Abstract
The ambisense S segment of Rift Valley fever (RVF) virus (a phlebovirus in the Bunyaviridae family) codes for two proteins: the viral complementary-sense RNA for the N nucleoprotein and the genomic-sense RNA for the nonstructural protein NSs. Except for the fact that the NSs protein is phosphorylated and forms filamentous structures in the nuclei of infected cells (R. Swanepoel and N. K. Blackburn, J. Gen. Virol. 34:557–561, 1977), its role is poorly understood, especially since the replication cycle of all these viruses takes place in the cytoplasm. To investigate the mechanisms involved in filament formation, we expressed NSs in mammalian cells via a recombinant Semliki Forest virus and demonstrated that the protein alone was able to form structures similar to those observed in RVF virus-infected cells, indicating that the presence of other RVF virus proteins is not required for filament formation. The yeast two-hybrid system was used to show that the protein interacts with itself and to map the interacting domains. Various deletion and substitution mutants were constructed, and the mutant proteins were analyzed by immunoprecipitation, Western blotting and immunofluorescence. These experiments indicated that the 10 to 17 amino acids of the carboxy-terminal domain were involved in self-association of the protein and that deletion of this acidic carboxy-terminal domain prevents the protein from forming filaments but does not affect its nuclear localization. The role of two phosphorylation sites present in this domain was also investigated, but they were not found to have a major influence on the formation of the nuclear filament.
In addition to the structural proteins incorporated into the viral particles, many viruses express a number of nonstructural proteins, whose function is not always understood. For Bunyaviridae, a family of spherical enveloped viruses with a trisegmented single-stranded RNA genome of negative or ambisense polarity (27), the role of the nonstructural proteins remains to be established. Although the five genera of this family, Bunyavirus, Phlebovirus, Nairovirus, Hantavirus, and Tospovirus, possess similar coding capacities (the L, M, and S segments, coding for the L protein, for the precursor to envelope glycoproteins G1 and G2, and for the N nucleoprotein, respectively), most of the diversity exists in the nonstructural proteins and their gene organization (for reviews, see references 11 and 31). Bunyaviruses, phleboviruses, and tospoviruses code for two nonstructural proteins called NSs and NSm (encoded by the S and M segments, respectively), whereas hantaviruses and possibly nairoviruses do not seem to possess a coding capacity for such nonstructural proteins. The NSm protein is generated by cleavage from the glycoprotein precursor in bunyaviruses and phleboviruses and is translated from a unique mRNA transcribed from the ambisense M segment in tospoviruses. Similarly, the NSs protein of tospoviruses and phleboviruses is encoded by a unique genome-sense mRNA transcribed from the ambisense S segment, whereas the small NSs protein of bunyaviruses is translated from a bicistronic mRNA which synthesizes both the NSs and N proteins from two overlapping reading frames. With regard to the NSs protein, not only does the strategy of expression vary within genera but also the primary sequence of this protein is poorly conserved among different members. For instance, the amino acid sequences of the phlebovirus NSs proteins could not be aligned (16). No function has so far been ascribed to the NSs protein of any member of the family. Within the Phlebovirus genus, NSs appears to vary with different representatives: in Punta Toro virus-infected cells, it localizes in the cytoplasm and is present in minute amounts in purified particles (28); in Uukuniemi virus-infected cells, it is associated with the 40S ribosomal subunit (32); in Karimabad virus-infected cells, it was found exclusively in the cytoplasm (33); and for Rift Valley fever (RVF) virus, NSs differs from its counterparts in other phleboviruses because it is phosphorylated and forms filamentous structures in the infected cell nuclei (34–36).
Early studies by Swanepoel and his group (34–36) and Ellis et al. (12) showed that the nuclear filament is composed of bundles of 50-nm-thick fibrils, which occupy half the length of the nucleus and are confined exclusively to the nuclei but not associated with nucleoli. The presence of a nonstructural protein in the nucleus of cells infected with RVF virus is unexpected, since this virus, like all the members of the family, utilizes only the cytoplasm as its site for multiplication. A possible role for this nonstructural protein was tested in a transcription reconstitution system, but the protein was not found to have any stimulatory or inhibitory activity (23).
RVF is a matter of public health concern in Africa; it is transmitted by mosquitoes and is responsible for large epidemics of hemorrhagic fevers with fatal cases. The causative virus was isolated for the first time in Kenya in 1931 during an epizootic affecting young animals, lambs, calves, and kids and causing an acute hepatitis. Since then, human infections, involving a variety of symptoms from benign fever to fatal hemorrhagic fever with hepatitis or encephalitis, have been described. The most recent epidemics occurred in 1977 and 1993 in Egypt, in 1987 in Mauritania, in 1990 to 1991 in Madagascar, and in 1997 to 1998 in Kenya, Somalia, and Tanzania (3).
As an approach to understanding the role of the nuclear filaments during RVF virus infection, we investigated the mechanisms involved in their formation and expressed the NSs protein via recombinant Semliki Forest viruses (SFV). The formation of the filamentous structures was dependent on the expression of the NSs protein alone, indicating that no other viral gene product is involved. Evidence for self-association of NSs was obtained with the yeast two-hybrid system, which was further used to map the interaction domain. Finally, by expressing various mutants of the NSs protein in mammalian cells, we demonstrated that the acidic amino acids representing the carboxy-terminal domain of NSs are required for self-association and are essential for filament formation but not for NSs transport into the nucleus.
MATERIALS AND METHODS
Cells and virus stocks.
BHK21 cells were cultured and maintained in Glasgow modified minimal essential medium (MEM) (Gibco-BRL) containing 10% tryptose phosphate, 10 mM HEPES, and 5% fetal calf serum (FCS) (Boehringer, Meylan, France). BSR cells were grown in Glasgow modified MEM containing 10% FCS. Vero (VC10) and CV1 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 5 and 10% FCS, respectively. Penicillin (200 U/ml) and streptomycin (200 μg/ml) were added to the media.
The MP12 strain of RVF virus (8) was grown in Vero (VC10) cells maintained in Dulbecco’s medium containing 2% FCS. Vaccinia virus vTF7-3 (kindly provided by B. Moss) was used for transient expression in CV1 cells transfected with plasmids expressing NSs by using DAC-30 (Eurogentec) as a transfecting agent.
