Abstract
Background:
Down syndrome, or Trisomy 21, is the leading genetic cause of cognitive disability in children and is associated with a high risk of several comorbidities, particularly congenital heart defects, early onset Alzheimer’s disease, leukaemia, and autoimmune disorders.
Objective:
This study describes the design, methods, and operational procedures employed to establish a biobank dedicated to Down syndrome that can support research projects investigating the effects of various genetic and environmental factors on this complex disease.
Methods:
Blood was collected from all recruited subjects, processed, aliquoted and immediately frozen at −80 °C in the Interinstitutional Multidisciplinary BioBank (BioBIM) facilities. A small aliquot of the sample was used to perform blood tests for which analysis would not be feasible at a later date, such as blood cell counts. Each biological sample was coded, assigned a Standard PREanalytical Code, and registered in the oloBIOBANK software connected to a medical card containing all the donor’s anamnestic data. All samples were stored under continuous real-time temperature recording using a freezer connected to a T-GUARD alarm system. In addition, a radiofrequency identification tracking system strictly monitored each cryopreservation operation performed throughout the sample lifecycle.
Results:
Biological samples were collected from 454 individuals with Down syndrome from 2007 to 2023. A total of 2233 biological samples were available for research purposes, including whole blood in different anticoagulants, serum, plasma, and frozen peripheral blood mononuclear cells. The quality of the nucleic acids obtained through specific standard operating procedures demonstrated that these samples were appropriate for clinical and basic research.
Conclusion:
By establishing this biobank, we have gathered a significant number of biological samples and clinical data from individuals with Down syndrome, thereby fostering collaboration between different research groups in an open and transparent manner. Sharing expertise and resources among scientists will ultimately facilitate the transfer of knowledge to clinical practice, leading to the development of more effective therapeutic treatments to improve the outcomes and quality of life of patients with Down syndrome.
Keywords: Down syndrome, biospecimens, biobank, genetics, Alzheimer’s disease, congenital heart disease
Introduction
Down Syndrome
Down syndrome (DS) (OMIM #190685) is the most common chromosomal abnormality in live births and the leading cause of cognitive disabilities in children. It was first defined in 1866 by London Hospital physician Langdon Down [1], who described the main features of the syndrome as epicanthal folds, wide face, macroglossia, cognitive disability, and shortened life span. In 1959, Lejeune et al. [2] demonstrated that DS was caused by the presence of three copies of chromosome 21, with 95% of cases caused by a free extra chromosome 21 which arises de novo [3,4]. About 3–4% of cases involve translocations in which the extra chromosome 21 material is translocated to another chromosome; these unbalanced translocations can be inherited from a normal parent carrying a balanced translocation [4]. Approximately 1% of other cases may be caused by genetic “mosaicism” [5], characterised by the presence of both euploid and trisomic cell populations and clinical presentation is usually attenuated [4,6]. DS occurs in less than 1 out of 700 live births in the USA and affects millions worldwide [4,7,8]. The prevalence in Europe is 10.12 per 10,000 births [9]. In addition to variable levels of cognitive disability, individuals with DS are at a higher risk (~50%) for congenital heart defects and almost inevitable early onset Alzheimer’s disease, as well as an increased risk of autoimmune disorders, leukaemia, autism, and other potential comorbidities [10-13].
The understanding of the biology of DS has greatly increased in recent years; however, the specific cellular impacts and molecular mechanisms responsible for DS-related cognitive impairment, as well as other comorbidities, are yet to be elucidated. DS mouse models have been commonly used for many years; however, recent initiatives have emphasised the need for larger clinical cohorts and genomic-scale resources to study this condition in humans. A high degree of variability among individuals has been described, reflecting the complexity and differences in genetic backgrounds of individuals with trisomy 21 [7,11]. Since 2000, significant results have been achieved in dissecting the genetic content of the long arm of chromosome 21 (HSA21), which was fully sequenced and catalogued in an international collaborative study [14]. However, whole-genome studies in large cohorts of clinically characterised individuals with DS have recently become a priority. These offer substantial opportunities to study the molecular pathogenic mechanisms from individual genetic and phenotypic profiles, thereby developing targeted therapeutic interventions. This is important for individuals with DS but is also relevant to their comorbidities that affect the non-DS population such as congenital heart disease and Alzheimer’s disease (AD).
Design and Development of a Dedicated Biobank
Planning and developing a dedicated biobank is crucial for research focused on genetic mechanisms, potential interactions with environmental factors, and their impact on the diverse phenotypes of individuals with DS [11]. Biological banks are defined by the Oviedo Convention as “operational units that provide a service for the preservation and management of biological material and associated clinical data in accordance with good laboratory practice, privacy law and ethics guidelines”. They constitute a fundamental resource, even many years after sample collection, and are essential prerequisites and fundamental supports for research and clinical trials of rare genetic diseases, especially from the current perspective of personalised medicine [15]. There are few biobanks dedicated to DS worldwide and none in Italy, therefore, our group decided to establish a biorepository dedicated to DS within the Interinstitutional Multidisciplinary BioBank (BioBIM) of the Scientific Institute for Research, Hospitalisation, and Healthcare (IRCCS), San Raffaele, Rome, Italy. BioBIM is a non-profit service unit for translational research in the medical field, configured as a Biological Resource Centre (BRC) and provided with a proprietary Data Lake for data matching, data mining, and integration. Its activities include obtaining, storing, processing, and sharing biological samples and their associated health data. Processing health data includes but is not limited to data storage, pseudo-anonymisation, anonymisation, curation, analysis, data transfer, and data sharing. BioBIM is well-established within the national health system and international research community. This has created a strong network of collaborations with other research institutions and biobanks; ultimately strengthening the possibility of promoting cultural exchange at a national and international level and supporting projects, multicentre protocols, and research agreements. BioBIM is ISO certified and accredited by the Italian Ministry of Health (https://directory.bbmri.it/#/board).
