4B]).
FIG. 4.
Characterizing interactions between aptamer 8-5 and the Rex protein. (A) RNA titration of the high-affinity aptamer 8-5. The Kapp of the high-affinity aptamer (30 nM) was determined by titrating increasing amounts of RNA against a constant amount of protein. Samples were filtered through modified nitrocellulose, and counts retained were quantitated with a PhosphorImager. (B) Gel mobility shift assay of the high-affinity aptamer 8-5. An increasing amount of Rex fusion protein was titrated against a constant amount of body-labeled aptamer (10 ng). Complexes were separated by electrophoresis in an 8% native acrylamide gel in 1× TBE at room temperature. The amount of complex formed as a function of protein concentration was determined with a PhosphorImager. (C) Competition assay of the high-affinity aptamer 8-5 with stem IID of the XRE. Competition for binding to stem IID of the XRE was measured by incubating increasing amounts of aptamer 8-5 with a constant amount of body-labeled stem IID (0.5 μM) in the presence of the Rex protein. As the concentration of unlabeled aptamer increased, a corresponding decrease in the amount of stem IID retained following filtration was observed. A least squares fit of the data indicated that 50% inhibition of complex formation occurred at an RNA concentration of 100 nM. In contrast, no consistent diminution in binding was observed with a nonspecific RNA competitor, tRNA.
In order to determine whether the aptamer interacted with Rex at the same site and in the same manner as the wild-type XBE, competition assays in which a constant amount of radiolabeled stem IID was incubated with increasing amounts of unlabeled aptamer 8-5 in the presence of Rex protein were set up. Complexes were allowed to equilibrate and were then separated from free RNA by filtration. As the aptamer concentration was increased, the fraction of stem IID that was bound was correspondingly reduced. At the highest aptamer concentration tested (5 μM), almost complete (>90%) inhibition of complex formation was observed; at an aptamer concentration of only 100 nM, 50% inhibition was observed (Fig. 4C).
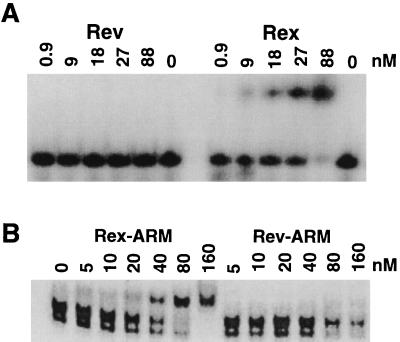
The selected anti-Rex aptamer was specific for the Rex protein. In a gel mobility shift assay, the anti-Rex aptamer did not form a stable complex with a different but functionally analogous ARM protein, HIV-1 Rev (Fig. 5A). Moreover, a minimal version of the anti-Rex aptamer could bind to the isolated Rex ARM but not the isolated Rev ARM (Fig. 5B). This result is especially significant given that the minimal XBE has not been shown to bind to the isolated Rex ARM.
FIG. 5.
The arginine-rich motif of Rex interacts specifically with aptamer 8-5. (A) The Rex protein interacts specifically with aptamer 8-5. Increasing concentrations of Rex or Rev were individually mixed with end-labeled clone 8-5, and complexes were separated on an 8% native polyacrylamide gel (19:1, 1× TBE, 5 W, ambient temperature). (B) The Rex ARM interacts specifically with a minimal version of aptamer 8-5. Peptides spanning the ARMs of Rex or Rev were titrated against a constant amount of an end-labeled, minimal version of aptamer 8-5. The minimal version of aptamer 8-5 was used in order to allow a larger electrophoretic separation between free and complexed RNA molecules. Complexes were separated on a 10% native polyacrylamide gel (59:1, 0.5× TBE, 350 V, 4°C).
Aptamers selected in vitro are functional in vivo.