Autographa californica nuclear polyhedrosis virus and recombinant baculovirus were grown in Spodoptera frugiperda cells (Sf9 cells) maintained in TC100 medium supplemented with 10% FCS and antibiotics.
Yeast and bacterial strains.
Saccharomyces cerevisae CG1945 and SFY526 were obtained from P. Legrain (Institut Pasteur, Paris, France). Strain CG1945 is MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 GAL4-542 gal80-538 cyhr2 LYS2::GAL1UAS-GAL1TATA-HIS3 URA3::GAL417-mers(x3)-CYC1TATA-lacZ, whereas strain SFY526 is MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112, canr GAL4-542 gal80-538 URA3::GAL1UAS-GAL1TATA-lacZ.
Manipulations of DNA, RNA, Escherichia coli XLI Blue (Promega), and yeasts were performed by previously described methods (4).
Construction of recombinant plasmids expressing the NSs protein and mutated forms.
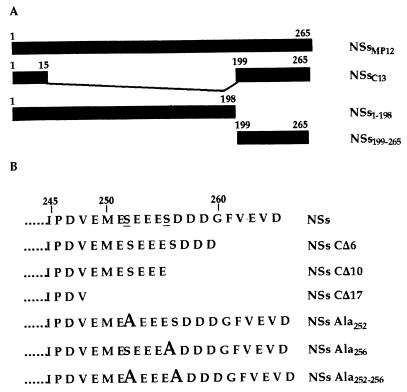
All the DNA fragments representing the open reading frame (ORF) coding for the NSs protein of MP12 (NSsMP12) or the deleted forms were generated by PCR with, as a template plasmid, pGem4Z-NSs containing the sequence of the ORF inserted into the unique HindIII site of pGem4Z (Promega). The sequence of the clone 13 virus protein was inserted into pBS-NSs-C13 (23). Deletion and substitution mutants are presented in Fig. 1. The oligonucleotides used for these constructions and their sequences are listed in Table 1. The sequence coding for NSsMP12 was amplified with the set of oligonucleotides NSFG 5′ and NSFAG 3′. The sets NSFG 5′ plus NSFAC1 3′, NSFG 5′ plus NSFAC2 3′, and NSFG 5′ plus NSFAC3 3′ were used to synthesize the genes of the deletion mutants NSs-CΔ6, NSs-CΔ10, and NSs-CΔ17, respectively. The sequences coding for the region corresponding to amino acids 1 to 198 and 199 to 265, were amplified with the sets NSFG 5′ plus NSpMP12 3′ and NSF2.5′ plus NSFAG 3′, respectively. Site-directed mutagenesis of the NSs gene was performed with the ExSite PCR-based mutagenesis kit (Stratagene, La Jolla, Calif.). Mutation of Ser252 or Ser256 to Ala were obtained with primer sets A2.1 plus A2.2 and A3.1 plus A3.2, respectively (Table 1). The double mutant containing Ser252 and Ser256 to Ala was obtained with primers A3.1 and A3-to-2 (Table 1). Primers A2.2, A3.2, and A3-to-2 were 5′ phosphorylated. All the mutations were checked by sequencing.
FIG. 1.
Schematic representation of NSs and the mutated forms. (A) Positions of the first and last amino acids of the complete and truncated NSs molecules. (B) Carboxy-terminal sequence of the NSs protein and mutated forms expressed in yeasts or in mammalian cells. The phosphoserines are underlined, and mutations to alanine are indicated in bold.
TABLE 1.
Oligonucleotides used for the construction of the NSs-expressing-plasmids
| Oligonucleotide | Sequence (5′-3′)a |
|---|---|
| NSFG 5′ | GGCCAAGCTTagatctTCATGGATTACTTTCCTGTGA |
| NSFAG 3′ | GGCCAAGCTTagatctTAACCTCTAATCAACCTCA |
| NSFAC1 3′ (∂17) | GGCCAAGCTTagatctTAAACATCTGGGATTGGAG |
| NSFAC2 3′ (∂10) | GGCCAAGCTTagatctTACTCCTCCTCTGATTCCAT |
| NSFAC3 3′ (∂6) | GGCCAAGCTTagatctTAATCATCATCACTCTCCTC |
| NSF2 5′ | GGCCAAGCTTagatctGCAGCAAGAGAG |
| NspMP12 3′ | GGCCAAGCTTagatctTACAGAAGCCGAACGCACTGT |
| A2.1 | GCAGAGGAGGAGAGTGATGATGATGGA |
| A2.2 | TTCCATCTCAACATCTGGGATTGG |
| A3.1 | GCTGATGATGATGGATTTGTTGAGGTT |
| A3.2 | CTCCTCCTCTGATTCCATCTCAAC |
| A3-to-2 | CTCCTCCTCTGCTTCCATCTCAAC |
The sequences corresponding to the HindIII and BglII sites are in boldface type; the HindIII site is in italics. The initiation and termination codons of the NSs gene are underlined. The nucleotides introducing mutations are in bold italic and underlined.
The amplified DNA fragments were digested with BglII for insertion into BamHI-cleaved pSFV1, pACTII, and pAS2 or with HindIII for insertion into pGem4Z previously digested with HindIII.
Yeast two-hybrid assays.
Yeast expression plasmids pAS2-1 and pACTII, as well as pAS2-prp11 and pACT-prp21, used for a positive control interaction (29), were kindly provided by P. Legrain (Institut Pasteur, Paris, France).
Transformations in CG1945 or SFY526 yeasts were performed by the lithium acetate method (6, 15, 19), and the transformants were plated on solid medium lacking leucine and tryptophan. To assay induction of the GAL4-driven HIS3 reporter gene, single colonies of transformed CG1945 cells were isolated and streaked on medium lacking leucine, tryptophan, and histidine. β-Galactosidase activity was assayed by using single colonies of SFY526-transformed cells as described previously (6, 18).