Herein, we present the procedures and pre-analytical, analytical, and cryopreservation processes used in the BioBIM DS biorepository which aims to provide high-quality biological samples and matched clinical-genetic data to the scientific community. The availability of both mutually correlated elements provides an important resource for studying DS and its clinical evolution and developing new diagnostic tests and targeted treatments [16,17]. We are confident that this resource, which features hundreds of well-characterised individuals with DS, will be of great interest and value to the DS research community.
Materials and Methods
Recruitment of Individuals with DS
Starting in 2007, individuals with DS included in the project were all admitted to the Paediatric Rehabilitation and Development Disabilities Department; instituted in the early 2000s by Prof. Giorgio Albertini of the IRCCS. This is a highly specialised reference centre that focuses on childhood disabilities, including clinical practice, rehabilitation, and scientific research. The only criterion for enrolment was written informed consent signed by a parent or legal guardian. All subjects with DS for which informed consent was available were enrolled and prospectively followed up in the outpatient clinic for treatment and periodic care. Exclusion criteria included lack of written informed consent, other chromosomal disorders, or incomplete clinical information. For all individuals with DS included in the biobank, complete clinical data were recorded and monitored during follow-up, together with additional sampling whenever possible by a highly specialised team. The diagnosis and management of comorbidities were handled using a multidisciplinary approach that included paediatricians, cardiologists, orthopaedic surgeons, endocrinologists, neurologists, and psychologists. Clinical information was coded according to the International Classification of Diseases, Ninth Revision (ICD-9) [18], a medical classification used worldwide in epidemiology, public health surveillance, health management, and clinical purposes to promote international comparability in the collection, processing, classification, and presentation of diseases.
All participants were evaluated using standardised and Italian-translated versions of the Brunette-Lezine, Griffiths, and Wechsler Preschool and Primary Scale of Intelligence assessments of psychomotor development in preschool children [19-21]. The intelligence quotient was assessed using Raven’s Coloured and Standard Progressive Matrices [22,23]. In addition, the Wechsler Adult Intelligence Scale and age-based Wechsler Intelligence Scale for Children were used [24,25]. Language abilities were evaluated by administering the Italian equivalents of the battery test for language assessment, the Rustioni test for language assessment in Italian, and the Cornoldi test for learning assessment [26-28].
Before participating in the project, all subjects and their parents or legal guardians were adequately informed by a paediatrician about the importance of biobanks for scientific research and the reasons for the collection of biological samples. A printed information sheet was provided to the family, and an informed consent form was signed by both the legal guardians and physicians. All participants were recruited and followed up under appropriate institutional ethics approval and in accordance with the principles of the World Medical Association Declaration of Helsinki.
Biospecimen Collection
Approximately 10 mL of venous peripheral blood was collected by a single venipuncture over 20–30 s into Vacutainer tubes (BD, Franklin Lakes, NJ, USA) containing different anticoagulants: K2 EDTA (3 mL tube; 367838); Na Citrate (2.7 mL tube; 363095); or SSTTM II Advance tubes Silica (3.5 mL tube; 366127). When feasible, an additional blood aliquot was collected into two lithium heparin vacutainer tubes (4 mL tube; 367883) for isolation of peripheral blood mononuclear cells (PBMCs). This method allowed the widest possible range of blood sample derivatives to be stored for future studies. The ambient temperature of the BioBIM building was maintained for all work environments at 18–22 °C. Immediately after collection, the samples were transferred to the BioBIM Laboratory, where they were processed using the most appropriate standard operating procedures [29].
After appropriate separation of plasma and serum by centrifugation, all samples were aliquoted into 1.8 mL cryotubes and immediately frozen at −80 °C [30,31]. A small aliquot of the sample was used to perform blood tests which would not be feasible at a later date such as blood cell counts. Information on the instruments used for routine laboratory tests is presented in Table 1.
Table 1.
Instruments and manufacturer information for the blood test.
| Blood test | Instrument | Manufacturer |
|---|---|---|
| Routine chemistry | ARCHITECT c8000 | Abbott Laboratories, Abbott Park, IL, USA |
| Complete and differential blood cell counts | Coulter LH 750 Hematology Analyzer | Beckman Coulter, Brea, CA, USA |
| Routine coagulation tests | ACL TOP automated coagulometer | Instrumentation Laboratory (IL) Co., Lexington, MA, USA |
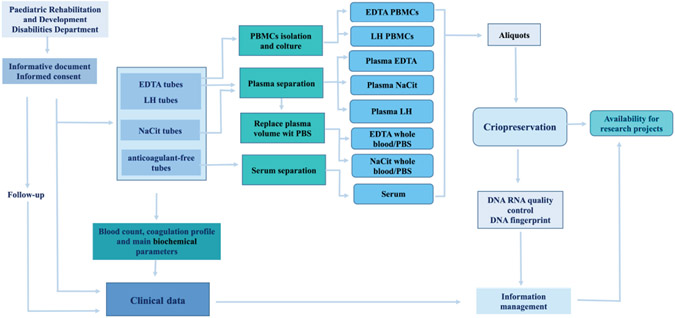
The sample processing workflow is shown in Fig. 1. PBMCs were isolated from blood from EDTA or lithium heparin collection tubes using phosphate-buffered saline and Lympholyte®-H (CL5010, Cedarlane, Hornby, ON, Canada; CL5010) before resuspending in 1 mL of denaturing solution and storing 1–3 aliquots at −80 °C [32,33]. In addition, the viable PBMCs required for immunopheno-typing and functional assays, such as chromosome-based analysis, detection of genotoxicity, or cytotoxicity assays, were processed as previously described and 1–3 vials of each stored in liquid nitrogen [33,34]
Fig. 1. Workflow of biosamples and clinical data acquisition within the general organization of the Down syndrome section of BioBIM, from recruitment of individuals with Down syndrome (DS) to sample cryopreservation (performed with Microsoft® Power Point® © 2013 Microsoft Corporation, Redmond, WA, USA).