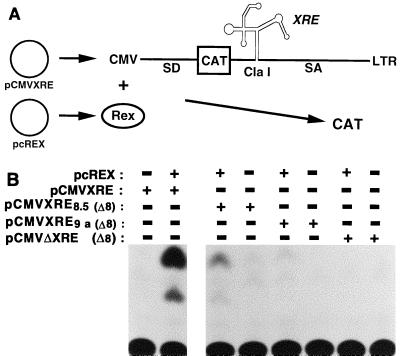
The high affinity, specificity, and stability of the selected aptamer-Rex complex and the fact that the aptamer effectively competes with the XBE for binding to Rex in vitro suggested that the aptamer might effectively mimic the function of the XBE in vivo. A reporter system that had previously been used to assay mRNA transport was adapted to assay anti-Rex aptamer function (Fig. 6A). In plasmid pCMV138, a reporter gene, CAT, is present in an HIV-1 intron and sequences responsible for Rev-dependent cytoplasmic transport (the Rev-responsive element) have been deleted from the intron. Thus, when pCMV138 is transfected into cells, mRNAs containing the HIV-1 intron are transcribed, the intron is spliced out, and the gene encoding CAT is degraded. However, when the XRE is introduced into the HIV-1 intron in place of the RRE and the plasmid is cotransfected with a Rex expression plasmid, unspliced mRNAs are transported to the cytoplasm and the gene encoding CAT is translated. In effect, expression of the CAT activity in pCMV138 is dependent on the dual presence of RNA elements and protein factors that can facilitate cytoplasmic transport (28, 37). This system therefore affords an opportunity to assay anti-Rex aptamers for the ability to bind Rex and support mRNA transport in vivo.
FIG. 6.
Anti-Rex aptamers mediate Rex responsiveness. (A) CAT reporter system and XRE chimeras. Plasmid pCMVXRE contains a strong CMV promoter that can generate an mRNA in which the sequences of the CAT protein and the XRE are embedded between splice donor (SD) and splice acceptor (SA) sites of an HIV-1 intron. In the absence of Rex, the intron is spliced out of the mRNA and degraded and little CAT activity can be observed in cell extracts. When pCMVXRE is transiently transfected into cells along with a Rex expression plasmid (pcREX), Rex mediates cytoplasmic transport of unspliced mRNA, more CAT is translated from the intact mRNAs, and the greater amounts of CAT activity can be observed in cell extracts. The amount of CAT activity observed is roughly proportional to the number of Rex-RNA complexes that are formed in the cell. (B) CAT assay of chimeric Rex-responsive elements. Reporter plasmids containing the wild-type XRE or XRE chimeras were cotransfected into CV-1 cells in the presence (+) or absence (−) of the Rex expression plasmid pcREX (24). Forty hours posttransfection, the cells were harvested and assayed for CAT activity. The identity of the aptamer that gave rise to the XRE chimera is indicated at the left. The positions of unacetylated and acetylated chloramphenicol on the TLC plate are shown.
To this end, a set of hybrid XRE sequences in which the XBE was replaced with aptamer sequences was generated (Fig. 6A). A deletion variant (REM 8) of the XRE which lacked the XBE was used as a starting point for the construction of chimeric XRE elements (2). Two different aptamer sequences were introduced into REM 8 in place of the wild-type XBE: the high-affinity aptamer that was isolated from round 8 (clone 8-5) and an aptamer that was isolated from round 4 but could not effectively compete for binding to stem IID of the XRE (clone 9A). The low-affinity aptamer serves as a negative control for the function of the high-affinity site. Plasmids bearing the hybrid XREs were transfected into tissue culture cells along with a Rex expression plasmid, and the Rex-dependent production of CAT activity was determined. The hybrid XRE containing aptamer 9A [Rex 9A (Δ8)] was not Rex responsive (less than 5% of wild-type activity). In contrast, aptamer 8-5 [Rex 8-5 (Δ8)] appeared to support Rex-responsive mRNA transport, although the hybrid XRE was less Rex responsive than the wild-type XRE (46% of wild-type activity [Fig. 6B]).
DISCUSSION
High-affinity aptamers that bind to a Rex fusion protein of HTLV-1 have been isolated from a short, conformationally constrained pool of RNA. While a longer pool could potentially have been used to derive aptamers, in order to make sure that anti-Rex aptamers were also Rex antagonists we wished to mimic as closely as possible the known sequence and structural requirements of the Rex-binding element. Moreover, we have previously observed that aptamers selected from both shorter (<30) and longer (>30) random sequence pools contained the same short consensus sequence and structural motifs (a phenomenon termed the “tyranny of short motifs”). Thus, it seemed likely that we might more fully cover sequence space with a shorter pool yet still extract the same high-affinity elements as with a longer random sequence pool. Finally, since the 79.9 pool had previously been used for selection of aptamers that could bind to HIV-1 Rev (16), it proved possible to directly compare the sequences, structures, and specificities of anti-Rex and anti-Rev aptamers.
