Abstract
Translation of mRNA encoding the L1 and L2 capsid proteins of papillomavirus (PV) is restricted in vivo to differentiated epithelial cells, although transcription of the L1 and L2 late genes occurs more widely. The codon composition of PV late genes is quite different from that of most mammalian genes. To test the possibility that PV late gene codon composition determines the efficiency of PV late gene expression in some cell types, synthetic bovine papillomavirus type 1 (BPV1) late genes were constructed with codon composition modified to resemble the typical mammalian gene. Expression of these genes from a strong promoter in Cos-1 cells was compared with expression of wild-type BPV1 late genes from the same promoter. Both unmodified and modified PV late genes were transcribed in Cos-1 cells, but only the codon-modified genes were translated. In vitro translation of wild-type but not synthetic BPV1 L1 mRNA was markedly enhanced by addition of aminoacyl-tRNAs. Codon composition thus limits BPV1 late gene translation in Cos-1 cells, and this limitation can be overcome by modification of the codon composition of the genes or by provision of excess tRNA. Replacement of codons in the green fluorescent protein (gfp) gene with those frequently used in PV late genes did not alter gfp transcription in Cos-1 cells but almost abolished translation, supporting the hypothesis that the observed differences in efficiency of translation of modified and unmodified PV capsid genes were related to codon usage rather than mRNA structure. As tRNA populations vary within and between tissues in the same eukaryotic organism, we speculate that matching of tRNA availability to codon usage may be one determinant of the restriction of expression of PV late genes to differentiated epithelium.
Papillomaviruses (PV) infect stratified squamous epithelia, and PV gene expression is linked to the state of differentiation of the epithelial cells. Early genes associated with plasmid replication and with regulation of host cell division and differentiation are expressed at low levels in the replicating basal cells of squamous epithelia, while the genes encoding immunogenic capsid proteins (L1 and L2) are transcribed predominantly in nonreplicating differentiated keratinocytes, and their protein product can be detected only in the most superficial layers of the epithelium (31). Transient translation of these genes follows infection of eukaryotic cells with viral vectors including vaccinia virus (10, 39) and baculovirus (18, 34), and stable transcription and translation of PV late genes from strong constitutive promoters can occur in yeast (14, 24). However, stable expression of authentic sequence late genes from plasmid eukaryotic expression vectors, as seen in yeast, has not been reported for replicating eukaryotic cells in vitro, although expression in nonreplicating differentiated cells in raft culture can be observed (22). Further, L1 translation is lost in vivo as cells dedifferentiate (31, 37). Lack of PV late gene expression in undifferentiated cells has been variously attributed to putative differentiation-specific transcriptional enhancers for late promoters (2, 9), to alternate message splicing in differentiated tissues (3), to message-processing sequences (16) or AU-rich instability-promoting sequences (7, 30) in the 3′ untranslated sequence of L1 mRNA, to RNA sequences within the late genes inhibitory to transcription (8, 25), or to transcriptional repressor proteins (33). However, these proposed mechanisms of regulation of PV late gene transcription or translation could not explain the absence of L1 protein observed in some tissues with abundant L1 mRNA (23).
The genetic code shows redundancy: several nucleotide triplets can encode the same amino acid, and several corresponding isoaccepting tRNAs are observed. Differences in the abundance of isoaccepting tRNAs are observed between organisms, and also between the tissues of a given organism, and are postulated as one explanation for why the same gene is more or less efficiently translated in different organisms (27). Virus genomes frequently have a G+C content significantly different from that of their host species, reflecting a different codon usage pattern of unknown significance (32). Analysis of codon usage in the PV genome shows that bovine papillomavirus type 1 (BPV1) late genes use 12 codons more than twice as frequently than the average for mammalian genes (Table 1). Similar observations apply to the late genes of other PV genotypes. We therefore hypothesize that as tRNA availability in a particular species is assumed to reflect codon usage in genes of that species (13, 27), PV late mRNAs may not be efficiently translated in undifferentiated cells in vitro or in vivo due to a mismatch of codon usage and tRNA availability. If this hypothesis were true, two alternate mechanisms could be proposed for the resolution in differentiated cells of the codon usage-tRNA mismatch in undifferentiated cells. tRNA availability might change with keratinocyte differentiation, or a reduction in the range and number of mRNAs competing for the rare tRNAs might allow more effective translation. This latter hypothesis could also explain why PV late gene expression can be induced in cell cultures infected with recombinant virus, such as vaccinia virus or baculovirus (10, 34), in which the virus infection switches off host mRNA synthesis. If our hypotheses are correct, conservative replacement of these codons in PV late genes less frequently used in mammalian genes should result in a gene encoding mRNA which can be efficiently expressed in dividing cells in culture. We therefore produced synthetic versions of BPV1 L1 and L2 genes, substituting codons preferentially used by mammalian genes for the codons in the original BPV1 L1 and L2 sequences which are less commonly used in mammalian genes (creating so-called humanized genes). We used these synthetic variants to examine the efficiency of transcription and translation of modified and unmodified BPV1 late genes expressed from strong constitutive and natural promoters in cultured cells in vitro. In further testing of the hypothesis, we examined whether replacing the major codons of gfp (encoding green fluorescent protein) with synonymous PV-preferred codons would reduce the level of translation of gfp mRNA in cultured cells.
TABLE 1.
