ABSTRACT
Context:
Dengue is one of the important vector-borne viral diseases affecting humans with diverse manifestations. Toll-like receptors (TLR) are pattern recognition receptors and play an important role in innate immunity against microbes. TLR3 plays a critical role in controlling the innate immune response mediated by flaviviruses such as dengue.
Aim:
We attempted to study the susceptibility of single nucleotide polymorphism of the TLR3 gene in dengue encephalitis (DE) patients and determine the association in terms of genotype, allele, and haplotype distribution along with the clinical outcome.
Settings and Design:
It was a case-controlled observational study in a tertiary care hospital.
Methods and Material:
We investigated the single nucleotide polymorphism in the TLR3 Leu412Phe gene using real-time polymerase chain reaction in 29 cases of DE and compared them with equal number of age- and sex-matched dengue patients without neurological features.
Statistical Analysis Used:
The genotype and allele frequencies were compared using a two-sided Chi-square or Fisher’s exact test.
Results:
The findings revealed that the genotypic distribution of TLR3 Leu412Phe polymorphism for the mutant genotype Phe/Phe (TT) demonstrated increased association of DE (31.03% vs 6.8%, P 0.019, odds ratio 6.075, 95% confidence interval 1.181–31.245). However, the number of heterozygous (H) genotype (Leu/Phe–CT) and mutant Phe allele (T) did not show any statistically significant association. TLR3 gene polymorphism did not show any correlation with mortality outcome at 1 month.
Conclusion:
The presence of mutant TLR3 Leu412Phe polymorphism may confer the propensity to have DE in patients with dengue infection in the Indian population. TLR3 polymorphism did not affect mortality outcome at 1 month.
Keywords: Dengue, encephalitis, genotype, mutant, polymorphism, toll-like receptors
Introduction
Dengue virus (DENV) is a ribonucleic acid (RNA) virus transmitted by the infected mosquito vector Aedes aegypti. Its clinical spectrum ranges from a mild dengue fever (DF) to a severe state of dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). Dengue encephalitis (DE) is considered to be another important severe manifestation of dengue infection. DE is considered a critical complication of the neurological manifestations of dengue infection.
Toll-like receptors (TLRs) belong to the family of pattern recognition receptors (PRRs). They act as detectors of the pathogen-associated molecular pattern (PAMR) to the invading pathogens. Out of all TLRs, the one that plays the most critical role in controlling the innate immune response mediated by viruses is TLR3. TLR3 (also named CD283 or IIAE2) is a receptor that senses nucleic acid, which helps in recognizing the product of the virus (double stranded ribonucleic acid, dsRNA) and synthetic legend polyinosinic–polycytidylic acid [poly (I: C)].[1] A study showed that out of all TLRs (1–9), TLR3 was most significantly expressed in the central nervous system. TLR3 has been found in B cells, microglia, T cells, macrophages, killer cells, and, especially in high concentrations, in human astrocytes.[2] There is an enhanced astrocytic expression of TLR3 upon activation by its known ligand, dsRNA. Unlike other TLR signaling that is mediated through adaptor MyD88, TLR3 mediates its signaling via TIR-domain-containing adapter-inducing interferon-beta (TRIF).[3] Different groups of viruses either have a dsRNA genome or produce dsRNA during replication. Therefore, the dsRNA-sensing property of TLR3 is attributed to its antiviral property. The TLR ligands activate the innate immunity signaling, and this property of TLR ligands can be used in preventing and treating infectious diseases.[4,5] TLR3 distinguishes dengue viruses after endosomal acidification.[6] The presence of heterotypic antibodies decreases the TLR stimulation, resulting in better replication of dengue viruses. In various studies, it has been noted that macrophages with TLR3 knocked out are more vulnerable to dengue viruses. The innate-immunity mediated by TLR3 has a defensive role toward dengue infection.[7] The expression of the TLR profile is directly proportional to the severity of dengue. The levels of TLR3 and TLR9 are observed to be higher in patients with DF than in DHF.[8] Other than the effects of TLR3 on the activation of dengue virus replications, few studies have also observed TLR4-modulated activation in dengue, but the reports are inconclusive.
