Summary Paragraph:
Gram-negative antibiotic development has been hindered by a poor understanding of the types of compounds that can accumulate within these bacteria.1–2 The presence of efflux pumps and substrate-specific outer membrane porins in Pseudomonas aeruginosa render this pathogen particularly challenging.3 As a result, there are few antibiotic options for P. aeruginosa infections4, and its many porins has made the prospect of discovering general accumulation guidelines seem unlikely.5 Here, we assess the whole-cell accumulation of 345 diverse compounds in P. aeruginosa and E. coli. While certain positively charged compounds permeate both bacterial species, P. aeruginosa is more restrictive as compared to E. coli. Computational analysis identified distinct physicochemical properties of small molecules that specifically correlate with P. aeruginosa accumulation, such as formal charge, positive polar surface area, and hydrogen bond donor surface area. Surprisingly, mode of uptake studies revealed that most small molecules permeate P. aeruginosa using a porin-independent pathway, thus enabling discovery of general P. aeruginosa accumulation trends with important implications for future antibiotic development. Retrospective antibiotic examples confirmed these trends, and these discoveries were then applied to expand the spectrum of activity of a gram-positive-only antibiotic, fusidic acid, into a version that demonstrates a dramatic improvement in antibacterial activity against P. aeruginosa. We anticipate that these discoveries will facilitate the design and development of high-permeating antipseudomonals.
Introduction:
Antibiotic discovery for P. aeruginosa has proven to be challenging, as achieving drug accumulation in these bacteria is particularly difficult compared to many other gram-negative pathogens.3 This lower permeability of P. aeruginosa is in part due to the absence of non-specific porins, such as OmpF in E. coli, which facilitate the diffusion of small, hydrophilic compounds across the outer membrane. Instead, P. aeruginosa possesses ~40 monomeric channels for nutrient transport with size restrictions of ~200 Da.6 Additionally, P. aeruginosa can express a broad range of tripartite efflux pumps, resulting in highly efficient antibiotic efflux.7 This combination confers high intrinsic resistance to many antibacterials, including some that are commonly used to treat infections caused by other gram-negative pathogens; for example, while antibiotics such as chloramphenicol, tetracyclines, and trimethoprim/sulfamethoxazole have clinical activity against other gram-negative organisms, they are not effective against P. aeruginosa.4 Notably, the lack of clinical efficacy of these drugs is attributed to poor intracellular accumulation in P. aeruginosa, not to a lack of target engagement.8 This is also true for antibiotics that are ‘gram-positive-only’ (e.g. erythromycin, linezolid, fusidic acid, rifampin, etc.) and have no activity against any gram-negative pathogens.8 Ultimately, only four classes of antibiotics (β-lactams, fluoroquinolones, aminoglycosides, and polymyxins) are used clinically to treat P. aeruginosa infections, and with resistance rampant and significant toxicity of certain active classes (e.g. aminoglycosides and polymyxins),9–10 novel antibacterials for P. aeruginosa infections are an area of significant need.
The identification of physicochemical properties that promote intracellular compound accumulation would greatly aid in the development of new anti-pseudomonal antibiotics.1–2, 11–12 For example, an understanding of compound accumulation in E. coli has led to actionable guidelines11, 13 that have been used to imbue gram-negative antibacterial activity into multiple different classes of compounds.14–25 However, despite important retrospective and prospective analyses of antibiotic activity and accumulation in P. aeruginosa,8, 26–28 general rules for compound accumulation in this bacterium have remained elusive. And, with multiple sources of potential uptake and efflux in P. aeruginosa, the possibility for discovery of generalizable accumulation guidelines for P. aeruginosa has been questioned.5 Here we utilize a whole-cell accumulation assay to assess the ability of non-antibiotic, structurally diverse small molecules to accumulate in this pathogen, ultimately leading to principles that will be helpful in the design of novel antipseudomonals.
Results:
Validation of assay in P. aeruginosa
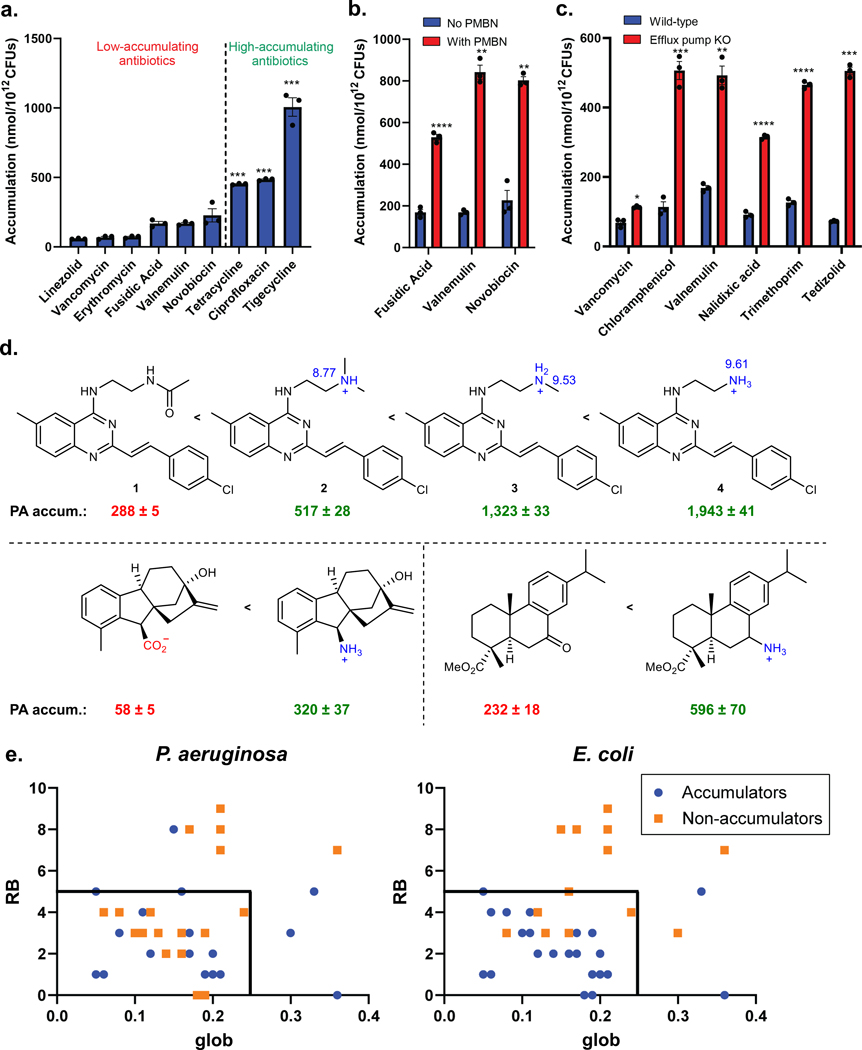
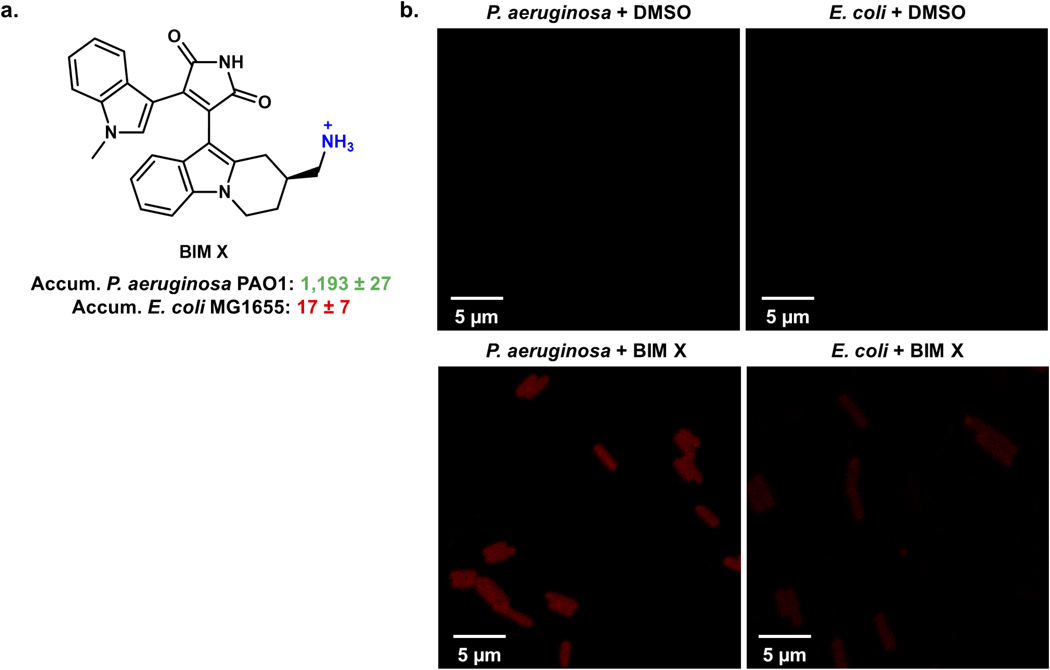
To study compound accumulation in P. aeruginosa, an LC-MS/MS-based assay was utilized.13, 29 Although predominately used to examine compound accumulation in E. coli,13–15, 30 this assay has been used with P. aeruginosa.15, 28 To validate the assay, a selection of antibiotic controls were evaluated for whole-cell accumulation in P. aeruginosa PAO1. This panel (Extended Data Table 1a) includes antibiotics with activity (minimum inhibitory concentration (MIC) ≤16 μg/mL) against P. aeruginosa in culture and antibiotics inactive against P. aeruginosa (MIC ≥256 μg/mL). The inactive antibiotics all showed minimal accumulation in PAO1, whereas the active antibiotics all demonstrated statistically significant levels of accumulation relative to the inactive compounds (Fig. 1a). To further evaluate this assay in P. aeruginosa, bacteria were treated with the membrane permeabilizer polymyxin B nonapeptide (PMBN).31 The low-accumulating antibiotics fusidic acid, valnemulin, and novobiocin all showed a significant increase in accumulation upon membrane permeabilization, consistent with the assay not simply reporting on non-specific interactions with the outer membrane (Fig. 1b, Extended Data Table 1a). As an additional control, antibacterial activity and accumulation of low-accumulating antibiotics were also evaluated in a genetically modified strain of P. aeruginosa PAO1, which has six efflux pumps knocked out (PAΔ6).26 MIC ratios were used to identify efflux substrates (Extended Data Table 1b); an increase in accumulation in the PAΔ6 strain was observed for the antibiotics that were highly potentiated upon efflux pump knockout (chloramphenicol, valnemulin, nalidixic acid, trimethoprim, and tedizolid), while vancomycin, which shows no potentiation upon efflux pump knockout,8 showed minimal change in accumulation in this experiment (Fig. 1c). Finally, to compare the results obtained by LC-MS/MS analysis to those obtained with fluorescence imaging, the accumulation of bisindolylmaleimide X (BIM X), an intrinsically fluorescent non-antibiotic, was evaluated in both P. aeruginosa PAO1 and E. coli MG1655. The LC-MS/MS assay revealed that BIM X accumulates in P. aeruginosa but not in E. coli (as described later), and confocal fluorescence imaging validated these results (Extended Data Fig. 1a, b).
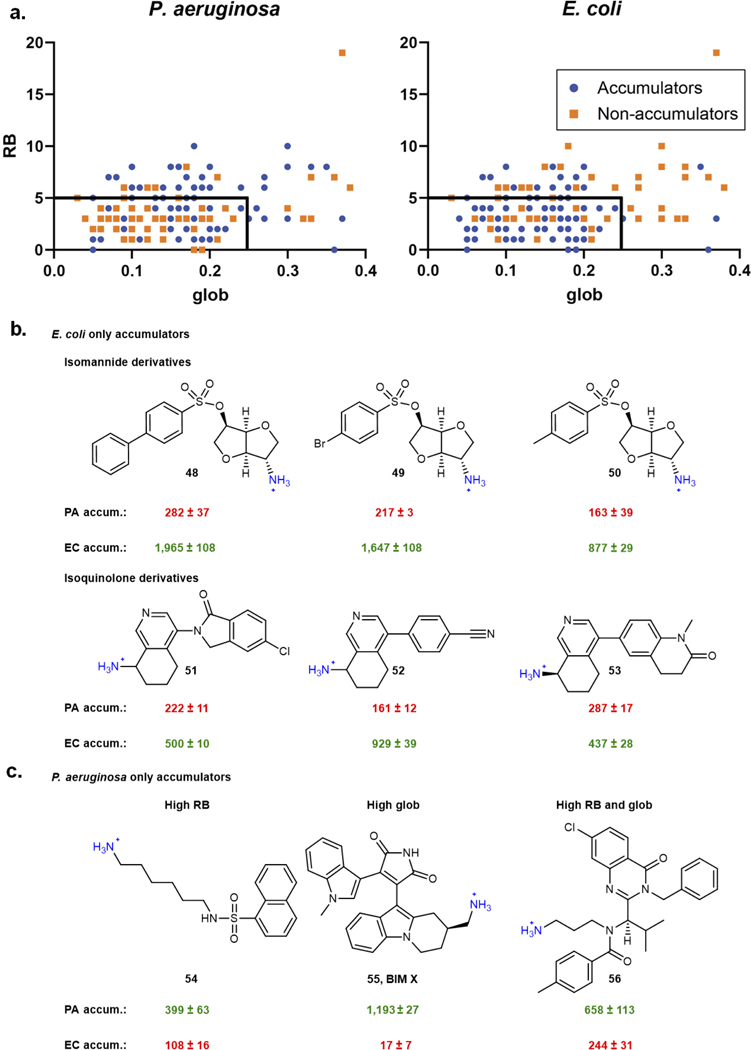
Fig. 1. Assessment of antibiotic controls in P. aeruginosa accumulation assay and evaluation of eNTRy rules predictability in P. aeruginosa.
a) Inactive antibiotics show low accumulation in P. aeruginosa PAO1, while active antibiotics are high accumulators. Statistically significant accumulation over the average of the low-accumulating controls is indicated with asterisks (***P<0.001; tetracycline (P=7.6E-5), ciprofloxacin (P=3.1E-5), tigecycline (P=1.6E-8).). b) Low-accumulating antibiotics show an increase in accumulation with treatment of polymyxin B nonapeptide (PMBN, 8 μg/mL). Statistically significant accumulation differences for low accumulating compounds in permeabilized and non-permeabilized PAO1 are indicated with asterisks (**P < 0.01, ****P< 0.0001; fusidic acid (P=5.8E-5), valnemulin (P=1.8E-3), novobiocin (P=3.0E-3)). c) Efflux substrates show increased accumulation in the efflux pump knockout strain, P. aeruginosa PAΔ6, whereas non-substrate vancomycin shows no significant (P=1.9E-2) increase. Statistically significant accumulation differences for low accumulating compounds in wild-type P. aeruginosa PAO1 and efflux deficient P. aeruginosa are indicated with asterisks (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; chloramphenicol (P=7.6E-4), valnemulin (P=4.5E-3), nalidixic acid (P=9E-6), trimethoprim (P=2E-6), and tedizolid (P = 5.3E-4)). d) The influence of amines on accumulation in P. aeruginosa PAO1 for three different series of compounds. pKa values calculated using Chemaxon. e) The influence of globularity and rotatable bonds on accumulation in P. aeruginosa PAO1 versus E. coli MG1655 for 40 primary amines. Low globularity and low rotatable bonds are predictive for ~80% of compounds tested in E. coli, but only ~50% for P. aeruginosa. Structures of all compounds are in Supplementary Table 1 and Compound Master Table, and data for E. coli is taken from Richter and co.13 For all figure panels, the average and s.e.m. are reported for accumulation values (nmol/1012 CFUs). n=3 biologically independent samples. Statistical significance was determined using a two-sample Welch’s t-test (one-tailed test, assuming unequal variance).