Frequency of codon usage for individual organismsa
| Amino acid | Codon | Frequency (10−3) of codon usage
|
||||
|---|---|---|---|---|---|---|
| Human | Cow | Yeast | Wheat | BPV L1 and L2 | ||
| Arg | CGA | 5.4 | 5.5 | 2.3 | 2.3 | 7.2 |
| CGC | 11.3 | 12.2 | 2.0 | 7.5 | 4.1 | |
| CGG | 10.4 | 11.2 | 1.1 | 4.6 | 5.1 | |
| CGU | 4.7 | 3.7 | 7.5 | 1.1 | 10.4 | |
| AGA | 9.9 | 9.9 | 24.0 | 4.1 | 14.4 | |
| AGG | 11.1 | 11.4 | 7.5 | 7.1 | 9.3 | |
| Leu | CUA | 6.2 | 4.9 | 11.8 | 12.1 | 18.6 |
| CUC | 19.9 | 21.2 | 4.1 | 18.6 | 6.2 | |
| CUG | 42.5 | 46.6 | 8.3 | 15.5 | 15.5 | |
| CUU | 10.7 | 10.6 | 9.6 | 6.5 | 20.7 | |
| UUA | 5.3 | 4.0 | 24.5 | 1.8 | 14.5 | |
| UUG | 11.0 | 9.6 | 32.1 | 15.3 | 15.5 | |
| Ser | UCA | 9.3 | 7.6 | 15.6 | 14.6 | 16.6 |
| UCC | 17.7 | 17.6 | 14.4 | 10.1 | 11.4 | |
| UCG | 4.2 | 4.5 | 6.5 | 9.6 | 6.2 | |
| UCU | 13.2 | 11.2 | 24.6 | 14.8 | 15.5 | |
| AGC | 18.7 | 18.7 | 7.1 | 12.8 | 12.4 | |
| AGU | 9.4 | 8.6 | 11.7 | 12.9 | 21.7 | |
| Thr | ACA | 14.4 | 11.4 | 15.6 | 4.6 | 37.3 |
| ACC | 23.0 | 21.1 | 13.9 | 15.9 | 19.7 | |
| ACG | 6.7 | 7.8 | 6.7 | 4.5 | 4.1 | |
| ACU | 12.7 | 9.6 | 22.0 | 11.8 | 28.0 | |
| Pro | CCA | 14.6 | 12.0 | 21.4 | 71.2 | 22.8 |
| CCC | 20.0 | 19.2 | 5.9 | 11.1 | 15.5 | |
| CCG | 6.5 | 7.9 | 4.1 | 19.4 | 0.0 | |
| CCU | 15.5 | 14.6 | 12.8 | 10.3 | 33.1 | |
| Ala | GCA | 14.0 | 13.1 | 15.3 | 11.2 | 33.1 |
| GCC | 29.1 | 35.8 | 15.5 | 19.5 | 17.6 | |
| GCG | 7.2 | 9.3 | 5.1 | 13.8 | 4.1 | |
| GCU | 19.6 | 19.1 | 28.3 | 9.6 | 13.5 | |
| Gly | GGA | 17.1 | 16.2 | 8.9 | 25.9 | 22.8 |
| GGC | 25.4 | 28.1 | 8.9 | 28.0 | 12.4 | |
| GGG | 17.3 | 19.2 | 5.1 | 28.5 | 22.8 | |
| GGU | 11.2 | 11.8 | 34.9 | 9.6 | 18.6 | |
| Val | GUA | 5.9 | 5.1 | 10.0 | 4.4 | 15.5 |
| GUC | 16.3 | 18.4 | 14.9 | 14.8 | 6.2 | |
| GUG | 30.9 | 32.9 | 9.5 | 12.9 | 23.8 | |
| GUU | 10.4 | 9.9 | 26.6 | 11.6 | 16.6 | |
| Lys | AAA | 22.2 | 21.6 | 37.7 | 4.5 | 37.2 |
| AAG | 34.9 | 37.1 | 35.2 | 17.4 | 13.5 | |
| Asn | AAC | 22.6 | 22.4 | 25.8 | 14.2 | 10.3 |
| AAU | 16.6 | 12.5 | 31.4 | 6.7 | 24.8 | |
| Gln | CAA | 11.1 | 9.7 | 29.8 | 171.8 | 22.8 |
| CAG | 33.6 | 34.4 | 10.4 | 79.4 | 17.6 | |
| His | CAC | 14.2 | 14.0 | 8.2 | 8.2 | 6.2 |
| CAU | 9.3 | 7.5 | 12.3 | 7.1 | 13.4 | |
| Glu | GAA | 26.8 | 24.4 | 48.9 | 7.8 | 36.2 |
| GAG | 41.4 | 45.4 | 16.9 | 19.7 | 21.7 | |
| Asp | GAC | 29.0 | 31.5 | 22.3 | 13.0 | 18.6 |
| GAU | 21.7 | 19.2 | 37.0 | 4.0 | 33.1 | |
| Tyr | UAC | 18.8 | 20.3 | 16.5 | 24.5 | 17.6 |
| UAU | 12.5 | 10.5 | 16.5 | 12.5 | 18.6 | |
| Cys | UGC | 14.5 | 13.9 | 3.7 | 14.8 | 5.2 |
| UGU | 9.9 | 9.4 | 7.6 | 4.9 | 5.2 | |
| Phe | UUC | 22.6 | 25.5 | 20.0 | 14.1 | 7.2 |
| UUU | 15.8 | 17.0 | 23.2 | 15.0 | 23.8 | |
| Ile | AUA | 5.8 | 5.2 | 12.8 | 5.4 | 22.7 |
| AUC | 24.3 | 25.8 | 18.4 | 19.7 | 8.2 | |
| AUU | 14.9 | 13.1 | 31.1 | 10.7 | 20.7 | |
The codon usage data for human, cow, yeast, and wheat proteins are derived from published results (20). The BPV1 data are from the sequences in the GenBank database. Usage of triplets in boldface is more than twice as frequently observed in PV L1 and L2 genes as mammalian consensus codons.
MATERIALS AND METHODS
Codon replacements in the BPV L1, L2, and gfp genes.
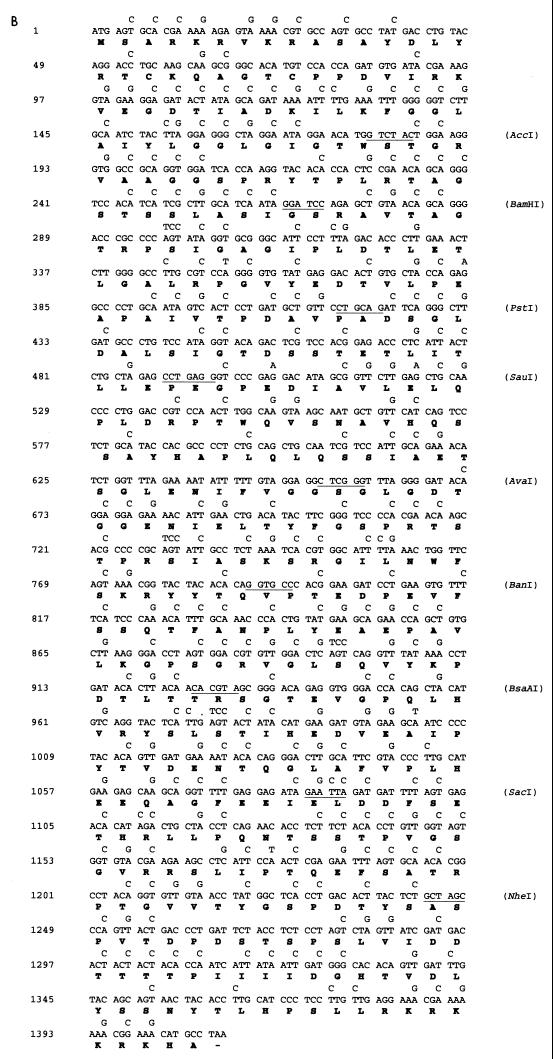
The DNA and amino acid sequences of the L1 and L2 genes are shown in Fig. 1. To determine whether the presence in the L1 and L2 genes of codons less frequently used in mammalian genes (Table 1) could in part explain the reduced translation of the L1 and L2 genes in some mammalian cells, we synthesized codon-modified L1 and L2 genes by using synonymous substitutions as shown. To construct the synthetic sequences, we synthesized 11 pairs of oligonucleotides for L1 and 10 pairs of oligonucleotides for L2. Each pair of oligonucleotides has restriction sites incorporated to facilitate subsequent cloning (Fig. 1). The degenerate oligonucleotides were used to amplify L1 and L2 sequences by PCR using a plasmid with the BPV1 genome as the template. The amplified fragments were cut with appropriate enzymes and sequentially ligated to the pUC18 vector, producing pUCHBL1 and pUCHBL2. The synthetic L1 and L2 sequences were sequenced and found to be error free; they were then subcloned into the mammalian expression vector pCDNA3 containing simian virus 40 (SV40) ori (Invitrogen), giving expression plasmids pCDNA/HBL1 and pCDNA/HBL2. To compare expression of L1 and L2 with that of the original sequences, the wild-type (wt) L1 and L2 genes were cloned into the pCDNA3 vector, resulting in pCDNA/BPVL1wt and pCDNA/BPVL2wt. To construct Pgfp, a modified gfp gene with PV-preferred codons, six pairs of oligonucleotides were synthesized. Each pair of oligonucleotides has restriction sites incorporated and was used to amplify gfp by using a humanized gfp gene (GIBCO) as the template. The PCR fragments were ligated into the pUC18 vector to produce pUCPGFP. The Pgfp gene was sequenced and cloned into the BamHI site of the same mammalian expression vector, pCDNA3, under the cytomegalovirus (CMV) promoter.
