SUMMARY
Resident cardiac macrophages are critical mediators of cardiac function. Despite their known importance to cardiac electrophysiology and tissue maintenance, there are currently no stem-cell-derived models of human engineered cardiac tissues (hECTs) that include resident macrophages. In this study, we made an induced pluripotent stem cell (iPSC)-derived hECT model with a resident population of macrophages (iM0) to better recapitulate the native myocardium and characterized their impact on tissue function. Macrophage retention within the hECTs was confirmed via immunofluorescence after 28 days of cultivation. The inclusion of iM0s significantly impacted hECT function, increasing contractile force production. A potential mechanism underlying these changes was revealed by the interrogation of calcium signaling, which demonstrated the modulation of β-adrenergic signaling in +iM0 hECTs. Collectively, these findings demonstrate that macrophages significantly enhance cardiac function in iPSC-derived hECT models, emphasizing the need to further explore their contributions not only in healthy hECT models but also in the contexts of disease and injury.
In brief
Lock and Graney et al. develop a human engineered cardiac tissue with an incorporated iPSC-derived macrophage population to better mimic the complex cell landscape of the native myocardium. Macrophage inclusion leads to increased contractile function of the tissue, which is attributed to macrophage stimulation of the cardiomyocyte β-adrenergic signaling pathway.
Graphical Abstract
INTRODUCTION
In recent years, human engineered cardiac tissues (hECT) have emerged as sophisticated, versatile platforms for studying the human heart in a simplified and highly controllable manner.1–5 As the adult human heart is extraordinarily complex, engineered models aim to recapitulate particular subsets of the heart’s structural and functional characteristics that can then be utilized to investigate specific biological questions of interest. While all hECT designs use combinations of cells, biomaterials, and bioactive factors, the resulting tissue models vary widely and are often designed toward specific applications in modeling disease, injury, and drug testing.6–9
The development and adoption of these tissue models for specific applications has been greatly advanced by the advent of induced pluripotent stem cell (iPSC) technology, which has provided a means to sustainably generate and study the cardiomyocytes and supporting cells of the human heart in an accessible and ethically non-controversial manner.10 However, it is well known that iPSC-derived cardiomyocytes are immature compared to cardiomyocytes native to the adult human heart.11,12 Historically, many engineered tissue designs have been dedicated to finding ways to advance the tissue to more faithfully capture the heart’s unique characteristics or more comprehensively characterize the tissue’s structure and function. Often, the focus is on the design of the tissue itself, by changing the materials used,13–15 the tissue architecture,3,5,16 or the readouts to better quantify tissue function.17–19 In contrast, modulation of the cellular composition of the tissues is less explored, with most of the focus centered on the cardiomyocytes and fibroblasts. Although they are the most prevalent cell types in the myocardium, they are far from the only cell types present in the cellular landscape, which includes populations like endothelial cells, pericytes, natural killer cells, lymphocytes, and dendritic cells, among others.20 Notably, no existing entirely iPSC-derived 3D cardiac tissue model, to our knowledge, has incorporated immune cells, despite their importance to the heart’s function and their central roles during disease and injury.1,2,21–23 In the native myocardium, immune cells account for 10.4% of the atrial and 5.3% of the ventricular cell landscapes.20 While there are many different cardiac immune cells, we opted to focus on macrophages, as myeloid cells represent the most abundant cardiac immune cells.24,25
Macrophages are highly heterogeneous cells that play pivotal roles in the human heart during both health and disease.26–28 Classified as part of the innate immune system, macrophages were originally considered to be mainly present in and derived from the blood, performing phagocytosis, chemotaxis, cytokine secretion, and antigen presentation for immune defense and tissue healing. More recent studies have revealed that there are also separate macrophage populations that reside within most tissues of the body. These tissue-resident macrophages not only differ in origin and self-renew independently from blood monocyte-derived macrophages but also have distinct phenotypic profiles and functions specific to the tissue microenvironment in which they reside.27,29,30
Recent studies of resident macrophages in the heart have demonstrated that under steady-state conditions, murine resident macrophages electrically modulate cardiomyocytes via connexin-43 gap junctions and maintain cardiac homeostasis by clearing dead cells and debris.31 Resident macrophages also play critical cardioprotective roles, as their depletion inhibits adaptive cardiomyocyte growth in response to hypertensive stress32 and promotes aberrant remodeling following myocardial infarction,33 leading to cardiac dysfunction in mice. While these discoveries have laid the groundwork for establishing the physiological importance of cardiac resident macrophages to the function of the myocardium, the reliance on murine models may limit translation to human health and disease. However, studies of human cardiac resident macrophages remain challenging due to the inaccessibility of human myocardium.
Recently developed protocols have established that CD14+CD68+ macrophages can be successfully derived from iPSCs (iM0s).34,35 iM0s are fully functional, capable of exerting phagocytosis and efferocytosis, as well as exhibiting the trademark phenotypic plasticity toward pro-inflammatory, M1-like and pro-regenerative, M2-like phenotypes in response to directed chemokine stimulation. While these macrophages are monocyte derived, they have a shared ontogeny with tissue-resident macrophages,36 making them a suitable population to use as a “resident” macrophage population in a hECT model.
In this work, an all-iPSC model of human cardiac tissue was engineered to create an in vitro model that is more recapitulative of the native heart and to provide a way to probe the impact of resident macrophages on human cardiac tissue phenotype and function. Bulk RNA sequencing was performed to evaluate changes in hECT molecular phenotypes. We further assessed the impact of iM0s on cardiac function through the assessment of inflammatory cytokine secretion from the hECT and by quantifying changes in contractility and force generation via bright-field imaging and calcium handling.