Comparison with eNTRy rules
To identify physicochemical properties that promote accumulation in P. aeruginosa, accumulation of a collection of diverse, non-antibiotic compounds was evaluated, including natural product-like compounds generated through the Complexity-to-Diversity strategy,32 and some commercially available compounds. As a major interest was defining how accumulation trends for P. aeruginosa compared to E. coli, a set of 67 compounds (Supplementary Table 1) whose accumulation was previously assessed in E. coli13 was evaluated in P. aeruginosa PAO1.
In this set of 67 compounds, positive charge appeared to correlate with compound accumulation in P. aeruginosa PAO1, with all accumulating compounds being positively charged (Supplementary Table 1), as was observed for this same compound set in E. coli (Supplementary Table 1).13 The importance of positive charge for accumulation in P. aeruginosa was further demonstrated via side-by-side comparisons of similar compounds differing only in the nature of the charge. As shown with a series of 6-methylquinazolines, amine-containing versions accumulate in PAO1 much higher than the corresponding neutral amide, with the primary amine leading to the highest levels of accumulation among the set, likely due to both the increased steric accessibility and increased pKa of the primary amine relative to the secondary and tertiary amine comparators (Fig. 1d; 1–4). These data mirror trends seen in E. coli with the same compounds.33 Other matched compound pairs (5 and 6, 7 and 8 in Fig. 1d) also demonstrate how a primary amine can facilitate compound accumulation in P. aeruginosa in certain contexts.
The importance of positive charge for compound accumulation, observed here for P. aeruginosa, invites comparison to the eNTRy rules (formulated for E. coli) which state that compounds with a positive charge (primary amine being best), low globularity, and a low number of rotatable bonds are most likely to accumulate in E. coli.11, 13 To assess the applicability of the eNTRy rules to P. aeruginosa, data for the compounds bearing primary amines in the set of 67 compounds were plotted according to their number of rotatable bonds and globularity scores; for this analysis amines that did not meet the other criteria identified previously as important for accumulation (steric accessibility and low amphiphilic moment)13 were removed, leaving a total of 40 compounds (see Compound Master Table). This analysis revealed that low globularity (glob) and rotatable bonds (RB) were not predictive for accumulation in P. aeruginosa, as these cutoffs include many compounds that are non-accumulators in P. aeruginosa; further, four compounds outside the eNTRy rules accumulate well in P. aeruginosa, with ultimately only ~50% of compounds binned correctly in this data set (Fig. 1e). For E. coli, these cutoffs bin ~80% of all compounds correctly (Fig. 1e), consistent with previous results in E. coli for a large compound set.13, 33 These exceptions to the eNTRy rules in both directions suggest that, besides the beneficial introduction of a primary amine, the eNTRy rules do not predict compound accumulation in P. aeruginosa.
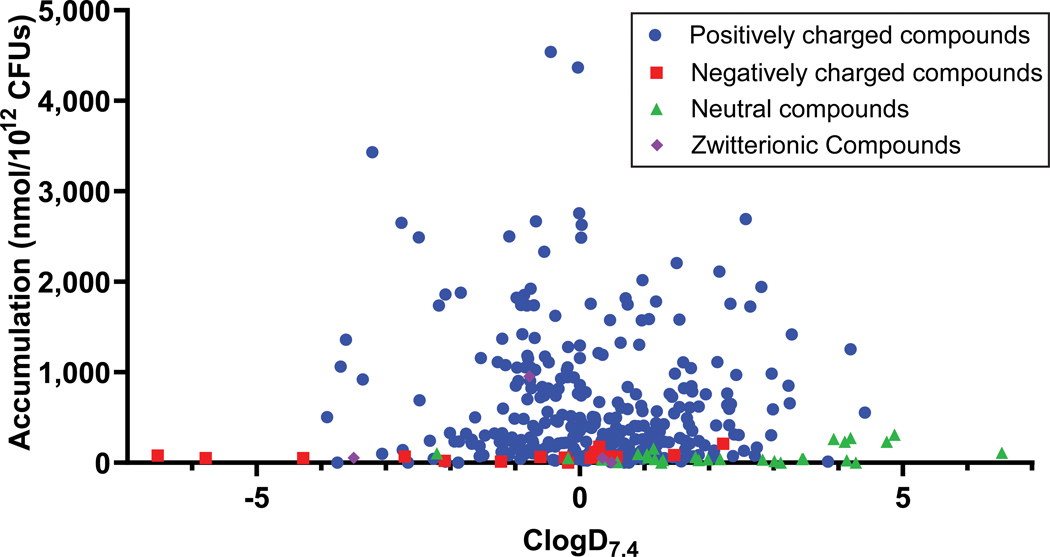
As not all compounds that possess a primary amine accumulate in P. aeruginosa, the compound test set was expanded to identify other properties that facilitate accumulation. In all, 240 primary amines (Supplementary Table 2) and 105 other compounds with varying charge states (Supplementary Table 3) were evaluated for accumulation in P. aeruginosa PAO1. Data gathered with this collection of 345 compounds confirmed the findings from the smaller compound set. It was observed that many positively charged compounds accumulate (Fig. 2a) and accumulators span a broad range of ClogD7.4 values (Fig. 2a). To make explicit head-to-head comparisons, accumulation of a large set of primary amines was assessed in both P. aeruginosa and E. coli. Analysis of these results for 154 compounds shows that while ~80% of compounds meeting the eNTRy rule cutoffs accumulate in E. coli, in agreement with the original report,13 the number of rotatable bonds and globularity are not useful for predicting accumulation in P. aeruginosa, with only ~40% of the compounds being sorted correctly as accumulators or non-accumulators in P. aeruginosa by the eNTRy rules (Extended Data Fig. 2a). Extended Data Fig. 2 also shows examples of compounds that accumulate in E. coli but not P. aeruginosa (Extended Data Fig. 2b), and vice versa (for example BIM X, Extended Data Fig. 2c).
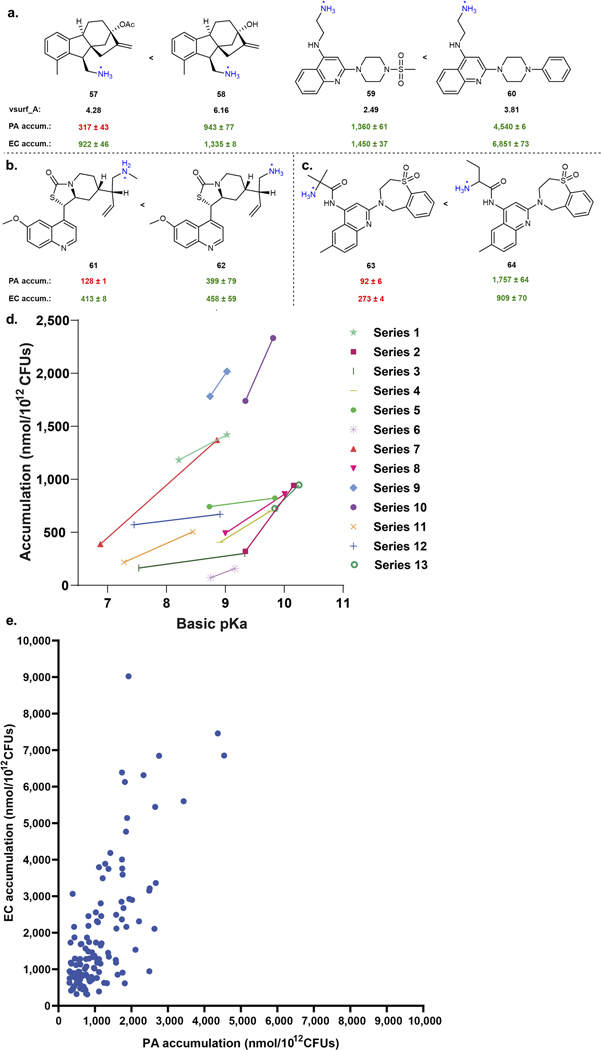
Fig. 2. Importance of ClogD7.4, Hydrogen Bond Donor Surface Area, and positive charge descriptors for accumulation in P. aeruginosa PAO1.
a) A set of 345 compounds, including 240 primary amines, was evaluated for accumulation in P. aeruginosa PAO1. All accumulating compounds have a positive charge. Structures, properties, and accumulation are reported in Supplementary Tables 2 and 3 and includes all 67 compounds from Supplementary Table 1. b) Analysis of 240 primary amines, correlation of Formal Charge or Positive Polar Surface Area and HBD Surface Area with compound accumulation in P. aeruginosa PAO1. In this analysis, compounds with HBD Surface Area ≥23 and either Positive Polar Surface Area ≥ 80 or Formal Charge ≥ 0.98 were most likely to accumulate. >80% of compounds that met these criteria were accumulators, shown in the green box. 113 compounds met the criteria, and 92 of them were accumulators, while 21 were non-accumulators. 127 compounds did not meet the criteria, and 55 of them were accumulators and 72 were non-accumulators. c) Increasing HBD Surface Area leads to an increase in accumulation in P. aeruginosa PAO1. d) Increasing the Positive Polar Surface Area positively correlates with accumulation in P. aeruginosa PAO1. e) Increasing the Formal Charge of a molecule through pKa modulation of amines or other ionizable atoms increases accumulation in P. aeruginosa PAO1. Calculated pKa values for basic functionalities are shown in blue, and calculated pKa values for acidic functionalities are shown in red. Accumulation units are reported in nmol/1012 CFUs. n=3 biologically independent samples. The average and s.e.m. are reported for accumulation values. ClogD7.4 was calculated using the online compound property calculation software FAFdrugs. Formal charge (FCH), HBD Surface Area (vsa_don in MOE), and Positive Polar Surface Area (Q_vsa_PPos in MOE) were calculated in MOE. pKa values were calculated using Chemaxon.
Beyond the three eNTRy rule parameters, two other properties were previously identified as important for accumulation in E. coli, amphiphilic moment and amine steric accessibility.13 Consistent with the observation in E. coli, increasing amphiphilic moment correlated with an increase in compound accumulation in P. aeruginosa (Extended Data Fig. 3a). The steric accessibility of amines also made a significant difference in accumulation values. Like E. coli, compounds with primary amines accumulated to a greater extent in P. aeruginosa than those with a secondary amine (Extended Data Fig. 3b). Additionally, compounds with primary amines on secondary carbons accumulated to a greater extent than compounds with primary amines on tertiary carbons (Extended Data Fig. 3c).