For immunoblotting, single colonies of transformed yeast CG1945 were grown in selective medium lacking leucine and tryptophan. Cells pelleted by low-speed centrifugation were lysed as described previously (37), and the proteins were analyzed by electrophoresis in a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel and immunoblotting with antibodies against GAL4-DNABD or GAL4-AD (Clontech) and anti-NSs ascitic fluid.
Generation of recombinant SFV.
Recombinant pSFV1-NSs plasmids digested at the unique SpeI site were transcribed in vitro by the SP6 RNA polymerase in the presence of cap analog as recommended by the supplier (Promega). Capped RNA transcripts were electroporated into BHK21 cells, together with an equal amount of RNA synthesized from the pSFV-helper 2 coding for the structural proteins of SFV and enabling packaging of the recombinant genomic RNA (7, 22). The progeny virus was then used for nonproductive infection in BSR cells and analysis of the recombinant protein.
Radiolabeling and cell fractionation.
Monolayers of approximately 106 BSR cells were infected with MP12, recombinant SFV, or vaccinia virus vTF7-3 at a multiplicity of infection (MOI) of 5 PFU per cell. After adsorption for 1 h at 37°C, the infected cells were incubated at 37°C in BHK-21 medium (Gibco-BRL) supplemented with 2% FCS. At 1 h before labeling, the cells were starved in methionine- and cysteine-free MEM and then labeled for 1 h with 200 μCi of a mixture of [35S]methionine and [35S]cysteine (Promix; Amersham) per ml and fractionated into cytoplasmic and nuclear extracts by the procedure described in reference 5. For protein analysis, the nuclear fraction was resuspended in 0.5 volume of Laemmli buffer, boiled, and clarified by centrifugation.
Production of antibodies against the RVF virus NSs protein.
A recombinant baculovirus expressing the NSs protein of the MP12 strain was generated after insertion of the DNA fragment into the YM1 vector and by using classical procedures to produce recombinant baculovirus (25). Sf9 cells infected at a MOI of 2 to 5 PFU per cell were incubated for 48 h at 28°C, harvested, washed in phosphate-buffered saline (PBS), and lysed in 0.5 mM Tris-HCl (pH 7.5) (approximately 107 cells per ml). After centrifugation at 10,000 × g for 30 min, the cytoplasmic fraction was discarded and the nuclear extract, corresponding to approximately 106 cells, was inoculated intraperitoneally into outbred mice (OF1 mice; Iffa Credo). Complete Freund’s adjuvant (Sigma) was added to the mixture for the first injection, and incomplete Freund’s adjuvant was added to the following ones on days 21, 42, and 63. On day 74, the ascitic TG180 cells (106 cells) were injected intraperitoneally, and ascitic fluid was withdrawn 11 days later.
Immunofluorescence staining.
BSR cells grown to subconfluency on coverslips were infected with MP12 or recombinant SFV at a MOI of 5 PFU per cell. At 24 h postinfection, the cells were washed twice with PBS, fixed with 3.7% formaldehyde in PBS for 30 min at 4°C, washed with PBS, and permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature. After being washed and treated with RNase A (250 μg/ml in PBS) for 10 min at room temperature, the cells were incubated for 30 min at 37°C with the anti-NSs mouse ascitic fluid at a 1:200 dilution in PBS-Tween 20 containing 10% FCS. Bound antibodies were visualized after incubation for 30 min at 37°C with fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G at a dilution of 1:100 in PBS-Tween 20 containing 10% FCS. Then the nuclei were stained with propidium iodine (0.5 μg/ml in PBS) for 10 min at room temperature. After the cells were washed extensively with PBS-Tween 20 containing 10% FCS, the coverslips were mounted for examination under a confocal microscope (Zeiss).
Immunoprecipitation.
Prior to immunoprecipitation, a 50-μl aliquot of cytoplasmic or nuclear extract was diluted 10- or 20-fold, respectively, with solubilization buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40) and incubated with 5 μl of nonimmune ascitic fluid to eliminate nonspecific complexes, and the supernatant was incubated overnight at 4°C with 10 μl of the anti-NSs ascitic fluid as described previously (23). The immunoprecipitates were eluted from the protein A-Sepharose beads in 25 μl of 2× Laemmli buffer and analyzed after electrophoresis in an SDS–12% polyacrylamide gel.
Cross-linking of NSs in cellular extracts.
VC10 cells (106) infected with MP12 virus at a MOI of 5 PFU per cell were harvested at 24 h postinfection and lysed in 100 μl of buffer A (10 mM HEPES [pH 8.0], 50 mM NaCl, 0.5 M sucrose, 1 mM EDTA, 0.2% Triton X-100). After the cells were centrifuged at 1,500 × g for 10 min at 4°C, the pellet containing membranes and nuclei was resuspended with gentle agitation in 100 μl of buffer B (10 mM HEPES [pH 8.0], 500 mM NaCl, 25% glycerol, 0.1 mM EDTA) containing 10 μl of a cocktail of protease inhibitors (Sigma). After incubation of the cells for 1 h at 4°C followed by centrifugation at 7,000 × g for 30 min, the supernatant was collected and incubated for 30 min at 25°C with freshly prepared dilutions of glutaraldehyde (25% solution; Merck). The proteins were analyzed after electrophoresis in an SDS–8% polyacrylamide gel.
RESULTS
The NSs protein expressed alone is able to form nuclear filaments.
Intranuclear inclusions were first detected in the hepatocytes of RVF virus-infected animals by Daubney et al. (10). Later, Swanepoel and his group (34–36) detected nuclear filaments in cells infected with various virulent strains and correlated this observation with the synthesis of a 31-kDa nonstructural phosphoprotein. To further characterize the NSs protein and the filaments, first we prepared monospecific polyclonal ascitic fluid from mice inoculated with a lysate of Sf9 cells infected with a recombinant baculovirus expressing the NSs of MP12 in large amounts. The hyperimmune ascitic fluid reacted positively in the enzyme-linked immunosorbent assay and in immunofluorescence tests involving mammalian cells infected with RVF virus but did not react with uninfected cells. We confirmed the distinct pattern of intranuclear fluorescence in various mammalian cells (BHK21, BSR, Vero, and CV1) and mosquito cells (AP61) infected with the MP12 attenuated strain of RVF virus.