RESULTS
Generation of hECT model
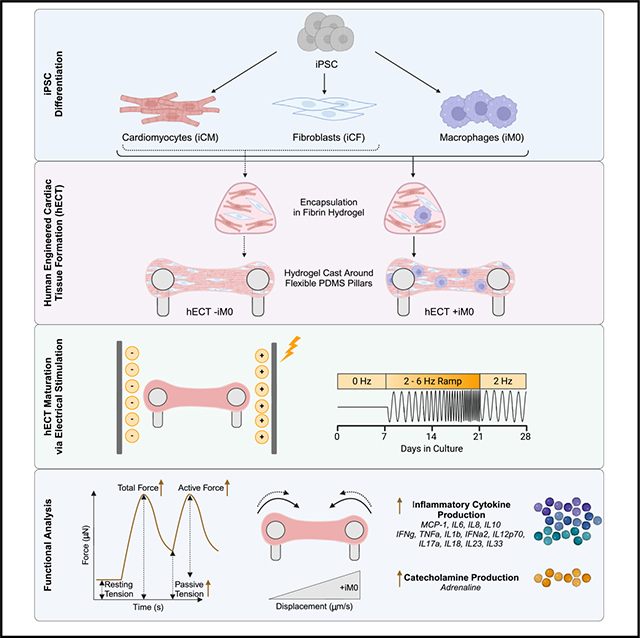
To establish a hECT model with an immune component, we leveraged our previously developed engineered tissue model design (milliPillar platform, Figure 1A).37 The cardiomyocytes (iCMs), cardiac fibroblasts (iCFs), and iM0s were independently differentiated from iPSCs using previously established protocols.34,38,39 iCM identity was confirmed by immunofluorescence markers of cardiac troponin T (cTNT) and α-actinin, and F-actin was used as a way to identify any non-myocyte cells present that were not positive for cTNT and α-actinin (Figure S1A). iCM function was confirmed by the spontaneous contraction, which typically began around day 10 of differentiation (Figure S1B). iCF identity was validated by immunofluorescence staining for GATA binding protein 4 (GATA4), transcription factor 21 (TCF21), and vimentin (Figure S1C) and gene expression (Figure S1D). Collagen secretion by iCFs increased significantly when stimulated by transforming growth factor β1, confirming iCF functionality (Figure S1E). Lastly, macrophages were differentiated from a CD68-YFP reporter iPSC line to enable real-time imaging of differentiated cells. Stable expression of CD68-YFP in iM0s relative to both unmodified and undifferentiated iPSCs was confirmed at the gene level using RT-qPCR, and immunofluorescence confirmed the expression of identity markers CD11b and CD68, as well as the expression of the endogenous CD68-YFP reporter (Figures S1F and S1G). The expression of myeloid markers (CD11b, CD14, human leukocyte antigen DR isotype [HLA-DR], and signal regulatory protein alpha [SIRPa]) was also confirmed by flow cytometry (Figure S1H). In addition, phagocytic uptake of latex beads confirmed iM0 functionality (Figure S1I).
Figure 1. Generation of iPSC-derived human engineered cardiac tissues containing macrophages.
(A) Cardiomyocytes (iCMs), cardiac fibroblasts (iCFs), and macrophages (iM0s) were derived from human iPSCs, encapsulated in a fibrin hydrogel, cast around PDMS pillars, and matured by ramped electrical stimulation over 28 days.
(B) Immunofluorescence of cardiac tissues prepared without (−iM0) and with (+iM0) macrophages. Scale bars are 100 μm.
(C) Quantification of macrophages retained in tissues after 28 days via CD45 staining.
(D) Dispersion of cTNT alignment.
(E) Dispersion of vimentin alignment.
(F) Size of iCMs with or without co-culture with iM0s.
(G) Circularity of iCMs with or without co-culture with iM0s.
Data represent mean ± SEM and were assessed via two-tailed t test; **p < 0.01.
Following iCM, iCF, and iM0 differentiation, the cells were mixed together in a fibrin hydrogel and suspended around flexible pillars to fabricate the engineered tissues. In tissues without macrophages (−iM0), cardiomyocytes were combined with fibroblasts in a 4:1 ratio to mimic the volumetric proportions observed in healthy tissue.40 In tissues with macrophages (+iM0), iM0s accounted for 7% of the total cell composition, since immune cells account for 10.4% of the atrial and 5.3% of the ventricular cell landscapes in the native myocardium.20 For the remainder of the tissue composition, the 4:1 ratio of cardiomyocytes to fibroblasts was preserved (resulting in 69.75% iCMs and 23.25% iCFs), as this ratio is known to affect tissue compaction, remodeling, and force generation.41,42 The formed hECTs were cultured over a period of 4 weeks and subjected to a frequency-ramped electrical stimulation regimen reported in our previous studies to improve tissue structure and function.37,43
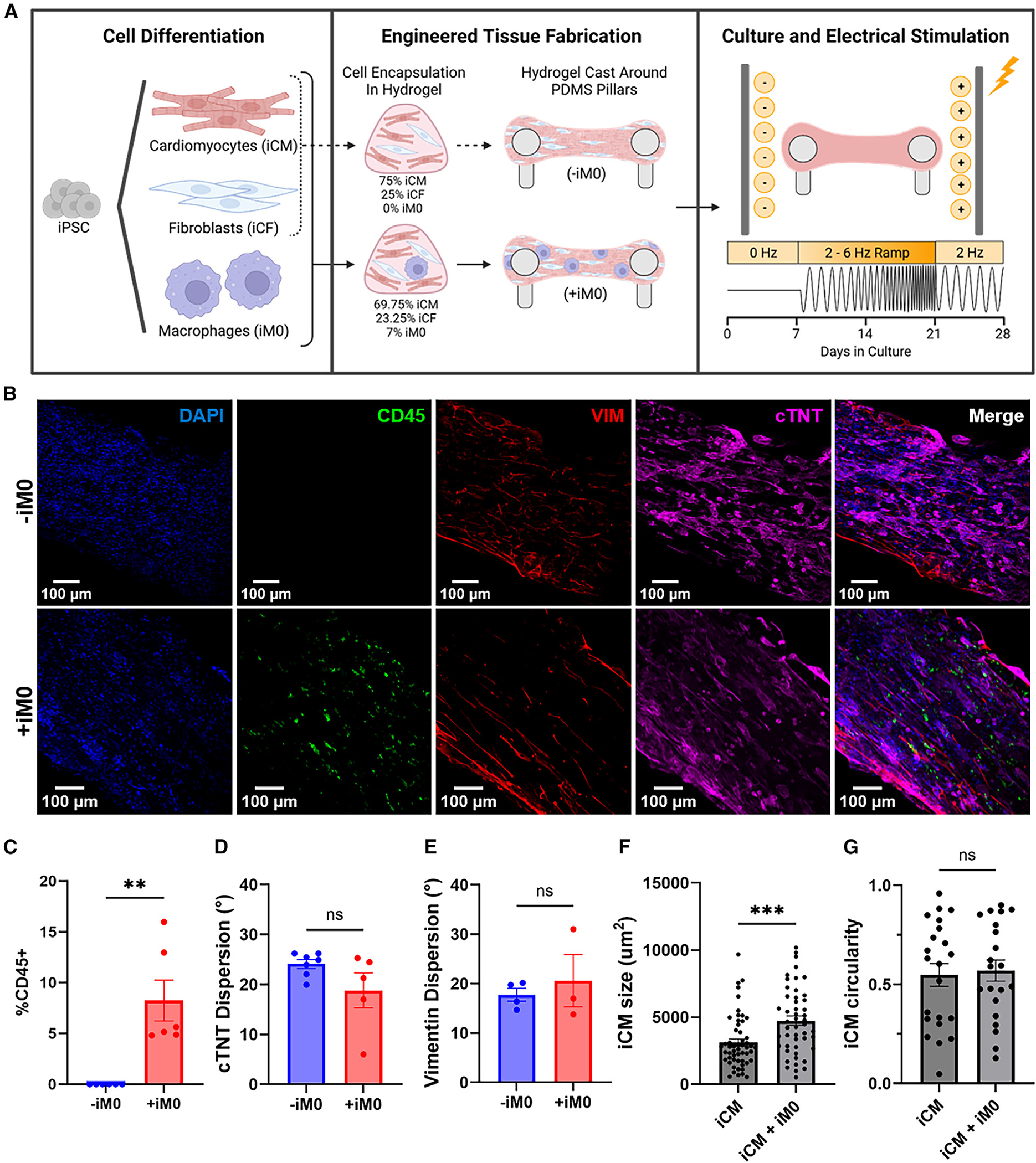
Importantly, we sought to determine whether the macrophage population integrated into the cardiac tissues during fabrication was retained throughout this extended culture period. The use of CD68-YFP-labeled macrophages enabled visualization of iM0s within the macrophage-seeded tissues (Figure S1J). At the end of the 28-day period, whole-mount immunofluorescence imaging of the tissues confirmed the maintained presence of an immune population using CD45, a pan-leukocyte antigen (Figure 1B). Based on quantitative analysis, CD45+ cells comprised approximately 8% of the cell population within hECTs seeded with iM0s (Figure 1C). Notably, iM0s lacked expression of the proliferation marker Ki-67 (Figure S1K), confirming that the macrophages were terminally differentiated and non-proliferative at the time of integration within hECTs. These data suggest that a resident macrophage population was retained in the hECTs for 28 days, with minimal change in the composition of the cellular landscape.