FIG. 1.
Nucleotide sequences of BPV1 L1 (A), L2 ORF (B) and gfp (C) and deduced amino acid sequences. Amino acids (in single-letter code) are presented below the second nucleotide of each codon. Mutations introduced into the genes are indicated above the corresponding nucleotides of the original sequence. The sites and enzymes used for cloning are indicated by horizontal lines. This replacement of nucleotides resulted in a polynucleotide sequence encoding BPV1 L1, L2, and GFP polypeptides with amino acid sequences identical to the original, but having preferred (for L1 and L2 genes) or PV (for the gfp gene) codons substituted for the synonymous codons of the wt nucleotide sequences.
To construct plasmids with the L2 gene of BPV1 in the context of the BPV1 sequence and with only BPV1 promoters, the BPV genome was cleaved at nucleotides (nt) 4450 and 6958 with BamHI/HindIII and cloned into pUC18. In the resultant plasmid, the original L1 (nt 4186 to 5595) and L2 (nt 5068 to 7095) open reading frames (ORFs) were removed. The synthetic humanized L2 gene, together with an SV40 poly(A) sequence to allow mRNA processing and the SV40 ori sequence to allow plasmid replication in eukaryotic cells, was cleaved from pCDNAHBL2 with HindIII/SmaI and inserted into the BamHI-cleaved and Klenow-blunted BPV genome lacking L1 and L2 ORF sequences. This plasmid was designated pCICR2. A similar plasmid was constructed with wt rather than synthetic humanized L2 and designated pCICR1.
Immunofluorescence and Western blot staining.
For immunoblotting assays, Cos-1 cells in six-well plates were transfected with 2 μg of L1 or L2 expression plasmid by using Lipofectamine (Gibco); 35 h after transfection, cells were washed with phosphate-buffered 0.15 M NaCl, pH 7.4 (PBS), and lysed in sodium dodecyl sulfate (SDS) loading buffer. The cellular proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) on a 10% gel blotted onto a nitrocellulose membrane. The L1 and L2 proteins were identified by enhanced chemiluminescence (Amersham, Little Chalfont, England), using BPV1 L1-specific (DAKO) or L2-specific (19) antiserum. For immunofluorescent staining, Cos-1 cells were grown on eight-chamber slides, transfected with plasmids, and fixed and permeabilized with 85% ethanol 36 h after transfection. The slides were blocked with 5% milk–PBS and probed with L1- or L2-specific antiserum, followed by fluorescein isothiocyanate (FITC)-conjugated anti-rabbit immunoglobulin G (IgG) (Sigma). For gfp or Pgfp plasmid-transfected cells, the cells were fixed with 4% buffered formaldehyde and viewed by epifluorescence microscopy. Data presented are representative of at least three experiments for each transfection.
In vitro translation assay.
One microgram of each plasmid was incubated with 20 μCi of [35S]methionine (Amersham) and 40 μl of T7-DNA polymerase coupled rabbit reticulocyte or wheat germ lysates (Promega). Translation was performed at 30°C and stopped by adding SDS loading buffer. The L1 proteins were separated by SDS-PAGE on a 10% gel and examined by autoradiography.
Northern blotting.
Cytoplasmic or total RNA was extracted from Cos-1 cells, transfected 24 h previously with various plasmids, using a Qiagen RNeasy mini kit as instructed by the supplier. Briefly, for cytoplasmic RNA purification, buffer RLN (50 mM Tris [pH 8.0], 140 mM NaCl, 1.5 mM MgCl2, and 0.5% Nonidet P-40) was directly added to monolayer cells, and cells were lysed at 4°C for 5 min. After the nuclei were removed by centrifugation, cytoplasmic RNAs were purified by column. For total RNA extraction, the monolayer cells were lysed in buffer RLT supplied with the kit, and RNA was purified on a spin column. Purified RNA (10 μg per lane) was separated on a 1.5% agarose gel in the presence of formaldehyde. The RNAs were then blotted onto nylon membrane and probed with (i) 1:1 mixed 5′-end-labeled L1 wt and HBL1 fragments, (ii) 1:1 mixed 5′-end-labeled L2 wt and HBL2 fragments, (iii) 1:1 mixed 5′-end-labeled gfp and Pgfp fragments, or (iv) randomly labeled glyceraldehyde-3-phosphate dehydrogenase GAPDH fragment. The blots were washed extensively at 65°C and exposed to X-ray films for 3 days.
Production of aminoacyl-tRNA.
tRNA (2.5 × 10−4 M; Boehringer) was added to a 20-μl reaction mixture containing 10 mM Tris-acetate (pH 7.8), 44 mM KCl, 12 mM MgCl2, 9 mM β-mercaptoethanol, 38 mM ATP, 0.25 mM GTP, and 7 μl of rabbit reticulocyte extract. The reaction was carried out at 25°C for 20 min, and 30 μl of H2O was added to the reaction to dilute the tRNAs to 10−4 M. The aminoacyl-tRNAs were then aliquoted and stored at −70°C.
RESULTS
Expression of synthetic L1 and L2 protein in cultured cells.