The stringency of selection was graded in order to efficiently drive the selection process. Protein concentration, competitor concentration, and equilibrium dynamics were systematically varied, and low-affinity or non-specifically binding RNA molecules were eliminated from the population within a few rounds. The selected pool showed a marked improvement over the unselected pool and bound Rex severalfold better than isolated stem IID of the XRE, which contains the XBE. The aggregate improvement in the binding ability of the pool was mirrored by improvements in the affinities of individual aptamers between rounds. Between rounds 4 and 8, low-affinity class III molecules had been eliminated from the population, only a minor set of class II sequences remained, and the highest-affinity binding species in class I predominated. When assayed in direct competition with stem IID, individual aptamers from round 8 bound either as well as or up to ninefold better than the XBE.
Although sequences and structures similar to the wild-type XBE would have been contained in the original random sequence pool, the sequences of the best anti-Rex aptamers (class I) do not closely resemble the sequence of the XBE. Moreover, although both class I aptamers and the XBE contain dinucleotide bulges separated by a central, base-paired triplet, the bulged nucleotides are on the same strand in class I aptamers but on opposing strands in the XBE. The isolation of such non-wild-type RNA-binding species from completely randomized pools has previously been observed. For example, RNA hairpins selected to bind phage T4 DNA polymerase bore little resemblance to the wild-type sequence (51), while RNA aptamers that bound HIV-1 RT were predicted to form a unique pseudoknot structure that differed from a tRNA cloverleaf (52). Sequence differences between natural and in vitro-selected RNA ligands may be the result of biological restrictions on natural selection, such as codon usage or RNA folding in the context of a viral genome, that have no counterpart in in vitro selection experiments.
Both class I aptamers and the XBE interact with the same site on the Rex protein, the ARM that lies between residues 1 and 16. Since the constant sequence helices in the original pool have been retained in the predicted structures of the anti-Rex aptamers, it is likely that selected residues in the formerly random region are not involved in structure presentation but instead directly participate in contacts with the Rex ARM. In support of this hypothesis, anti-Rev aptamers have previously been selected from the same random sequence pool and structural studies have revealed that many of the selected residues directly contact the Rev protein (16, 58).
Our results may also help to resolve conflicting reports in the literature over whether and how the wild-type Rex ARM interacts with the XBE or XRE. Hope and coworkers have shown when the Rev and Rex proteins are fused, the Rex ARM is dispensible for Rex-dependent mRNA transport (29). These results are supported in part by the work of Bohnlein, Hauber, and their coworkers, who have demonstrated that Rex deletion variants that lack an ARM can nonetheless mediate Rex-dependent mRNA transport, albeit at reduced levels (27). In contrast, Grassmann and coworkers (20) have shown that Rex deletion variants that lack an ARM do not bind well to the XRE in vitro. Nonconservative substitutions in the Rex ARM (Arg5-Arg6-Arg7 to Asp-Leu) disrupt the function of Rex (42) and the ability of Rex to bind to the XRE (8). Similarly, Hammes and Greene (23) have demonstrated that conservative lysine-for-arginine substitutions within the ARM can abrogate Rex-dependent mRNA transport. Since the anti-Rex aptamers bind the Rex ARM and also directly compete with the XBE for binding to Rex, our results strongly imply that Rex must also bind to the XBE via its ARM. In support of this conclusion, when the Rex ARM is fused to a heterologous proteins (β-galactosidase), weak binding to the XRE can be demonstrated (20). However, as a caveat to this discussion it should be noted that additional XBE binding sites within Rex may fall outside the ARM, as suggested by Bohnlein et al. (10) and Weichselbraun and coworkers (56).
Precisely because class I aptamers do not resemble the XBE yet bind to the same site on Rex, they should prove to be useful reagents for probing the role of RNA sequence and structure in Rex-mediated RNA transport. The highest-affinity aptamer, 8-5, was cloned into the XRE in place of the XBE and was found to efficiently support Rex-responsive mRNA transport, though not as well as the wild-type XRE. The diminished activity of the chimeric XRE may be a result of not knowing precisely how to substitute the aptamer for the XBE or how to position the aptamer relative to other Rex-binding sites on the XRE, such as an adjacent Rex-binding site mapped by Askjaer and Kjems (3). It is also possible that the binding affinities of aptamers in the in vivo cellular milieu are different than those calculated based on in vitro binding reactions. However, while the requirements for transport function or protein binding may be more stringent in vivo than in vitro, it is nonetheless impressive that an unnatural RNA element selected solely on the basis of its ability to bind Rex can support Rex-dependent RNA transport. Moreover, the fact that a high-affinity anti-Rex aptamer supports mRNA transport better than a low-affinity anti-Rex aptamer supports the contention that the aptamers are serving as functional substitutes for the XBE.