We subsequently assessed the impact of iM0 on the overall cell alignment within the tissues by quantifying the orientation of cTNT and vimentin within hECTs. There were no significant differences detected through whole-mount image analysis between tissues prepared with and without iM0 (Figures 1D and 1E); however, 2D co-culture of iCMs with iM0s led to increased iCM size, with no changes in iCM circularity (Figures 1F and 1G).
RNA sequencing confirms sustained macrophage presence
To further validate the presence of macrophages in the hECT model and provide insight into the functional differences caused by the macrophage population, we performed bulk RNA sequencing on the hECTs after 28 days of culture. Principal-component analysis demonstrated distinct separation of the hECTs prepared in the presence (+iM0) and absence (−iM0) of macrophages, and tight clustering of the replicates within each group, with principal component 1 accounting for 98% variance within the data (Figure 2A). Differential expression analysis (p < 0.01, log2 fold change >1) identified hundreds of differentially expressed genes, the bulk of which were upregulated in the +iM0 group compared to the −iM0 control group (Figure 2B). As expected, among the top upregulated genes were those associated with myeloid and macrophage identity, such as CD14 and CD68. Gene Ontology (GO) pathway analysis of biological processes also revealed an overwhelming upregulation of leukocyte-specific pathways and pathways involved in immune response, including “inflammatory response,” “innate immune response,” “cytokine-mediated signaling pathway,” “positive regulation of cytokine mediated production,” and “macrophage activation,” further confirming the presence of an immune population within the tissues (Figure 2C).
Figure 2. RNA-seq confirms iM0 presence within cardiac tissues after 4 weeks.
(A) Principal-component analysis of gene count data for hECTs prepared with (+iM0) and without (−iM0) macrophages.
(B) Enhanced volcano plot of differentially expressed genes, determined by p < 0.01, log2(fold change) > 1, including markers indicative of macrophage presence in CD14 and CD68.
(C) Chord diagram of the top 10 biological processes and corresponding differentially expressed genes generated by AdvaitaBio iPathwayGuide.
(D) Hierarchical clustering of gene counts for a panel of markers indicative of CCR2-HLA-DR+ tissue-resident macrophages in +iM0 and −iM0 tissues.
(E) Single-sample gene set enrichment analysis for gene sets describing macrophage populations found within the adult human heart. Analysis was not restricted to only differentially expressed genes. Data were analyzed via two-tailed t test.
(F) Production by +iM0 and −iM0 hECTs of inflammatory cytokines in tissue supernatant after 7 and 28 days.
Statistical significance was determined by two-way ANOVA with Tukey’s post hoc analysis; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Data represent mean ± SEM.
We aimed to further characterize the phenotype of iM0s within our hECT model after the 28-day cultivation period. Using the RNA sequencing data, we compared the expression of genes in hECTs containing iM0s against published lists of genes describing cardiac tissue-resident macrophages, as well as M1/M2 activation. Hierarchical clustering for genes indicative of CCR2-MERTK+ cardiac tissue-resident macrophages27 revealed the elevated expression of these markers in tissues prepared with iM0s (Figure 2D). To expand on these findings, we performed single-sample gene set enrichment analysis against transcriptomic signatures for macrophages in the adult human heart,20 including three populations of LYVE1+ macrophages, two populations of DOCK4+ macrophages, antigen-presenting macrophages, and monocyte-derived macrophages (Figures 2E and S2A). Interestingly, tissues prepared with iM0s were significantly enriched for LYVE1+MP-1 relative to control tissues without iM0s, whereas we did not observe a significant enrichment for any other profile of cardiac macrophages (Figure 2E). These data suggest that iM0s within the hECTs are a relatively homogeneous population of macrophages and may resemble a cardiac tissue-resident phenotype associated with cardiovascular remodeling. Given the monocytic origin of our iM0s, we further analyzed the tissue samples for enrichment against the transcriptomic profiles of circulating monocyte-derived pro-inflammatory (M1) and pro-regenerative (M2a, M2c) macrophages.44 Tissues prepared with iM0s were significantly enriched relative to control tissues for both M1 and M2a activation states (Figure S2B), suggesting an underlying inflammatory response of iM0.
Macrophages alter hECT cytokine production
Cytokine secretion plays an important role in influencing the microenvironment and directing cell behavior. Based on the bulk RNA-sequencing data, we chose to quantify the secretion of inflammatory mediators in conditioned supernatant collected from hECTs prepared with and without macrophages. Interestingly, only minimal differences were observed after 1 week of culture; however, by day 28, macrophages within hECTs significantly increased production of both pro-inflammatory cytokines, such as monocyte chemoattractant protein-1 (MCP-1), interleukin (IL)-6, IL-8, interferon γ, and tumor necrosis factor α (Figures 2F and S2C), and pro-regulatory cytokines, such as IL-10 (Figure 2F). While it could not be conclusively determined whether the macrophages are solely responsible for this phenomenon, it does suggest that the presence of the macrophages within the tissues drives cytokine production by directly producing cytokines and by inducing production by surrounding iCM and iCF cell populations.