To test the hypothesis that the codon composition of the genes encoding the L1 and L2 capsid proteins of PV contributes to their restricted expression in undifferentiated epithelial cells, we produced synthetic BPV1 L1 and L2 genes, substituting codons preferentially used in mammalian genes for the codons frequently present in the wt BPV1 L1 and L2 sequences (Fig. 1). Codons where the ratio of usage in BPV1 late genes to mammalian genes was >2, shown in boldface in Table 1, were replaced; additionally, most codons where the ratio was >1.5 were replaced. For the L1 gene, a total of 202 base substitutions were made in 196 codons, without changing the encoded amino acid sequence (Fig. 1A). This synthetic humanized BPV L1 gene was designated HBL1. In a similarly modified BPV1 L2 gene designated HBL2, 303 bases were changed to replace 290 less frequently used codons with the corresponding preferentially used codons. Using the synthetic HBL1 and HBL2 genes, we constructed two eukaryotic expression plasmids based on pCDNA3, designated pCDNA/HBL1 and pCDNA/HBL2, respectively. Similar expression plasmids, constructed with the wt BPV1 L1 and BPV1 L2 genes, were designated pCDNA/BPVL1wt and pCDNA/BPVL2wt, respectively. In each of these plasmids, the SV40 ori allowed replication in Cos-1 cells, and the L1 or L2 gene was driven by a strong constitutive CMV promoter.
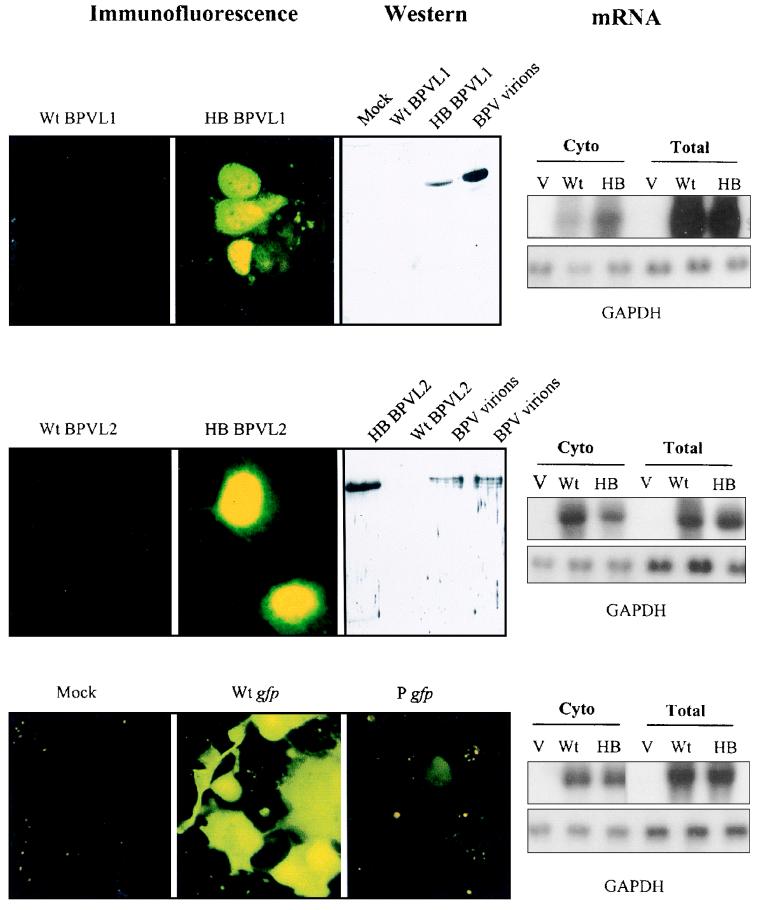
To compare the expression of the synthetic humanized and wt BPV1 L1 and BPV1 L2 genes, we separately transfected Cos-1 cells with each of the L1 and L2 plasmids described above. Transfected cells were analyzed for expression of L1 or L2 protein by immunofluorescence 36 h after transfection (Fig. 2). Cells transfected with the pCDNA3 expression plasmid containing the synthetic humanized L1 or L2 gene produced large amounts of the corresponding protein. About 80% of cells showed significant L1 or L2 staining, within the cell nucleus as expected from the nuclear localization of these proteins (38). Cells transfected with expression plasmids with the wt L1 or L2 sequences produced no detectable L1 or L2 protein (Fig. 2). To compare more accurately the expression of the different L1 and L2 constructs, L1 and L2 protein expression was assessed by immunoblot analysis of Cos-1 cells transfected with the wt or synthetic humanized BPV1 L1 or L2 pCDNA3 expression construct. Immunoreactive L1 and L2 proteins were expressed from the synthetic humanized L1 and L2 sequences, but no L1 or L2 protein was expressed from the wt L1 and L2 sequences (Fig. 2).
FIG. 2.
Expression of L1 and L2 proteins and GFP and of the corresponding mRNAs from wt or codon-modified genes. Cos-1 cells were transfected with wt or modified L1 and L2 expression plasmids pCDNA/BPVL1wt plus pCDNA/HBL1 (row 1), pCDNA/BPVL2wt plus pCDNA/HBL2 (row 2), and pWt gfp plus P gfp (row 3). Cells were fixed after 36 h and incubated with rabbit anti-BPV1 L1 (row 1) or L2 (row 2) antiserum, followed by FITC-conjugated goat anti-rabbit IgG antibody, and protein was visualized by immunofluorescence. L1 and L2 proteins were visible in the nuclei of cells transfected with HBL1 (row 1, panel 2) and HBL2 (row 2, panel 2) plasmids. The cells transfected with wt L1 (row 1, panel 1) or L2 (row 2, panel 1) sequences failed to produce detectable protein. Cells transfected with humanized gfp (row 3, panel 2) produced GFP, whereas cells transfected with the Pgfp produced little GFP (row 3, panel 3). Immunoblotting of cell lysates of HBL1-transfected cells, and of cell transfected with purified BPV1 but not wt BPV1 L1 (row 1 panel 3), was positive for L1 protein. L2 protein appeared as a 78-kDa band in cells transfected with HBL2 and BPV1 but not in cells transfected with wt L2 (row 2, panel 3). mRNA extracted from the cytoplasm (Cyto) or whole-cell preparation (total) of cells transfected with control pCDNA3 (V), with pCDNA with wt or humanized (HB) L1 or L2 gene inserts, or with wt gfp or Pgfp was probed with 32P-labelled probes made with wt L1 (row 1, panel 4) or L2 (row 2, panel 4) or gfp (row 3, panel 4) sequences. The total amount of mRNA in these samples was examined by hybridization of the mRNA sample with a GAPDH probe (panel 4, lower).