Features influencing accumulation in PA
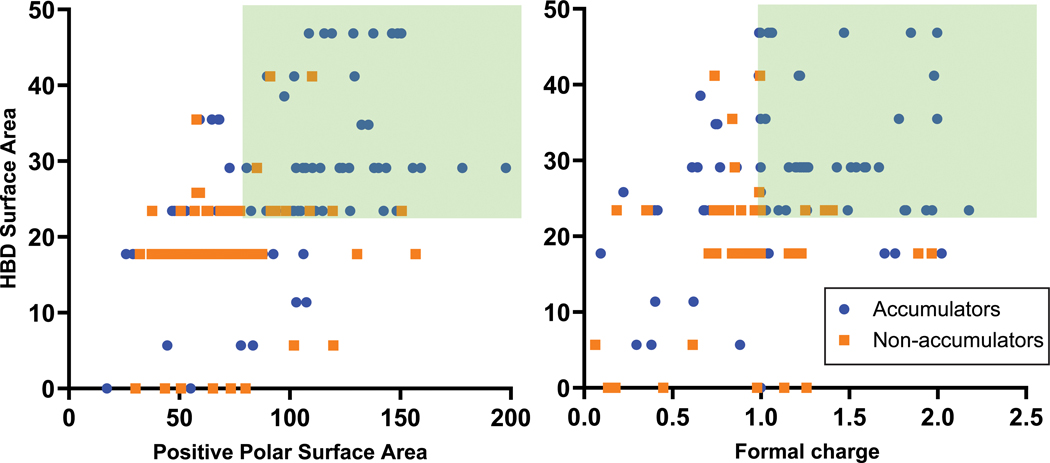
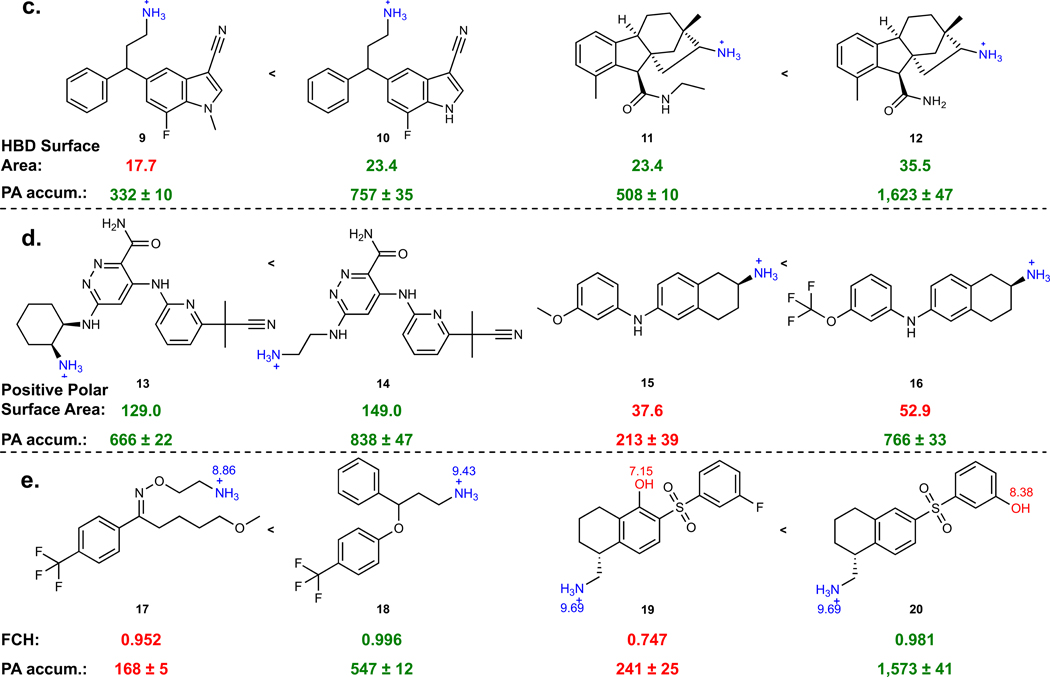
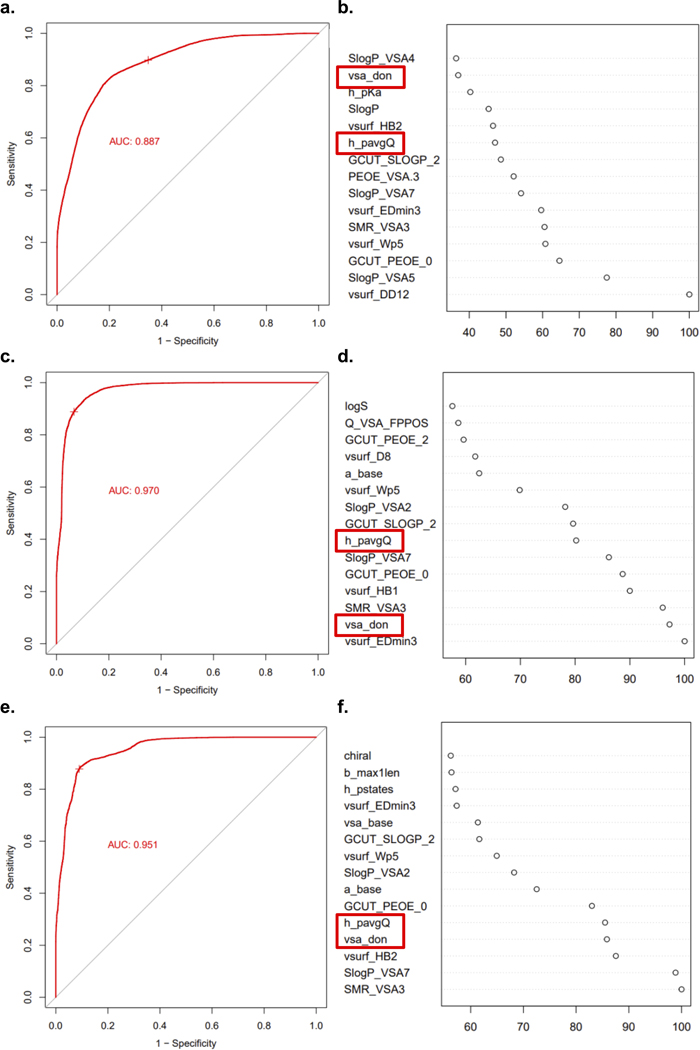
While the presence of a sterically unencumbered primary amine on a compound with a high amphiphilic moment correlates with accumulation in P. aeruginosa, these traits are not enough to form a useful set of guidelines. Thus, we investigated other physicochemical traits that might facilitate accumulation in P. aeruginosa. For this work a cheminformatic approach was utilized, starting with the calculation of 288 physicochemical properties for each of the 240 primary amines from Fig. 2a. These descriptors were used to train a random forest classification model to predict compound accumulation (Extended Data Fig. 4).13 Through this analysis, it was noted that Hydrogen Bond Donor (HBD) Surface Area and positive charge descriptors were positively correlated with accumulation.
The most predictive descriptors associated with positive charge were Positive Polar Surface Area and Formal Charge. While these two positive charge descriptors have commonalities, using both accounts for compounds that 1) may have a primary amine with a lower pKa and less localized charge, but have other partially charged functional groups that are beneficial for accumulation (low formal charge, high positive polar surface area), and that 2) are smaller compounds that may be highly charged, but have lower overall surface area (high formal charge, low positive polar surface area). Compounds with HBD Surface Area ≥23 and with high positive charge (Polar Positive Surface Area ≥80 and/or Formal Charge ≥0.98) were most likely to accumulate (green boxes in Fig. 2b), with >80% (92/113) of all compounds that meet these criteria accumulating in P. aeruginosa PAO1. Side-by-side comparisons demonstrate the benefit of increasing HBD Surface Area (Fig. 2c; 9–12), Positive Polar Surface Area (Fig. 2d; 13–16), and Formal Charge, either through modulation of amine pKa (Fig. 2e; 17 and 18) or through modulation of the pKa of other ionizable atoms (Fig. 2e; 19 and 20). Further analysis of matched molecular pairs demonstrates a clear correlation between increased amine pKa (and increased amine steric accessibility) and higher compound accumulation in P. aeruginosa (Extended Data Fig. 3d, Supplementary Table 8). It is notable that overall accumulation values in P. aeruginosa PAO1 are still ~50% lower on average compared to E. coli MG1655, consistent with the increased permeability barrier of P. aeruginosa relative to other gram-negative pathogens (Extended Data Fig. 3e).34
Mode of uptake
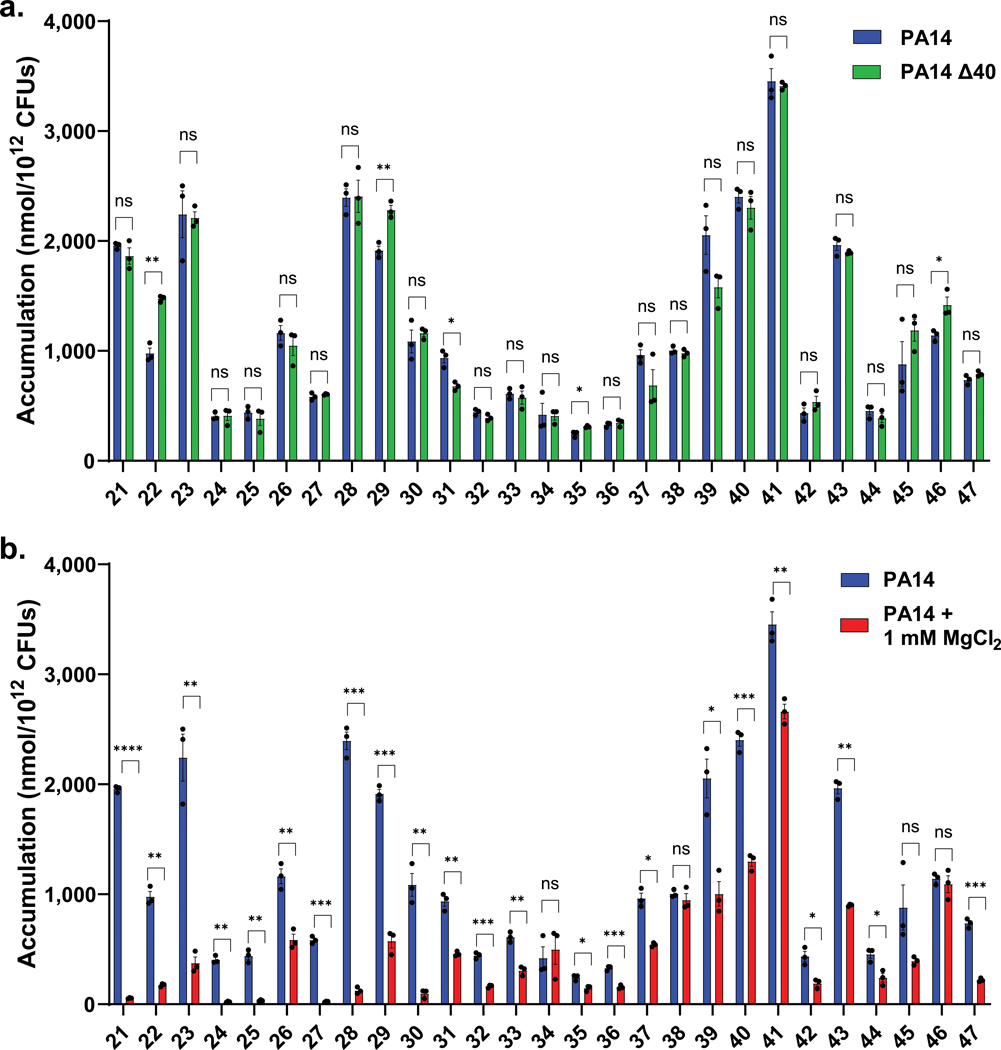
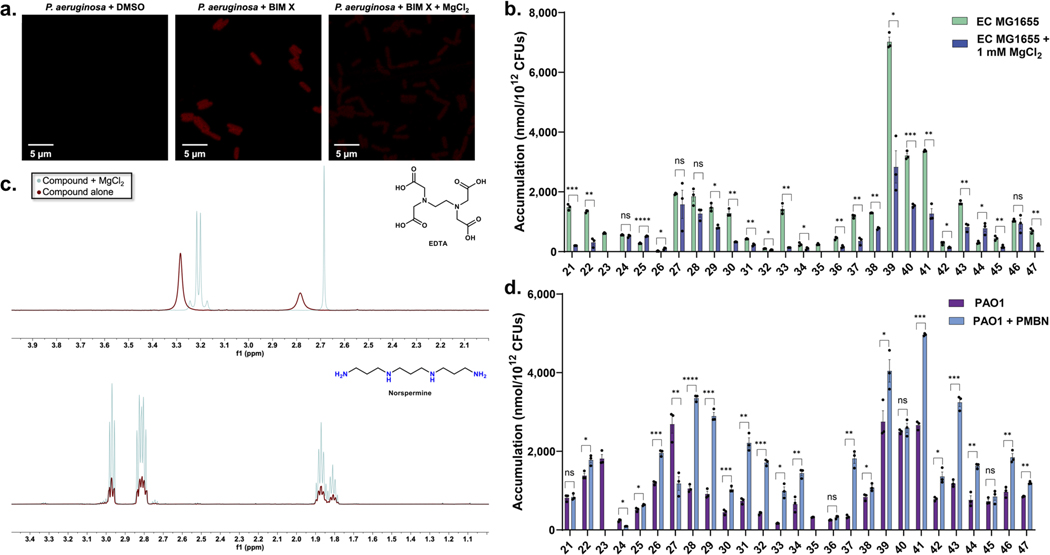
With the importance of positive charge for accumulation in P. aeruginosa established, and with a wide range of potential porins responsible for compound uptake in this organism, it was of interest to investigate the mode of uptake for these compounds. In an attempt to identify compounds that rely on substrate-specific channels of P. aeruginosa for uptake, accumulation was assessed for a subset of compounds in a strain of P. aeruginosa that has 40 putative porins knocked out (PA14 Δ40)35 and accumulation values were compared to the parental strain, PA14. Compounds ranged in molecular weight from less than 200 to greater than 500 Da, and included monoamines, diamines, and guanidiniums (Supplementary Table 4). Strikingly, most compounds showed no statistically significant difference in accumulation between the two strains, indicating that even the absence of 40 porins does not significantly alter accumulation (Fig. 3a). Only one compound (31) showed a statistically significant decrease in accumulation in the PA14 Δ40 strain, while three compounds (22, 29, 46) had slightly higher accumulation values. Suspecting the self-promoted uptake pathway36 as the mode of permeation, the same subset of compounds was evaluated for accumulation in P. aeruginosa in media supplemented with MgCl2. Magnesium ions compete for LPS binding sites with cationic species, and co-treatment is often used to identify compounds that use the self-promoted uptake pathway.37 Notably, in P. aeruginosa, monoamines, diamines, and guanidiniums showed substantially diminished accumulation in the presence on magnesium ions (Fig. 3b, and see Extended Data Fig. 5a for confocal images of BIM X in the presence and absence of MgCl2), suggesting a significant role for the self-promoted uptake pathway for these compounds. The same experiment was performed in E. coli MG1655, where both porin-mediated and self-promoted uptake are likely to be operative.37 While co-incubation with MgCl2 did lower accumulation levels in E. coli as expected, it was to a lesser extent than was observed for P. aeruginosa (Extended Data Fig. 5b). As a control, magnesium binding to a polycation (norspermine) was evaluated by NMR, revealing no/low binding, suggesting it is unlikely that magnesium is simply reducing compound availability (Extended Data Fig. 5c). Accumulation in P. aeruginosa PAO1 was also evaluated in media supplemented with PMBN to weaken the LPS layer. Most compounds showed a statistically significant increase in accumulation, further supporting entry via the self-promoted uptake pathway (Extended Data Fig. 5d). These accumulation results suggest that, in contrast to other gram-negative species, P. aeruginosa does not primarily rely on porins for most small-molecule uptake; instead, self-promoted uptake likely accounts for most permeation into the cell.
Fig. 3. Evaluation of accumulation in a porin deficient strain and in the presence of magnesium.
a) Compounds tested in P. aeruginosa PA14 with all 40 putative porins knocked out (PA14 Δ40) show minimal accumulation differences relative to the parental strain PA14, suggesting a porin-independent mode of uptake. b) The same set of compounds shows a significant decrease in accumulation upon co-treatment with MgCl2 (1 mM), suggesting self-promoted uptake as the primary mode of entry for these compounds. The same PA14 data is used in Fig. 3a and 3b. All structures and accumulation values are listed in Supplementary Table 4a. n=3 biologically independent samples. The average and s.e.m are reported for accumulation values. Statistical significance was determined using a two-sample Welch’s t-test (one-tailed test, assuming unequal variance). Statistically significant accumulation differences for compounds in P. aeruginosa PA14 versus P. aeruginosa PA14 with MgCl2 treatment or P. aeruginosa PA14 Δ40 are indicated with asterisks (n.s. not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Accumulation of diamines
The importance of self-promoted uptake and positive charge for accumulation led us to examine the impact of multiple positive charges on accumulation. In two examples it was observed that diamines exhibited higher accumulation values relative to their monoamine comparators (Extended Data Fig. 6a; 65–68). While diamines containing two primary amines are almost always high accumulators (Extended Data Fig. 6a,b), diamines with one primary amine and one secondary or tertiary amine less reliably provide an accumulation benefit in P. aeruginosa (Extended Data Fig. 6c).