A study revealed that inhibition of dengue infection in coenocytes occurs via the blocking of viral entry through lipopolysaccharides.[9] The response of innate immunity toward dengue infection has been modulated by TLR3/7/9 activation. Single nucleotide polymorphism (SNP) in TLR3/8 and TIR domain-containing adaptor protein (TIRAP) genes are thought to be associated with increasing the risk of developing DHF. TLR3 and TLR8 expression in HepG2 cells are increased by dengue infection.[10] Similarly, dengue virus multiplication is restricted by TLR3 activation.[11] In a study to find the relationship between the presence of SNP in TLR3/7/8 and TIRAP and its outcome in dengue infection, it was observed that TLR3 rs3775291 C/T genotype expression was lower in DHF compared to patients with dengue fever. Higher expression of TLR3 (rs3775291) is found to be protective against developing DHF via reducing inflammation. Among the many risk factors, the TLR3 and TIRAP gene variant is significantly associated with the risk of developing severe dengue infection.[12] Various viral infections and non-viral diseases/states have been linked with TLR3 (rs3775291: L412F) polymorphism.[13] With this knowledge of the effect of TLR3 on innate immunity, it could be postulated that modifications in TLR3 genes and its operational ability can make dengue virus-infected patients more likely to develop dengue encephalitis. In the current study, we aimed to detect TLR3 polymorphism with respect to genotype and allele polymorphism in patients with dengue encephalitis and analyzed mortality outcome.
Material and Methods
It was a case-controlled observational study done in a tertiary care hospital in northern India. This institute is situated in the eastern part of Uttar Pradesh, which is an endemic region for dengue virus infection. Ethical approval was obtained from the Institutional Ethics Committee of our university. Written informed consent was obtained for all the cases and controls. Patients were enrolled over 2 years and followed-up at 1 month.
Selection of cases and control
Patients with confirmed dengue infection were screened, and patients with dengue encephalitis were included in the study. The diagnosis of dengue encephalitis was made according to the criteria by Cristiane and Marzia.[14] It included 1) presence of fever; 2) acute signs of cerebral involvement such as altered consciousness or change in personality, seizures, and focal neurological signs; 3) reactive immunoglobulin M (IgM) dengue antibody, non-structural protein 1 (NS1) antigen, or positive dengue polymerase chain reaction (PCR) on serum or cerebrospinal fluid; and 4) exclusion of other causes of viral encephalitis and encephalopathy. Patients with co-infection with other brain infections causing acute meningo-encephalitis (bacterial, fungal, tubercular, rickettsia, and viruses like herpes simplex, varicella zoster, enterovirus, chikungunya, Ebstein–Barr virus, cytomegalovirus, Japanese encephalitis, mumps, and measles), metabolic encephalopathy, and acquired immunodeficiency syndrome-induced central nervous system disease were excluded from the study. Age and sex-matched dengue patients without neurological features were enrolled as controls from the same geographic area.
Clinical assessment, classification, and baseline characteristics of cases
In all patients, we analyzed the detailed history and performed complete neurological examination. The patients/attendants were asked about the duration of fever, altered mental status, seizures, limb weakness, and extra pyramidal symptoms. The baseline Glasgow coma scale (GCS) score and vitals were recorded at the time of enrollment. Entire neurological assessment including other system evaluation was recorded.
Laboratory investigations
We performed blood investigations, including complete blood count, liver and renal function test, blood sugar, and electrolytes. The human immunodeficiency virus ELISA was done in all patients. Serum NS1 and IgM were assessed to confirm dengue infection.
Cerebrospinal fluid (CSF) analysis
CSF analysis was done within the first week of admission in all the cases for determining total leukocyte and differential leukocyte counts, protein, and sugar with simultaneous measurement of plasma sugar. CSF IgM ELISA was performed using the National Institute of Virology Dengue MAC ELISA kit (Version No. 1.4). This kit is manufactured by National Institue of Virology, Pune, Maharashtra(State), India. In addition, all CSF samples were tested for the Cartridge-Based Nucleic Acid Amplification Test CBNAAT (for Mycobacterium Tuberculosis), gram stain, culture sensitivity, fungal stain, and ELISA for Japanese encephalitis and herpes simplex, varicella zoster, enterovirus, chikungunya, Ebstein–Barr virus, cytomegalovirus, mumps, and measles virus to rule out co-infections.
Neuroimaging
Computed tomography (CT) and/or magnetic resonance imaging (MRI) was done according to the clinical status of the patient. CT images were investigated for any hypodense or hyperdense areas, while MRI of the brain with contrast was especially evaluated for any area of hyperintensity on fluid-attenuated inversion recovery and T2-weighted images, restriction on diffusion-weighted imaging, presence of contrast enhancement, and blooming on gradient-recalled echo sequences.