Overall, these findings support and complement other mechanistic studies of viral mRNA transport. Current models for the mechanism of Rex responsiveness suggest that the cellular mRNA transport apparatus interacts with the Rex protein through an effector domain (7, 13, 45). The effector domains of Rex and Rev are similar, and when these domains are swapped for one another the chimeric proteins can still facilitate cytoplasmic mRNA transport (56). These ARM proteins therefore appear to function as simple connectors that transitively bind viral RNA to the cellular machinery. We now show that the XRE may contribute little to this connection beyond the ability to act as a handle for Rex binding. This interpretation accords with a previous study in which anti-Rev aptamers substituted for the RBE within the RRE were found to support Rev responsiveness (47).
Taken together, our results suggest that it may be possible to use class I aptamers as intracellular decoys for Rex and, thus, as inhibitors of HTLV-1 replication. First, since aptamer 8-5 can functionally substitute for the XBE, it likely interacts with the Rex protein in vivo. Second, since aptamer 8-5 binds specifically to the Rex protein and Rex ARM, eschewing interactions with the similarly charged, functionally analogous Rev protein and Rev ARM even at high peptide concentrations, it should not bind nonspecifically to other cellular or viral proteins. Third, the essential sequences and structures of aptamer 8-5 can be reduced to a synthetically accessible 22-nucleotide RNA by the incorporation of synthetic linker elements; the minimal construct shows no loss of binding activity (39a). Finally, a similar strategy based on intracellular expression of anti-Rev aptamers has proven successful at inhibiting HIV-1 replication (19, 33).
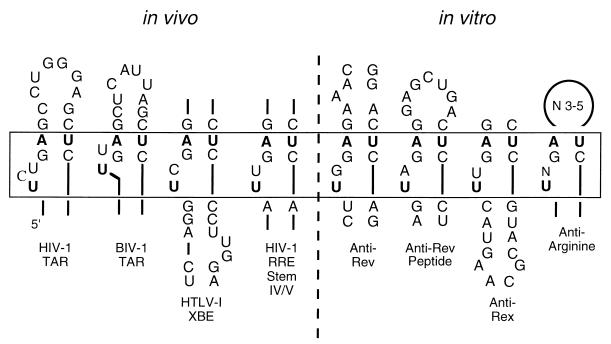
Other anti-Rex aptamers showed a greater resemblance to the wild-type XBE and to other RNA molecules that bind ARMs. Class II aptamers contained a conserved sequence motif, 5′ UUGAG … CUC 3′, that was predicted to form a short stem adjacent to a bulge loop (Fig. 7). A similar sequence is also found in the wild-type XBE and is also predicted to form a stem-bulge structure. In vitro selection experiments with partially randomized populations based on the wild-type XBE have previously shown that the 5′ UUGAG … CUC 3′ stem-bulge is important for Rex binding (5). Moreover, an XBE “half-site” that contains the stem-bulge and that resembles class II aptamers can bind Rex (5).
FIG. 7.
Arginine-binding sites in natural and in vitro-selected RNA molecules. The sequence 5′ UUGAG … CUC 3′ forms a stem-bulge structure that has been shown to be an arginine-binding pocket in HIV-1 TAR (1, 41). Variations on this sequence and structural motif that are nonetheless known to form arginine-binding pockets have also been identified in other natural RNA molecules (e.g., bovine immunodeficiency virus type 1 TAR [40, 59]) and RNA molecules generated by in vitro selection (e.g., an anti-Rev aptamer [16, 53, 58] and antiarginine aptamers [49]; see also Fig. 3B). Other natural (RRE stem IV/V) and selected (anti-Rev peptide aptamers [57]) RNAs contain similar sequences and predicted structural motifs and are hypothesized to recognize arginine in a similar manner. The residues that are known or postulated to be involved in arginine recognition are boxed.