To establish whether the alterations to the primary sequence of the L1 and L2 mRNAs which resulted from the codon alterations also affected steady-state expression of the corresponding message, total and cytoplasmic mRNAs were prepared from Cos-1 cells transfected with the various capsid protein gene constructs. Using GAPDH as an internal standard, we established by Northern blotting that two to three times more modified than wt L1 mRNA and similar levels of wt and modified L2 mRNAs were present in the cytoplasm of transfected cells (Fig. 2, right panel). The total amount of L1 or L2 protein expressed in Cos-1 cell cultures per arbitrary unit of L1 or L2 mRNA expressed was assessed by densitometry as at least 1,000-fold higher for the humanized gene constructs than for the natural gene constructs, confirming that the majority of the difference in level of expressed PV late protein between the two constructs was a consequence of increased L1 or L2 translation, rather than increased gene transcription.
PV late protein translation in vitro.
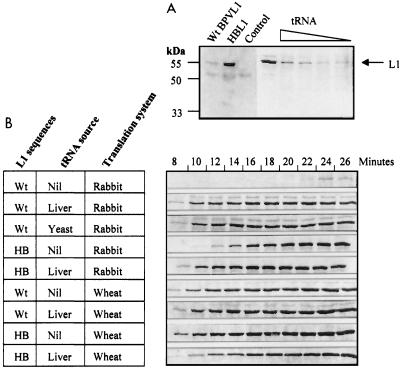
As the major limitation to expression of the wt BPV L1 and L2 genes in dividing cells in vitro appeared to be translational, we wished to test whether this limitation reflected a limited availability of the appropriate tRNA species for gene translation. As transient expression of the synthetic genes within intact cells may be regulated by many factors, we tested our hypothesis in a cell-free system using rabbit reticulocyte lysate (RRL) or wheat germ lysate to examine gene translation. Similar amounts of plasmids expressing the wt or synthetic humanized BPV1 L1 gene were added to a T7-DNA polymerase coupled RRL transcription-translation system in the presence of [35S]methionine. After 20 min, translated proteins were separated by SDS-PAGE and visualized by autoradiography. Efficient translation of the modified L1 gene was observed (Fig. 3A, lane 2), while translation of the wt BPV1 L1 sequence resulted in a weak 55-kDa L1 band (lane 1). We reasoned that although the wt sequence was not optimized for translation in RRL, some translation would occur since there would be no cellular mRNA species competing for the rare codons present in the wt L1 sequence. These data suggest that the observed difference in efficiency of translation of the wt and synthetic humanized L1 genes is a consequence of limited availability of the tRNAs required for translation of the rare codons present in the wt gene. We therefore expected that addition of excess tRNA to the in vitro translation system would enhance translation of the wt L1 gene to a greater extent than that of the synthetic gene. To address this question, 10−5 M aminoacyl-tRNA from yeast or bovine liver was added to the RRL translation system and L1 protein synthesis was assessed. Introduction of exogenous tRNAs resulted in a dramatic improvement in translation of the wt L1 sequence, which now gave a yield of L1 protein comparable to that observed with the synthetic humanized L1 sequence (Fig. 3A, right lanes). Enhancement of translation of the wt L1 gene by aminoacyl-tRNA was dose dependent, with an optimum efficiency at 10−5 M tRNA. As addition of exogenous tRNA improved the yield of L1 protein translated from the wt L1 gene sequence, we assessed the speed of translation of wt and humanized L1 mRNAs. Samples were collected from the translation mixture every 2 min, starting at min 8. Translation of L1 from the wt sequence was much slower than that from the humanized L1 sequence (Fig. 3B), and the retardation of translation could be completely overcome by adding exogenous liver tRNA. Addition of a comparable amount of exogenous liver tRNA to the humanized L1 gene expression mixture somewhat enhanced expression of this gene, as might be predicted from a mass action effect on the kinetics of protein translation. Translation of the wt gene in the absence of exogenous tRNA was too slow to allow exact determination of the increased rate of translation following addition of exogenous tRNA but was greater than 10-fold, in contrast to the 1.3-fold increase observed with the synthetic gene upon addition of exogenous tRNA (Fig. 3B). In separate experiments, we established that wt L1 translation could be similarly enhanced by yeast tRNA (Fig. 3B) and by tRNAs extracted from bovine skin epidermis, which presumably constitutes a mixture of tRNAs from differentiated and undifferentiated epithelial cells (data not shown).
FIG. 3.
For in vitro translation of BPV L1 sequences, wt BPV L1 (wt) or synthetic L1 (HB) sequences were translated in vitro, using RRL or wheat germ extract in the presence of [35S]methionine. (A) wt L1 or HB L1 plasmid DNA was added to the T7-DNA polymerase coupled in vitro translation system (Promega), and L1 protein was translated for 20 min at 30°C. L1 protein was detected by PAGE and autoradiography. The BPV L1 wt gene produced a weak L1 band at 55 kDa (lane 1). The synthetic HBL1 gene made a larger amount of L1 protein (lane 2). The translation reaction where only vector was added produced no visible band (lane 3). When 10−5 to 10−9 M (lanes 4 to 8) bovine liver aminoacyl-tRNA was added to the translation system, the protein translation efficiency from wt L1 sequence was improved in a tRNA dose-dependent manner. (B) Comparison of the translation efficiencies of wt L1 and HB L1 sequences in the presence and absence of tRNA. Translation was carried out in RRL (rabbit) or wheat germ extract (wheat), and samples were collected every 2 min starting from min 8. Addition of 10−5 M bovine liver or yeast tRNA is indicated at the left.
Translation of wt L1 is efficient in wheat germ extract.
To further test our hypothesis that tRNA availability is a determinant of expression of the wt BPV1 L1 gene, we examined the translation of L1 in a cell type in which a quite different set of tRNAs would be available. In a wheat germ translation system, wt L1 mRNA was translated as efficiently as humanized L1 mRNA, and addition of exogenous aminoacyl-tRNAs did not improve the translation efficiency of either wt or humanized sequences (Fig. 3B). This finding suggested that in wheat germ, in contrast to RRL, there are sufficient of the tRNAs which are limiting for translation of wt L1 sequence in RRL to allow efficient L1 translation.
Modified late genes can be expressed from constructs with only the natural BPV promoters.
While the data presented above indicate that translation is limiting for the production of BPV1 capsid proteins in our test system, these experiments were conducted in systems which are not truly representative of the viral late gene transcription from the BPV genome, in part because the genes were driven by a strong CMV promoter. We therefore wished to establish whether synthetic humanized BPV capsid protein mRNA would be translated more efficiently than the wt mRNA if transcribed from a plasmid containing only the natural BPV1 promoters. This would establish whether translation was indeed one of the limiting factors for expression of BPV1 late genes in the context of the BPV1 genome and in an undifferentiated cell. Cos-1 cells were transfected with plasmid pCICR1 or pCICR2 (Fig. 4, top panel), in which the wt or synthetic humanized L2 gene was inserted into the BPV1 genome, cloned into pUC18. L2 protein expression was examined by immunofluorescence of transfected cells and by immunoblotting. Synthetic humanized L2 was efficiently expressed, whereas the wt L2 sequence, driven from a similar construct, produced no immunoreactive L2 protein (Fig. 4, bottom panel). As undifferentiated cells supported the expression of the humanized L2 gene but not the wt L2 gene expressed in the context of the BPV1 genome without an additional promoter, the results confirmed our earlier observations from experiments using the CMV promoter. The plasmids tested here contained the SV40 ori, designed to replicate the DNA in Cos-1 cells. The increased copy number of the BPV1 L2 plasmids or the transcription-enhancing activity of the SV40 ori might explain in part the increased efficiency of expression of L2 in this experimental system compared with infected skin. However, the marked difference in expression between the natural and humanized genes seen with a CMV promoter construct was still observed with a plasmid containing only the natural promoter, suggesting that the variation in L2 expression efficiency observed was independent of the transcriptional promoter used in our system.