Analysis of TLR3 polymorphism
Genomic DNA extraction
Peripheral blood was collected in ethylene diamine tetra acetic acid (EDTA) vials from the study subjects. The genomic DNA was extracted from total blood using the salting-out method and purified using the Genomic DNA extraction kit (Thermo Fisher Scientific, USA). DNA samples were stored at -80°C until used.
TLR 3 Leu412Phe (L412F) genotyping polymorphism assay
A single set of primer sequences was designed for the TLR3 Leu412Phe polymorphism: forward primer sequence of 5′-TCACAGGATTGATAAACCTGAAAT-3′ and reverse primer sequence of 5′-AAAGACAGATGCTGGAAGTGTG-3′ (Integrated DNA technology, USA). In the TaqMan Single nucleotide polymorphism (SNP) genotyping assay, PCR amplification was performed in a 25 μl reaction mixture containing 5 μl of genomic DNA, 12.5 μl of TaqMan SNP Genotyping Master Mix (Thermo Fisher Scientific, USA), and TaqMan SNP Genotyping Assay probe for rs3775291 (Cat No. 4351379) (Thermo Fisher Scientific, USA) SNP. The quantitative polymerase chain reaction (QPCR) thermal cycling program was 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Of all samples, 10% of the samples were confirmed by using PCR direct sequencing.
Treatment
The patients were managed conservatively with intravenous fluids, nasogastric tube feeding, antibiotics, antipyretics, anticonvulsants, and decongestants (mannitol, glycerol, and acetazolamide).
Statistical analysis
The genotype and allele frequencies were compared using a two-sided Chi-square or Fisher’s exact test to confirm the Hardy–Weinberg equilibrium. Simple gene counting was used to determine combined genotype and allele frequencies, and data was analyzed using Microsoft Excel 2007 (Microsoft, Seattle, WA, USA). Graph Pad Prism (version 5.01, GraphPad Software, Inc. La Jolla, CA, USA) and IBM SPSS were used for statistical analysis (20.0 version). All categorical variables were expressed as percentages and continuous variables were expressed as mean ± standard deviation. Chi square test, odds ratio, and 95% confidence interval were calculated as applicable. Yates correction was done wherever required. A value of P < 0.05 was considered to be statistically significant.
Results
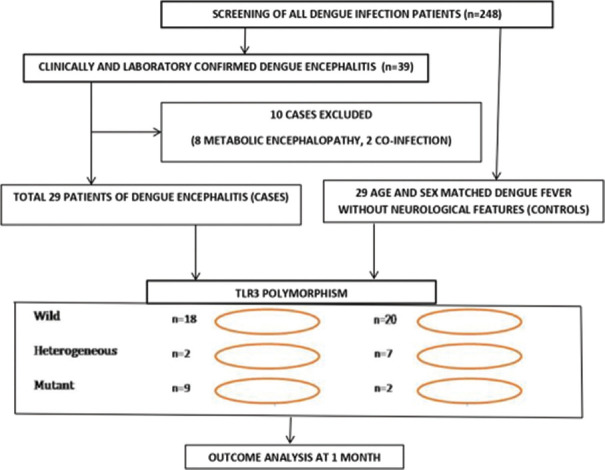
A total of 248 patients with dengue infection were screened during the period of enrolment. Out of them, 39 patients with dengue encephalitis were observed, among which 29 patients were included as cases after excluding 10 patients (eight metabolic encephalopathy and two co-infection). An equal number (n = 29) of age- and sex-matched patients with dengue fever not having neurological manifestations from the same geographical area were recruited as controls [Figure 1].
Figure 1.
Flow diagram of the study schemata and genotypic distribution of patients
Demographic, clinical, and laboratory parameters
The baseline demographic, clinical, and laboratory features of cases and controls are shown in Table 1.
Table 1.