Accumulation of guanidiniums/pyridiniums
Guanidiniums and pyridiniums provide positively-charged alternatives that are more lipophilic relative to primary amines and have previously been demonstrated to improve accumulation in E. coli to differing extents.33 To assess the accumulation benefit of these functional groups in P. aeruginosa, a set of 16 amine, guanidinium, and pyridinium side-by-side comparisons (48 compounds total) were evaluated for accumulation in P. aeruginosa PAO1. Guanidiniums had comparable accumulation values and classification relative to their primary amine comparators, whereas most pyridiniums led to a substantial decrease in accumulation and were not classified as accumulators (Extended Data Fig. 6d and e, Supplementary Table 5); this result matches observations in E. coli.33
Accumulation trends across PA strains
P. aeruginosa has a large pan-genome, leading to significant heterogeneity between strains.38 To test the generality of the accumulation trends observed in P. aeruginosa PAO1, additional representative strains of P. aeruginosa were selected. P. aeruginosa PA14 is a highly virulent strain that is a part of the most common clonal group and contains pathogenicity islands in its genome that are absent in PAO1.39 Additionally, a clinical isolate from an acute infection, P. aeruginosa PA1280, was evaluated.40 MICs for antibiotics against each of these strains were determined and intracellular accumulation was measured. Accumulation of these antibiotic controls correlated well with the observed activity (Extended Data Fig. 6f and Extended Data Table 1c).
Additional compounds were selected for evaluation of accumulation in PA14 and PA1280, comprising 27 non-antibiotics with a range of accumulation levels in PAO1. While some variance in overall values occurred, and accumulation was on average lower in PA1280, very few compounds changed classification from “accumulator” to “non-accumulator” or vice versa in either strain, suggesting that the accumulation model will be robust across different P. aeruginosa strains (Extended Data Fig. 6g, full data set reported in Supplementary Table 6).
Retrospective examples
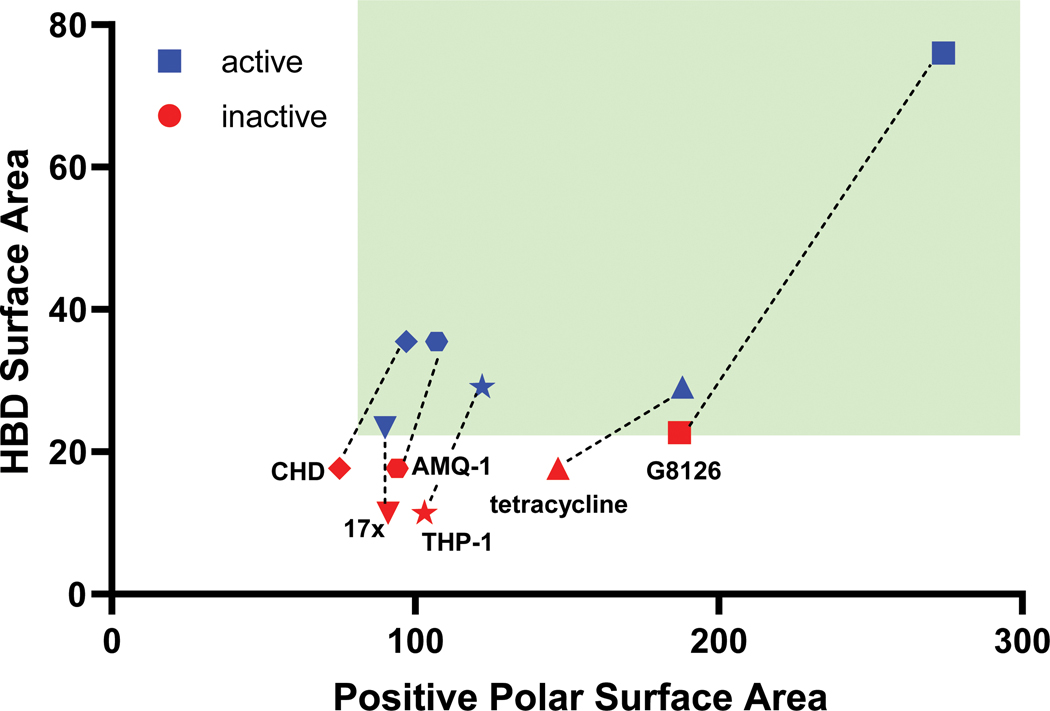
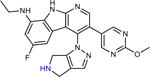
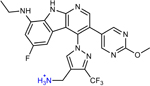
A retrospective analysis of the literature was performed to assess whether previous serendipitous incorporation of these physicochemical properties had proved beneficial in historical drug discovery campaigns. Six examples of antibiotic pairs were identified in which one derivative had low whole-cell activity against P. aeruginosa, but another derivative was reported with improved activity. These antibiotics are from diverse structural classes and engage a variety of biological targets, including DNA gyrase and topoisomerase (17x and 17r16; AMQ-1 and AMQ-219; THP-1 and THP-241), the ribosome (CHD and CDCHD18; tetracycline and tigecycline42), and the bacterial type 1 signal peptidase (G8126 and G0775, all structures in Extended Data Table 2).43 As biochemical potency and accumulation both contribute to whole-cell activity, where possible we aimed to select side-by-side comparisons of compounds with similar target engagement. MIC values in efflux pump deficient strains were used as a surrogate for target engagement (Extended Data Table 2), and many of the reported pairs demonstrated similar activity in efflux pump deficient bacterial strains. The HBD Surface Area and Formal Charge/Positive Polar Surface Area were calculated for these antibiotic pairs. Notably, increasing HBD Surface Area to ≥23 and Positive Polar Surface Area to ≥80 led to an increase in antibacterial activity against P. aeruginosa, ranging from 4- >32-fold improvements (data plotted in Fig. 4; structures and activity reported in Extended Data Table 2),16, 18–19, 41, 43 suggesting that increased accumulation is a significant contributor to the observed improvement in antibacterial activity. Indeed, as shown in Fig. 1a, we find tigecycline accumulates to a higher extent than tetracycline. Further, when considering antibiotics used for the treatment of P. aeruginosa infections, both classes of positively charged antibiotics (aminoglycosides and polymyxins) match these guidelines for P. aeruginosa accumulation. These results suggest these guidelines as a generalizable means to increase accumulation in P. aeruginosa across a variety of scaffolds and targets. β-lactam and fluoroquinolone antibiotics meet the guidelines less consistently, likely due to their zwitterionic or negatively charged states.
Fig. 4. Six retrospective examples where an antibiotic derivative is more active (≥4-fold) against P. aeruginosa than the parent.
Analysis shows that the derivatives with improved antibacterial activity meet the HBD surface area and charge requirements for accumulation in P. aeruginosa (as shown by the green box). Target engagement (inferred from MIC values against efflux-deficient strains) is similar for most of the compound pairs (for structures of each antibiotic, see Extended Data Table 2), suggesting that the improved antibiotic activity against P. aeruginosa is due to an increase in accumulation; this has been explicitly shown tetracycline/tigecycline pair (Fig. 1a).
A PA-active fusidic acid derivative
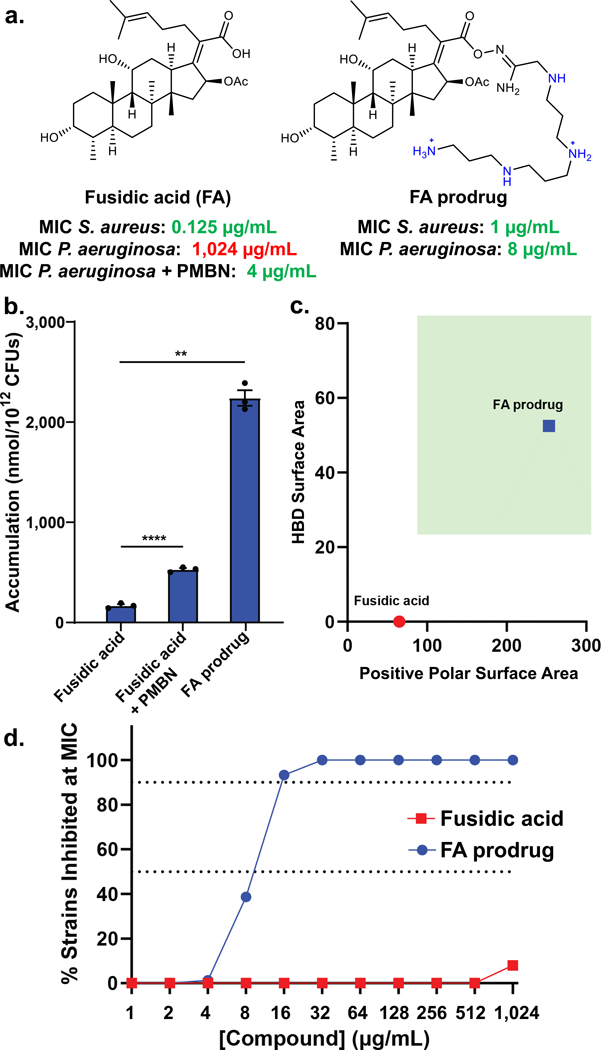
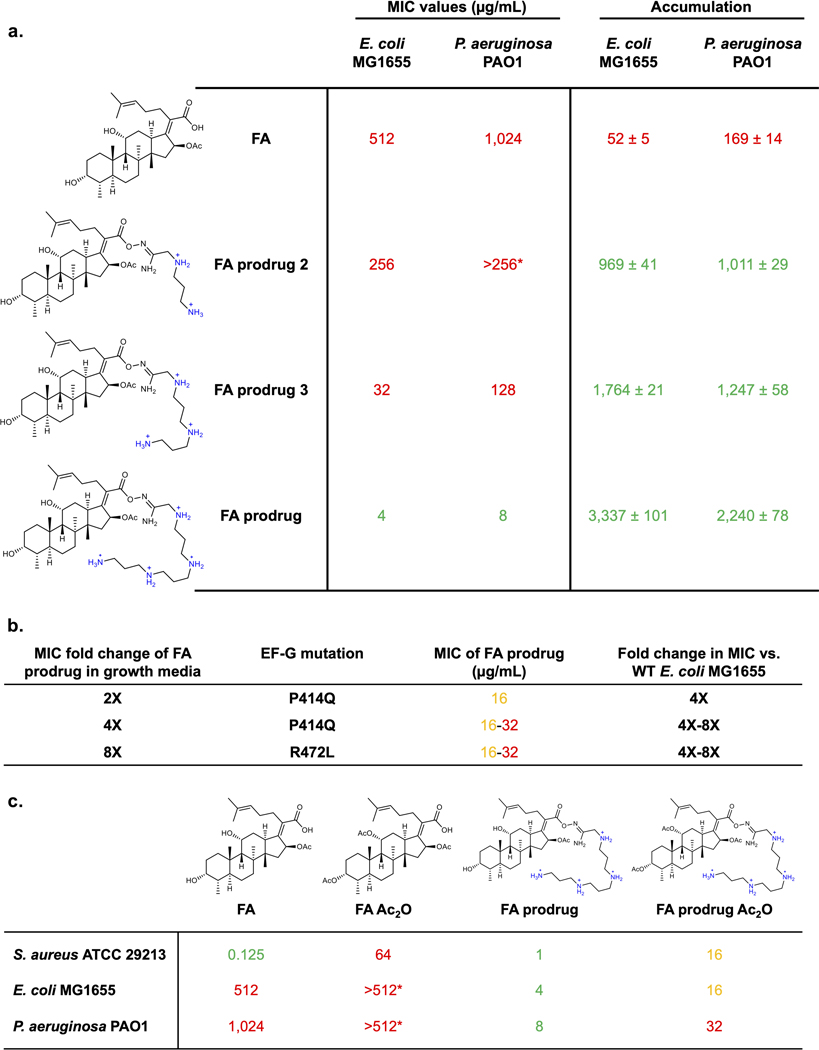
With an improved understanding of the physicochemical properties that promote intracellular accumulation, as well as the porin-independent mode of uptake of these compounds, we were interested in applying these findings to develop a novel antipseudomonal. Our discoveries suggest that increasing the HBD Surface Area and the Formal Charge and/or Positive Polar Surface Area of an antibiotic candidate should lead to improved whole-cell antibacterial activity against P. aeruginosa if target binding is not disrupted. As stated previously, there are many antibiotics (Extended Data Table 1d) that would have activity against P. aeruginosa if they could achieve higher intracellular accumulation. Fusidic acid (FA), a potent gram-positive-only antibiotic, is one example. FA has negligible whole-cell antibacterial activity in P. aeruginosa (MIC = 1024 μg/mL versus PAO1, Fig. 5a) and poor intracellular accumulation (Fig. 5b). Notably, FA has an MIC of 4 μg/mL against P. aeruginosa PAO1 when co-administering PMBN (Fig. 5a), as well as elevated accumulation in the presence of PMBN (Fig. 5b), suggesting that its biological target (elongation factor G, EF-G44) can be engaged in P. aeruginosa, and that if FA could achieve higher intracellular concentrations, it would exhibit whole-cell antibacterial activity.
Fig. 5. Development of PA-active fusidic acid (FA) derivative.
a) FA has potent activity against gram-positive bacteria, but no activity against P. aeruginosa PAO1. However, when co-administering the membrane permeabilizer PMBN (8 μg/mL), FA has good antibacterial activity, indicating that the target (EF-G) can be exploited, and if FA could accumulate, it would be active against wild-type P. aeruginosa. To improve accumulation using the predictive guidelines for PA, while maintaining the necessary acid for FA activity, an amidoxime prodrug moiety was generated with a polyamine linker. The prodrug is cleaved through hydrolysis to release FA inside the cell, leading to an MIC value of 8 μg/mL, a 128-fold improvement in activity against wild-type P. aeruginosa. b) FA accumulation in P. aeruginosa PAO1 is enhanced in the presence of PMBN (8 μg/mL). FA prodrug shows >20x higher accumulation levels that FA. As the prodrug hydrolyzes under assay conditions, the accumulation of FA is reported for FA prodrug. c) FA does not meet the predictive guidelines for accumulation in P. aeruginosa, while FA prodrug fits the described physicochemical properties, highlighted in the green box. d) FA prodrug possesses 64–256x improved activity relative to FA against a panel of 75 clinical isolates of P. aeruginosa. MICs were performed in LB Lennox broth according to the CLSI guidelines in biological triplicate. Accumulation units are reported in nmol/1012 CFUs. n=3 biologically independent samples. The average and s.e.m are reported for accumulation values. Statistical significance was determined using a two-sample Welch’s t-test (one-tailed test, assuming unequal variance). Statistically significant accumulation differences for fusidic acid in the presence of PBMN or with the prodrug linker are indicated with asterisks (**P < 0.01, ****P < 0.0001)
With a HBD Surface Area of 0 and a Positive Polar Surface Area of 65.3 (Fig. 5c), FA does not meet our guidelines for accumulation in P. aeruginosa and was selected as a challenging starting point for conversion to a version active against this pathogen. Using the established structure-activity relationship of FA along with the accumulation guidelines for P. aeruginosa, FA derivatives were designed and synthesized. As the carboxylic acid is a convenient handle for structural modifications, but necessary for target engagement,45 we envisioned using a prodrug approach to increase permeation into the cell, while maintaining on-target activity against EF-G through the release of free FA inside the cell. An amidoxime prodrug moiety was ultimately selected as it was hydrolytically cleaved within a biologically relevant timeframe, liberating FA (Extended Data Fig. 8a). Amidoxime prodrug derivatives containing functional groups whose presence increases overall HBD Surface Area and Positive Polar Surface Area were synthesized and evaluated. Data collected in both P. aeruginosa PAO1 and E. coli MG1655 demonstrate a substantial increase in accumulation for all FA prodrug derivatives tested (Extended Data Fig. 7a). Accumulation values increase with the number of amines, with the version containing four ionizable nitrogens (FA prodrug, Fig. 5a) demonstrating the highest accumulation (Extended Data Fig. 7a). FA prodrug meets the HBD Surface Area and Positive Polar Surface Area requirements for accumulation in P. aeruginosa (Fig. 5c) and does indeed achieve very high accumulation levels in PAO1 (Fig. 5b; accumulation reported for free FA). Importantly, FA prodrug has an MIC of 8 μg/mL against P. aeruginosa PAO1, a 128-fold increase relative to FA (Fig. 5a) and has MIC values of 8–16 μg/mL against a panel of 75 clinical isolates of P. aeruginosa; FA has essentially no antibacterial activity against this clinical isolate panel (Fig. 5d, MIC values in Supplementary Table 7).