FIG. 4.
Expression of L2 protein from the native PV promoter. (Top) Schematic representation of plasmids used to determine L2 expression from the BPV cryptic promoter(s). The original L1 and most of the L2 sequences were deleted from the BPV1 genome by BamHI and HindIII digestion, and the remaining BPV1 sequence was cloned into pUC18. Wild-type or synthetic humanized L2 sequences were inserted into the BamHI site of the BPV1 genome. The position of the inserted SV40 ori sequence is indicated. The plasmid where modified L2 was used but without the SV40 ori sequence was also used as a control. (Middle) The plasmids were used to transfect Cos-1 cells, and the expression of L2 protein was determined by using BPV1 L2-specific polyclonal antiserum followed by FITC-linked anti-rabbit IgG. Expression of L2 was observed only in cells transfected with a plasmid (pCICR2) containing the SV40 ori and a synthetic humanized L2 gene. Cells transfected with the plasmid with the wt L2 sequence (pCICR1) with SV40 or a modified L2 sequence without the SV40 ori (pCICR3) failed to produce detectable L2 protein. A mock transfection in which the cells did not receive a plasmid was used as a control. (Bottom) Transfected cells were subjected to SDS-PAGE and immunoblotting with a rabbit polyvalent antibody to L2, and L2 was detected only in cells transfected with pCICR2.
Substitution of PV-preferred codons prevents translation but not transcription of a non-PV gene in undifferentiated cells.
To further confirm that codon usage can alter gene expression in mammalian cells, we made a further variant of a synthetic gfp gene modified for optimal expression in eukaryotic cells (40). In our variant, codons optimized for expression in eukaryotic cells were replaced with those preferentially used in PV late genes. Of 240 codons in the humanized gfp gene, which expresses high levels of GFP in cultured cells, 156 were changed to the corresponding PV late gene-preferred codons to produce a new gfp gene designated Pgfp. Expression of Pgfp in cells in vitro was compared with that of humanized gfp. Cos-1 cells transfected with the humanized gfp produced a bright fluorescent signal after 24 h, while cells transfected with Pgfp produced only a faint fluorescent signal (Fig. 2). Thus, modification of codon usage in other genes to match that of PV late genes can reduce expression of those genes in cell lines, as is observed for PV late genes.
DISCUSSION
In this study, we have confirmed that one determinant of the efficiency of expression of three different genes in dividing mammalian cells in vitro is their codon composition. This observation has commonly been made when genes from prokaryotic organisms have been expressed in eukaryotic cells (27), and these data are now extended to apply to two PV genes and one eukaryotic gene modified to resemble a PV gene in codon composition. The differential expression of genes utilizing different codons to express the same protein appears predominantly to reflect different efficacy of gene translation, and tRNA availability is apparently rate limiting for translation when genes use codons rarely used by consensus genes from the expression host.
Alterations to mRNA secondary structure or protein binding (29) as a consequence of the changes to the primary sequence of the genes might contribute to the observed differences in efficiency of translation between natural and modified genes in cultured cells. However, the marked enhancement of translation of the natural but not the modified mRNA that was observed after addition of tRNA in a mammalian in vitro translation system strengthens the argument that tRNA availability is rate limiting for translation of the natural gene in mammalian cells. A shortage of critical tRNAs could result in slowed elongation of the nascent peptide or premature termination of translation (21). Slowed elongation appears to be the major consequence for the PV late gene. Analysis of codon usage in the PV genome shows that PV late genes use many codons that mammalian cells rarely use. For example, PV frequently uses UUA for leucine, CGU for arginine, ACA for threonine, and AUA for isoleucine, whereas these codons are significantly less often used in mammalian genes. In contrast, PV late genes can be translated efficiently when stably expressed in yeast (14, 24), and the codon compositions of yeast and PV genes are similar (Table 1). The correlation of codon usage in yeast genes and PV late genes is puzzling and may simply represent chance convergent evolution. An apparent exception to the inefficient expression of PV late genes in dividing cells is that PV L1 genes can be expressed in insect cell lines (18) by using recombinant baculovirus or in mammalian cell lines by using recombinant vaccinia virus (39). As infection with these viruses down regulates cellular protein synthesis, the efficient expression of the L1 capsid proteins under these circumstances may occur because less cellular mRNA is available in a virus-infected cell to compete with the L1 mRNA for the rarer tRNAs.
Codon composition could be a more general determinant of gene expression within different stages of differentiation of the same tissue. Although the genetic code is essentially universal, different organisms show differences in codon composition of their genes, while the codon compositions of genes tend to be relatively similar for all genes within each organism and matched to the population of isoaccepting tRNAs for that organism (13). However, populations of tRNAs in differentiating and neoplastic cells are different (15, 35, 36), and the tRNA populations also vary in cells growing under different growth conditions (6). Together these observations suggest that codon composition and tRNA availability may provide a primitive mechanism for spatial or temporal regulation of gene expression. It is well recognized that the G+C content of many double-stranded DNA viruses, a crude marker for viral gene codon composition, is markedly different from the G+C content of the DNA of the cells they infect (32). Viruses may therefore have evolved to take advantage of codon composition to regulate their own program of gene expression, perhaps to avoid expression of lethal quantities of viral proteins in undifferentiated cells where the virus utilizes the cellular machinery to replicate its genome. Similar alteration of efficiency of expression of at least one gene from human immunodeficiency virus type 1 upon alteration of codon usage has recently been described (1), although the mechanism of increased efficacy of expression was related in that study to release of translation from dependence on human immunodeficiency virus type 1 rev. The early genes of PV, with the notable exception of E4, have codon usage patterns similar to those of the late genes, which could explain the very low levels of expression of these proteins observed in dividing cells in vivo and in vitro (26, 28). In keeping with our hypotheses, it has recently been shown that the translation of at least one PV early gene, E7, can be enhanced in vitro by addition of exogenous tRNA (5).