Clinical profile of dengue encephalitis (n=29) and dengue fever without neurological features (n=29)
| Parameters | Case (n=29) | Control (n=29) |
|---|---|---|
| 1. Mean age (years) | 28.32±15.81 | 28.32±15.81 |
| <20 Years | 11 | 11 |
| >20 Years | 18 | 18 |
| 2. Male:female ratio | 24:5 (4.8:1) | 24:5 (4.8:1) |
| 3. Fever | 29 (100%) | 29 (100%) |
| 4. Headache | 15 (51.72%) | 22 (75.86%) |
| 5. Vomiting | 9 (31.03%) | 15 (51.72%) |
| 6. Altered sensorium | 16 (58.62%) | 0 |
| 7. Altered behavior | 6 (20.69%) | 0 |
| 8. Seizure | 3 (10.35%) | 0 |
| 9. Focal neurological deficit | 5 (17.24%) | 0 |
| 10. Mean duration of illness (days) | 7.85±7.49 | 5.62±3.16 |
| 11. Mean glassgow coma scale | 9.1±1.87 | 15 |
| 12. Glassgow coma scale | ||
| <8 | 8 (27.59%) | 0 |
| >8 | 21 (72.41%) | 15 |
| 13. Mortality | 2 (6.89%) | 1 (3.45%) |
Note: Case, dengue encephalitis; Control, dengue fever without neurological features; n, number
Analysis of TLR3 polymorphism
The genotypic frequency distribution was found to be consistent with the Hardy–Weinberg equilibrium. The genotypic analysis of the TLR3 Leu412Phe polymorphism showed a statistically significantly higher number of mutant (M) genotype (Phe/Phe–TT) in cases compared to that in the control group (31.03% vs 6.8%, odds ratio = 6.075, 95% confidence interval = 1.181–31.245, P = 0.019). On comparing cases with control with respect to the number of heterozygous (H) genotype (Leu/Phe–CT), the difference was not found to be statistically significant (6.8% vs 24.13%%, odds ratio = 0.233, 95% confidence interval = 0.043–1.279, P = 0.07). We adopted a recessive model to calculate the association of DE due to lesser frequency of the variant genotype. A comparison was made between the frequency of mutant (M) + heterozygous (H) genotype in both cases and controls, but the difference was not statistically significant (37.93% vs 31.03%, odds ratio = 1.36, 95% confidence interval = 0.458–4.027, P = 0.65). On further analysis of the mutant Phe allele (T), mutant allelic frequency was found to be higher in cases than in controls but the difference was also statistically insignificant (34.48% vs 18.97%, odds ratio = 2.290, 95% confidence interval = 0.98–5.30, P = 0.052) [Table 2]. The electropherogram denoting the different genotypes (wild, heterozygous, and mutant) are shown in Figure 2.
Table 2.
Analysis of TLR3 Leu412Phe polymorphism in dengue encephalitis (n=29) and dengue fever without neurological features (n=29)
| Type of mutation TLR 3 - Leu412Phe | Genotype | Case (n=29), % | Control (n=29), % | P | Odd’s ratio | 95% C.I. |
|---|---|---|---|---|---|---|
| Wild (W) | CC | 18 (29), (62%) | 20 (29), (68%) | 0.581 | 0.736 | (0.248-2.184) |
| Heterozygous (H) | CT | 2 (29), (6.8%) | 7 (29), (24.13%) | 0.070 | 0.232 | (0.043-1.279) |
| Mutant (M) | TT | 9 (29), (31.03%) | 2 (29), (6.8%) | 0.019 | 6.075 | (1.181-31.245) |
| H+M | CT + TT | 11 (29), (37.93%) | 9 (29), (31.03%) | 0.650 | 1.360 | (0.458-4.027) |
| Phe allele | T | 20 (58), (34.48%) | 11 (58), (18.97%) | 0.052 | 2.290 | (0.98-5.300) |
Note: TLR, Toll-Like Receptor; Case, dengue encephalitis; Control, dengue fever without neurological features; n, number; P, probability value; CI, confidence interval
Figure 2.