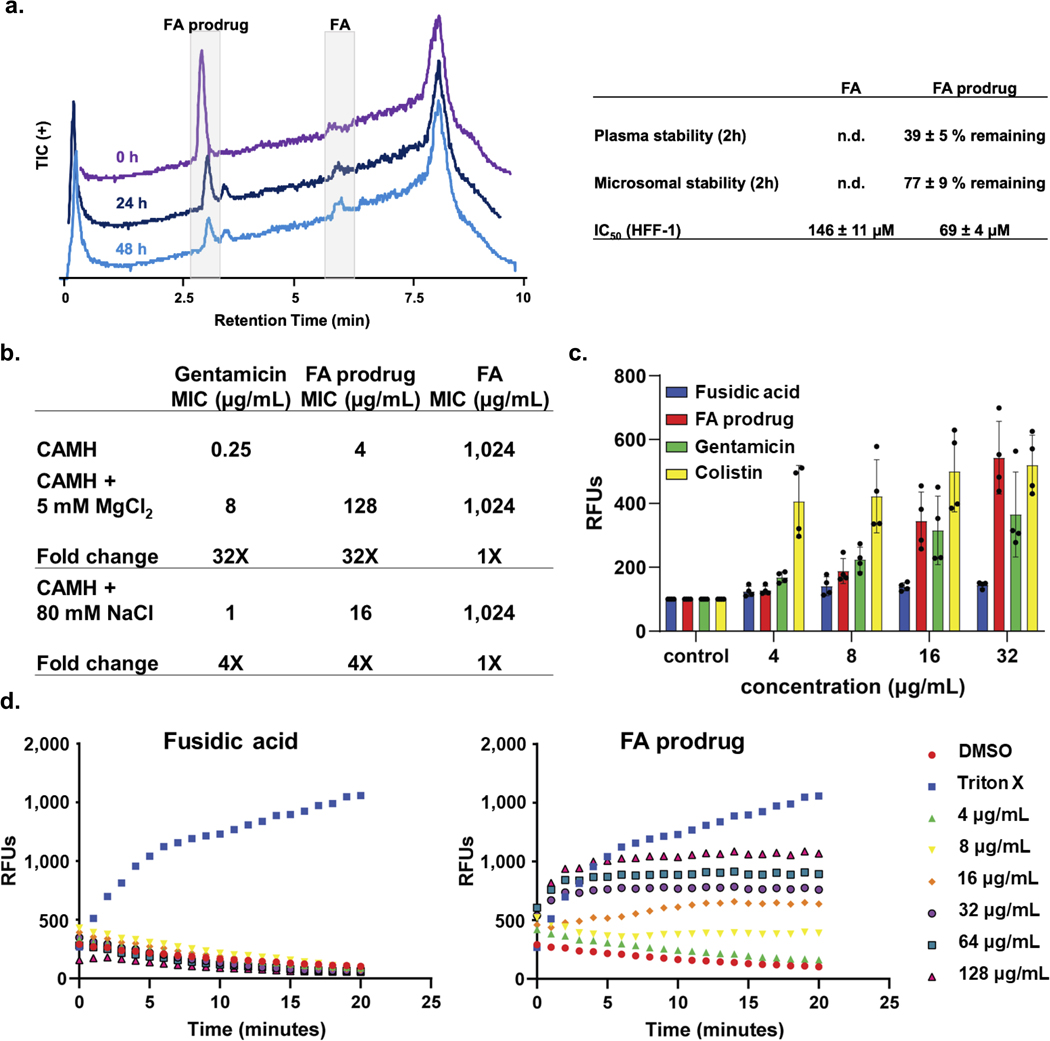
To explore the antibacterial mode of action of FA prodrug, E. coli MG1655 mutants resistant to FA prodrug were generated using a serial exposure method. Mutants that were viable at 2X, 4X, and 8X the MIC concentration were isolated. Resistant colonies showed a 4–8 fold increase in MIC values against FA prodrug, and mutations map back to the FA binding pocket in EF-G indicating that liberated FA can engage its intracellular target and exert antibacterial activity (Extended Data Fig. 7b). Generation of P. aeruginosa PAO1 mutants resistant to FA prodrug was achieved using the large inoculum method, with a frequency of resistance at 2.67 × 10-7. When sequenced, these mutants did not have mutations in fusA, indicating that FA prodrug likely has more than one mechanism of action.
The mode of action of FA prodrug in P. aeruginosa was then probed using structural derivatives. It is known that diacylation of FA significantly reduces antibacterial activity, and indeed this compound (FA Ac2O) has an MIC = 64 μg/mL in S. aureus (Extended Data Fig. 7c). To evaluate the dependence of FA prodrug activity on inhibition of EF-G, a diacetylated version, FA prodrug Ac2O, was synthesized (Extended Data Fig. 7c). This compound loses antibacterial activity against P. aeruginosa relative to FA prodrug, suggesting that some antibacterial activity of FA prodrug is derived from the inhibition of EF-G, with prodrugs with more amines providing more antibacterial activity through membrane depolarization (Extended Data Fig. 7a).
It is apparent that FA prodrug has a dual mode of antibacterial activity. Based on the polyamine moiety we hypothesized that membrane-based interactions may be contributing to the antibacterial activity, and a variety of experiments were performed to characterize such interactions. Co-treatment with MgCl2 led to a 32-fold shift in MIC for FA prodrug and gentamicin in P. aeruginosa PAO1, while FA showed no change in antibacterial activity, suggesting that FA prodrug entry into P. aeruginosa occurs via the self-promoted uptake pathway (Extended Data Fig. 8b); NaCl had minimal effects in the same experiment (Extended Data Fig. 8b). The fluorescent probe N-phenylnapthylamine (NPN) was used to assess outer membrane permeability changes upon treatment with FA, gentamicin, colistin, and FA prodrug. NPN is membrane impermeable and is only weakly fluorescent in aqueous solutions. Upon entry to the cell and interacting with phospholipids, significant fluorescence is observed. While treatment of P. aeruginosa PAO1 with FA for ten minutes did not lead to any change in fluorescence, treatment with gentamicin, colistin, and FA prodrug led to a dose-dependent increase (Extended Data Fig. 8c). Finally, using the potentiometric dye DiSC3 (5), it was demonstrated that treatment with FA prodrug leads to dose-dependent inner membrane depolarization, monitored by an increase in fluorescence, while treatment with FA did not impact membrane polarization (Extended Data Fig. 8d). All this data is consistent with FA prodrug relying on membrane interactions to enter the cell; in addition to the liberated FA engaging EF-G to kill the bacterial cells, inner membrane depolarization by FA prodrug likely plays a role in the observed antibacterial activity. This residual membrane depolarizing antibacterial activity can be observed in the diacylated compound (Extended Data Fig. 7c). This effect can also be observed amongst the different versions of the FA prodrug, where potency is not solely impacted by high accumulation values, as extending the polyamine moiety increases membrane-disrupting ability in addition to increasing accumulation (Extended Data Fig. 7a).
Discussion
Comparison of results from the unbiased accumulation experiments between E. coli and P. aeruginosa highlights the benefit of ionizable nitrogens (with minimal steric encumbrance) and high amphiphilic moment to promote intracellular accumulation in both E. coli and P. aeruginosa. HBD Surface Area and positive charge parameters were found to positively correlate with accumulation in P. aeruginosa, and ~80% of compounds that meet the appropriate cutoffs accumulate. As evidenced by this study, the unbiased evaluation of diverse non-antibiotic compounds can identify properties that are not apparent in the assessment of antibiotics.
An interesting and surprising result is the observed porin-independent uptake of compounds in P. aeruginosa. Assessing compound accumulation in a strain of P. aeruginosa with 40 putative porins knocked out (Δ40) demonstrates few significant accumulation differences in the absence of porins. Consistent with our result, a similar trend was observed in assessing MIC values for antibiotics in the wild-type and Δ40 strain; aside from the carbapenem antibiotics, no antibiotics had an MIC fold change in this study.35 Collectively, these results suggest that in contrast to E. coli, where the general porin OmpF facilitates uptake for some monoamines,13, 46 in P. aeruginosa the self-promoted uptake pathway may be the primary mode of uptake for all positively charged compounds. Multicharged antibiotics, such as the aminoglycosides and colistin, have been suggested to utilize this pathway for uptake into bacteria, but the utility of this pathway for monoamine uptake had not yet been disclosed. Evidence does suggest the importance of porin-mediated uptake for some negatively charged compounds into P. aeruginosa, most recently demonstrated by the rational design of ETX0462, which was chemically optimized for uptake through P. aeruginosa porins.47 The development of a prodrug moiety that can facilitate self-promoted uptake into P. aeruginosa for fusidic acid may prove generalizable for other antibiotics where antibacterial activity is limited due to poor intracellular accumulation; of course, as polyamine functionality can introduce liabilities (toxicity or off-target membrane interactions), optimization and exploration of other prodrugs will be beneficial.
Historical data and trends from the literature begin to make more sense in light of the results presented herein. For example, the impenetrability of P. aeruginosa, even relative to other gram-negative bacteria, has been noted by others and is powerfully shown (in comparison to E. coli) in Extended Data Fig. 3e. As certain small molecules can enter E. coli through porins, the general impenetrability of P. aeruginosa appears to be linked to the squelching of most porin-dependent uptake pathways. The inability to exploit porin-mediated uptake in P. aeruginosa has stymied antibiotic development for this pathogen, but data presented herein suggests that purposeful design of antibiotics to leverage porin-independent mechanisms can be fruitful in the generation of novel antibiotics for this dangerous pathogen.
Methods:
Bacterial Strains.
P. aeruginosa PAO1 and E. coli MG1655 were obtained from the American Type Culture Collection (ATCC). P. aeruginosa AR strains were obtained from the Centers for Disease Control and Prevention and FDA Antibiotic Resistance Isolate Bank. P. aeruginosa Cubist strains were obtained from Cubist Pharmaceuticals in 2008. P. aeruginosa PA14 and PA14 Δ40 were a generous gift from Prof. Dirk Bumann, Beatrice Claudi, and co-workers.35 P. aeruginosa PAO1 Δ6 and P. aeruginosa PAO1 Δ6•pore were generous gifts from Prof. Helen Zgurskaya and co-workers.26 P. aeruginosa 1113 and 1727 were a generous gift from Catherine Llanes.48 P. aeruginosa Carle strains were obtained from Carle Hospital in Urbana, IL.
Antimicrobial Susceptibility Tests.
Susceptibility testing was performed in biological triplicate, using the microdilution broth method as outlined by the Clinical and Laboratory Standards Institute. Bacteria were cultured with cation-adjusted Meuller-Hinton (CAMH) broth (Sigma-Aldrich; catalogue number: 90922) or Luria-Bertani (LB) broth (Lennox) in round-bottom 96-well plates (Corning; catalogue number: 3788). To probe the magnesium-dependence of antibacterial activity, CAMH was supplemented with 5 mM MgCl2.
Accumulation Assay.
The accumulation assay was performed in triplicate in batches of ten samples, with each batch containing tetracycline or ciprofloxacin as a positive control. P. aeruginosa PAO1, PAO1 Δ6, PA14, PA14 Δ40, and PA1280, and E. coli MG1655 were used in these experiments. For each replicate, 2.5 mL (E. coli) or 5 mL (P. aeruginosa) of an overnight culture was diluted into 250 mL of fresh Luria−Bertani broth (Lennox) or Tryptic Soy Broth (TSB) and grown at 37 °C with shaking to an optical density (OD600) of 0.55–0.60. The bacteria were pelleted at 3220 r.c.f. for 10 min at 4 °C, and the supernatant was discarded. The pellets were resuspended in 40 mL of phosphate buffered saline (PBS) and pelleted as before, and the supernatant was discarded. The pellets were resuspended in 8.8 mL of fresh PBS and aliquoted into ten, 1.5 mL Eppendorf tubes (875 μL each). The number of colony-forming units (CFUs) was determined by a calibration curve. The samples were equilibrated at 37 °C with shaking for 5 min; compound was added (final concentration = 50 μM), and then the samples were incubated at 37 °C with shaking for either 10 min. These time points were short enough to minimize metabolic and growth changes (no changes in OD600 or CFU count observed). After incubation, 800 μL of the cultures was carefully layered on 700 μL of silicone oil (9:1 AR20/Sigma High Temperature, cooled to −78 °C). Bacteria were pelleted through the oil by centrifuging at 13 000 r.c.f. for 2 min at room temperature (with the supernatant remaining above the oil); the supernatant and oil were then removed by pipetting. To lyse the samples, each pellet was dissolved in 200 μL of water, and then, they were subjected to three (E. coli) or four (P. aeruginosa) freeze−thaw cycles of 3 min in liquid nitrogen followed by 3 min in a water bath at 65 °C. P. aeruginosa samples were then treated with 50 μM DNase and RNase and incubated at 37 °C for 15 minutes. The lysates were pelleted at 13 000 r.c.f. for 2 min at room temperature, and the supernatant was collected (180 μL). The debris was resuspended in 100 μL of methanol and pelleted as before. The supernatants were removed and combined with the previous supernatants collected. Finally, the remaining debris was removed by centrifuging at 20 000 r.c.f. for 10 min at room temperature. Supernatants were analyzed by LC−MS/MS.