Macrophages impact cardiac tissue function
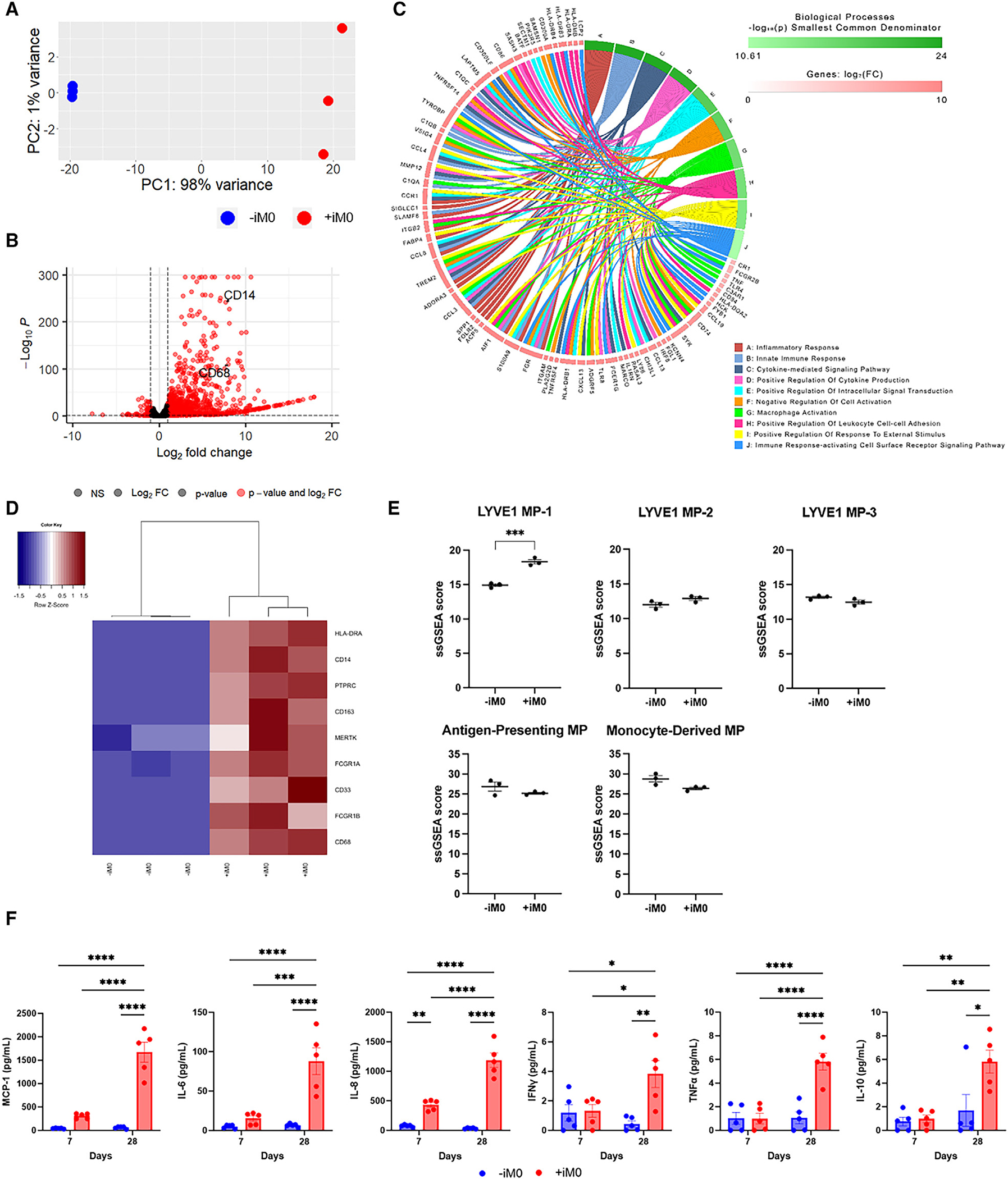
To assess the impact of macrophages on the overall contractile function of the cardiac muscle, video-enabled microscopy was utilized to track and analyze the displacement of the flexible pillars that move as the tissue contracts (Figure S3). Importantly, the mechanical properties of the pillars were measured, enabling force generation of the tissue to be calculated from the pillar displacement measurements. Furthermore, by taking into account (1) the distance between the pillars at rest, (2) the pillar displacement during tissue contraction, and (3) the cross-sectional area of each tissue, several metrics (e.g., force, stress, and contraction and relaxation velocity of the tissues) can be calculated to comprehensively characterize tissue functionality.
After 4 weeks of culture, we compared the final cross-sectional areas of each tissue to their original cross-sectional areas measured after the first 7 days of formation to assess tissue compaction over time and observed that +iM0 tissues tended to compact more than their −iM0 counterparts (Figure 3A), though the final cross-sectional areas of the tissues did not significantly differ between groups (Figure 3B). This indicates that macrophages may facilitate increased remodeling of the tissue during its initial formation and throughout the culture period. At rest, the passive tension, which is the force the tissues exert when at maximum relaxation, was higher in tissues prepared with macrophages compared to −iM0 controls (Figure 3C). However, the passive stress, which accounts for the cross-sectional area of the tissue, indicated no significant differences between the two groups (Figure 3D). This was not the case for the active force produced during contraction. Surprisingly, the tissues containing a macrophage population demonstrated a significantly increased active force production that was, on average, twice as high as the force exerted by the −iM0 tissues (Figure 3E), and this disparity became even more apparent when taking the cross-sectional area into account in the active stress metric (Figure 3F). This difference was also reflected in the total force generation (Figure 3G) and total stress of the tissues (Figure 3H), which denote the sum of their respective passive and active components. Macrophages also induced pronounced differences in the contraction and relaxation velocities (Figures 3I and 3J) compared to tissues without macrophages, likely caused by the increased active force production, as the pillar displacement for each contraction is larger. Collectively, these data suggest that macrophage inclusion in hECT facilitates more robust tissue contractility.
Figure 3. iM0s alter cardiac tissue function.
(A and B) Quantification of (A) tissue compaction and (B) tissue cross-sectional area at 28 days.
(C–J) Quantification of metrics of contractile function at 1 Hz stimulation including (C) passive tension, (D) passive stress, (E) mean active force, (F) mean active stress, (G) mean force, (H) mean stress, (I) mean contraction velocity, and (J) mean relaxation velocity.
Data represent mean ± SEM and were assessed via two-tailed t test; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Tissue-resident macrophages modulate β-adrenergic responses
To investigate the underlying reasons behind this increase in force generation, we referred back to the RNA sequencing data, aiming to discover affected pathways related to cardiomyocyte contractile function. By excluding the immune-related terms, the GO analysis revealed that there were also many upregulated genes in the +iM0 tissues that impact other relevant biological processes. Notably, many of these were related to calcium signaling, including “positive regulation of cytosolic calcium ion concentration,” “calcium ion homeostasis,” and “calcium iron transport” (Figure 4A). This result was particularly interesting, as calcium is the central element of excitation-contraction coupling, serving as a main regulator of force production in the cardiac tissue. Furthermore, among the affected pathways listed by KEGG analysis on the differentially expressed gene set was the “adrenergic signaling in cardiomyocytes” pathway (p < 0.05). While not the most significantly affected pathway listed, adrenergic signaling was of considerable interest because of its regulation by calcium signaling and known effects on cardiomyocyte contractile function, matching some of the functional changes identified between groups.
Figure 4. iM0s alter calcium and β-adrenergic pathways.