BioBIM, Interinstitutional Multidisciplinary BioBank; PBMCs, peripheral blood mononuclear cells.
All freezers were monitored, continuous real-time temperature recorded, and fitted with a T-GUARD alarm system (Biomed-Consulting, Milan, Italy).
Information Management
One of the most crucial aspects of setting up a biobank is the implementation of automated processes for the integrated traceability of samples and their storage and faster sample retrieval; improving sample integrity, traceability, and safety and increasing efficiency.
To this end, each biological sample was coded and registered in the oloBIOBANK software (https://www.olomedia.com/biobank-biobanca/) (Olomedia, Palermo, Italy). This is a web-based system that provides a host server that stores donor data and information on samples and aliquots. The oloBIOBANK software allows the management of individual sample preservation and traceability, regardless of the storage method or location, by connecting it to a donor card which represents a medical record containing all the donor’s anamnestic data.
oloBIOBANK uses the Standard PREanalytical Code (SPREC) designed by the International Society for Biological Environment Repositories to identify samples [35-37]. This assigns a 7-element code to each sample corresponding to seven pre-analytical variables, with different strings of letters for fluids or solid tissues.
BioBIM was validated and a radio-frequency identification (RFID) tracking system was introduced. This is a data identification and storage technology that allows the tracing of the specific paths of each biological sample until freezing. The RFID system designed and developed at the biobank consists of a device that provides a chronological report of the processes involved in each cryopreservation operation, such as door opening/closing, rack insertion/extraction, sample insertion, and the identification of the laboratory operator [29,38].
Nucleic Acid Extraction Quality Control
Pre-analytical variables, including storage time and ambient temperature, may influence the quality and quantity of the isolated nucleic acids. Considering the large number of samples collected, optimising standard procedures for storing whole blood prior to DNA extraction is a crucial step in a biological repository.
Quality assessment of the cryopreserved biological samples was performed by extracting nucleic acids from randomly selected samples in accordance with the planned timelines. Standard operating procedures from pre-analytical steps to DNA and RNA extraction were previously validated and described [30,32].
Briefly, DNA was extracted from whole blood using the QIAamp DNA Blood Mini Kit (51104, QIAGEN, Hilden, Germany), and RNA was extracted from PBMCs using the QIAamp RNA Blood Mini Kit (52304, QIAGEN, Hilden, Germany). The concentration and quality of the DNA and RNA were assessed using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). DNA integrity was assessed by PCR to obtain DNA fragments of various lengths, whereas RNA integrity was determined using an Agilent 2100 Bio-analyzer with RNA 6000 Nano and RNA 6000 Pico assays (Agilent Technologies, Santa Clara, CA, USA) [30,32].
Finally, a low-cost and easy-to-use protocol for DNA fingerprinting capable of ensuring DNA identity in a biorepository was validated and introduced within the biobank standard operating procedures [39].
Results
Clinical Database
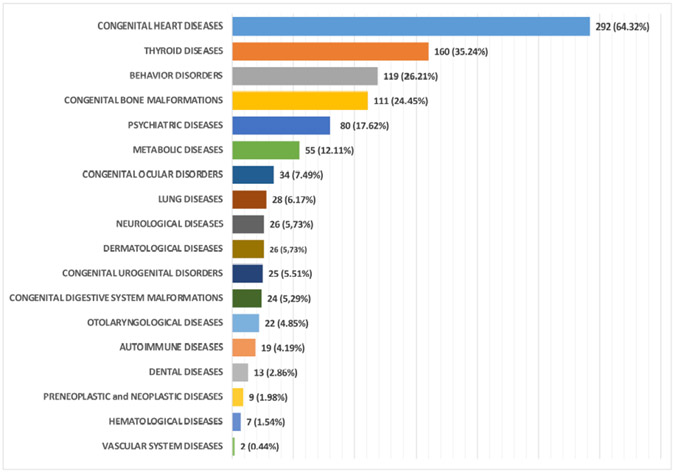
To date, a total of 454 subjects, 253 males (55.73%) and 201 females (44.27%) with an average age of 7 years, ranging from 1–55 years, have been enrolled. The majority of participants were referred to our centre from southern Italy (51.10 %) and central Italy (44.05 %); only 3.96% came from Northern Italy and 0.88% from other areas. The chromosomal assessment found a non-disjunction in 418 (92.07%), translocation in 11 (2.42%), and mosaicism in 5 (1.10%) individuals with DS, however, the result of the cytogenetic analysis was not available for 20 (4.41%) cases. Table 2 provides the basic demographic information and age distribution of the enrolled individuals with DS. A summary of the various patient comorbidities associated with the syndrome is shown in Fig. 2, and additional details on the phenotypic subtypes are listed in Fig. 3 (Ref. [18]) and Supplementary Table 1. For each case, all diagnosed conditions were coded using the International Classification of Diseases, Ninth Revision (ICD-9) [18].