Samples were analyzed with the 5500 QTRAP LC/MS/MS system (AB Sciex) with a 1200 series HPLC system (Agilent Technologies) including a degasser, an autosampler, and a binary pump. Liquid chromatography separation was performed on an Agilent SB-Aq column (4.6 × 50 mm, 5 μm) (Agilent Technologies) with mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile). The flow rate was 0.3 mL/min. The linear gradient was as follows: 0−3 min, 100% mobile phase A; 10−15 min, 2% mobile phase A; 15.5−21 min, 100% mobile phase A. The autosampler was set at 5 °C. The injection volume was 15 μL. Mass spectra were acquired with both positive electrospray ionization at the ion spray voltage of 5500 V and negative electrospray ionization at the ion spray voltage of −4500 V. The source temperature was 450 °C. The curtain gas, ion source gas 1, and ion source gas 2 were 33, 50, and 65, respectively. Multiple reaction monitoring was used to quantify the metabolites. Power analysis was not used to determine the number of replicates. Error bars represent the standard error of the mean of three biological replicates. The statistical significance of the accumulation was determined using a two sample Welch’s t test (one-tailed test, assuming unequal variance) relative to the negative controls. All compounds evaluated in the biological assays were ≥95% pure.
PMBN assays.
Assays measuring permeabilization by polymyxin B nonapeptide (PMBN) were performed as above, with the addition of 8 μg/mL PMBN immediately before the compound of interest was added.
Accumulation assay with magnesium.
Assays measuring impact of magnesium treatment were performed as above, with the addition of 1 mM MgCl2 immediately before the compound of interest was added.
Assays with PAO1 Δ6•pore.
Assays were performed exactly as described above, with the exception that outgrowth media (LB) was supplemented with IPTG to a final concentration of 0.1 mM to induce the pore.
NPN outer membrane permeabilization assay.
An overnight culture of P. aeruginosa PAO1 was diluted into 10 mL fresh TSB and grown to mid-log phase. Cells were collected by centrifugation at 3,220xg for 10 minutes at room temperature. The supernatant was discarded, and the pellet was resuspended in 5 mM HEPES (pH=7.2), 5 mM glucose. Cells were collected again by centrifugation, supernatant discarded, 125 and pellet resuspended to an OD600 = 1. Then, 100 μL of washed cells and 100 μL of assay buffer containing 20 μM NPN were mixed together and added to a 96-well optical-bottom black plate. Either 2 μL of a fusidic acid, FA prodrug, gentamicin, or DMSO, was added to each well and fluorescence was immediately monitored at an excitation wavelength of 350 nm and emission wavelength of 420 nm for 10 min at 30 sec intervals. Fluorescence values were normalized to the control wells.
Membrane depolarization assay.
100 μL of an overnight culture of P. aeruginosa PAO1 was diluted into 10 mL fresh TSB and grown to mid-log phase. Cells were collected by centrifugation at 3,220xg for 10 minutes at room temp. The supernatant was discarded, and the pellet was resuspended in 5 mM HEPES (pH=7.8). Cells were collected again by centrifugation and supernatant discarded. The pellet was then resuspended in 5 mM HEPES with 100 mM KCl and diluted to an OD600 of 0.1. 0.2 M EDTA was added to the bacterial solution to a final concentration of 0.2 mM EDTA to permeabilize the bacteria to the dye. Into a black 96-well plate, 192 μL of the bacterial/EDTA solution and 8 μL of 10 μM DiSC dye was added to the wells. The fluorescence of the plate was monitored for 30 minutes, with reads every 5 minutes at the excitation wavelength of 622 nm and emission wavelength of 670 nm. To re-establish the proton gradient, 2 μL of 10 M KCl solution (final concentration of 100 mM KCl) was added to each well. 2 μL of compound was added at each concentration and fluorescent monitored for 60 minutes. Triton X (1%) was used as the positive control and DMSO was the negative control.
Selection of resistant mutants.
Resistant mutants were selected using the large inoculum method. Briefly, P. aeruginosa Δ6 (1.8 × 109 c.f.u.) was plated on 100 mm plates of Luria Bertani agar containing 64, 32 or 16 μg ml−1 fusidic acid with 10 μg/mL gentamicin. Colonies were visible after incubating at 37 °C for 72 h. Resistant colonies were confirmed by streaking on selective media with the same concentration of fusidic acid.
Confocal fluorescence microscopy and sample preparation.
Accumulation was performed as described above for either PAO1 or EC MG1655 with either Bisindolylmaleimide X (± MgCl2) or DSMO up until the point immediately after removing the oil and supernatant and before the freeze-thaw lysis. The pellet was then resuspended in 1 mL 3.7 % Formaldehyde in PBS, transferred to a new tube, and allowed to sit on a nutator for 30 minutes at room temperature. 3 μL of the now fixed bacterial cells were placed on a clean microscope cover slip (22 mm x 40 mm, VWR), followed by an agarose pad prepared as described previously49, followed by a second cover slip. Fluorescence images were taken with a Zeiss 710 multiphoton confocal microscope through a 63x/1.4 RI oil objective. All samples were excited at 488 nm with an argon laser, and fluorescence emission was recorded from 599 nm- 689 nm. Images were acquired and processed using Zen Black (Zen 2.3).
Hydrolysis of FA prodrug.
The hydrolysis of FA prodrug (10 μg/mL) in PBS at 37 °C (shaking incubation) was monitored using LC-MS over a period of 48 hours.
Microsomal stability.
To a 1.7 mL Eppendorf tube was added PBS (442.5 μL), 20 X NADPH regenerating solution A (Corning Gentest) (25 μL), and 100 X NADPH regenerating solution B (Corning Gentest) (5 μL). This was placed in a shaking incubator (250 RPM) for 5 minutes at 37 °C. To the solution was then added the compound of interest (2.5 μL of a 10 mM stock) and finally the mouse liver microsomes (Gibco Mouse (CD-1)) (25 μL, 20 mg/mL). The assay was then mixed via pipet and a 50 μL aliquot was removed and added to 50 μL cold acetonitrile. This was taken to be T = 0 h. The remaining assay was placed back into the incubator and allowed to stand for an additional 2 hours before repeating this same process. Quenched samples were centrifuged at 13,000 rcf for 3 minutes, and 75 μL of the supernatant was taken and submitted for LC-MS/MS analysis.
Plasma stability.
To a 1.7 mL Eppendorf tube was added PBS (39 μL) followed by bovine plasma (160 μL) for each assay. This was placed in a shaking incubator (250 RPM) for 5 minutes at 37 °C. To the warm sample was then added compound of interest (1 μL of a 10 mM stock). The samples were vortexed and a 50 μL aliquot was removed and added to 200 μL cold acetonitrile. This was taken to be T = 0 h. The remaining assay was placed back into the incubator and allowed to stand for an additional 2 hours before repeating this same process. Quenched samples were centrifuged at 16,000 rcf for 5 minutes, and 100 μL of the supernatant was taken and submitted for LC-MS/MS analysis.
Cytotoxicity.
IC50 values for FA and FA prodrug were recorded in HFF-1 cells (male, newborn; obtained from ATCC). Cells were seeded in a 96 well plate at 3,000 cells/well and allowed to grow for 24 hours. Cells were then treated with compound and allowed to incubate for another 72 hours before assessing cell viability with Alamar blue assay. The cells were not tested for mycoplasma beforehand.
Calculation of Physiochemical Properties.
Data sets of chemical structures were created and managed using Canvas (Schrödinger, LLC). Initial structure preparation and 3D minimization was performed with LigPrep (Schrödinger, LLC) using OPLS_2005 force fields Generation of ensembles of conformations was performed using Conformational Search in MOE 2015.10 using the LowModeMD method with default. Physiochemical descriptors (both 2D- and 3D-based) were calculated using MOE 2015.1049 for each conformation. Descriptors were averaged (unweighted mean) across all conformations for each molecule. Data with descriptors were used to train a random forest classification prediction model using the R package caret. The random forest model offers many advantages for this application including resistance to over-fitting and the ability to measure descriptor importance44. Preprocessing of data removed descriptors with near-zero variance or high co-correlation with other descriptors. Source code for data analysis and model training can be found at https://github.com/HergenrotherLab/GramNegAccum. ClogD7.4 values were calculated using the online compound property calculation software FAFdrugs.
Statistical analyses.
GraphPad Prism 8.2.1.441 was used for data analysis and figure generation. Data are shown as means ± s.e.m. Statistical significance was determined by one-way ANOVA (with Tukey’s multiple comparisons test) for two groups at a single time point, two-way ANOVA (with Sidak’s multiple comparison’s test) for two groups at multiple time points, or two-way ANOVA (with Tukey’s multiple comparisons test) for three or more groups at multiple time points. P < 0.05 was considered statistically significant. In this study, no statistical methods were used to predetermine the sample size. The experiments were not randomized, and the investigators were not blinded to allocation during the experiments and outcome assessments.
Data availability:
The authors declare that all data supporting the findings of this study are available within the paper and the Supplementary Information. Source data is provided for all Figures and Extended Data Figures.
Code availability:
Source code for data analysis and model training can be found at https://github.com/HergenrotherLab/GramNegAccum.
Extended Data
Extended Data Fig. 1. Confocal fluorescent images of an intrinsically fluorescent compound, Bisindolylmaleimide X (BIM X).
a) The structure of BIM X along with accumulation values. The LC-MS/MS assay reveals BIM X as a poor accumulator in E. coli MG1655, while it is a good accumulator in P. aeruginosa PAO1. Accumulation units are in nmol/1012 CFU and the error is reported as the s.e.m. b) The standard accumulation assay was performed in P. aeruginosa (PAO1) and E. coli (MG1655) with either BIM X (50 μM) or DMSO until just after the oil removal step. Cells were fixed in 3.7 % formaldehyde in PBS, and fluorescence images were taken with a Zeiss 710 multiphoton confocal microscope through a 63x/1.4 oil objective. All samples were excited at 488 nm with an argon laser, and fluorescence emission was recorded from 599–689 nm. Images were acquired using Zen Black (Zen 2.3). DMSO controls for both cell types tested indicate no autofluorescence. Images are representative of n = 3 biologically independent samples.
Extended Data Fig. 2. Comparison of species-specific accumulation trends.
a) In the expanded set of primary amines, many compounds that fit the eNTRy rules do not accumulate in P. aeruginosa PAO1, and many compounds that do not fit the eNTRy rules do accumulate in P. aeruginosa PAO1. Low globularity (glob) and low rotatable bonds (RB) are predictive for ~80% of compounds tested in E. coli MG1655, but only 41% of compounds in P. aeruginosa PAO1. Compounds with poor amine steric accessibility, low amphiphilic moment, and multiple charges were removed from this analysis; 154 compounds total are included in the plots, see Compound Master Table for compounds. b) Compounds that accumulate in E. coli, but not in P. aeruginosa, can often be grouped according to structural class, but no additional trends were identified. c) Compounds that accumulate in P. aeruginosa, but not E. coli, are primarily compounds that have high rotatable bonds, high globularity, or both. Accumulation units are reported in nmol/1012 CFUs. n=3 biologically independent samples. The s.e.m. is reported for accumulation values.
Extended Data Fig. 3. Accumulation trends, matched molecular pairs, and comparison of P. aeruginosa and E. coli accumulation.
a) Amphiphilic moment (vsurf_A) positively correlates with accumulation in both P. aeruginosa and E. coli. b) Amine steric accessibility is important for accumulation in both P. aeruginosa and E. coli. c) Similarly, compounds with primary amines on secondary carbons tend to accumulate higher than compounds with primary amines on tertiary carbons. d) Matched molecular pairs show a correlation between amine pKa and accumulation in P. aeruginosa. For structures, see Supporting Table 8. The pKa estimation was calculated using the software MoKa (v3.2.2) from Molecular Discovery suite. e) Distribution of accumulation values in P. aeruginosa vs. E. coli for compounds that accumulate in both bacterial strains (131 compounds plotted, see Compound Master Table for structures). Accumulation levels were lower on average in P. aeruginosa PAO1 relative to E. coli MG1655. Accumulation units are reported in nmol/1012 CFUs. n=3 biologically independent samples. The s.e.m. is reported for accumulation values. Strains used: E. coli MG1655, P. aeruginosa PAO1.
Extended Data Fig. 4. Random forest prediction modeling.
a) Random forest prediction model results on data set of all 240 primary amines, structures reported in Supplementary Table 2. ROC plot with 10 repeated cross-validations in the training classification models. b) Relative importance of top 15 descriptors for all 240 primary amines. c) Random forest prediction model results on data set of 50 highest primary amine accumulators and 50 lowest primary amine accumulators (100 amines total; compounds 2.1–2.50; 2.191–2.240 in Supplementary Table 2). d) Relative importance of top 15 descriptors for 100 primary amines. e) Random forest prediction model results on data set of 30 highest primary amine accumulators and 30 lowest primary amine accumulators (60 amines total; compounds 2.1–2.30; 2.211–2.240 in Supplementary Table 2). f) Relative importance of top 15 descriptors for 60 primary amines. Formal charge (h_pavgQ) and hydrogen bond donor surface area (vsa_don) are boxed in red. Early iterations of the model with the incomplete compound library always identified these properties within the top 15 most important.