As our current observations may represent an apparently novel mechanism of regulation of gene translation within a single tissue, it is relevant to consider how this relates to previously proposed hypotheses for the restriction of expression of PV late genes to differentiated epithelium. It has been suggested that reduced late gene expression may reflect dependence of expression from the late promoter on transcription factors expressed only in differentiated epithelium or may alternatively be due to suppression of late promoter transcription by viral (33) or cellular gene products expressed in undifferentiated cells. The late promoters of human papillomavirus types 31b and 5 (11, 12) are described as differentiation dependent, although the search for relevant transcription control factors in differentiated keratinocytes by conventional footprinting and DNA binding studies has been unrewarding. Our data show that capsid proteins are not translated from PV L1 and L2 mRNAs in cells transfected with CMV promoter-based expression vectors (Fig. 2), suggesting that in addition to any transcriptional controls that may exist, there is also a posttranscriptional block to capsid protein synthesis in undifferentiated cells. Within L1 or L2 mRNA or within flanking untranslated message are sequences resembling 5′ splice donor sites which are inhibitory to transcription of genes with which they are associated (8, 17). Other AU-rich sequences in L1 or L2 mRNA promote mRNA degradation (30). Consistently moderately decreased levels of mRNA for the wt L1, compared with the sequence of modified L1, were observed in the present study following expression of these genes in Cos-1 cells. This observation confirms that transcription or stability of wt L1 may be diminished in comparison with other genes. However, the overall contribution of reduced transcriptional efficacy to PV L1 expression in our systems was small in comparison to the effect of codon usage on translational efficiency, and no similar effect on transcription was seen for L2 although the translational difference with the modified L2 gene was equally large. Because untranslated inhibitory sequences were not included in our test system, we cannot determine the roles of inhibitory sequences and codon mismatch in suppression of PV late gene expression in vivo. However, regulatory sequences promoting RNA degradation or inhibiting translation are presumed to act through interaction with nuclear or cytoplasmic proteins (29), and inefficient translation of native sequence L1 mRNA was observed in a cell-free translation system from anucleate cells, demonstrating that codon composition of the PV late genes must play some role in regulation of PV late gene translation. Further, the mechanisms shown to inhibit L1 and L2 mRNA production or increase message instability in undifferentiated cells have yet to be shown to be inactive in differentiated epithelium, as would be required to explain by these mechanisms the successful translation of PV late genes in this tissue.
Further evidence supporting the hypothesis that codon composition may be an important determinant of PV capsid gene expression was gathered from an analysis of the 84 PV L1 sequences currently available in GenBank. The codon composition of the L1 genes, and particularly the frequency of usage of the rarer codons, was essentially the same across all the published sequences (data not shown), as would be expected given the similar G+C contents of the PV genomes. The PV L1 gene is relatively conserved at the amino acid level, showing 60 to 80% amino acid homology between PV genotypes, as might be expected given the constraints on capsid protein function. There are, however, no obvious constraining influences on the codon composition of the PV late genes beyond those of our hypothesis, as the late gene region does not code for other genes, either in other reading frames or on the complementary DNA strand, and has no known cis-acting regulatory functions other than those intrinsic to L1 or L2 gene expression. If codon composition of the capsid genes were not important for PV function, a considerable heterogeneity of codon usage might therefore be expected, given the evolutionary diversity of PVs (4).
Taken together, the data presented in this report makes a case that codon usage should be considered as a possible determinant of differential expression of PV late genes between undifferentiated and differentiated epithelial cells, in addition to many proven regulation mechanisms, and suggests that this observation may be generalizable to other genes. The relative role of message instability and codon mismatch in determining PV late gene expression in differentiated tissues will require comparisons of transcriptional activity and translation of the L1 and L2 genes driven from strong constitutive promoters in differentiated and undifferentiated epithelium. Such work using either transgenic technology or keratinocyte raft cultures should now be feasible. The hypothesis that expression of other genes may be regulated by tRNA availability in vivo is now testable, as methods for fingerprinting tRNA distributions in tissues have recently been developed (15).
ACKNOWLEDGMENTS
We thank Mark Williams for helpful discussions throughout the study.
This work was funded in part by grant RO1-CA 57789 from NIH and by grants from the NH&MRC, the Queensland Cancer Fund, the Mayne Bequest, and the Princess Alexandra Hospital Research and Development Foundation.
REFERENCES
- 1.Andre S, Seed B, Eberle J, Schraut W, Bultmann A, Haas J. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J Virol. 1998;72:1497–1503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker C C, Howley P M. Differential promoter utilization by the bovine papillomavirus in transformed cells and productively infected wart tissues. EMBO J. 1987;6:1027–1035. doi: 10.1002/j.1460-2075.1987.tb04855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barksdale S K, Baker C C. Differentiation-specific alternative splicing of bovine papillomavirus late mRNAs. J Virol. 1995;69:6553–6556. doi: 10.1128/jvi.69.10.6553-6556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan S-Y, Delius H, Halpern A L, Bernard H-U. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J Virol. 1995;69:3074–3083. doi: 10.1128/jvi.69.5.3074-3083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Pasquale C, Kandue D. Modulation of HPV16 E7 translation by tRNAs in eukaryotic cell-free translation systems. Biochem Mol Biol Int. 1998;45:1005–1009. doi: 10.1002/iub.7510450518. [DOI] [PubMed] [Google Scholar]
- 6.Doi R H, Kaneko I, Igarashi R T. Pattern of valine transfer ribonucleic acid of Bacillus subtilis under different growth conditions. J Biol Chem. 1968;243:945–951. [PubMed] [Google Scholar]