Table 2.
Demographic information and age distribution of individuals with Down syndrome included in BioBIM (N = 454).
| Cases number | % | |
|---|---|---|
| Age group, years* | ||
| 1–3 | 22 | 4.85 |
| 4–6 | 24 | 5.29 |
| 7–9 | 42 | 9.25 |
| 10–12 | 61 | 13.44 |
| 13–15 | 57 | 12.56 |
| 16–18 | 63 | 13.88 |
| 19–21 | 40 | 8.81 |
| 22–24 | 44 | 9.69 |
| 25–27 | 30 | 6.61 |
| 28–30 | 24 | 5.29 |
| 31–33 | 15 | 3.30 |
| 34–36 | 11 | 2.42 |
| 37–39 | 6 | 1.32 |
| 41–43 | 7 | 1.54 |
| 46–50 | 5 | 1.10 |
| 51–55 | 3 | 0.66 |
| Gender | ||
| Female | 201 | 44.27 |
| Male | 253 | 55.73 |
| Chromosomal assessment | ||
| Non-disjunction | 418 | 92.07 |
| Translocation | 11 | 2.42 |
| Mosaicism | 5 | 1.10 |
| NA | 20 | 4.41 |
| Geographical origin | ||
| Southern Italy | 232 | 51.10 |
| Central Italy | 200 | 44.05 |
| Northern Italy | 18 | 3.96 |
| Other | 4 | 0.88 |
Age at the time of recruitment to the biobank.
Fig. 2. Graphic representation of the incidence of specific comorbidities categorized according to specific disease groups in the 454 Down syndrome afferent to BioBIM.
At the end of the bars is the number of instances the type of comorbidity was diagnosed in total and the parentheses indicate the frequency of the presence of the specific condition in the 454 subjects.
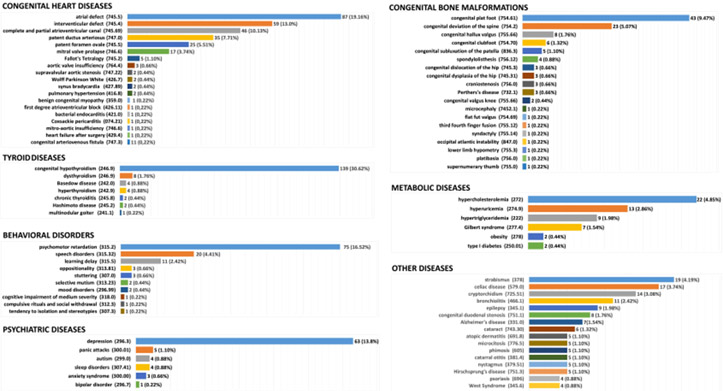
Fig. 3. Graphic representation of comorbidities associated with the 454 individuals with Down syndrome in BioBIM.
For each disease group, the specific disease and the code according to the International Classification of Diseases, Ninth Revision (ICD-9) [18] is shown on the left, and the right of the bars shows the absolute number and frequency.
As summarised in Table 3, 206 of 454 individuals with DS (45.4%) had congenital heart defects, with a slightly higher percentage in males (52.9%) than in females (47.1%). This was the most common comorbidity and was consistent with previous evidence. Some individuals with DS had more than one diagnosed heart defect, therefore, 292 heart defects were observed, which corresponded to 64.32% of these comorbidities in the 454 subjects. (Fig. 2, Supplementary Table 1). As shown in Fig. 3, atrial defects (ICD-9 745.5), intraventricular defects (ICD-9 745.4), and complete and partial atrioventricular canals (ICD-9 745.7) were the most common heart defects and were detected in 87 (19.16%), 59 (13.0%), and 46 (10.13%) patients, respectively. Other defects observed were 35 (7.71%) patent ductus arteriosus (ICD-9 747.0), 25 (5.51%) patent foramen ovale (ICD-9 745.5), and 17 (3.74%) mitral valve prolapses (ICD-9 746.6). In addition, five (1.10%) patients with tetralogy of Fallot (ICD-9 745.2) and three (0.66%) patients with aortic valve insufficiency (ICD-9 764.4) were included. Other diseases (15 [5.1%]) showed individual frequencies of less than 1% (Fig. 3, Supplementary Table 1).
Table 3.
Demographic information and age distribution of congenital heart defects in individuals with Down syndrome included in the BioBIM (n = 206).
| Cases number | % | |
|---|---|---|
| Age group, years* | ||
| 1–3 | 12 | 5.83 |
| 4–6 | 17 | 8.25 |
| 7–9 | 24 | 11.65 |
| 10–12 | 39 | 18.93 |
| 13–15 | 25 | 12.14 |
| 16–18 | 28 | 13.59 |
| 19–21 | 20 | 9.71 |
| 22–24 | 12 | 5.83 |
| 25–27 | 11 | 5.34 |
| 29–31 | 7 | 3.40 |
| 32–37 | 6 | 2.91 |
| 41–51 | 5 | 2.43 |
| Gender | ||
| Female | 97 | 47.09 |
| Male | 109 | 52.91 |
Age at the time of recruitment to the biobank.