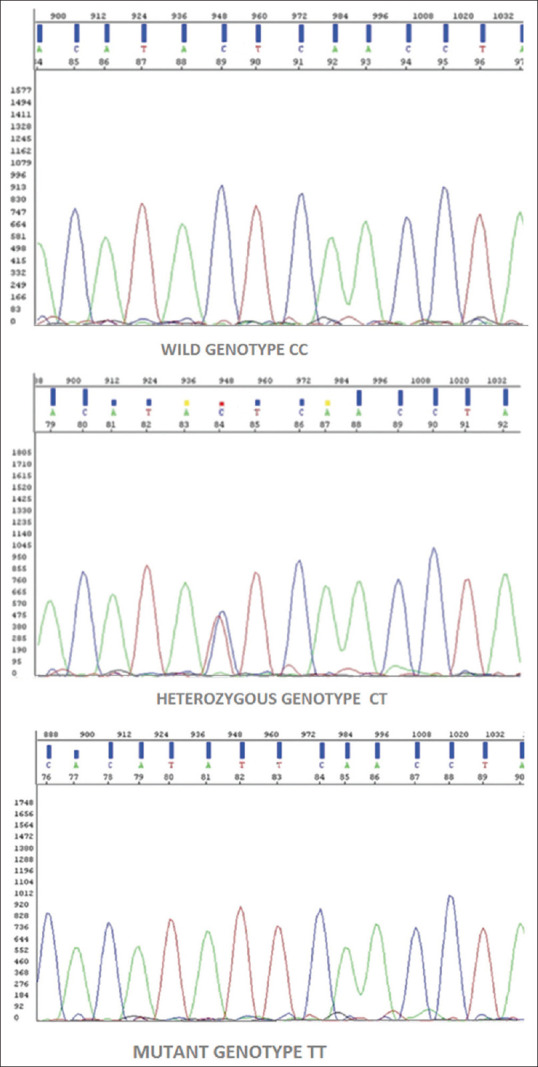
Prototype electropherograms of patients with dengue encephalitis depicting the wild genotype, heterozygous genotype, and mutant genotype
Analysis of outcome and mortality
Out of the 29 patients with dengue encephalitis, mortality was observed in two cases, and the remaining 27 patients recovered completely at the 1-month follow-up. On univariate analysis of age <20 years, GCS ≤8, seizures, severe thrombocytopenia, and mutant TLR3 genotype (TT), none of the parameters was found to be significantly associated with mortality at 1 month in dengue encephalitis [Table 3].
Table 3.
Outcome analysis of dengue encephalitis (n=29) with respect to mortality
| Parameters | Chi-square test | P | Odd’s ratio | 95% C I |
|---|---|---|---|---|
| Age <20 years | 0.133* | 0.71 | 1.7 | 0.1-30.28 |
| Gcs <8 | 0.54* | 0.46 | 2.85 | 0.16-52.05 |
| Seizure | 0.497* | 0.48 | 12.5 | 0.55-284.12 |
| Severe thrombocytopenia | 0.066 | 0.79 | 1.45 | 0.08-25.81 |
| Mutant TLR3 genotype | 0.361* | 0.54 | 2.37 | 0.13-42.8 |
Note: P, probability value; C I, confidence interval; GCS, Glassgow Coma Scale; *, with Yates correction; TLR, Toll-Like Receptor
Discussion
In this study, we observed a significantly higher frequency of mutant genotype (Phe/Phe) of TLR3 Leu142Phe polymorphism in DE cases as compared to controls.
Dengue virus has been associated with various neurological complications. The dengue illness manifests neurologically by different mechanisms. It can have direct involvement in the form of acute meningo-encephalitis, myelitis, and myositis. Various post-infectious immune-mediated disorders such as acute disseminated encephalomyelitis, myelitis, neuromyelitis optica, optic neuritis, and Guillain–Barré syndrome are found to be associated with dengue.[15,16] In the 2013 data recording, dengue has been accounted for approximately 1.14 million (0.73–1.98 million) disability-adjusted life-years, with the majority arising from southeast Asia (52%), while India contributed to 34% of the 96 million DENV cases.[17] Multiple human infective viruses like herpes viruses, arboviruses, Epstein–Barr virus, enterovirus, human immunodeficiency virus, rabies virus, measles virus, and mumps virus have neurotropic activity implicating the nervous system.[18] Dengue encephalitis (DE) is an important neurological entity with an incidence ranging from 0.5% to 6.2% in the Indian population.[19] Immune surveillance of central nervous system (CNS) is an important element in determining the fate of neurotropic viral infections to brain. The exact mechanism for susceptibility to DE in dengue patients is not precisely known. Susceptibility to DE may be attributed to the genetic alterations of the host DNA, leading to altered immune mechanism required for effective clearance of dengue virus.
Toll-like receptors (TLR) are germline-encoded pattern recognition receptors (PRRs), which identify conserved molecular structures better known as pathogen-associated molecular patterns (PAMPs) on pathogenic microbes. It activates multiple intracellular signaling pathways as well as induces genes that are involved in immune responses and inflammation. By mediating dendritic cell maturation and activation of T lymphocytes specific to pathogens, it serves as a link between the innate and adaptive pathways of the immune system. TLR3 significantly contributes in virus-mediated innate immune reactions. TLR3 has high expression in human astrocytes. TLR3 affects clinical outcome through protective (antiviral) or harmful (immunoregulatory) properties. The effect of the genetic variation of TLR3 on the outcome of an infectious disease is complex. It includes possible effects of ethnicity, gender, mutation in other genes, and environmental factors. The underlying molecular mechanisms by which TLR3 gene variation affects innate immune responses are yet to be known. Single nucleotide polymorphism (SNP) of genes coding for cytokines has been observed with altered cytokine reactions. SNP in TLR3 is reported with nervous system infection with various neurotropic viruses.[20] TLR3 Leu412Phe genotype may have a relevant role in determining the susceptibility to DE.