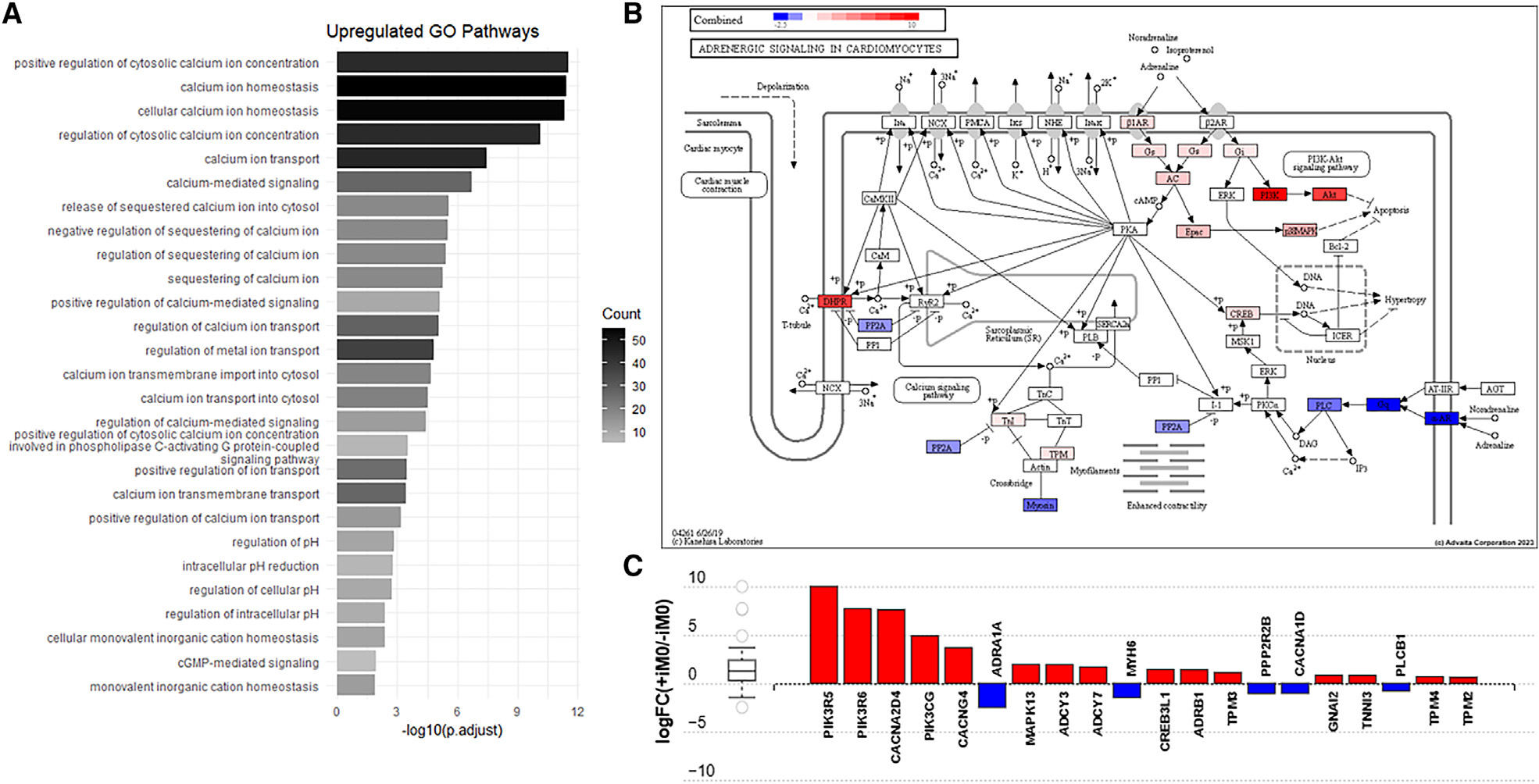
(A) Upregulated GO pathways in +iM0 compared to −iM0 tissues related to calcium signaling.
(B) KEGG pathway diagram of adrenergic signaling in cardiomyocytes showing up- and downregulated genes in +iM0 compared to −iM0 tissues combined with predicted downstream effects generated by AdvaitaBio iPathwayGuide.
(C) Quantification of differentially expressed pathway genes shown in the KEGG pathway diagram of adrenergic signaling in cardiomyocytes generated by AdvaitaBio iPathwayGuide.
The KEGG pathway diagram for adrenergic signaling in cardiomyocytes (Figure 4B) shows the computed perturbation of each gene in the pathway, accounting for the measured expression of differentially expressed genes from our RNA-seq dataset (Figure S4A), combined with the computed accumulated perturbation propagated across the pathway topology from upstream genes (Figure S4B), to show how the β-adrenergic signaling pathway is impacted. Assessment of the pathway diagram reveals that the +iM0 hECTs significantly increased the expression of adrenergic receptor-β1 (ADRB1) and adenylyl cyclase (ADCY3, ADCY7), as well as upregulation along the PI3K-Akt pathway (PIK3R5, PIK3R6, PIK3CG), both of which are involved in cardiomyocyte survival, growth, hypertrophy, and contractile function (Figure 4C). However, we also observed downregulated expression of ADRA1A, which encodes for the α-1A adrenergic receptor found in myocytes, as well as several downstream genes, including phospholipase C (PLCB1). Interestingly, macrophage integration also induced changes in the expression of genes responsible for the dihydropyridine receptor, an L-type Ca2+ channel complex, with significant upregulation of CACNA2D4 and CACNG4 and a slight downregulation in the expression of CACNA1D. Finally, we also noted changes in genes that code for sarcomeric proteins, including downregulation of myosin heavy chain 6 (MYH6) and upregulation of cardiac type troponin I3 (TNNI3) and tropomyosin-encoding genes TPM4 and TPM2. The differential expression of these genes may explain the altered force production observed between tissues prepared with and without macrophages.
To better understand how differences in gene expression affect calcium signaling and adrenergic signaling in hECTs, we utilized cardiomyocytes genetically encoded with a fluorescent calcium indicator (GCaMP) and subjected the tissues to an isoproterenol challenge. Isoproterenol is a well-established inotropic and chronotropic β-adrenergic agonist, which enhances local Ca2+ release in cardiomyocytes.45,46 As expected, both groups exhibited the characteristic dose-dependent increase in beat frequency (Figure 5A). However, an analysis of tau (Figure 5B), the calcium transient decay constant, revealed that at low concentrations of isoproterenol, the presence of macrophages led to a shorter tau compared to −iM0 controls; however, this difference was abolished at high isoproterenol concentrations. Both the period of contraction during the release of intracellular Ca2+ (Figure S5A) and the period of relaxation during the reuptake of intracellular Ca2+ (Figure S5B) were equally affected. At low concentrations of isoproterenol, iM0s consistently reduced the period of contraction and period of relaxation for the release and reuptake of intracellular Ca2+, respectively, indicating that this difference likely is not caused by a change to any specific step of the calcium cycling. To further test this, we subjected the tissues to 10 μM forskolin, which activates adenylyl cyclase, the protein directly downstream of the β-adrenergic receptor. The response to forskolin was similar to that observed for isoproterenol (Figures 5C, 5D, S5C, and S5D).
Figure 5. Macrophages modulate cardiac tissue β-adrenergic response.