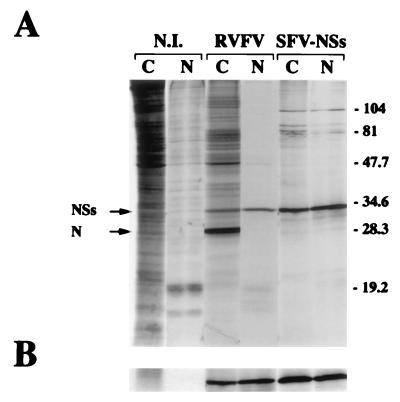
The sequence of the S segment of MP12 had already been reported (16), and the theoretical molecular weight of the NSs protein was estimated to be 29,900 with a net charge of 7.4. Expression of the recombinant NSs protein was analyzed after infection of confluent monolayers of BSR cells with the recombinant SFV at a MOI of 5 PFU per cell. Cells infected with the MP12 strain, or mock infected were run as controls. After incubation for 24 h at 37°C, the cells were labeled for 1 h in the presence of [35S]methionine and [35S]cysteine and fractionated into nuclear and cytoplasmic extracts. The proteins were analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography, before (Fig. 2A) and after (Fig. 2B) immunoprecipitation with the NSs-monospecific mouse polyclonal ascitic fluid. The recombinant NSs protein and the authentic protein were detected in both the cytoplasmic and nuclear compartments, in the same proportion (approximately 50% as estimated by PhosphoImager analysis [Molecular Dynamics Inc.]) (Fig. 2A). It should be noted that, as expected, the nucleoprotein N was clearly visible in the cytoplasmic fraction of MP12 virus-infected cells but not in the nucleus (Fig. 2A), providing a control for the fractionation procedure.
FIG. 2.
Distribution of the NSs protein between the cytoplasmic and nuclear fractions. (A) Confluent monolayers of BSR cells were infected with the MP12 strain of RVF virus or SFV-NSs or mock infected (N.I.). At 6 h postinfection, the cells were labeled with 200 μCi of a mixture of [35S]methionine and [35S]cysteine per ml for 1 h and fractionated into cytoplasmic (C) and nuclear (N) extracts. The proteins were analyzed by electrophoresis in SDS–12% polyacrylamide gels and autoradiography. The positions of the N and NSs proteins and the molecular mass markers (in kilodaltons) are shown on the left and right, respectively. (B) Nuclear and cytoplasmic extracts were immunoprecipitated with the mouse polyclonal antibodies against the NSs protein. The immune complexes were analyzed by SDS–12% polyacrylamide gel electrophoresis followed by autoradiography.
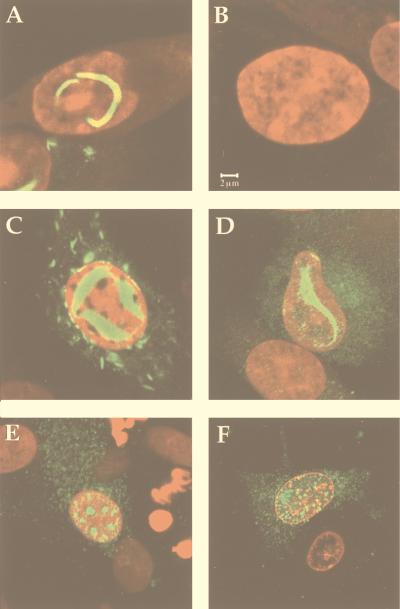
To determine whether the recombinant NSs protein assembled into filamentous structures in the nucleus, BSR cells grown on coverslips were infected with SFV-NSs (or with MP12 virus as a control), collected 24 h postinfection, and processed for immunofluorescence tests. Examination by confocal microscopy showed the presence of filaments in the nuclei of cells infected with MP12 virus (Fig. 3A) as well as with SFV-NSs (Fig. 3C). In the SFV-NSs-infected cells, NSs was expressed to a high level, forming a thick intranuclear ribbon-like filament. The similarity between the patterns of intranuclear fluorescence in cells infected with MP12 and SFV-NSs (Fig. 3A and C) clearly demonstrates that NSs is able to form filaments in the absence of other RVF virus proteins. Although the purpose of this paper is to focus on the nuclear filaments, it is worthwhile noting that fluorescence was also visible in the cytoplasm of SFV-NSs- and MP12 virus-infected cells. In fact, work still in progress in our laboratory showed that a fraction of NSs remains in the cytoplasm and associates with viral and cellular structures (39).
FIG. 3.
Localization of NSs in BSR cells infected with RVF virus, recombinant SFV expressing NSs, or carboxy-terminally deleted mutants. BSR cells were infected with RVF virus (A), SFV-NSs (C), SFV-NSsCΔ6 (D), SFV-NSsCΔ10 (E), or SFV-NSsCΔ17 (F) or not infected (B). At 24 h postinfection, the cells were fixed and stained with mouse polyclonal antibodies against NSs protein followed by fluorescent goat anti-mouse immunoglobulin antibodies.
The NSs protein expressed in yeast interacts with itself.