The stem-bulge sequence and structural motif seen in class II aptamers and in the XBE is similar to a stem-bulge that has been predicted to form in HIV-1 TAR (Fig. 3A, Fig. 7). Structural studies of TAR have revealed that the stem-bulge forms a compact pocket that can interact with one of the arginines in the ARM of HIV-1 Tat (41). In a Tat-TAR complex, the guanidino group of arginine is thought to form a pseudo-Hoogsteen pair with the G:C base pair adjacent to the bulge, while the bulged U residue forms a base triple with A:U that pulls the negatively charged phosphate backbone into apposition with the positively charged arginine.
A number of groups have previously found that Rex can recognize the RRE and functionally substitute for HIV-1 Rev (2, 8, 10, 24, 43, 54, 56). The primary Rex-binding site on the RRE appears to be different from the primary Rev-binding site: Rev binds first to stem II of the RRE, while Rex binds to a region that includes stems IV and V (2, 54). Since the anti-Rex aptamers we have identified serve as examples of individual Rex-binding sites, we compared the sequences and structures of the anti-Rex aptamers with the sequence and structure of the RRE. Surprisingly, a paired 5′ GAG … CUC 3′ triplet is found adjacent to a U-rich single-stranded region in stem IV/V of the RRE. This putative arginine-binding pocket differs from those observed previously in that a homopurine base pair may separate paired and single-stranded regions. However, arginine-binding pockets based on the 5′ GAG … CUC 3′ motif have previously been shown to be structurally diverse: mutational analysis of the TAR element indicates that two- to four-base bulges are functional and bovine immunodeficiency virus TAR bisects the bulge-loop with a base pair (Fig. 7). To the extent that the putative arginine-binding pocket in RRE stem IV/V and the arginine-binding pockets predicted to form in class II anti-Rex aptamers and in the XBE are functionally equivalent, our selection results may provide a basis for understanding how Rex recognizes the RRE.
REFERENCES
- 1.Aboul-ela F, Karn J, Varani G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J Mol Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed Y F, Hanly S M, Malim M H, Cullen B R, Greene W C. Structure-function analyses of the HTLV-I Rex and HIV-1 Rev RNA response elements: insights into the mechanism of Rex and Rev action. Genes Dev. 1990;4:1014–1022. doi: 10.1101/gad.4.6.1014. [DOI] [PubMed] [Google Scholar]
- 3.Askjaer P, Kjems J. Mapping multiple RNA binding sites of human T-cell lymphotropic virus type I Rex protein within 5′- and 3′-Rex response elements. J Biol Chem. 1998;273:11463–11471. doi: 10.1074/jbc.273.19.11463. [DOI] [PubMed] [Google Scholar]
- 4.Bartel D P, Zapp M L, Green M R, Szostak J W. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell. 1991;67:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 5.Baskerville S, Zapp M, Ellington A D. High-resolution mapping of the human T-cell leukemia virus type 1 Rex-binding element by in vitro selection. J Virol. 1995;69:7559–7569. doi: 10.1128/jvi.69.12.7559-7569.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beraud C, Lombard-Platet G, Michal Y, Jalinot P. Binding of the HTLV-1 Tax 1 transactivator to the inducible 21 bp enhancer is mediated by the cellular factor HEB1. EMBO J. 1991;10:3795–3803. doi: 10.1002/j.1460-2075.1991.tb04949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogerd H P, Fridell R A, Madore S, Cullen B R. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 8.Bogerd H P, Huckaby G L, Ahmed Y F, Hanly S M, Greene W C. The type I human T-cell leukemia virus (HTLV-I) Rex trans-activator binds directly to the HTLV-I Rex and the type 1 human immunodeficiency virus Rev RNA response elements. Proc Natl Acad Sci USA. 1991;88:5704–5708. doi: 10.1073/pnas.88.13.