In a study by Barkhash et al.,[21] it was found that there was an increased expression of the TLR3 SNP (rs3775291) G allele and G/G homozygote in Tick–Borne encephalitis patients compared to healthy controls in the Russian population. A recent study by Mickienė et al.[22] stated that TLR3 gene polymorphism plays a role in the development of clinical tick borne encephalitis (TBE) in the Lithuanian population and may be associated with disease severity. In an Indian study by Biyani et al.,[23] it was found that mutant (TLR3 Leu412Phe polymorphism) and heterozygous (Leu/Phe–CT) TLR3 polymorphism have statistically significantly increased frequency in cases of Japanese encephalitis (JE). The study concluded that persons with these two polymorphisms have higher predisposition for Japanese encephalitis. The study by Lim et al.[24] revealed that the pathophysiology of herpes simplex encephalitis may be influenced by anomalies of intrinsic TLR3 immunity in the central nervous system. TLR3 deficiency leads to herpes simplex virus-2-related Mollaret’s meningitis. The literature revealed a case description of recurrent HSV-2 meningitis with the presence of homozygous polymorphic TLR3 allele.[25] TLR3 deficiency causes HSV-2-associated Mollaret’s meningitis. According to a case study by Willman and colleagues, a patient with severe and repeated episodes of HSV-2 meningitis possessed the homozygous polymorphic TLR3 variant (Mollaret’s meningitis).[26] A new missense mutation (F303S) in the TLR3 gene was discovered by Hidaka et al.[27] in a single patient with influenza-associated encephalopathy. In an experimental study by Daffis et al.[28] to determine the protective effect of TLR3 against West Nile virus in the brain, they found that TLR3-deficient mice died frequently when there was a high viral load in the cerebral neurons.
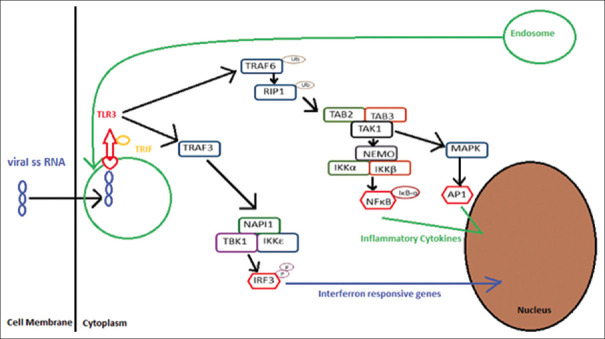
DENV is known to up-regulate TLR 3 within HepG2 cells. A study by Torres et al.[8] revealed that TLR3 blocks the replication of DENV with the help of interferon-β in vitro cultured hepatoma cells. TLR3 polymorphism Leu412Phe is associated with reduced activation of IRF-3 (interferon regulatory factor 3) and NF-ĸB (nuclear factor- kappa B). It leads to reduced production of type I interferon (IFN), and this phenomenon can promote viral proliferation. “Leucine” 412 is situated just next to glycosylated “asparagine” residue, and its glycan moiety contacts dsRNA. TLR3 polymorphism (Leu412Phe) leads to replacement of the amino acid “leucine” by “phenylalanine” at the 412 position. This can affect the process of glycosylation of asparagine or prevent the N-glycosyl moiety-dsRNA interaction, resulting in reduced signaling activity of the Phe-412 TLR3 gene[29,30] [Figure 3]. As per published literature and our experience, we have tried to explore the genetic polymorphism of TLR3 gene susceptibility in dengue encephalitis. Furthermore, in a recent Taiwanese study, an association of autoimmune disorder has been associated with dengue infection, especially encephalomyelitis.[31] The importance of newer advancements in genetics and inflammation is crucial to the better understanding of tropical diseases. Newer studies into genetic interplay of inflammation can substantiate the scope for future research in dengue infection.
Figure 3.