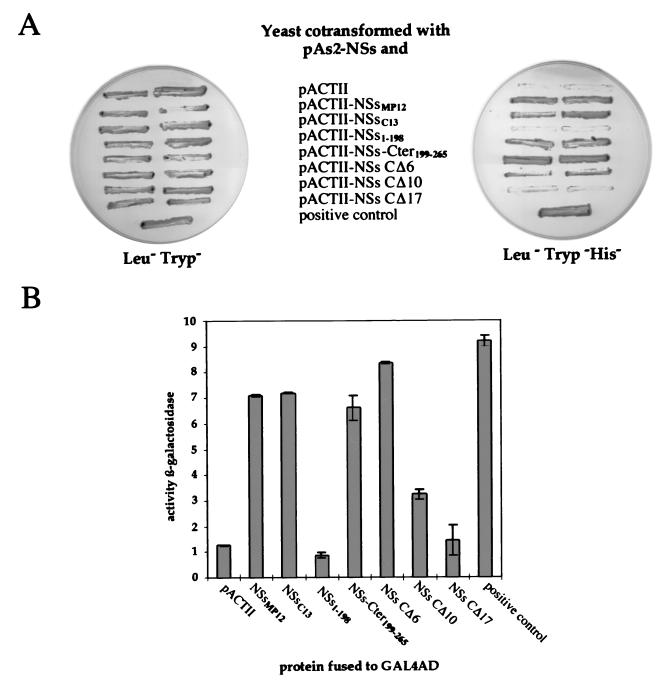
To determine the process involved in filament formation, we investigated whether the NSs protein associates with itself. To this end, we tested possible protein-protein interactions by using the yeast GAL4-based two-hybrid system (13). The complete ORF coding for the NSs protein of the MP12 strain was cloned into the yeast protein expression vectors pAS2 and pACTII, as fusion proteins, in frame with the GAL4 DNA-binding and transcription activation domain, respectively, of the GAL4 transactivator. Figure 4A shows that yeast colonies cotransformed with pAS2-NSs and pACTII-NSs did grow in this medium, indicating that the NSs protein associated with itself. The absence of growth of yeasts transformed with pAS2-NSs and the pACTII control plasmid confirmed the specificity of the interaction. Qualitative and quantitative assays were also carried out after cotransformation of strain SFY526, which contains the lacZ reporter gene. Interaction was assessed by the appearance of blue colonies in the presence of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (results not shown) and by assaying the β-galactosidase activity of yeast cells grown in liquid medium (Fig. 4B). These assays confirmed that NSs interacts with itself.
FIG. 4.
Analysis of the ability of NSs to self-associate in the yeast two-hybrid system. (A) Analysis of interactions between NSs and itself or truncated forms in the yeast two-hybrid assay with the HIS3 gene. Yeast CG1945 cotransformed with the pAS2-NSs and pACTII plasmids expressing the indicated proteins were selected on plates containing minimal medium lacking leucine and tryptophan (Leu−Tryp−) or lacking leucine, tryptophan, and histidine (Leu−Tryp−His−) to assay the induction of the GAL4-driven HIS3 reporter gene. The interaction between prp11 and prp21 was used as a positive control. (B) Quantitative analysis of interactions by induction of the lacZ reporter gene. The yeast cells SFY526 cotransformed by the same combination of plasmids as described for panel A were assayed for β-galactosidase activity. The results are the mean β-galactosidase activity determined from two independent yeast cotransformants assayed in triplicate with o-nitrophenyl-β-d-galactopyranoside as a substrate. Error bars indicate standard deviation.
The C-terminal acidic domain of the NSs protein is involved in the interaction of the protein with itself.
To map the NSs interaction domain involved in self-association, deletion mutants were constructed and expressed in the yeast two-hybrid system. All the deletion mutants depicted in Fig. 1A were cloned into both pAS2 and pACTII plasmids, but for clarity and because the results were similar, only one combination is shown.
Clone 13 is a naturally attenuated isolate of RVF virus which does not produce nuclear filaments and possesses a large deletion of 70% of the NSs ORF, conserving in frame the 15 and 67 amino acids of the amino and carboxy termini, respectively, of the protein (26). One of the obvious deletion mutants to test was the clone 13 NSs protein. The ORF encoding this 82-amino-acid protein was cloned into the yeast plasmids and expressed in frame with the GAL4 activation domain. Yeast cotransformed with pAS2-NSsMP12 and pACTII-NSsC13 were able to grow in the absence of histidine (Fig. 4A), demonstrating that this internally deleted NSs associated with the complete NSs. Other experiments also showed that NSsC13 can interact with itself (results not shown).
To determine the region involved in the interaction, we designed and tested two deletion mutants, NSs1–198, containing the region of MP12 virus NSs from amino acids 1 to 198, and NSs-Cter199–265, containing the carboxy terminus encoded by the ORFs of both MP12 and clone 13 and corresponding to the 67 terminal amino acids (Fig. 1A). Expression of the DNA-binding domain–NSs and activation domain–NSs fusion proteins in extracts of yeasts cotransformed with pAS2-NSs and PACTII-NSs was confirmed by Western blotting with monoclonal antibodies against GAL4 DNA-binding or activation domain or polyclonal antibodies against the NSs protein (results not shown). Figure 4 showed that the fusion protein containing NSs-Cter199–265 but not NSs1–198 activated the transcription of the HIS3 and lacZ genes, demonstrating that the interaction domain localized within the carboxy-terminal region of the protein.
To investigate the amino acids involved in the interaction, three additional deletion mutants were constructed, i.e., NSs-CΔ6, NSs-CΔ10, and NSs-CΔ17, in which the very last 6, 10, or 17 terminal amino acids were deleted (Fig. 1B). The carboxy terminus of NSs is rich in acidic amino acids, which are conserved in NSs-CΔ6 and partially and completely deleted in NSs-CΔ10 and NSs-CΔ17, respectively. As shown in Fig. 4A, yeasts coexpressing NSsMP12 and NSs-CΔ6 or NSs-CΔ10 grew in medium lacking histidine whereas those coexpressing NSsMP12 and NSs-CΔ17 did not. These results were confirmed in assays involving activation of lacZ (Fig. 4B), except that the interaction between NSs and the mutant NSs-CΔ10 was found negative for activation of the lacZ gene. Considering that activation of the HIS3 gene is more sensitive than that of lacZ, this would indicate that the interaction between the NSs and NSs-CΔ10 proteins is weak. Taken together, these data suggest that most of the 17 amino acids of the carboxy terminus and probably the acidic ones from 249 to 259 are responsible for the self-association of the NSs protein.
The carboxy terminus of NSs protein is involved in filament formation but is not necessary for nuclear localization.