Extended Data Fig. 5. Influence of magnesium on accumulation, evaluation of its binding by NMR, and influence of magnesium and PMBN on compound accumulation.
a) Confocal fluorescent images of an intrinsically fluorescent compound, Bisindolylmaleimide X (BIM X) in P. aeruginosa with or without MgCl2. The standard accumulation assay was performed in P. aeruginosa (PAO1) with either DMSO, BIM X (50 μM), or BIM X (50 μM) + MgCl2 (1 mM), until just after the oil removal step. Cells were fixed in 3.7 % formaldehyde in PBS, and fluorescence images were taken with a Zeiss 710 multiphoton confocal microscope through a 63x/1.4 oil objective. All samples were excited at 488 nm with an argon laser, and fluorescence emission was recorded from 599–689 nm. Images were acquired using Zen Black (Zen 2.3). The images for P. aeruginosa + DMSO and P. aeruginosa + BIM X are the same as in Extended Data Fig. 1. DMSO controls indicate no autofluorescence. Images are representative of n = 3 biologically independent samples. b) Evaluation of accumulation of compounds in E. coli MG1655 upon co-treatment with MgCl2 (1 mM). c) Determination of Mg2+ interactions with polyamines. Ethylenediaminetetraacetic acid (EDTA), a compound known to chelate to Mg2+ in solution, clearly shows a marked chemical shift change in the NMR as well as changes in coupling when comparing EDTA alone to EDTA plus Mg2+. When this same experiment was performed with norspermine, there were no observable changes in coupling or chemical shift in the presence of MgCl2, suggesting a lack of perturbation of any electronic environment on the molecule, thus, a lack of interaction between Mg2+ and norspermine. Compounds were evaluated at a final concentration of 1 mM, and MgCl2 was at a final concentration of 20 mM to maintain the same Mg2+-to-compound ratio as in the accumulation experiments. The pD of all solutions was adjusted to 7.2 prior to analysis. d) The same set of compounds as in Extended Data Fig. 5b showed a statistically significant increase in accumulation in P. aeruginosa PAO1 upon co-treatment with the permeabilizer PMBN (8 μg/mL). All structures and accumulation values are listed in Supplementary Table 4b. For samples with additives in Extended Data Fig. 5b & d, compounds 23 and 35 did not meet the mass spec standards and are thus excluded from this analysis. n=3 biologically independent samples. The average and s.e.m are reported for accumulation values. Statistical significance was determined using a two-sample Welch’s t-test (one-tailed test, assuming unequal variance). Statistically significant accumulation differences for compounds in P. aeruginosa PA14 versus P. aeruginosa PA14 with PMBN treatment or E. coli MG1655 versus E. coli MG1655 with MgCl2 treatment are indicated with asterisks (n.s. not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Extended Data Fig. 6. Multiple amines and alternative positive charges often aid in accumulation in P. aeruginosa PAO1.
a) Diamines accumulate to significantly higher levels intracellularly in P. aeruginosa relative to their monoamine comparators. b) Diamines containing two primary amines consistently accumulate to a significant extent. c) Diamines containing one primary amine and one secondary or tertiary amine have more variable accumulation levels in P. aeruginosa, depending on hydrogen bond donor ability. d) Accumulation summary of 16 amine, guanidinium, and pyridinium containing compounds (48 compounds total, structures in Supplementary Table 5) in P. aeruginosa PAO1. Compounds are classified as accumulators or non-accumulators based on statistical significance relative to the negative antibiotic controls. e) Examples of side-by-side amine (A), guanidinium (G), and pyridinium (P) comparators and their relative accumulation values in P. aeruginosa PAO1. Amine and guanidinium comparators tend to accumulate to a very similar extent, while pyridiniums often do not accumulate to a significant extent. f) Accumulation of antibiotic controls in three P. aeruginosa strains. Accumulation is consistent with antibacterial activity reported in Extended Data Table 1c. g) Accumulation of representative non-antibiotics in three P. aeruginosa strains. While there is some variance in accumulation levels between strains, high concordance of accumulation classification was observed. Compounds are classified as accumulators or non-accumulators based on statistical significance relative to the negative antibiotic controls. Structures and accumulation values are reported in Supplementary Table 6. Accumulation units are reported in nmol/1012 CFUs. n=3 biologically independent samples. The average and s.e.m. are reported for accumulation values. Accumulation units are reported in nmol/1012 CFUs. All compounds were tested in biological triplicate. The average and s.e.m. are reported for accumulation values. Formal charge (FCH) was calculated using MOE.
Extended Data Fig. 7. Accumulation and activity of FA and various derivatives, and resistance generation in E. coli.
a) Introducing a hydrolyzable amidoxime linker onto FA provides a strategy to increase gram-negative activity and accumulation. Increasing the number of amines on the linker results in improved antibacterial activity and accumulation in two gram-negative species, E. coli MG1655 and P. aeruginosa PAO1, with the 4-amine linker-containing FA derivative (FA prodrug) demonstrating the most potent activity and highest absolute accumulation values. Accumulation values are reported in nmol/1012 CFUs and represent the concentration of free FA in the cell. n=3 biologically independent samples. The average and s.e.m. are reported for accumulation values. b) E. coli MG1655 mutants resistant to FA prodrug were generated using a serial passage method and exhibit amino acid mutations mapping back to the FA binding pocket of EF-G. Bacteria (1 × 108 CFU/mL) were first inoculated in a standard MIC experiment against both sub- and supra-MIC antibiotic concentrations. The well containing the highest concentration of FA prodrug where bacterial growth was still observed was then isolated and re-subjected to this experiment. c) Acetylating both alcohols of FA greatly disrupts target engagement of FA with EF-G as inferred from MIC values against S. aureus. The diacetylated version of FA prodrug (FA prodrug Ac2O) loses 4–16x activity against both gram-positive and gram-negative bacterial strains, suggesting that inhibition of EF-G contributes to the observed antibacterial activity of FA prodrug. MICs were performed in MH or LB broth per Clinical and Laboratory Standards Institute (CLSI) guidelines. Accumulation values are reported in nmol/1012 CFUs and represent the concentration of free FA in the cell. n=3 biologically independent samples. The average and s.e.m. are reported for accumulation values. * Indicates compound solubility limit.
Extended Data Fig. 8. Mode of uptake and membrane interactions of FA prodrug.
a) (RIGHT) In PBS at 37°C, FA prodrug (10 μg/mL) hydrolyzes to fusidic acid (FA) over a period of 48 hours, as monitored via LCMS. TIC LCMS traces showing the disappearance of FA prodrug and the appearance of FA. Traces are all normalized to 2E6 ion counts. (LEFT) Plasma stability, microsomal stability, and 72-hour IC50 values for FA and FA prodrug. Average percentage errors are reported as s.e.m. IC50 errors reported as s.d. n=3 biologically independent samples. b) Co-treatment with magnesium ions leads to a 32x increase in MIC for gentamicin and FA prodrug, while FA shows no change. Treatment with 80 mM NaCl shows minimal change when compared to the MgCl2 treated samples. MICs performed in P. aeruginosa PAO1 according to CLSI guidelines, and values are reported in μg/mL. n=3 biologically independent samples c) FA prodrug, gentamicin, and colistin all permeabilize the outer membrane of P. aeruginosa PAO1 to the membrane impermeable fluorophore NPN at a 10-minute time point, while treatment with FA shows no effect. Error bars represent the standard deviation from the average of RFUs. n=3 biologically independent samples d) Treatment with FA prodrug leads to dose-dependent inner membrane depolarization in P. aeruginosa PAO1, quantified using the potentiometric dye DiSC3 (5), while treatment with FA shows no effect. Concentrations of FA and FA prodrug are listed in μg/mL. 1% Triton X was used as the positive control, while 2% DMSO was used as the negative control. n=3 biologically independent samples.
Extended Data Table 1.
a) MICs of antibiotic controls used in the accumulation assay against P. aeruginosa PAO1 and with co-treatment of 8 μg/mL of PMBN. b) MIC ratios between P. aeruginosa PAO1 and P. aeruginosa PAΔ6 indicate whether a compound is subject to efflux. Vancomycin shows no potentiation in the efflux pump KO strain, indicating it is not liable to significant efflux, whereas chloramphenicol, valnemulin, trimethoprim, nalidixic acid, and tedizolid show substantial improvements in antibacterial activity in the efflux pump KO strain, indicating these compounds have considerable efflux liabilities. c) MICs of antibiotic controls in three different strains of P. aeruginosa. d) Many broad-spectrum antibacterials have limited efficacy against P. aeruginosa due to poor accumulation, not due to a lack of target engagement. In an efflux pump deficient strain of P. aeruginosa, PAΔ626, antibiotics demonstrate significantly improved activity compared to the wild-type (WT) strain P. aeruginosa PAO1, along with increased intracellular accumulation. MICs were performed in MH or LB broth per Clinical and Laboratory Standards Institute (CLSI) guidelines. Accumulation units are reported in nmol/1012 CFUs. The s.e.m. is reported for accumulation values.. n.v. = not viable. MICs were performed in MH or LB broth per Clinical and Laboratory Standards Institute (CLSI) guidelines. n=3 biologically independent samples
| a. | ||
|---|---|---|
|
| ||
| Compound | PA01 MIC (|jg/mL) | PA01 + PMBN MIC (|jg/mL) |
| Inactive antibiotics | ||
| Linezolid | >512 | n.d. |
| Vancomycin | >512 | n.d. |
| Erythromycin | 256 | n.d. |
| Fusidic acid | >512 | 4 |
| Vainemuiin | 256 | 1 |
| Novobiocin | >512 | 1 |
| Active antibiotics | ||
| Tetracycline | 16 | n.d. |
| Ciprofloxacin | 0.5 | n.d. |
| Tigecycline | 1 | n.d. |
| b. | |||
|---|---|---|---|
|
| |||
| Compound | PA01 MIC (pg/mL) | PAA6 MIC (Hg/mL) | Fold change |
| Poor Efflux Substrate | |||
| Vancomycin | >512 | >512 | 1 |
| Good Efflux Substrates | |||
| Chloramphenicol | >512 | 4 | >128 |
| Vainemuiin | 256 | 0.125 | 2048 |
| Trimethoprim | >512 | 2 | >256 |
| Nalidixic acid | >128 | 2 | >64 |
| Tedizolid | >256 | 16 | >16 |
| c. | |||
|---|---|---|---|
|
| |||
| Compound | PA01 MIC (ug/mL) | PA14 MIC (ug/mL) | PA1280 MIC (ug/mL) |
| Inactive antibiotics | |||
| Linezolid | >512 | >512 | >512 |
| Vancomycin | >512 | >512 | >512 |
| Erythromycin | 256 | 256 | 256 |
| Novobiocin | >512 | >512 | >512 |
| Active antibiotics | |||
| Tetracycline | 16 | 8 | 32 |
| Ciprofloxacin | 0.5 | 0.125 | 0.5 |
| Tigecycline | 1 | 2 | 16 |
| d. | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Compound | WT MIC (ug/mL) | PAA6 MIC (ug/mL) | Fold change | WT Accum | PAA6 Accum | Fold change |
| Chloramphenicol | >512 | 4 | >128 | 114 ± 14 | 537 ± 11 | 4.7 |
| Trimethoprim | >512 | 2 | >256 | 127 ±6 | 465 ±6 | 3.6 |
| Sulfamethoxazole | >64 | 4 | >16 | 101 ±26 | 420 ± 15 | 4.2 |
| Tetracycline | 16 | 0.25 | 64 | 452 ±3 | n.v. | - |
Extended Data Table 2.
Biological data on compounds included in the retrospective analysis comparing the properties of PA-active compounds versus less active derivatives. MICs reported in μg/mL. n.r. = not reported. * indicates that the MIC of compounds was determined in the E. coli ΔacrB efflux mutant instead of the ΔtolC efflux mutant.
| Structure | Compound ID | WT PA MIC | PA efflux KO MIC | WT EC MIC | AtolC EC MIC |
|---|---|---|---|---|---|
|
17x | 10 | n.r. | 0.81 | n.r. |
|
17r | 1.3 | n.r. | 0.31 | n.r. |
|
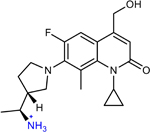
AMQ-1 (11 in original manuscript) | 16 | 4 | 16 | 0.25* |
|
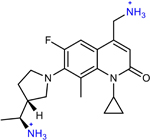
AMQ-2 (10 in original manuscript) | 2 | 0.5 | 1 | 0.06* |
|
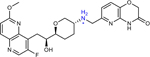
THP-1 (4 in original manuscript) | 4 | n.r. | 0.5 | 0.004 |
|
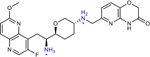
THP-2 (21 in original manuscript) | 1 | n.r. | 0.063 | 0.008 |

|
CHD | 16 | n.r. | n.r. | 0.5 |
|
CDCHD | 1 | n.r. | n.r. | 0.25 |
|
Tetracycline | 16 | 0.25 | 1 | 0.25 |
|
|
Tigecycline | 1 | 0.125 | 0.5 | 0.125 |
|
G8126 | >64 | n.r. | 32 | n.r. |

|
G0775 | 2 | n.r. | 0.125 | 0.063 |
Supplementary Material
Acknowledgements:
We thank the NIH (AI136773), the University of Illinois, and Roche for support of this work. M.K.G. is a member of the NIH Chemistry-Biology Interface Training Grant (T32-GM136629). We thank Lucas Li (Metabolomics Center, Roy J. Carver Biotechnology Center, UIUC and Duke University School of Medicine Proteomics and Metabolomics Core Facility) for all LC-MS/MS analysis. We thank Dean Olson, Lingyang Zhu, and Nikki Duay at the School of Chemical Sciences NMR Laboratory at UIUC for NMR services. We thank Austin Cyphersmith and the Core Facilities at the Carl Woese Institute for Genomic Biology for assistance with confocal imaging. We are grateful to all past Hergenrother lab members who have contributed compounds to the Complexity-to-Diversity collection, and to Dr. Bryon Drown, who wrote the code used for the random forest analysis. We thank Prof. Helen Zgurskaya for the generous donation of the P. aeruginosa PA01 Δ6 and PA01 Δ6-pore strains, and Prof. Dirk Bumann for the generous donation of the P. aeruginosa PA14 and PA14 Δ40 strain. We are further grateful to additional collaborators at Roche, including Christian Kramer who helped establish this collaboration early on, and Guillaume Marc Daniel for his guidance of the collaboration as alliance manager.
Footnotes
Competing Interests Statement: The University of Illinois has filed patents on some of the compounds described herein on which P.J.H. and M.G.C are inventors.