Pathophysiological hypothesis of TLR3 Leu412Phe susceptibility in dengue encephalitis
Conclusion
Dengue encephalitis is an important neurological manifestation of dengue illness. Immune surveillance of CNS is important in determining the fate of viral brain infection. The effect of genetic variation in TLR3 on the outcome of an infectious disease is complex and multifactorial. The underlying molecular mechanisms by which TLR3 gene variation affects innate immune responses and influences host susceptibility to DE in dengue illness are largely unknown. Our study suggests that TLR3 Leu412Phe polymorphism has a higher propensity to develop DE in dengue illness. There is a need for larger multi-corpus studies to consolidate the association of TLR3 polymorphism with susceptibility to different viral infections of the brain.
Future Perspectives
This is a prototype study performed on dengue encephalitis across the world. The novelty of this study is the effect of the TLR3 Leu412Phe polymorphism in dengue encephalitis in the Indian population. The limitation of our study is that the sample size of dengue encephalitis was 29 due to less CNS involvement observed in dengue illness. In view of the smaller sample size and lack of healthy controls, polymorphism studies may not have adequate power. There may be an association of genetic alterations in the host DNA with specific disease manifestations as observed in this study for the development of encephalitis. In the future, there is a need for larger multi-centric collaborated case-controlled studies in prevalent infections such as dengue to explore further genetics and validate such genetic effects.
Highlights
Toll-like receptors (TLRs) are pattern recognition receptors contributing to the innate response against microbial infections.
TLR3 and its polymorphism play an important role in viral infections of the nervous system.
The presence of mutant TLR3 genotype polymorphism may increase the susceptibility for dengue encephalitis.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-?B by Toll-like receptor 3. Nature. 2001;413:732–8. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 2.Carpentier PA, D’Anne SD, Miller SD. Glial toll-like receptor signaling in central nervous system infection and autoimmunity. Brain Behav Immunity. 2008;22:140–7. doi: 10.1016/j.bbi.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–15. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Uematsu S, Akira S. Toll-like receptors and Type I interferons. J Biol Chem. 2007;282:15319–23. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- 5.Wu MH, Zhang P, Huang X. Toll-like receptors in innate immunity and infectious diseases. Front Med China. 2010;4:385–93. doi: 10.1007/s11684-010-0600-x. [DOI] [PubMed] [Google Scholar]
- 6.Tsai YT, Chang SY, Lee CN, Kao CL. Human TLR3 recognizes dengue virus and modulates viral replication in vitro. Cell Microbiol. 2009;11:604–15. doi: 10.1111/j.1462-5822.2008.01277.x. [DOI] [PubMed] [Google Scholar]
- 7.Nasirudeen AM, Wong HH, Thien P, Xu S, Lam KP, Liu DX. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl Trop Dis. 2011;5:e926. doi: 10.1371/journal.pntd.0000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torres S, Hernández JC, Giraldo D, Arboleda M, Rojas M, Smit JM, et al. Differential expression of Toll-like receptors in dendritic cells of patients with dengue during early and late acute phases of the disease. PLoS Negl Trop Dis. 2013;7:e2060. doi: 10.1371/journal.pntd.0002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen YC, Wang SY, King CC. Bacterial lipopolysaccharide inhibits dengue virus infection of primary human monocytes/macrophages by blockade of virus entry via a CD14-dependent mechanism. J Virol. 1999;73:2650–7. doi: 10.1128/jvi.73.4.2650-2657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conceição C, El-Bacha T, Villas-Bôas CS, Coello G, Ramírez J, Montero-Lomeli M, et al. Gene expression analysis during dengue virus infection in HepG2 cells reveals virus control of innate immune response. J Infect. 2010;60:65–75. doi: 10.1016/j.jinf.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Liang Z, Wu S, Li Y, He L, Wu M, Jiang L, et al. Activation of Toll-like receptor 3 impairs the dengue virus serotype 2 replication through induction of IFN-? in cultured hepatoma cells. PloS One. 2011;6:e23346. doi: 10.1371/journal.pone.0023346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alagarasu K, Bachal RV, Memane RS, Shah PS, Cecilia D. Polymorphisms in RNA sensing toll like receptor genes and its association with clinical outcomes of dengue virus infection. Immunobiology. 2015;220:164–8. doi: 10.1016/j.imbio.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 13.Kindberg E, Vene S, Mickiene A, Lundkvist Å, Lindquist L, Svensson L. A functional Toll-like receptor 3 gene (TLR3) may be a risk factor for tick-borne encephalitis virus (TBEV) infection. J Infect Dis. 2011;203:523–8. doi: 10.1093/infdis/jiq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cristiane S, Marzia PS. Diagnosis criteria of dengue encephalitis. Arq Neuropsiquiatr. 2014;72:263. doi: 10.1590/0004-282x20130251. [DOI] [PubMed] [Google Scholar]