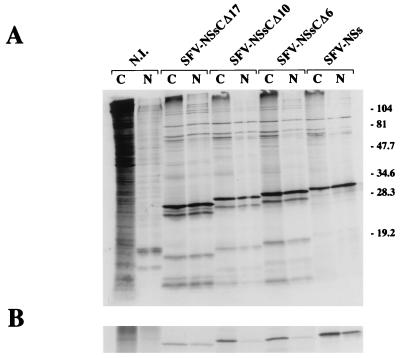
To determine whether the carboxy-terminal domain necessary for self-association of NSs is responsible for filament formation and nuclear localization, recombinant SFV expressing the truncated forms NSs-CΔ6, NSs-CΔ10, and NSs-CΔ17 were constructed after cloning the complete or deleted ORF of the NSs protein into plasmid pSFV1. Expression of mutant proteins was analyzed by radiolabeling and immunoprecipitation and by immunofluorescence tests. Figure 5 shows the analysis of nuclear and cytoplasmic proteins from BSR cells infected with the recombinant viruses before (Fig. 5A) or after (Fig. 5B) immunoprecipitation with the NSs-monospecific antibodies. Although the nuclear truncated forms appeared to be immunoprecipitated less efficiently than the cytoplasmic forms, all the mutated NSs proteins were immunoprecipitated and detected in both compartments. Other experiments showed that the truncated forms of NSs were poorly recognized in Western blotting, suggesting that a significant fraction of the polyclonal antibodies was raised against linear epitopes present in the carboxy-terminal region and that the remaining part of the protein contained conformational epitopes. It should be noted that the pattern of migration of the proteins in Fig. 5A suggested that the recombinant protein, deleted or not, were sensitive to degradation even in the presence of high concentrations of protease inhibitors or that incomplete translation products were generated.
FIG. 5.
Distribution of the deletion mutants of NSs protein between the cytoplasmic and nuclear fractions. BSR cells were infected with SFV expressing NSs, NSsCΔ6, NSsCΔ10, or NSsCΔ17 or not infected (NI). At 6 h postinfection, the cells were labeled with 200 μCi of a mixture of [35S]methionine and [35S]cysteine per ml for 1 h. After cytoplasmic (C) and nuclear (N) fractionation, proteins were analyzed on SDS–12% polyacrylamide gels before (A) and after (B) immunoprecipitation with mouse polyclonal antibodies against the NSs protein. The positions of molecular size markers in kilodaltons are shown on the right.
Confocal microscopy of BSR cells infected with SFV recombinants expressing the truncated forms NSs-CΔ17, NSs-CΔ10, and NSs-CΔ6 indicated that like the NSsMP12 protein expressed by RVF virus or by recombinant SFV (Fig. 3A and C), the mutant protein NSs-CΔ6 (Fig. 3D) was still able to form filamentous structures in the nuclei of infected BSR cells. In contrast, in cells expressing NSs-CΔ10 (Fig. 3E) or NSs-CΔ17 (Fig. 3F), no filament was visible but the mutated protein was detected in the nuclei, forming aggregates with a punctate pattern.
Although the SFV system was chosen because its replication cycle occurs in the cytoplasm, one of the nonstructural protein, nsP2, was shown to localize in the nuclear compartment (30). To exclude the possibility that the SFV nsP2 biased the analysis of the NSs protein and its mutants, we used the transient-expression system based on plasmids transfected in cells expressing the T7 RNA polymerase via the vaccinia virus vTF7-3 (14). After transfection of pGem4Z-NSs into CV1 cells infected with vTF7-3, labeling, and immunoprecipitation or immunofluorescence assay, all the mutant proteins were detected in the nucleus and the cytoplasm (data not shown), indicating that the SFV expression system did not bias the results.
The carboxy-terminal sequence depicted in Fig. 1B shows that the domain for self-association contains two potential sites of phosphorylation by casein kinase II at positions 252 and 256. Experiments in progress in our laboratory clearly indicate that these two serine residues are phosphorylated during infection of Vero cells with MP12 (20). To assess the role of the phosphoserines in the self-association and filament formation, the two serine residues were mutated to alanine by site-directed mutagenesis. Thus, we generated plasmids pGem4Z-NSs-Ala252 and pGem4Z-NSNs-Ala256, each containing a single substitution, and pGem4Z-NSs-Ala252&256, containing a double mutation. Expression of these mutated proteins was analyzed after transfection of these T7 promoter-driven plasmids in cells infected with the vaccinia virus vTF7-3. The proteins from the cytoplasmic and nuclear extracts were analyzed by Western blotting and immunofluorescence (results not shown). Each of the single- and double-mutant protein was detected in both compartments, and nuclear filaments were observed in cells transfected with each of the three plasmids.
Together, these results clearly indicated that the domain for self-association is required for filament formation but not for nuclear localization and that the acidic amino acids present in the domain play a major role. In addition, the two phosphorylation sites present in this acidic region do not appear to have a major influence on filament formation and transport of the protein into the nucleus.
The nuclear filament is composed of NSs dimers.
In an attempt to purify the filament, the nucleoplasmic proteins were extracted from the nuclear fraction by treatment with a high concentration of NaCl (500 mM). Chromatin, DNA, and membranes were pelleted by centrifugation, and most of the NSs protein was detected in the supernatant (results not shown), strongly suggesting that the filament is not tightly associated with chromatin.
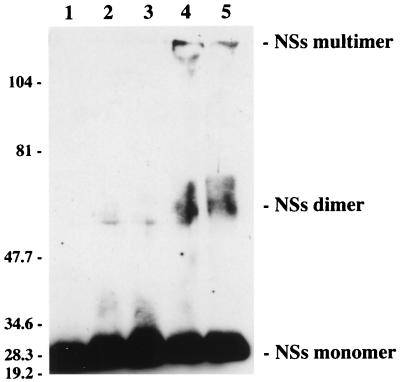
To confirm by more direct means that NSs associates with itself to form the filament, we performed cross-linking experiments with glutaraldehyde as a cross-linker. The nucleoplasmic proteins were treated with various concentrations of glutaraldehyde and analyzed in an 8% polyacrylamide gel. Western blotting indicated that concentrations of glutaraldehyde as low as 0.1% modified the migration of NSs: in addition to the monomer, two bands were detected corresponding to the size of dimers and to a very high molecular mass visible at the top of the gel, probably representing the stabilized form of the filament (Fig. 6). When the cytoplasmic extract was treated similarly, NSs was found associated with several high-molecular-mass proteins but the dimeric form was not observed (results not shown), suggesting that dimerization occurs only in the nucleus.