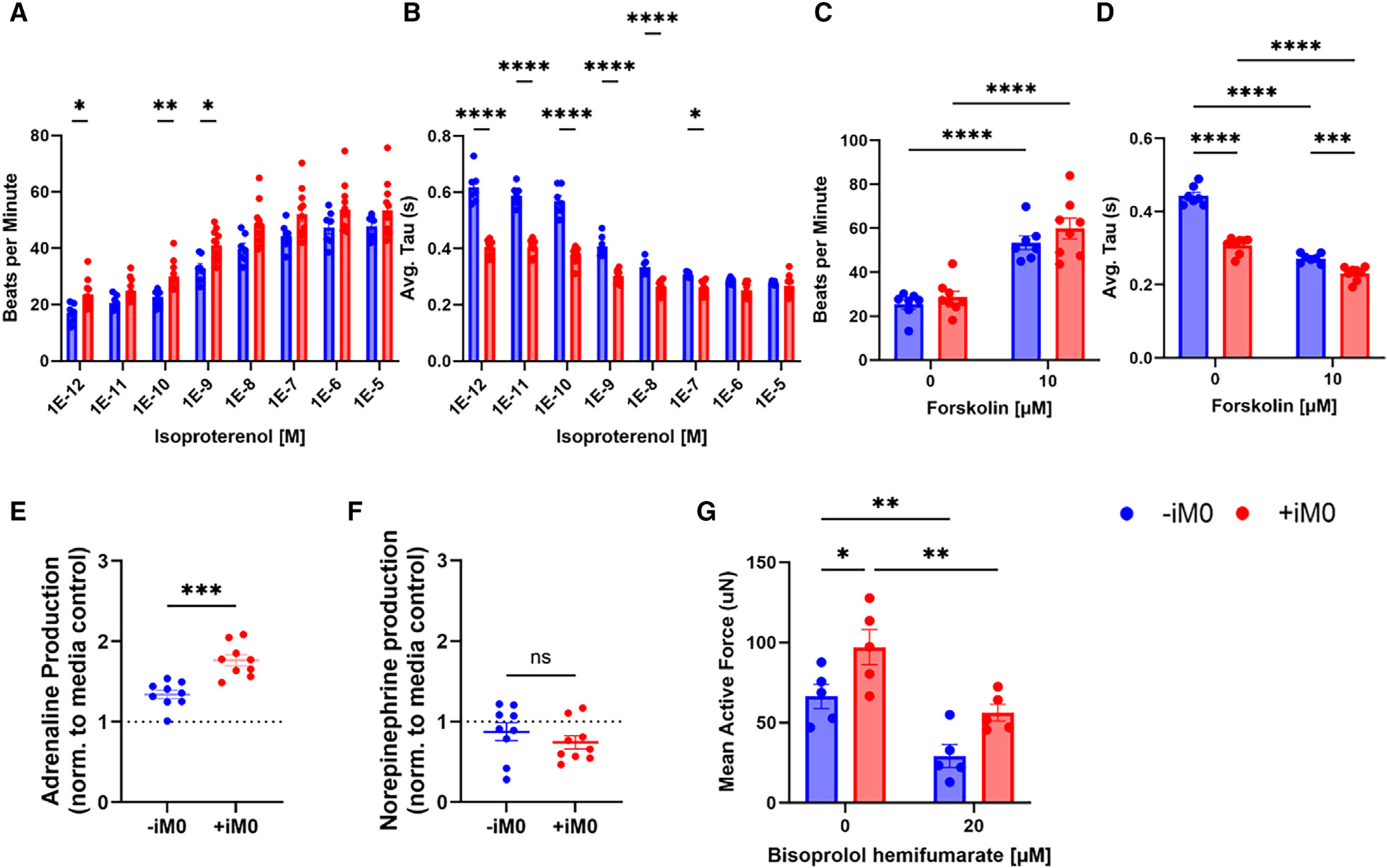
(A and B) Effect of isoproterenol challenge on hECT calcium transients shown through quantification of (A) beat rate and (B) average tau.
(C and D) Effect of 10 mM forskolin on hECT calcium transients shown through quantification of (C) beat rate and (D) average tau.
(E) Adrenaline and (F) norepinephrine production in −iM0 versus +iM0 hECTs.
(G) Mean active force production of tissues in response to treatment with β1-adrenergic receptor blocker bisoprolol hemifumarate.
Data represent mean ± SEM and were assessed via two-tailed t test for assessment of adrenaline and norepinephrine, or via repeated-measures two-way ANOVA with multiple comparisons comparing between conditions at each concentration of isoproterenol, forskolin, or bisoprolol respectively; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
The main catecholamines responsible for activating the β-adrenergic signaling pathway are adrenaline (epinephrine) and noradrenaline (norepinephrine).47 Quantification of catecholamine levels in the supernatant of hECTs confirmed that tissues containing iM0s produced significantly more adrenaline compared to hECTs without iM0s (Figure 5E). Additionally, among the three individual cell types, iM0s displayed the highest adrenaline production (Figure S5E). In contrast, we did not observe significant production of norepinephrine by hECTs, irrespective of macrophage presence (Figure 5F). Consistent with this finding, iM0s, iCMs, and iCFs did not produce significant quantities of norepinephrine when cultured in isolation (Figure S5F). These results suggest that iM0s are a source of adrenaline, which in turn stimulates the β-adrenergic signaling pathway in cardiomyocytes, leading to altered calcium signaling dynamics and the increased contractile function observed in the +iM0 hECTs.
To confirm this, we inhibited the β-adrenergic receptor 1 within hECTs using the β-blocker bisoprolol hemifumarate and quantified contractile force production. Indeed, β-blocker inhibition significantly reduced the mean active force production by the tissues and abrogated differences between hECTs prepared with and without macrophages (Figure 5G). Similar trends were observed in the mean contraction and relaxation speeds (Figures S5G and S5H). Together, these results suggest that iM0s enhance cardiac tissue contractile function through the stimulation of the β-adrenergic signaling pathway.
DISCUSSION
Here, we report the first all-iPSC in vitro model of engineered human myocardium containing a population of resident macrophages. We demonstrate that the macrophage population is retained across 28 days of cultivation and results in significantly altered contractile tissue function, increasing the active force and stress, as well as the contraction and relaxation velocities. Subsequent interrogation of the calcium signaling within the hECTs revealed that iM0s’ presence alters calcium signaling along the β-adrenergic pathway, which may contribute to the observed differences in contractile force.