Various forms of thyroid disease were diagnosed in 160 (35.24%) individuals. Congenital hypothyroidism (ICD-9 243) was by far the most common, representing 139 cases (30.62%), whereas only eight cases of dysthyroidism (ICD-9 246.9) (1.76%) and four cases (0.88%) with both hyperthyroidism (ICD-9 242.9) and Basedow disease (ICD-9 242.0) were identified (Fig. 3, Supplementary Table 1).
In view of the underlying diagnosis of DS, which includes the presence of global developmental delay, only cases in which the delay was greater than “mild” on the specific scale are reported in Supplementary Table 1 and Fig. 3. Psychomotor retardation (ICD-9 315.2), speech disorders (ICD-9 315.32), and learning delay (ICD-9 315.5) were the most frequent disorders, with 75 (16.52%), 20 (4.41%), and 11 (2.42%) cases, respectively, from a total of 119 (26.21%) individuals with behavioural disorders.
Individuals with DS were also evaluated for the presence of bone malformations, which were identified in 111 (24.25%) cases, of which the most frequent pathologies were congenital plantar foot (ICD-9 754.61) in 43 (38.74%) and congenital deviation of the spine (ICD-9 754.2) in 23 (20.72%) individuals (Fig. 3, Supplementary Table 1).
Of the 454 individuals with DS that were recruited, 80 (17.62%) were categorised as having psychiatric disorders. Notably, this percentage was mostly attributable to 63 individuals (78.75%) with a diagnosis of depression (ICD-9 396.3) (Fig. 3, Supplementary Table 1). Older individuals with DS had a general increase in the frequency of depression.
Other metabolic conditions were diagnosed in 55 (12.11%) of the 454 individuals with DS. Hypercholesterolaemia (ICD-9 272), hyperuricaemia (ICD-9 274.9), hypertriglyceridaemia (ICD-9 222), and Gilbert syndrome (ICD-9 277.4) were found in 22 (4.85%), 13 (2.86%), 9 (1.98%), and 7 (1.54%) patients, respectively (Fig. 3, Supplementary Table 1).
Other clinical features such as congenital eye, urogenital and digestive system, lung, dermatologic, otolaryngological, autoimmune, neoplastic, vascular system, and neurological diseases were present in 235 subjects, with a frequency ranging of 0.44–7.49% (Fig. 3, Supplementary Table 1).
Since individuals with DS are now typically raised in families and live longer, we have unfortunately learned that trisomy 21 causes an extremely high frequency (approximately 80%) of early-onset AD. Since the 454 individuals represented in BioBank were mostly children or younger than the age at which AD manifests, the AD frequency in our population was lower than expected. Only seven (1.54%) cases of AD (ICD-9 331.0) were included among the neurological comorbidities; strikingly, these were diagnosed at ages 31, 32, 34, 36, 37 (n = 2), and 46 years. Therefore, of the 47 individuals with DS over the age of 30, with the oldest being 55, seven have been identified with AD and most have been diagnosed at a younger age than expected. Of the 15 cases of age 40 or older, only one was diagnosed with AD at the time of recruitment to the biobank, highlighting the importance of following individuals longitudinally whenever possible.
Sample Resource
To date, 2233 samples from the 454 individuals with DS enrolled into the biobank have been cryopreserved relative to the time of recruitment, including 454 whole blood samples in EDTA; 428 serum samples; 370 plasma citrated samples, 370 plasma EDTA samples, 370 plasma lithium heparin samples and 241 frozen PBMCs. In multiple instances, the collection of biological samples was repeated during clinical follow-up, on average every three years. Sampling for the second, third, fourth, and fifth times has been performed for 154, 56, 18, 5, and 1 subjects, respectively. There are 850 repeated samples, including 218 whole blood samples; 189 serum samples; 375 plasma samples, citrated, EDTA, and lithium heparin; 46 PBMCs; and 22 urine samples.
An additional 69 samples of whole blood, EDTA and citrated; serum; plasma, citrated, EDTA, and lithium heparin; and live PBMCs obtained from EDTA- and lithium heparin-anticoagulated blood (maintained in liquid nitrogen) were collected from members of the 23 families with DS recruited to the biobank, resulting in a total of 552 biological samples.
The frozen PBMCs had an average cell count of 11.8 × 106/mL, with a range of 4.37–19.95 × 106/mL. Quality checks of nucleic acid extractions indicated that DNA concentrations averaged 35 ng/μL, with a range of 15–5050 ng/μL, and an acceptable absorbance ratio at 260 and 280 nm. Similarly, RNA extraction had an average concentration of 60 ng/μL, with a range of 40–170 ng/μL, and an absorbance ratio at 260/280 nm and 260/230 always higher than 1.9 and 1.8, respectively [30,31].
Discussion
In this study, we report the creation of a DS biobank within the BioBIM BRC that can provide a freely accessible resource for translational research studies on this population. Our focus on this condition stems from the large number of patients with disabilities who visit our centre, including individuals with DS, and the strong commitment of the IRCCS; whose institutional mission is to guarantee a high-quality and specialised integrated assistance and rehabilitation network. To the best of our knowledge, this is the first dedicated DS biobank in Italy. An advantage of this biobank is its coexistence with a highly specialised laboratory, which has made it possible to carry out verification and analytical tests on the samples to establish the characteristics and criteria for collection and preservation, as demonstrated by scientific papers published as part of the project [29-32,37]. To date, BioBIM has recruited 454 and 80 individuals with DS and AD, respectively. Each biological sample was linked to the detailed sociodemographic and clinical characteristics of the donor.