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogerd H P, Tiley L S, Cullen B R. Specific binding of the human T-cell leukemia virus type I Rex protein to a short RNA sequence located within the Rex response element. J Virol. 1992;66:7572–7575. doi: 10.1128/jvi.66.12.7572-7575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohnlein E, Berger J, Hauber J. Functional mapping of the human immunodeficiency virus type 1 Rev RNA binding domain: new insights into the domain structure of Rev and Rex. J Virol. 1991;65:7051–7055. doi: 10.1128/jvi.65.12.7051-7055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen B R. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol Rev. 1992;56:375–394. doi: 10.1128/mr.56.3.375-394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellington A D, Conrad R. Aptamers as potential nucleic acid pharmaceuticals. Biotechnol Annu Rev. 1995;1:185–214. doi: 10.1016/s1387-2656(08)70052-8. [DOI] [PubMed] [Google Scholar]
- 13.Fritz C C, Zapp M L, Green M R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 14.Fu L, White K N. Enhancement of nucleocytoplasmic export of HTLV-I Rex mRNA through cis and trans interactions of the mRNA with the complex of Rex protein and Rex-responsive element. FEBS Lett. 1996;396:47–52. doi: 10.1016/0014-5793(96)01062-9. [DOI] [PubMed] [Google Scholar]
- 15.Gessain A, Barin F, Vernant J C, Gout O, Maurs L, Calender A, de The G. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet. 1985;ii:407–410. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 16.Giver L, Bartel D, Zapp M, Pawul A, Green M, Ellington A D. Selective optimization of the Rev-binding element of HIV-1. Nucleic Acids res. 1993;21:5509–5516. doi: 10.1093/nar/21.23.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giver L, Bartel D P, Zapp M I, Green M R, Ellington A D. Selection and design of high-affinity RNA ligands for HIV-1 Rev. Gene. 1993;137:19–24. doi: 10.1016/0378-1119(93)90246-y. [DOI] [PubMed] [Google Scholar]
- 18.Gold L. Conformational properties of oligonucleotides. Nucleic Acids Symp Ser. 1995;33:20–22. [PubMed] [Google Scholar]
- 19.Good P D, Krikos A J, Li S X, Bertrand E, Lee N S, Giver L, Ellington A, Zaia J A, Rossi J J, Engelke D R. Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther. 1997;4:45–54. doi: 10.1038/sj.gt.3300354. [DOI] [PubMed] [Google Scholar]
- 20.Grassmann R, Berchtold S, Aepinus C, Ballaun C, Boehnlein E, Fleckenstein B. In vitro binding of human T-cell leukemia virus rex proteins to the rex-response element of viral transcripts. J Virol. 1991;65:3721–3727. doi: 10.1128/jvi.65.7.3721-3727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grone M, Hoffmann E, Berchtold S, Cullen B R, Grassmann R. A single stem-loop structure within the HTLV-1 Rex response element is sufficient to mediate Rex activity in vivo. Virology. 1994;204:144–152. doi: 10.1006/viro.1994.1518. [DOI] [PubMed] [Google Scholar]
- 22.Grone M, Koch C, Grassmann R. The HTLV-1 Rex protein induces nuclear accumulation of unspliced viral RNA by avoiding intron excision and degradation. Virology. 1996;218:316–325. doi: 10.1006/viro.1996.0200. [DOI] [PubMed] [Google Scholar]
- 23.Hammes S R, Greene W C. Multiple arginine residues within the basic domain of HTLV-I Rex are required for specific RNA binding and function. Virology. 1993;193:41–49. doi: 10.1006/viro.1993.1101. [DOI] [PubMed] [Google Scholar]
- 24.Hanly S M, Rimsky L T, Malim M H, Kim J H, Hauber J, Duc Dodon M, Le S Y, Maizel J V, Cullen B R, Greene W C. Comparative analysis of the HTLV-I Rex and HIV-1 Rev trans-regulatory proteins and their RNA response elements. Genes Dev. 1989;3:1534–1544. doi: 10.1101/gad.3.10.1534. [DOI] [PubMed] [Google Scholar]