- 15.Sahu R, Verma R, Jain A, Garg RK, Singh MK, Malhotra HS, et al. Neurologic complications in dengue virus infection: A prospective cohort study. Neurology. 2014;83:1601–9. doi: 10.1212/WNL.0000000000000935. [DOI] [PubMed] [Google Scholar]
- 16.Chan DP, Teoh SC, Tan CS, Nah GK, Rajagopalan R, Prabhakaragupta MK, et al. Ophthalmic complications of dengue. Emerg Infect Dis. 2006;12:285–9. doi: 10.3201/eid1202.050274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murhekar MV, Kamaraj P, Kumar MS, Khan SA, Allam RR, Barde P, et al. Burden of dengue infection in India, 2017: A cross-sectional population based serosurvey. Lancet Glob Health. 2019;7:e1065–73. doi: 10.1016/S2214-109X(19)30250-5. [DOI] [PubMed] [Google Scholar]
- 18.Akira S. TLR signaling. Curr Top Microbiol Immunol. 2006;311:1–16. doi: 10.1007/3-540-32636-7_1. [DOI] [PubMed] [Google Scholar]
- 19.Mehta VK, Verma R, Jain A, Sharma N, Mahdi AA. A study of dengue encephalitis with laboratory and clinical parameters in Tertiary Center of North India. J Family Med Prim Care. 2021;10:4041–6. doi: 10.4103/jfmpc.jfmpc_632_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moumad K, Lascorz J, Bevier M, Khyatti M, Ennaji MM, Benider A, et al. Genetic polymorphisms in host innate immune sensor genes and the risk of nasopharyngeal carcinoma in North Africa. G3 Genes Genom Genet. 2013;3:971–7. doi: 10.1534/g3.112.005371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barkhash AV, Voevoda MI, Romaschenko AG. Association of single nucleotide polymorphism rs3775291 in the coding region of the TLR3 gene with predisposition to tick-borne encephalitis in a Russian population. Antivir Res. 2013;99:136–8. doi: 10.1016/j.antiviral.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Mickienė A, Pakalnienė J, Nordgren J, Carlsson B, Hagbom M, Svensson L, et al. Polymorphisms in chemokine receptor 5 and Toll-like receptor 3 genes are risk factors for clinical tick-borne encephalitis in the Lithuanian population. PLoS One. 2014;9:e106798. doi: 10.1371/journal.pone.0106798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biyani S, Garg RK, Jain A, Malhotra HS, Kumar R, Prakash S, et al. Toll-like receptor-3 gene polymorphism in patients with Japanese encephalitis. J Neuroimmunol. 2015;286:71–6. doi: 10.1016/j.jneuroim.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Lim HK, Seppänen M, Hautala T, Ciancanelli MJ, Itan Y, Lafaille FG, et al. TLR3 deficiency in herpes simplex encephalitis: High allelic heterogeneity and recurrence risk. Neurology. 2014;83:1888–97. doi: 10.1212/WNL.0000000000000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Y, Audry M, Ciancanelli M, Alsina L, Azevedo J, Herman M, et al. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J Exp Med. 2011;208:2083–98. doi: 10.1084/jem.20101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willmann O, Ahmad-Nejad P, Neumaier M, Hennerici MG, Fatar M. Toll-like receptor 3 immune deficiency may be causative for HSV-2-associated mollaret meningitis. Eur Neurol. 2010;63:249–51. doi: 10.1159/000287585. [DOI] [PubMed] [Google Scholar]
- 27.Hidaka F, Matsuo S, Muta T, Takeshige K, Mizukami T, Nunoi H. A missense mutation of the Toll-like receptor 3 gene in a patient with influenza-associated encephalopathy. Clin Immunol. 2006;119:188–94. doi: 10.1016/j.clim.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Daffis S, Samuel MA, Suthar MS, Gale M, Jr, Diamond MS. Toll-like receptor 3 has a protective role against West Nile virus infection. J Virol. 2008;82:10349–58. doi: 10.1128/JVI.00935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang SY, Jouanguy E, Ugolini S, Smahi A, Elain G, Romero P, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–7. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 30.Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–81. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shih HI, Chi CY, Tsai PF, Wang YP, Chien YW. Re-examination of the risk of autoimmune diseases after dengue virus infection: A population-based cohort study. PLoS Negl Trop Dis. 2023;17:e0011127. doi: 10.1371/journal.pntd.0011127. [DOI] [PMC free article] [PubMed] [Google Scholar]