A notable advantage of our model is that it is entirely iPSC derived, providing a uniform genetic background and patient specificity. Primary human cardiomyocytes and cardiac tissue-resident macrophages are very difficult to source, and while primary peripheral blood macrophages are more easily accessible, there is significant variation between donors, and their lifetime in culture is limited, making them less reproducible. In comparison, iPSC-derived cell types are both accessible and reproducible and can be scaled for higher-throughput studies. Furthermore, prior studies have demonstrated that for models containing multiple cell populations, the inclusion of iCFs rather than dermal fibroblasts as a stromal cell population significantly enhances the maturity and physiological relevance of the tissue model due to the increased cell-cell crosstalk.23 Recently, other groups have shown the potential for tissue-resident macrophage populations (e.g., resident macrophages in colonic organoids, microglia in brain organoids) to help mature iPSC-derived organoids past early fetal stages of development.48,49 In addition, fully iPSC-derived constructs have great potential for various future applications. These models can be genetically manipulated as desired to create isogenic controls or used to investigate inter-cellular interactions and cell-type-specific mechanisms in disease and injury modeling. With the inclusion of a resident macrophage population, this becomes even more pertinent, as the inflammatory response to pathophysiological stressors is often central to determining injury progression and outcome. This tissue engineered model could be particularly beneficial for investigating pathophysiological states involving the immune system, such as myocardial infarction and heart failure, cytotoxic injury, and viral and autoimmune myocarditis.50–52
We observed a relatively constant proportion of macrophages within +iM0 hECTs over 28 days that matched the initial cell composition used during fabrication. Considering the absence of Ki-67 expression by iM0s, and that iCMs are terminally differentiated, these findings suggest that iCFs within hECTs are also not proliferative. This is consistent with a recent report by Giacomelli et al., which suggests that COL1A+ iCFs have low levels of Ki-67 after 21 days of culture within engineered microtissues,23 leading to a relatively constant cellular landscape.
Given the plasticity of macrophages, we next sought to understand the changes in phenotype to iM0s following integration within our hECT model. The iM0s in this work were generated from hematopoietic progenitors originating from hemogenic endothelium; as a result, the ontogeny of iM0s is expected to be more similar to tissue-resident macrophages than peripheral blood monocyte-derived macrophages.34,36 Prior work has further demonstrated that iPSC-derived primitive macrophages can be differentiated into specialized tissue-resident-like macrophages in the presence of organ-specific stimuli.53 A comparison of our bulk transcriptomic data against immune populations in the adult human heart suggests that after 28 days, iM0s may begin to resemble a CCR2-HLA-DR+ cardiac tissue-resident phenotype. Although immune cells were absent from −iM0 hECTs, we did not observe enrichment in +iM0 hECTs for all transcriptomic profiles of macrophages. Importantly, these profiles define diverse macrophage subsets within cardiac tissues and contain genes that are not macrophage specific but are also expressed by other cells in hECTs. While our data suggest that iM0s resemble a single population of LYVE1+ cardiac macrophages, due to the known diversity of resident macrophages within cardiac tissue,20 single-cell transcriptomic analysis is needed to more precisely assess the macrophage landscape within hECTs at steady state.
During our characterization of the macrophages, we also observed an upregulation in the secretion of mostly pro-inflammatory cytokines from the hECT, especially during the later time points, suggesting that the cells are responding to a stressor. This is most likely caused by electrical stimulation applied to the cardiac tissues. Electrical signals can directly affect cell behavior and function, and while the electrical stimulation regimen used to develop this model has been shown to increase force production and cell organization within a tissue composed of cardiomyocytes and fibroblasts,43,54 it is less understood how the electrical stimulation affects the resident macrophage population. Primary macrophages display voltage-sensitive receptors and regulate their intracellular Ca2+ signaling pathways for functions like proliferation or polarization through different ion channels, suggesting that they could be impacted by electrical fields.55,56 Furthermore, studies of macrophage responses to an electrical field have found increased cytokine production, which supports the notion that it is the electrical stimulus causing the increase in cytokine production.57 Future work will explore the application of this model to injury and disease for a comprehensive analysis of the heterogeneity of and dynamic changes in resident cardiac macrophages. Notably, the macrophage-seeded hECT platform described in this work can be coupled with organ-chip systems to delineate the crosstalk between resident macrophages and circulating iPSC-derived monocytes, which remains incompletely understood.58,59
Importantly, we aimed to identify how the incorporation of an immune component into the hECT model would impact the function of the cardiac muscle as a whole. The most striking finding was that macrophage-seeded tissues had significantly increased force production compared to control tissues without macrophages. This result was surprising given that, by adding iM0s, the total numbers of cardiomyocytes and fibroblasts in the tissues were lower than their −iM0 counterparts. Force production is one of the hallmark characteristics by which hECT models are assessed with respect to determining the functionality and maturity of the tissue. While the force production by this +iM0 tissue model still falls short compared to that of the native human myocardium, it still demonstrates a marked improvement over the tissues containing only cardiomyocyte and fibroblast populations.
To uncover the mechanism by which macrophages facilitate this increased contractile force, we interrogated the β-adrenergic signaling pathway. Adrenergic signaling is known to have a variety of downstream effects on cardiac contractile function, and many gene expression differences, including the upregulation of β-adrenergic receptor 1 expression in +iM0 hECTs, were observed along this pathway. This was further supported by the observed increase in iCM size in 2D co-culture with iM0s, as cardiomyocyte hypertrophy is linked to adrenergic signaling.60 Isoproterenol challenge and forskolin treatment revealed differences in calcium transients of the hECTs at low versus high concentrations, with the shorter tau in +iM0 hECTs suggesting that the presence of macrophages in the tissue leads to altered calcium signaling by stimulating the β-adrenergic signaling pathway in +iM0 hECTs. Evaluation of the major catecholamines that stimulate the adrenergic pathway confirmed that +iM0 hECTs have higher adrenaline levels than −iM0 hECTs, and iM0s specifically produced the highest levels of adrenaline compared to iCMs and iCFs. This is consistent with existing literature of catecholamine production by primary immune cells, including lymphocytes and phagocytes.61–63 There is evidence that macrophages contain intracellular levels of catecholamines, such as dopamine and norepinephrine, and can release these as signals to surrounding cells.64 However, whether this is a de novo synthesis or the cells uptake and store catecholamines from their surrounding microenvironment is still unclear. Finally, to further confirm whether stimulation of the β-adrenergic pathway underlies the differences in contractile function between groups, a β-blocker was used to inhibit activation of the pathway. Administration of the β-blocker reduced the mean active force generation and abrogated differences between +iM0 and −iM0 hECTs. Together, these data suggest that the enhanced contractile function of the +iM0 cardiac tissues is facilitated by iM0 secretion of adrenaline, which in turn activates the β-adrenergic signaling pathway, leading to the downstream observed effects in cardiac tissue function.
In summary, we report a reproducible, patient-specific, iPSC-derived in vitro model of the human myocardium with a functional resident macrophage component. Under steady-state conditions, the inclusion of an additional cell population not only increased the complexity of the tissue’s cellular landscape and consequently the biological fidelity of the model but also significantly improved force generation and altered calcium signaling. Importantly, interrogation of this model revealed that macrophage presence modulates the β-adrenergic signaling pathway, potentially contributing to the observed changes in hECT function. Ultimately, this model can serve as a platform for future investigation of the interplay between resident macrophages and other cardiac cell populations under (patho)physiological conditions, leading to an improved understanding of disease and injury progression and the identification of potential therapeutic targets for translational applications.