For some years, several authors and research agencies have promoted the need to establish biological banks dedicated to DS, recognising its importance and that the number of individuals with DS has increased [17,40-42]. Many women over 35 years now experience pregnancies, which significantly increases the risk of DS. Despite the wide availability, they do not always undergo prenatal tests, or after a positive test, choose to continue the pregnancy [43].
Furthermore, owing to better healthcare and preventive medicine, as well as improved socioeconomic integration of individuals with DS, their life expectancy is progressively increasing [17,34]. Recent data indicate that individuals with DS now have a life expectancy of more than 55 years compared to only 25 years in the 1980s [44]. This evolution creates new and more demanding challenges in the field of healthcare for individuals with DS and overcomes the “culture of intractability”, which has characterised this disorder for years [15,41]. A longer life expectancy has increased the need to understand the causes of various comorbidities and their consequent diagnostic, prognostic, and therapeutic implications. An important example is the association between the aging of this population and a greater risk of developing dementia similar to AD, about which little is known and whose diagnosis is particularly challenging [11,13,14,17].
Recently, the LuMind IDSC Foundation and the National Down Syndrome Society, two leading DS research and advocacy organisations in the USA, formulated recommendations to the National Institutes of Health for a research strategy focused on the quality of life and care priorities for individuals with DS by 2030. As part of this health programming plan, which focuses on medical research as a tool to improve the health of individuals with DS, one of the overall priority recommendations was to establish specific centralised DS biobanks [44,45].
In this context, a dedicated biobank with serum, plasma, nucleic acids, and PBMCs linked to continually updated clinical data is the best strategy for conducting translational research studies to identify novel therapeutic approaches for individuals with DS [17]. A biobank is essential for the rapid application of “omics” scientific tools to translational and clinical trials to identify new biomarkers for therapeutic targets and response predictivity [46], guide the decision making process, and optimize the therapeutic strategy; as suggested by the “precision medicine” of DS [15,17].
Conclusion
The establishment of a biobank dedicated to DS within the BioBIM BRC aims to enhance and develop scientific research by studying different types of biological samples that can be stored and made available for high-quality research projects aimed at improving knowledge about this syndrome. All these samples allow us to study various aspects of the disease, such as genetic, molecular, biochemical, and cellular characteristics, and compare individuals with DS who show substantially different outcomes, particularly comorbidities. Individuals with trisomy 21 may also be compared to euploid controls to better understand the pathogenetic aspects of co-occurring conditions that impact populations with and without DS and identify new therapeutic options.
Supplementary Material
Acknowledgment
The authors express their deep gratitude to all the patients and their families for providing the opportunity to conduct the DS BioBIM project. The authors also wish to thank Gina Bonanni and all the nursery staff of the participating clinical centers who have contributed and supported the researchers in the overall success of the project. Special thanks are due to the Rotary Club Latina San Marco for having been awarded the biobank project. Finally, the authors wish to thank Luigi Narducci and Danilo Sayed Ibrahim for their excellent technical assistance in biobanking activities.
Funding
This research was partially funded by the Italian Ministry of Health (Ricerca Corrente).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Ethics Approval and Consent to Participate
The study protocol was reviewed and approved by the IRCCS San Raffaele Ethic Committee on November 15th, 2006, approval number: ISR/DMLBA/405. Written informed consent to participate in the study was obtained from all recruited individuals (or their parent/legal guardian/next of kin). All activities related to this project were performed in accordance with the national regulations of the EU General Data Protection Regulation (GDPR) (Reg. EU 2016/679), and the EU Charter of Fundamental Rights, both with respect to the security and protection of personal data and the technical requirements for storing these data. Individuals with DS are prospectively recruited and followed-up under the appropriate Institutional ethics approval and in accordance with the principles embodied in the World Medical Association Declaration of Helsinki. Furthermore, to guarantee the protection of patients’ personal information, the methods of consent collection, sample storage, and quality control were reviewed annually by qualified external auditors.
Dedication
This paper is dedicated to the memory of Prof. Giorgio Albertini, who committed his life to children with Down syndrome. He was a truly inspirational human being, not only a dedicated neurologist but a visionary in multiple ways, whose constant support has made possible the establishment of our DS biobank.
Availability of Data and Materials
The data that support the findings of this study are available from the BioBIM Scientific Coordinator but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of Scientific Direction and Administrative Department, taking into account all issues related to patient privacy and consent, as well as the correct use of data to comply with the EU GDPR.
Supplementary Material
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.24976/Discov.Med.202436184.85.