FIG. 6.
Analysis of nucleoplasmic proteins after cross-linking with various concentrations of glutaraldehyde. Proteins from the nucleoplasmic fraction were treated with 0% (lane 1), 0.05% (lane 2), 0.1% (lane 3), 0.5% (lane 4), or 1% (lane 5) glutaraldehyde, electrophoresed in an 8% polyacrylamide gel, and analyzed by Western blotting with NSs-specific antibodies. The positions of the molecular size markers in kilodaltons are shown on the left.
DISCUSSION
Among phleboviruses, RVF virus is unique in forming filamentous structures in the nuclei of infected cells, and the reason why NSs is transported to the nucleus even though all the steps of the viral cycle are localized in the cytoplasmic compartment is still unclear. Together, our results established a clear correlation between self-association of NSs and filament formation. Among other members of the family, tospoviruses are the only viruses to exhibit similar structures in infected plant cells, but, in contrast to the RVF virus-induced NSs filaments, these inclusions were located in the cytoplasm (reviewed in reference 17). Baculoviruses and entomopoxviruses (EPV) also induce specific filaments in infected cells during the late phase of infection. In the case of the well-studied insect poxvirus Amsacta moorei EPV, large bundles of filaments composed of a phosphoprotein called FALPE (filament-associated late protein of EPVs) accumulate exclusively in the cytoplasm. The role of these filaments is not known. In baculovirus-infected cells, filaments composed of the p10 protein were found in both the cytoplasm and nucleus (1). Like the RFV virus NSs protein, p10 and FALPE were shown to interact with themselves to form polymers. Self-association of these proteins appeared to be due to the presence of amphipathic alpha-helices engaged in coiled-coil interactions (reference 2 and references therein). Coiled coils are present in numerous fibrous proteins such as keratin, myosin, and fibrinogen and were identified as the dimerization element of leucine zipper proteins (reviewed in reference 24). When analyzed with the Robinson-Garnier and Chou-Fasman computer programs for secondary-structure prediction, the NSs protein was shown to contain helices at its carboxy terminus but no heptad repeat specific for coiled-coil structures. However, such structures may have escaped detection by the program, since coiled-coil domains identified in the crystal structures of several proteins were not predicted by the programs.
Analysis of the primary sequence of NSs of MP12 did not reveal the presence of the classical nuclear localization signal or its bipartite version (reviewed in reference 38). Although this 30-kDa molecule can, in principle, pass through the nuclear pore complex, it is probably actively transported. If this is the case, the mechanism and the signal for its import will have to be determined. The presence of the NSs protein of MP12 and all the deleted mutants in the nucleus indicated that the carboxy-terminal domain was not responsible for nuclear localization. This was confirmed by the observation that the small truncated clone 13 NSs protein of 8.5 kDa was found only in the cytoplasm (21). Also, this seems to indicate that the internal sequence specific for the NSs of MP12 contains the import signal(s).
As to the cytoplasmic form of NSs, it appears that neither the MP12 protein nor the clone 13 protein forms filaments in this compartment. The absence of filaments in the cytoplasm was already reported for cell infection with other RVF virus strains (12). If filament formation results from dimerization of NSs through its carboxy-terminal domain, it is possible that in the cytoplasm, this region interacts strongly with viral or cellular proteins and therefore is not accessible for dimerization. It might also be that filament formation requires a cellular protein present only in the nucleus. For the p10 protein of baculovirus, it was shown that once phosphorylated, the protein associated with microtubules, which probably play a role in process formation (9). Further studies are necessary to determine the mechanism of filament formation and to characterize the cytoplasmic form of NSs, its phosphorylation status, and the cellular and viral proteins with which it interacts with the aim of understanding the role of this protein in the biology and pathogenicity of the virus.
ACKNOWLEDGMENTS
We thank P. Legrain and M. Froment for their help in setting up the yeast two-hybrid system, R. Hellio for his excellent work in confocal microscopy, Claude Leclerc and Marie-Françoise Saron for their advice on immunizing the mice, and J. Foulon for antibody production. The use of the SFV replicon was initiated in collaboration with H. Garoff.
Fellowships to F.Y. were financed in part by La Direction Générale de l’Armement and by la Fondation Mérieux, and that to A.K. was financed in part by Ministère de l’éducation nationale du Grand Duché du Luxembourg. The confocal microscope was purchased with a donation from Marcel and Liliane Pollack.