- 25.Hidaka M, Inoue J, Yoshida M, Seiki M. Post-transcriptional regulator (rex) of HTLV-1 initiates expression of viral structural proteins but suppresses expression of regulatory proteins. EMBO J. 1988;7:519–523. doi: 10.1002/j.1460-2075.1988.tb02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinuma Y, Nagata K, Hanaoka M, Nakai M, Matsumoto T, Kinoshita K I, Shirakawa S, Miyoshi I. Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc Natl Acad Sci USA. 1981;78:6476–6480. doi: 10.1073/pnas.78.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofer L, Weichselbraun I, Quick S, Farrington G K, Bohnlein E, Hauber J. Mutational analysis of the human T-cell leukemia virus type I trans-acting rex gene product. J Virol. 1991;65:3379–3383. doi: 10.1128/jvi.65.6.3379-3383.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hope T J, Bond B L, McDonald D, Klein N P, Parslow T G. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex are functionally interchangeable and share an essential peptide motif. J Virol. 1991;65:6001–6007. doi: 10.1128/jvi.65.11.6001-6007.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hope T J, McDonald D, Huang X J, Low J, Parslow T G. Mutational analysis of the human immunodeficiency virus type 1 Rev transactivator: essential residues near the amino terminus. J Virol. 1990;64:5360–5366. doi: 10.1128/jvi.64.11.5360-5366.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue J, Yoshida M, Seiki M. Transcriptional (p40x) and post-transcriptional (p27x-III) regulators are required for the expression and replication of human T-cell leukemia virus type I genes. Proc Natl Acad Sci USA. 1987;84:3653–3657. doi: 10.1073/pnas.84.11.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen K B, Green L, MacDougal-Waugh S, Tuerk C. Characterization of an in vitro-selected RNA ligand to the HIV-1 Rev protein. J Mol Biol. 1994;235:237–247. doi: 10.1016/s0022-2836(05)80030-0. [DOI] [PubMed] [Google Scholar]
- 32.Klug S J, Famulok M. All you wanted to know about SELEX. Mol Biol Rep. 1994;20:97–107. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- 33.Lee S W, Gallardo H F, Gilboa E, Smith C. Inhibition of human immunodeficiency virus type 1 in human T cells by a potent Rev response element decoy consisting of the 13-nucleotide minimal Rev-binding domain. J Virol. 1994;68:8254–8264. doi: 10.1128/jvi.68.12.8254-8264.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Y S, Green M R. Similarities between prokaryotic and eukaryotic cyclic AMP-responsive promoter elements. Nature. 1989;340:656–659. doi: 10.1038/340656a0. [DOI] [PubMed] [Google Scholar]
- 35.Luo Y, Yu H, Peterlin B M. Cellular protein modulates effects of human immunodeficiency virus type 1 Rev. J Virol. 1994;68:3850–3856. doi: 10.1128/jvi.68.6.3850-3856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marriott S J, Lindholm P F, Brown K M, Gitlin S D, Duvall J F, Radonovich M F, Brady J N. A 36-kilodalton cellular transcription factor mediates an indirect interaction of human T-cell leukemia/lymphoma virus type I TAX1 with a responsive element in the viral long terminal repeat. Mol Cell Biol. 1990;10:4192–4201. doi: 10.1128/mcb.10.8.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDonald D, Hope T J, Parslow T G. Postranscriptional regulation by the human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex proteins through a heterologous RNA binding site. J Virol. 1992;66:7232–7238. doi: 10.1128/jvi.66.12.7232-7238.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy E L, Hanchard B, Figueroa J P, Gibbs W N, Lofters W S, Campbell M, Goedert J J, Blattner W A. Modelling the risk of adult T-cell leukemia/lymphoma in persons infected with human T-lymphotropic virus type I. Int J Cancer. 1989;43:250–253. doi: 10.1002/ijc.2910430214. [DOI] [PubMed] [Google Scholar]
- 39.Osame M, Usuku K, Izumo S, Ijichi N, Amitani H, Igata A, Matsumoto M, Tara M. HTLV-I associated myelopathy, a new clinical entity [letter] Lancet. 1986;i:1031–1032. doi: 10.1016/s0140-6736(86)91298-5. [DOI] [PubMed] [Google Scholar]
- 39a.Osborne, S. E. Unpublished results.