References
- [1].Down JLH. Observations on an ethnic classification of idiots. London Hospital Records. 1866; 3: 259–262. [Google Scholar]
- [2].Lejeune J, Turpin R, Gautier MM. Le mongolisme premier exemple d’aberration autosomique humaine. Ann Genet. 1959; 1: 41–49. [Google Scholar]
- [3].Hook EB, Cross PK, Schreinemachers DM. Chromosomal abnormality rates at amniocentesis and in live-born infants. JAMA. 1983; 249: 2034–2038. [PubMed] [Google Scholar]
- [4].Ivan DL, Cromwell P. Clinical practice guidelines for management of children with Down syndrome: Part I. Journal of Pediatric Health Care: Official Publication of National Association of Pediatric Nurse Associates & Practitioners. 2014; 28: 105–110. [DOI] [PubMed] [Google Scholar]
- [5].Pangalos C, Avramopoulos D, Blouin JL, Raoul O, deBlois MC, Prieur M, et al. Understanding the mechanism(s) of mosaic trisomy 21 by using DNA polymorphism analysis. American Journal of Human Genetics. 1994; 54: 473–481. [PMC free article] [PubMed] [Google Scholar]
- [6].Mutton D, Alberman E, Hook EB. Cytogenetic and epidemiological findings in Down syndrome, England and Wales 1989 to 1993. National Down Syndrome Cytogenetic Register and the Association of Clinical Cytogeneticists. Journal of Medical Genetics. 1996; 33: 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Palminiello S, Jarząbek K, Kaur K, Walus M, Rabe A, Albertini G, et al. Upregulation of phosphorylated alphaB-crystallin in the brain of children and young adults with Down syndrome. Brain Research. 2009; 1268: 162–173. [DOI] [PubMed] [Google Scholar]
- [8].Whooten R, Schmitt J, Schwartz A. Endocrine manifestations of Down syndrome. Current Opinion in Endocrinology, Diabetes, and Obesity. 2018; 25: 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Santoro M, Coi A, Spadoni I, Bianchi F, Pierini A. Sex differences for major congenital heart defects in Down Syndrome: A population based study. European Journal of Medical Genetics. 2018; 61: 546–550. [DOI] [PubMed] [Google Scholar]
- [10].Arumugam A, Raja K, Venugopalan M, Chandrasekaran B, Kovanur Sampath K, Muthusamy H, et al. Down syndrome-A narrative review with a focus on anatomical features. Clinical Anatomy (New York, N.Y.). 2016; 29: 568–577. [DOI] [PubMed] [Google Scholar]
- [11].Antonarakis SE. Down syndrome and the complexity of genome dosage imbalance. Nature Reviews. Genetics 2017; 18: 147–163. [DOI] [PubMed] [Google Scholar]
- [12].Capone GT, Chicoine B, Bulova P, Stephens M, Hart S, Crissman B, et al. Co-occurring medical conditions in adults with Down syndrome: A systematic review toward the development of health care guidelines. American Journal of Medical Genetics. Part A 2018; 176: 116–133. [DOI] [PubMed] [Google Scholar]
- [13].DeTure MA, Dickson DW. The neuropathological diagnosis of Alzheimer’s disease. Molecular Neurodegeneration. 2019; 14: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park HS, et al. The DNA sequence of human chromosome 21. Nature. 2000; 405: 311–319. [DOI] [PubMed] [Google Scholar]
- [15].McCabe LL, McCabe ERB. Personalized medicine for individuals with Down syndrome. Molecular Genetics and Metabolism. 2011; 104: 7–9. [DOI] [PubMed] [Google Scholar]
- [16].Steinberg K, Beck J, Nickerson D, Garcia-Closas M, Gallagher M, Caggana M, et al. DNA banking for epidemiologic studies: a review of current practices. Epidemiology (Cambridge, Mass.). 2002; 13: 246–254. [DOI] [PubMed] [Google Scholar]
- [17].McCabe LL. McCabe ERB. Down syndrome: issues to consider in a national registry, research database and biobank. Molecular Genetics and Metabolism. 2011; 104: 10–12. [DOI] [PubMed] [Google Scholar]
- [18].International Classification of Diseases, Ninth Revision (ICD-9). 2007. Available at: https://archive.cdc.gov/#/details?archive_url=https://archive.cdc.gov/www_cdc_gov/nchs/icd/icd9cm.htm (Accessed: 20 April 2023).
- [19].Brunet O, Lezine I, Ponzo E. Scala di sviluppo psicomotorio della prima infanzia: manuale. Organizzazioni Speciali: Firenze, Italy. 1991. [Google Scholar]
- [20].Lanfranchi S, Rea M, Vianello R, Ferri R. Griffiths Scales of Child Development. Hogrefe Editore: Firenze, Italy. 2017. [Google Scholar]
- [21].Onnivello S, Pulina F, Locatelli C, Marcolin C, Ramacieri G, Antonaros F, et al. Cognitive profiles in children and adolescents with Down syndrome. Scientific Reports. 2022; 12: 1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pueyo R, Junqué C, Vendrell P, Narberhaus A, Segarra D. Raven’s Coloured Progressive Matrices as a measure of cognitive functioning in Cerebral Palsy. Journal of Intellectual Disability Research. 2008; 52: 437–445. [DOI] [PubMed] [Google Scholar]
- [23].Bürkner PC. Analysing Standard Progressive Matrices (SPM-LS) with Bayesian Item Response Models. Journal of Intelligence. 2020; 8: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ryan JJ, Gontkovsky ST, Kreiner DS, Tree HA. Wechsler Adult Intelligence Scale-Fourth Edition performance in relapsing-remitting multiple sclerosis. Journal of Clinical and Experimental Neuropsychology. 2012; 34: 571–579. [DOI] [PubMed] [Google Scholar]
- [25].Na SD, Burns TG. Wechsler Intelligence Scale for Children-V: Test Review. Applied Neuropsychology: Child. 2016; 5: 156–160. [DOI] [PubMed] [Google Scholar]
- [26].Marini A, Marotta L, Bulgheroni S, Fabbro F. BVL: Battery Test for Language Assessment (Batteria per la Valutazione del Linguaggio). Organizzazioni Speciali: Firenze, Italy. 2015. [Google Scholar]