REFERENCES
- 1.Alaoui-Ismaili M H, Richardson C D. Identification and characterization of a filament-associated protein encoded by Amsacta moorei entomopoxvirus. J Virol. 1996;70:2697–2705. doi: 10.1128/jvi.70.5.2697-2705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alaoui-Ismaili M H, Richardson C D. Insect virus proteins (FALPE and p10) self-associate to form filaments in infected cells. J Virol. 1998;72:2213–2223. doi: 10.1128/jvi.72.3.2213-2223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. An outbreak of Rift Valley fever, Eastern Africa, 1997–98. Weekly Epidemiol Rep. 1998;73:105–112. [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kinsington R E, Moore D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 5.Avalos R T, Yu Z, Nayak D P. Association of influenza virus NP and M1 proteins with cellular cytoskeletal elements in influenza virus-infected cells. J Virol. 1997;71:2947–2958. doi: 10.1128/jvi.71.4.2947-2958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker D M, Lumblad V. Manipulations of yeast genes. Introduction of DNA into yeast cells. In: Ausubel F M, Brent M, Kingston R E, Moore D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. pp. 13.7.1–13.7.10. [Google Scholar]
- 7.Berglund P, Sjöberg M, Garoff H, Atkins G J, Sheahan B J, Lilljeström P. Semliki Forest virus expression system: production of conditionally infectious recombinant particles. Bio/Technology. 1993;11:916–920. doi: 10.1038/nbt0893-916. [DOI] [PubMed] [Google Scholar]
- 8.Caplen H, Peters C J, Bishop D H. Mutagen-directed attenuation of Rift Valley fever virus as a method for vaccine development. J Gen Virol. 1985;66:2271–2277. doi: 10.1099/0022-1317-66-10-2271. [DOI] [PubMed] [Google Scholar]
- 9.Cheley S, Kosik K S, Paskevich P, Bakalis S, Bayley H. Phosphorylated baculovirus p10 is a heat-stable microtubule-associated protein associated with process formation in Sf9 cells. J Cell Sci. 1992;102:739–752. doi: 10.1242/jcs.102.4.739. [DOI] [PubMed] [Google Scholar]
- 10.Daubney R J, Hudson J R, Garnham P C. Enzootic hepatitis or Rift Valley fever. An undescribed virus disease of sheep, cattle, and man from East Africa. J Pathol Bacteriol. 1931;34:545–579. [Google Scholar]
- 11.Elliott R M. The Bunyaviridae. New York, N.Y: Plenum Press; 1996. [Google Scholar]
- 12.Ellis D S, Shirodaria P V, Fleming E, Simpson D I H. Morphology and development of Rift Valley fever virus in Vero cell cultures. J Med Virol. 1988;24:161–174. doi: 10.1002/jmv.1890240205. [DOI] [PubMed] [Google Scholar]
- 13.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 14.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giorgi C, Accardi L, Nicoletti L, Gro M C, Takehara K, Hilditch C, Morikawa S, Bishop D H. Sequences and coding strategies of the S RNAs of Toscana and Rift Valley fever viruses compared to those of Punta Toro, Sicilian sandfly fever, and Uukuniemi viruses. Virology. 1991;180:738–753. doi: 10.1016/0042-6822(91)90087-r. [DOI] [PubMed] [Google Scholar]
- 17.Goldbach R, Peters D. Molecular and biological aspects of tospoviruses. In: Elliott R M, editor. The Bunyaviridae. New York, N.Y: Plenum Press; 1996. pp. 129–153. [Google Scholar]
- 18.Guarente L. Strategies for the identification of interacting proteins. Proc Natl Acad Sci USA. 1993;90:1639–1641. doi: 10.1073/pnas.90.5.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohl, A. 1998. Unpublished data.
- 21.Kohl, A., and A. Billecocq. 1998. Unpublished data.
- 22.Liljestrom P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 23.Lopez N, Muller R, Prehaud C, Bouloy M. The L protein of Rift Valley fever virus can rescue viral ribonucleoproteins and transcribe synthetic genome-like RNA molecules. J Virol. 1995;69:3972–3979. doi: 10.1128/jvi.69.7.3972-3979.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 25.Matsuura Y, Possee R D, Overton H A, Bishop D H. Baculovirus expression vectors: the requirements for high level expression of proteins, including glycoproteins. J Gen Virol. 1987;68:1233–1250. doi: 10.1099/0022-1317-68-5-1233. [DOI] [PubMed] [Google Scholar]
- 26.Muller R, Saluzzo J-F, Lopez N, Dreir T, Turell M, Smith J, Bouloy M. Characterization of clone 13, a naturally attenuated avirulent isolate of Rift Valley fever virus, which is altered in the small segment. Am J Trop Med Hyg. 1995;53:405–411. doi: 10.4269/ajtmh.1995.53.405. [DOI] [PubMed] [Google Scholar]
- 27.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D. Virus taxonomy. Sixth report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. pp. 300–315. [Google Scholar]
- 28.Overton H A T, Ihara T, Bishop D H L. Identification of the N and NSs proteins coded by the ambisense S RNA of Punta Toro phlebovirus using monospecific antisera raised to baculovirus expressed N and NSs proteins. Virology. 1987;157:338–350. doi: 10.1016/0042-6822(87)90276-5. [DOI] [PubMed] [Google Scholar]
- 29.Rain J C, Tartakoff A M, Kramer A, Legrain P. Essential domains of the PRP21 splicing factor are implicated in the binding to PRP9 and PRP11 proteins and are conserved through evolution. RNA. 1996;2:535–550. [PMC free article] [PubMed] [Google Scholar]
- 30.Rikkonen M. Functional significance of the nuclear-targeting and NTP-binding motifs of Semliki Forest virus nonstructural protein nsP2. Virology. 1996;218:352–361. doi: 10.1006/viro.1996.0204. [DOI] [PubMed] [Google Scholar]
- 31.Schmaljohn C. Bunyaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1447–1471. [Google Scholar]
- 32.Simons J F, Persson R, Pettersson R F. Association of the nonstructural protein NSs of Uukuniemi virus with the 40S ribosomal subunit. J Virol. 1992;66:4233–4241. doi: 10.1128/jvi.66.7.4233-4241.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith J F, Pifat D Y. Morphogenesis of sandfly fever virus (Bunyaviridae family) Virology. 1982;121:61–81. doi: 10.1016/0042-6822(82)90118-0. [DOI] [PubMed] [Google Scholar]
- 34.Struthers J K, Swanepoel R. Identification of a major non-structural protein in the nuclei of Rift Valley fever virus-infected cells. J Gen Virol. 1982;60:381–384. doi: 10.1099/0022-1317-60-2-381. [DOI] [PubMed] [Google Scholar]
- 35.Struthers J K, Swanepoel R, Shepherd S P. Protein synthesis in Rift Valley fever virus-infected cells. Virology. 1984;134:118–124. doi: 10.1016/0042-6822(84)90277-0. [DOI] [PubMed] [Google Scholar]
- 36.Swanepoel R, Blackburn N K. Demonstration of nuclear immunofluorescence in Rift Valley fever infected cells. J Gen Virol. 1977;34:557–561. doi: 10.1099/0022-1317-34-3-557. [DOI] [PubMed] [Google Scholar]
- 37.Transy C, Legrain P. The two-hybrid: an in vivo protein-protein interaction assay. Mol Biol Rep. 1995;21:119–127. doi: 10.1007/BF00986502. [DOI] [PubMed] [Google Scholar]
- 38.Whittaker G R, Helenius A. Nuclear import and export of viruses and virus genomes. Virology. 1998;246:1–23. doi: 10.1006/viro.1998.9165. [DOI] [PubMed] [Google Scholar]
- 39.Yadani, F. 1998. Unpublished data.