References:
- 1.Silver LL, A Gestalt approach to Gram-negative entry. Bioorg. Med. Chem. 2016, 24 (24), 6379–6389. [DOI] [PubMed] [Google Scholar]
- 2.Payne DJ; Gwynn MN; Holmes DJ; Pompliano DL, Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug. Discov. 2007, 6 (1), 29–40. [DOI] [PubMed] [Google Scholar]
- 3.Yoshimura F; Nikaido H, Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. J. Bacteriol. 1982, 152 (2), 636–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassetti M; Vena A; Croxatto A; Righi E; Guery B, How to manage Pseudomonas aeruginosa infections. Drugs Context 2018, 7, 212527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tommasi R; Iyer R; Miller AA, Antibacterial Drug Discovery: Some Assembly Required. ACS Infect. Dis. 2018, 4 (5), 686–695. [DOI] [PubMed] [Google Scholar]
- 6.Tamber S; Ochs MM; Hancock RE, Role of the novel OprD family of porins in nutrient uptake in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188 (1), 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aeschlimann JR, The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other gram-negative bacteria. Insights from the Society of Infectious Diseases Pharmacists. Pharmacotherapy 2003, 23 (7), 916–24. [DOI] [PubMed] [Google Scholar]
- 8.Krishnamoorthy G; Leus IV; Weeks JW; Wolloscheck D; Rybenkov VV; Zgurskaya HI, Synergy between Active Efflux and Outer Membrane Diffusion Defines Rules of Antibiotic Permeation into Gram-Negative Bacteria. mBio 2017, 8 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falagas ME; Kasiakou SK, Toxicity of polymyxins: a systematic review of the evidence from old and recent studies. Crit. Care 2006, 10 (1), R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mingeot-Leclercq MP; Tulkens PM, Aminoglycosides: nephrotoxicity. Antimicrob. Agents Chemother. 1999, 43 (5), 1003–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richter MF; Hergenrother PJ, The challenge of converting Gram-positive-only compounds into broad-spectrum antibiotics. Ann. N. Y. Acad. Sci. 2019, 1435 (1), 18–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zgurskaya HI; Rybenkov VV, Permeability barriers of Gram-negative pathogens. Ann. N. Y. Acad. Sci. 2020, 1459 (1), 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter MF; Drown BS; Riley AP; Garcia A; Shirai T; Svec RL; Hergenrother PJ, Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 2017, 545 (7654), 299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motika SE; Ulrich RJ; Geddes EJ; Lee HY; Lau GW; Hergenrother PJ, A Gram-Negative Antibiotic Active Through Inhibition of an Essential Riboswitch. J. Am. Chem. Soc. 2020, 142 (24), 10856–10862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker EN; Drown BS; Geddes EJ; Lee HY; Ismail N; Lau GW; Hergenrother PJ, Implementation of permeation rules leads to a FabI inhibitor with activity against Gram-negative pathogens. Nat. Microbiol. 2020, 5 (1), 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y; Shi H; Zhou M; Ren Q; Zhu W; Zhang W; Zhang Z; Zhou C; Liu Y; Ding X; Shen HC; Yan SF; Dey F; Wu W; Zhai G; Zhou Z; Xu Z; Ji Y; Lv H; Jiang T; Wang W; Xu Y; Vercruysse M; Yao X; Mao Y; Yu X; Bradley K; Tan X, Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV. J. Med. Chem. 2020, 63 (17), 9623–9649. [DOI] [PubMed] [Google Scholar]
- 17.Andrews LD; Kane TR; Dozzo P; Haglund CM; Hilderbrandt DJ; Linsell MS; Machajewski T; McEnroe G; Serio AW; Wlasichuk KB; Neau DB; Pakhomova S; Waldrop GL; Sharp M; Pogliano J; Cirz RT; Cohen F, Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase. J. Med. Chem. 2019, 62 (16), 7489–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukežič T; Fayad AA; Bader C; Harmrolfs K; Bartuli J; Groß S; Lešnik U; Hennessen F; Herrmann J; Pikl Š; Petković H; Müller R, Engineering Atypical Tetracycline Formation in Amycolatopsis sulphurea for the Production of Modified Chelocardin Antibiotics. ACS Chem. Biol. 2019, 14 (3), 468–477. [DOI] [PubMed] [Google Scholar]
- 19.Skepper CK; Armstrong D; Balibar CJ; Bauer D; Bellamacina C; Benton BM; Bussiere D; De Pascale G; De Vicente J; Dean CR; Dhumale B; Fisher LM; Fuller J; Fulsunder M; Holder LM; Hu C; Kantariya B; Lapointe G; Leeds JA; Li X; Lu P; Lvov A; Ma S; Madhavan S; Malekar S; McKenney D; Mergo W; Metzger L; Moser HE; Mutnick D; Noeske J; Osborne C; Patel A; Patel D; Patel T; Prajapati K; Prosen KR; Reck F; Richie DL; Rico A; Sanderson MR; Satasia S; Sawyer WS; Selvarajah J; Shah N; Shanghavi K; Shu W; Thompson KV; Traebert M; Vala A; Vala L; Veselkov DA; Vo J; Wang M; Widya M; Williams SL; Xu Y; Yue Q; Zang R; Zhou B; Rivkin A, Topoisomerase Inhibitors Addressing Fluoroquinolone Resistance in Gram-Negative Bacteria. J. Med. Chem. 2020, 63 (14), 7773–7816. [DOI] [PubMed] [Google Scholar]
- 20.Brem J; Panduwawala T; Hansen JU; Hewitt J; Liepins E; Donets P; Espina L; Farley AJM; Shubin K; Campillos GG; Kiuru P; Shishodia S; Krahn D; Lesniak RK; Schmidt Adrian J; Calvopina K; Turrientes MC; Kavanagh ME; Lubriks D; Hinchliffe P; Langley GW; Aboklaish AF; Eneroth A; Backlund M; Baran AG; Nielsen EI; Speake M; Kuka J; Robinson J; Grinberga S; Robinson L; McDonough MA; Rydzik AM; Leissing TM; Jimenez-Castellanos JC; Avison MB; Da Silva Pinto S; Pannifer AD; Martjuga M; Widlake E; Priede M; Hopkins Navratilova I; Gniadkowski M; Belfrage AK; Brandt P; Yli-Kauhaluoma J; Bacque E; Page MGP; Bjorkling F; Tyrrell JM; Spencer J; Lang PA; Baranczewski P; Canton R; McElroy SP; Jones PS; Baquero F; Suna E; Morrison A; Walsh TR; Schofield CJ, Imitation of beta-lactam binding enables broad-spectrum metallo-beta-lactamase inhibitors. Nat. Chem. 2022, 14 (1), 15–24. [DOI] [PubMed] [Google Scholar]
- 21.Schumacher CE; Rausch M; Greven T; Neudörfl J-M; Schneider T; Schmalz H-G, Total Synthesis and Antibiotic Properties of Amino-Functionalized Aromatic Terpenoids Related to Erogorgiaene and the Pseudopterosins. Eur. J. Org. Chem. 2022, e202200058. [Google Scholar]
- 22.Parker EN; Cain BN; Hajian B; Ulrich RJ; Geddes EJ; Barkho S; Lee HY; Williams JD; Raynor M; Caridha D; Zaino A; Shekhar M; Muñoz KA; Rzasa KM; Temple ER; Hunt D; Jin X; Vuong C; Pannone K; Kelly AM; Mulligan MP; Lee KK; Lau GW; Hung DT; Hergenrother PJ, An Iterative Approach Guides Discovery of the FabI Inhibitor Fabimycin, a Late-Stage Antibiotic Candidate with In Vivo Efficacy against Drug-Resistant Gram-Negative Infections. ACS Cent. Sci. 2022, 8 (9), 1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang K-J; Pantua H; Diao J; Skippington E; Volny M; Sandoval W; Tiku V; Peng Y; Sagolla M; Yan D; Kang J; Katakam AK; Michaelian N; Reichelt M; Tan M-W; Austin CD; Xu M; Hanan E; Kapadia SB, Deletion of a previously uncharacterized lipoprotein lirL confers resistance to an inhibitor of type II signal peptidase in Acinetobacter baumannii. Proc. Natl. Acad. Sci. U.S.A. 2022, 119 (38), e2123117119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onyedibe KI; Nemeth AM; Dayal N; Smith RD; Lamptey J; Ernst RK; Melander RJ; Melander C; Sintim HO, Re-sensitization of Multidrug-Resistant and Colistin-Resistant Gram-Negative Bacteria to Colistin by Povarov/Doebner-Derived Compounds. ACS Infect. Dis. 2023, 9 (2), 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goethe O; DiBello M; Herzon SB, Total synthesis of structurally diverse pleuromutilin antibiotics. Nat. Chem. 2022, 14 (11), 1270–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper CJ; Krishnamoorthy G; Wolloscheck D; Walker JK; Rybenkov VV; Parks JM; Zgurskaya HI, Molecular Properties That Define the Activities of Antibiotics in Escherichia coli and Pseudomonas aeruginosa. ACS Infect. Dis. 2018, 4 (8), 1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehla J; Malloci G; Mansbach R; Lopez CA; Tsivkovski R; Haynes K; Leus IV; Grindstaff SB; Cascella RH; D’Cunha N; Herndon L; Hengartner NW; Margiotta E; Atzori A; Vargiu AV; Manrique PD; Walker JK; Lomovskaya O; Ruggerone P; Gnanakaran S; Rybenkov VV; Zgurskaya HI, Predictive Rules of Efflux Inhibition and Avoidance in Pseudomonas aeruginosa. mBio 2021, 12 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leus Inga V; Adamiak J; Chandar B; Bonifay V; Zhao S; Walker Scott S; Squadroni B; Balibar Carl J; Kinarivala N; Standke Lisa C; Voss Henning U; Tan Derek S; Rybenkov Valentin V; Zgurskaya Helen I, Functional Diversity of Gram-Negative Permeability Barriers Reflected in Antibacterial Activities and Intracellular Accumulation of Antibiotics. Antimicrob. Agents Chemother. 2023, 67 (2), e01377–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geddes EJ; Li Z; Hergenrother PJ, An LC-MS/MS assay and complementary web-based tool to quantify and predict compound accumulation in E. coli. Nat. Protoc. 2021, 16 (10), 4833–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallace MJ; Dharuman S; Fernando DM; Reeve SM; Gee CT; Yao J; Griffith EC; Phelps GA; Wright WC; Elmore JM; Lee RB; Chen T; Lee RE, Discovery and Characterization of the Antimetabolite Action of Thioacetamide-Linked 1,2,3-Triazoles as Disruptors of Cysteine Biosynthesis in Gram-Negative Bacteria. ACS Infect. Dis. 2020, 6 (3), 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaara M, Agents that increase the permeability of the outer membrane. Microbiol. Rev. 1992, 56 (3), 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huigens RW 3rd; Morrison KC; Hicklin RW; Flood TA Jr.; Richter MF; Hergenrother PJ, A ring-distortion strategy to construct stereochemically complex and structurally diverse compounds from natural products. Nat. Chem. 2013, 5 (3), 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perlmutter SJ; Geddes EJ; Drown BS; Motika SE; Lee MR; Hergenrother PJ, Compound Uptake into E. coli Can Be Facilitated by N-Alkyl Guanidiniums and Pyridiniums. ACS Infect. Dis. 2021, 7, 162–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hancock RE; Woodruff WA, Roles of porin and beta-lactamase in beta-lactam resistance of Pseudomonas aeruginosa. Rev. Infect. Dis. 1988, 10 (4), 770–5. [DOI] [PubMed] [Google Scholar]
- 35.Ude J; Tripathi V; Buyck JM; Soderholm S; Cunrath O; Fanous J; Claudi B; Egli A; Schleberger C; Hiller S; Bumann D, Outer membrane permeability: Antimicrobials and diverse nutrients bypass porins in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 2021, 118 (31). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loh B; Grant C; Hancock RE, Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1984, 26 (4), 546–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hancock RE; Farmer SW, Mechanism of uptake of deglucoteicoplanin amide derivatives across outer membranes of Escherichia coli and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1993, 37 (3), 453–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kung VL; Ozer EA; Hauser AR, The accessory genome of Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2010, 74 (4), 621–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mikkelsen H; McMullan R; Filloux A, The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS One 2011, 6 (12), e29113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams JJ; Halvorsen EM; Dwyer EM; DiFazio RM; Hergenrother PJ, Toxin-antitoxin (TA) systems are prevalent and transcribed in clinical isolates of Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 2011, 322 (1), 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surivet JP; Zumbrunn C; Bruyere T; Bur D; Kohl C; Locher HH; Seiler P; Ertel EA; Hess P; Enderlin-Paput M; Enderlin-Paput S; Gauvin JC; Mirre A; Hubschwerlen C; Ritz D; Rueedi G, Synthesis and Characterization of Tetrahydropyran-Based Bacterial Topoisomerase Inhibitors with Antibacterial Activity against Gram-Negative Bacteria. J. Med. Chem. 2017, 60 (9), 3776–3794. [DOI] [PubMed] [Google Scholar]
- 42.Sum PE; Lee VJ; Testa RT; Hlavka JJ; Ellestad GA; Bloom JD; Gluzman Y; Tally FP, Glycylcyclines. 1. A new generation of potent antibacterial agents through modification of 9-aminotetracyclines. J. Med. Chem. 1994, 37 (1), 184–8. [DOI] [PubMed] [Google Scholar]
- 43.Smith PA; Koehler MFT; Girgis HS; Yan D; Chen Y; Chen Y; Crawford JJ; Durk MR; Higuchi RI; Kang J; Murray J; Paraselli P; Park S; Phung W; Quinn JG; Roberts TC; Rougé L; Schwarz JB; Skippington E; Wai J; Xu M; Yu Z; Zhang H; Tan M-W; Heise CE, Optimized arylomycins are a new class of Gram-negative antibiotics. Nature 2018, 561 (7722), 189–194. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka N; Kinoshita T; Masukawa H, Mechanism of protein synthesis inhibition by fusidic acid and related antibiotics. Biochem. Biophys. Res. Commun. 1968, 30 (3), 278–83. [DOI] [PubMed] [Google Scholar]
- 45.Garcia Chavez M; Garcia A; Lee HY; Lau GW; Parker EN; Komnick KE; Hergenrother PJ, Synthesis of Fusidic Acid Derivatives Yields a Potent Antibiotic with an Improved Resistance Profile. ACS Infect. Dis. 2021, 7 (2), 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haloi N; Vasan AK; Geddes EJ; Prasanna A; Wen PC; Metcalf WW; Hergenrother PJ; Tajkhorshid E, Rationalizing the generation of broad spectrum antibiotics with the addition of a positive charge. Chem. Sci. 2021, 12 (45), 15028–15044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durand-Reville TF; Miller AA; O’Donnell JP; Wu X; Sylvester MA; Guler S; Iyer R; Shapiro AB; Carter NM; Velez-Vega C; Moussa SH; McLeod SM; Chen A; Tanudra AM; Zhang J; Comita-Prevoir J; Romero JA; Huynh H; Ferguson AD; Horanyi PS; Mayclin SJ; Heine HS; Drusano GL; Cummings JE; Slayden RA; Tommasi RA, Rational design of a new antibiotic class for drug-resistant infections. Nature 2021, 597 (7878), 698–702. [DOI] [PubMed] [Google Scholar]
- 48.Llanes C; Hocquet D; Vogne C; Benali-Baitich D; Neuwirth C; Plesiat P, Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 2004, 48 (5), 1797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skinner SO; Sepulveda LA; Xu H; Golding I, Measuring mRNA copy number in individual Escherichia coli cells using single-molecule fluorescent in situ hybridization. Nat Protoc 2013, 8 (6), 1100–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and the Supplementary Information. Source data is provided for all Figures and Extended Data Figures.