- 40.Puglisi J D, Chen L, Blanchard S, Frankel A D. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science. 1995;270:1200–1203. doi: 10.1126/science.270.5239.1200. [DOI] [PubMed] [Google Scholar]
- 41.Puglisi J D, Tan R, Calnan B J, Frankel A D, Williamson J R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 42.Rimsky L, Dodon M D, Dixon E P, Greene W C. Trans-dominant inactivation of HTLV-I and HIV-1 gene expression by mutation of the HTLV-I Rex transactivator. Nature. 1989;341:453–456. doi: 10.1038/341453a0. [DOI] [PubMed] [Google Scholar]
- 43.Rimsky L, Hauber J, Dukovich M, Malim M H, Langlois A, Cullen B R, Greene W C. Functional replacement of the HIV-1 rev protein by the HTLV-1 rex protein. Nature. 1988;335:738–740. doi: 10.1038/335738a0. [DOI] [PubMed] [Google Scholar]
- 44.Seiki M, Inoue J, Hidaka M, Yoshida M. Two cis-acting elements responsible for posttranscriptional trans-regulation of gene expression of human T-cell leukemia virus type I. Proc Natl Acad Sci USA. 1988;85:7124–7128. doi: 10.1073/pnas.85.19.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 46.Sullenger B A, Gallardo H F, Ungers G E, Gilboa E. Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell. 1990;63:601–608. doi: 10.1016/0092-8674(90)90455-n. [DOI] [PubMed] [Google Scholar]
- 47.Symensma T L, Giver L, Zapp M, Takle G B, Ellington A D. RNA aptamers selected to bind human immunodeficiency virus type 1 Rev in vitro are Rev responsive in vivo. J Virol. 1996;70:179–187. doi: 10.1128/jvi.70.1.179-187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan R, Chen L, Buettner J A, Hudson D, Frankel A D. RNA recognition by an isolated alpha helix. Cell. 1993;73:1031–1040. doi: 10.1016/0092-8674(93)90280-4. [DOI] [PubMed] [Google Scholar]
- 49.Tao J, Frankel A D. Arginine-binding RNAs resembling TAR identified by in vitro selection. Biochemistry. 1996;35:2229–2238. doi: 10.1021/bi951844b. [DOI] [PubMed] [Google Scholar]
- 50.Toyoshima H, Itoh M, Inoue J-I, Seiki M, Takaku F, Yoshida M. Secondary structure of the human T-cell leukemia virus type 1 rex-responsive element is essential for rex regulation of RNA processing and transport of unspliced RNAs. J Virol. 1990;64:2825–2832. doi: 10.1128/jvi.64.6.2825-2832.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 52.Tuerk C, MacDougal S, Gold L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tuerk C, MacDougal-Waugh S. In vitro evolution of functional nucleic acids: high-affinity RNA ligands of HIV-1 proteins. Gene. 1993;137:33–39. doi: 10.1016/0378-1119(93)90248-2. [DOI] [PubMed] [Google Scholar]
- 54.Unge T, Solomin L, Mellini M, Derse D, Felber B K, Pavlakis G N. The Rex regulatory protein of human T-cell lymphotropic virus type I binds specifically to its target site within the viral RNA. Proc Natl Acad Sci USA. 1991;88:7145–7149. doi: 10.1073/pnas.88.16.7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weeks K M, Crothers D M. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell. 1991;66:577–588. doi: 10.1016/0092-8674(81)90020-9. . (Erratum, 70:1069, 1992.) [DOI] [PubMed] [Google Scholar]
- 56.Weichselbraun I, Farrington G K, Rusche J R, Bohnlein E, Hauber J. Definition of the human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex protein activation domain by functional exchange. J Virol. 1992;66:2583–2587. doi: 10.1128/jvi.66.4.2583-2587.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu W, Ellington A D. Anti-peptide aptamers recognize amino acid sequence and bind a protein epitope. Proc Natl Acad Sci USA. 1996;93:7475–7480. doi: 10.1073/pnas.93.15.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye X, Gorin A, Ellington A D, Patel D J. Deep penetration of an alpha-helix into a widened RNA major groove in the HIV-1 rev peptide-RNA aptamer complex. Nat Struct Biol. 1996;3:1026–1033. doi: 10.1038/nsb1296-1026. [DOI] [PubMed] [Google Scholar]
- 59.Ye X, Kumar R A, Patel D J. Molecular recognition in the bovine immunodeficiency virus Tat peptide-TAR RNA complex. Chem Biol. 1995;2:827–840. doi: 10.1016/1074-5521(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc Natl Acad Sci USA. 1982;79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao L J, Giam C Z. Interaction of the human T-cell lymphotrophic virus type I (HTLV-I) transcriptional activator Tax with cellular factors that bind specifically to the 21-base-pair repeats in the HTLV-I enhancer. Proc Natl Acad Sci USA. 1991;88:11445–11449. doi: 10.1073/pnas.88.24.11445. [DOI] [PMC free article] [PubMed] [Google Scholar]