- 7.Furth P A, Baker C C. An element in the bovine papillomavirus late 3′ untranslated region reduces polyadenylated cytoplasmic RNA levels. J Virol. 1991;65:5806–5812. doi: 10.1128/jvi.65.11.5806-5812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furth P A, Choe W-T, Rex J H, Byrne J C, Baker C C. Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol Cell Biol. 1994;14:5278–5289. doi: 10.1128/mcb.14.8.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grassmann K, Rapp B, Maschek H, Petry K U, Iftner T. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J Virol. 1996;70:2339–2349. doi: 10.1128/jvi.70.4.2339-2349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagensee M E, Yaegashi N, Galloway D A. Self-assembly of human papillomavirus type I capsids by expression of the L1 protein alone or by coexpression of the L1 and L2 capsid proteins. J Virol. 1993;67:315–322. doi: 10.1128/jvi.67.1.315-322.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haller K, Stubenrauch F, Pfister H. Differentiation-dependent transcription of the epidermodysplasia verruciformis-associated human papillomavirus type 5 in benign lesions. Virology. 1995;214:245–255. doi: 10.1006/viro.1995.0028. [DOI] [PubMed] [Google Scholar]
- 12.Hummel M, Hudson J B, Laimins L A. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J Virol. 1992;66:6070–6080. doi: 10.1128/jvi.66.10.6070-6080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- 14.Jansen K U, Rosolowsky M, Schultz L D, Markus H Z, Cook J C, Donnelly J J, Martinez D, Ellis R W, Shaw A R. Vaccination with yeast-expressed cottontail rabbit papillomavirus (CRPV) virus-like particles protects rabbits from CRPV-induced papilloma formation. Vaccine. 1995;13:1509–1514. doi: 10.1016/0264-410x(95)00103-8. [DOI] [PubMed] [Google Scholar]
- 15.Kanduc D. Changes of tRNA population during compensatory cell proliferation: differential expression of methionine-tRNA species. Arch Biochem Biophys. 1997;342:1–6. doi: 10.1006/abbi.1996.9869. [DOI] [PubMed] [Google Scholar]
- 16.Kennedy I M, Haddow J K, Clements J B. Analysis of human papillomavirus type 16 late mRNA 3′ processing signals in vitro and in vivo. J Virol. 1990;64:1825–1829. doi: 10.1128/jvi.64.4.1825-1829.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy I M, Haddow J K, Clements J B. A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J Virol. 1991;65:2093–2097. doi: 10.1128/jvi.65.4.2093-2097.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirnbauer R, Booy F, Cheng N, Lowy D R, Schiller J T. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci USA. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W J, Gissmann L, Sun X Y, Kanjanahaluethai A, Müller M, Doorbar J, Zhou J. Sequence close to the N-terminus of L2 protein is displayed on the surface of bovine papillomavirus type 1 virions. Virology. 1997;227:474–483. doi: 10.1006/viro.1996.8348. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura Y, Wada K, Doi H, Kanaya S, Gojobori T, Ikemura T. Codon usage tabulated from the international DNA sequence databases. Nucleic Acids Res. 1996;24:214–215. doi: 10.1093/nar/24.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oba T, Andachi Y, Muto A, Osawa T. Translation in vitro of codon UGA as tryptophan in Mycoplasma capricolum. Biochimie. 1991;73:1109–1112. doi: 10.1016/0300-9084(91)90153-r. [DOI] [PubMed] [Google Scholar]
- 22.Ozhun M A, Meyers C. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J Virol. 1997;71:5161–5172. doi: 10.1128/jvi.71.7.5161-5172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruesch M N, Stubenrauch F, Laimins L A. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J Virol. 1998;72:5016–5024. doi: 10.1128/jvi.72.6.5016-5024.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasagawa T, Pushko P, Steers G, Gschmeissner S E, Hajibagheri M A N, Finch J, Crawford L, Tommasino M. Synthesis and assembly of virus-like particles of human papillomaviruses type 6 and type 16 in fission yeast Schizosaccharomyces pombe. Virology. 1995;206:126–135. doi: 10.1016/s0042-6822(95)80027-1. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz S. Cis-acting negative RNA elements on papillomavirus late mRNAs. Semin Virol. 1998;8:291–300. [Google Scholar]
- 26.Selvey L A, Dunn L A, Murray B, Tindle R W, Frazer L H. An ELISA capture assay for the E7 transforming proteins of HPV16 and HPV18. J Virol Methods. 1992;37:119–127. doi: 10.1016/0166-0934(92)90039-g. [DOI] [PubMed] [Google Scholar]
- 27.Smith D W. Problems of translating heterologous genes in expression systems: the role of tRNA. Biotechnol Prog. 1996;12:417–422. doi: 10.1021/bp950056a. [DOI] [PubMed] [Google Scholar]
- 28.Smotkin D, Wettstein F O. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc Natl Acad Sci USA. 1986;83:4680–4684. doi: 10.1073/pnas.83.13.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sokolowski M, Tan W, Jeline M, Schwartz S. mRNA instability elements in the human papillomavirus type 16 L2 coding region. J Virol. 1998;72:1504–1515. doi: 10.1128/jvi.72.2.1504-1515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sokolowski M, Zhao C P, Tan W, Schwartz S. AU-rich mRNA instability elements on human papillomavirus type 1 late mRNAs and c-fos mRNAs interact with the same cellular factors. Oncogene. 1997;15:2303–2319. doi: 10.1038/sj.onc.1201415. [DOI] [PubMed] [Google Scholar]
- 31.Stoler M H, Rhodes C R, Whitbeck A, Wolinsky S M, Chow L T, Broker T R. Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum Pathol. 1992;23:117–128. doi: 10.1016/0046-8177(92)90232-r. [DOI] [PubMed] [Google Scholar]
- 32.Strauss E G, Strauss J H, Levine A J. Virus evolution. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1995. pp. 153–171. [Google Scholar]
- 33.Stubenrauch F, Leigh I M, Pfister H. E2 represses the late gene promoter of human papillomavirus type 8 at high concentrations by interfering with cellular factors. J Virol. 1996;70:119–126. doi: 10.1128/jvi.70.1.119-126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volpers C, Schirmacher P, Streeck R E, Sapp M. Assembly of the major and the minor capsid protein of human papillomavirus type 33 into virus-like particles and tubular structures in insect cells. Virology. 1994;200:504–512. doi: 10.1006/viro.1994.1213. [DOI] [PubMed] [Google Scholar]
- 35.Yang S S, Comb D G. Distribution of multiple forms of lysyl transfer RNA during early embryogenesis of sea urchin, Lytechinus variegatus. J Mol Biol. 1968;31:138–142. doi: 10.1016/0022-2836(68)90062-4. [DOI] [PubMed] [Google Scholar]
- 36.Yang W K, Novelli G D. Isoaccepting +RNA’s in mouse plasma cell tumors that synthesize different myeloma protein. Biochem Biophys Res Commun. 1968;31:534–539. doi: 10.1016/0006-291x(68)90510-x. [DOI] [PubMed] [Google Scholar]
- 37.Zeltner R, Borenstein L A, Wettstein F O, Iftner T. Changes in RNA expression pattern during the malignant progression of cottontail rabbit papillomavirus-induced tumors in rabbits. J Virol. 1994;68:3620–3630. doi: 10.1128/jvi.68.6.3620-3630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou J, Doorbar J, Sun X Y, Crawford L V, McLean C S, Frazer I H. Identification of the nuclear localization signal of human papillomavirus type 16 L1 protein. Virology. 1991;185:625–632. doi: 10.1016/0042-6822(91)90533-h. [DOI] [PubMed] [Google Scholar]
- 39.Zhou J, Sun X Y, Stenzel D J, Frazer I H. Expression of vaccinia recombinant HPV 16 L1 and L2 ORF proteins in epithelial cells is sufficient for assembly of HPV virion-like particles. Virology. 1991;185:251–257. doi: 10.1016/0042-6822(91)90772-4. [DOI] [PubMed] [Google Scholar]
- 40.Zolotukhin A S, Potter M, Hauswirth W W, Guy J, Muzyezka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]