- [27].Daniela Rustioni D, Lancaster M, Associazione “La nostra Famiglia”. Rustioni Test for Language Assessment (Prove di Valutazione della Comprensione Linguistica). Organizzazioni Speciali: Firenze, Italy. 1994. [Google Scholar]
- [28].Cornoldi C, Carretti B, Cesaretto J, Viola F. Prove MT avanzate-3-clinica: la valutazione delle abilità di lettura e comprensione per la scuola primaria e secondaria di 1. grado: manuale. Giunti Edu: Firenze, Italy. 2016. [Google Scholar]
- [29].Bonizzi G, Capra M, Cassi C, Taliento G, Pala O, Sajjadi E, et al. Biobank for Translational Medicine: Standard Operating Procedures for Optimal Sample Management. Journal of Visualized Experiments. 2022; 189: e63950. [DOI] [PubMed] [Google Scholar]
- [30].Palmirotta R, Ludovici G, De Marchis ML, Savonarola A, Leone B, Spila A, et al. Preanalytical Procedures for DNA Studies: The Experience of the Interinstitutional Multidisciplinary BioBank (BioBIM). Biopreservation and Biobanking. 2011; 9: 35–45. [DOI] [PubMed] [Google Scholar]
- [31].Welsh S, Peakman T, Sheard S, Almond R. Comparison of DNA quantification methodology used in the DNA extraction protocol for the UK Biobank cohort. BMC Genomics. 2017; 18: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Palmirotta R, De Marchis ML, Ludovici G, Leone B, Savonarola A, Ialongo C, et al. Impact of preanalytical handling and timing for peripheral blood mononuclear cells isolation and RNA studies: the experience of the Interinstitutional Multidisciplinary BioBank (BioBIM). The International Journal of Biological Markers. 2012; 27: e90–e98. [DOI] [PubMed] [Google Scholar]
- [33].Betsou F, Gaignaux A, Ammerlaan W, Norris PJ, Stone M. Biospecimen Science of Blood for Peripheral Blood Mononuclear Cell (PBMC) Functional Applications. Current Pathobiology Reports. 2019; 7: 17–27. [Google Scholar]
- [34].Kofanova OA, Davis K, Glazer B, De Souza Y, Kessler J, Betsou F, et al. Viable mononuclear cell stability study for implementation in a proficiency testing program: impact of shipment conditions. Biopreservation and Biobanking. 2014; 12: 206–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Betsou F, Lehmann S, Ashton G, Barnes M, Benson EE, Coppola D, et al. Standard preanalytical coding for biospecimens: defining the sample PREanalytical code. Cancer Epidemiology, Biomarkers & Prevention: a Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2010; 19: 1004–1011. [DOI] [PubMed] [Google Scholar]
- [36].Lehmann S, Guadagni F, Moore H, Ashton G, Barnes M, Benson E, et al. Standard preanalytical coding for biospecimens: review and implementation of the Sample PREanalytical Code (SPREC). Biopreservation and Biobanking. 2012; 10: 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Skoworonska M, Blank A, Centeno I, Hammer C, Perren A, Zlobec I, et al. Real-life data from standardized preanalytical coding (SPREC) in tissue biobanking and its dual use for sample characterization and process optimization. The Journal of Pathology. 2023; 9: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nanni U, Spila A, Riondino S, Valente MG, Somma P, Iacoboni M, et al. RFID as a new ICT tool to monitor specimen life cycle and quality control in a biobank. The International Journal of Biological Markers. 2011; 26: 129–135. [DOI] [PubMed] [Google Scholar]
- [39].Palmirotta R, De Marchis ML, Ludovici G, Ialongo C, Leone B, Lopez N, et al. A reliable and reproducible technique for DNA fingerprinting in biorepositories: a pilot study from BioBIM. The International Journal of Biological Markers. 2013; 28: e398–404. [DOI] [PubMed] [Google Scholar]
- [40].Oster-Granite ML, Parisi MA, Abbeduto L, Berlin DS, Bodine C, Bynum D, et al. Down syndrome: national conference on patient registries, research databases, and biobanks. Molecular Genetics and Metabolism. 2011; 104: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].McCabe LL, McCabe ERB. Down syndrome and personalized medicine: changing paradigms from genotype to phenotype to treatment. Congenital Anomalies. 2013; 53: 1–2. [DOI] [PubMed] [Google Scholar]
- [42].Lavigne J, Sharr C, Ozonoff A, Prock LA, Baumer N, Brasington C, et al. National down syndrome patient database: Insights from the development of a multi-center registry study. American Journal of Medical Genetics. Part A 2015; 167A: 2520–2526. [DOI] [PubMed] [Google Scholar]
- [43].Einfeld SI, Brown R. Down syndrome— new prospects for an ancient disorder. The Journal of the American Medical Association. 2010; 303: 2525–2526. [DOI] [PubMed] [Google Scholar]
- [44].Hendrix JA, Amon A, Abbeduto L, Agiovlasitis S, Alsaied T, Anderson HA, et al. Opportunities, barriers, and recommendations in down syndrome research. Translational Science of Rare Diseases. 2021; 5: 99–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Baumer NT, Becker ML, Capone GT, Egan K, Fortea J, Handen BL, et al. Conducting clinical trials in persons with Down syndrome: summary from the NIH INCLUDE Down syndrome clinical trials readiness working group. Journal of Neurodevelopmental Disorders. 2022; 14: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bardhan S, Li H, Tarver E, Schramm C, Brown M, Garcia L, et al. The National Institutes of Health INvestigation of Co-occurring conditions across the Lifespan to Understand Down syndromE (INCLUDE) Project: Accelerating research discoveries for people with Down syndrome across the lifespan. American Journal of Medical Genetics. Part C, Seminars in Medical Genetics. 2024; 196: e32081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.