Limitations of the study
There are several limitations to this study. The primary limiting factor of this work was due to the use of bulk RNA sequencing to determine transcriptomic changes within a complex tissue. Single-cell sequencing would allow for the delineation of differentially expressed genes specific to each cell population. Single-cell sequencing is of great interest for future studies, as it will enable greater insight into how the macrophage transcriptome changes within the cardiac microenvironment and whether it introduces heterogeneity within the macrophage population during culture. Additionally, in this study, we limited the scope to focus primarily on the impact of macrophages on the cardiomyocyte population given the differences we found in contractile function. However, we expect that iCFs are also affected by the presence of iM0s. Although we did not explore the impact of iM0s on the stromal cell population or the surrounding extracellular matrix of the tissue, this is an interesting avenue that would be greatly enabled by single-cell sequencing and proteomics. In this study, we show that iM0s increase the contractile function of hECTs, in part due to increased activation of the β-adrenergic pathway in cardiomyocytes. While our data suggest that β-adrenergic signaling contributes to these functional changes, our mechanistic understanding of this response is not comprehensive, and there are likely other factors that contribute. For example, we observed that +iM0 hECTs have increased production of several inflammatory cytokines that are known to influence many different cell types. Currently, our understanding of how the differences in inflammatory signal profiles may affect the hECT is limited. It is plausible that these cytokines impact tissue contractility and warrants further investigation. Finally, this study was only conducted using one cell line. It will be important in future studies to repeat this work using tissues fabricated from multiple iPSC lines from different patient donors to confirm and strengthen these initial findings.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Gordana Vunjak-Novakovic (gv2131@columbia.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Bulk RNA-sequencing data have been deposited at Gene Expression Omnibus (GEO) (GEO: GSE266797) and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Alexa Fluor 488 Phalloidin | ThermoFisher | Cat# A12379 |
| Cardiac Troponin T | Abcam | Cat# ab45932; RRID: AB_956386 |
| TCF21 | Sigma Aldrich | Cat# HPA013189; RRID: AB_10601215 |
| Vimentin | Abcam | Cat# ab24525; RRID: AB_778824 |
| CD11b | ThermoFisher | Cat# MA5-32793; RRID: AB_2810069 |
| CD68 | Abcam | Cat# ab955; RRID: AB_307338 |
| Alpha actinin | Sigma | Cat# A7811; RRID: AB_476766 |
| CD45 | Abcam | Cat# ab8216; RRID: AB_306361 |
| DAPI | BD Biosciences | Cat# 564907; RRID: AB_2869624 |
| AlexaFluor 700-conjugated anti-human CD45 | BioLegend | Cat# 304024; RRID: AB_493761 |
| Brilliant Violet 420-conjugated anti-human CD11b | BioLegend | Cat# 301324; RRID: AB_11219589 |
| Brilliant Violet 605-conjugated anti-human CD14 | BioLegend | Cat# 301834; RRID: AB_2563798 |
| PE/Cyanine7-conjugated anti-human HLA-DR | BioLegend | Cat# 307615; RRID: AB_493589 |
| APC-conjugated anti-human CD172/SIRPa | BioLegend | Cat# 372105; RRID: AB_2650862 |
| Ki-67 | Abcam | Cat# ab15580; RRID: AB_443209 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| (pLV(Exp)-Neo-CD68(short)>EYFP | VectorBuilder | Cat# VB211109-1377wwb |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Lenti-X Accelerator | Takara | Cat# 631257 |
| Isoproterenol | Sigma | Cat# I6504-500mg |
| Forskolin | Tocris | Cat# 1099 |
| Bisoprolol Hemifumarate | MedChemExpress | Cat# HY-B0076 |
| Y-27632 dihydrochloride | Tocris | Cat# 1254 |
| CHIR 99021 | Tocris | Cat# 4423 |
| Wnt-C59 | Tocris | Cat# 5148 |
| 6-aminocaproic acid | Sigma-Aldrich | Cat# A7824 |
| Fibrinogen | Sigma-Aldrich | Cat# F3879-100mg |
| Thrombin | Sigma-Aldrich | Cat# T6884 |
| PDMS/Sylgard 184 | Dow | Cat# 1024001 |
| Human Albumin | Sigma-Aldrich | Cat# A9731-1G |
| L-ascorbic acid 2-phosphate | Sigma-Aldrich | Cat# A8960 |
| Retinoic acid | Sigma-Aldrich | Cat# R1514 |
| FGF2 | Peprotech | Cat# 100-18B |
| SB35421 | Selleck Chemicals | Cat# S1067 |
| GlutaMAX | Invitrogen | Cat# 35050061 |
| Recombinant human macrophage colony stimulating factor | Peprotech | Cat# 300-25 |
| Human interleukin 3 | Peprotech | Cat# 200-03 |
|
| ||
| Critical commercial assays | ||
|
| ||
| STEMdiff Hematopoietic Kit | Stem Cell Technologies | Cat# 05310 |
| CD14 Microbeads, human | Miltenyi Biotec | Cat# 130-050-201 |
| LEGENDplex Human Inflammation Panel 1 | Biolegend | Cat# 740809 |
| RNeasy Micro Kit | Qiagen | Cat# 74004 |
| High-Capacity cDNA Reverse Transcription kit | Applied Biosystems | Cat# 4368813 |
| Sirius red total collagen detection kit | Chondrex | Cat# 9062 |
| Fluorescent red Latex beads | Sigma | Cat# L3030 |
| Adrenaline ELISA kit | Abcam | Cat# ab287788 |
| Norepinephrine ELISA kit | Abcam | Cat# ab287789 |
|
| ||
| Deposited data | ||
|
| ||
| Bulk RNA-sequencing | This paper | GEO: GSE266797 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| WTC11 | Material Transfer Agreements from Bruce Conklin, Gladstone Institute | Huebsch et al.65 |
| WTC11-GCAMP6f | Material Transfer Agreements from Bruce Conklin, Gladstone Institute | Huebsch et al.65 |
| WTC11-CD68-EYFP | Generated in this study | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S1 for list of RNA sequences used for RT-qPCR. | N/A | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| R Studio (version 4.3.1) | Posit | https://www.rstudio.com/ |
| GraphPad Prism 10.2.1 | GraphPad | https://www.graphpad.com/ |
| iPathwayGuide | Advaita Bioinformatics | https://advaitabio.com/ |
| Gene Ontology Resource | Global Core Biodata Resource | https://geneontology.org/ |
| REVIGO | REVIGO | http://revigo.irb.hr/ |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
hiPSC sourcing
hiPSCs were sourced from the previously generated WTC11 and WTC11-GCaMP6f hiPSC lines, gifted from Dr. Bruce Conklin.65 The parental hiPSC line, WTC11 (GM25256, Coriell Institute), was generated from an apparently healthy donor (Male, Asian, 30 years old at sampling) with no history of cardiovascular diseases. Additionally, to enable CD68 tracking within hiPSC-derived macrophages, a CD68-EYFP lentiviral vector (pLV(Exp)-Neo-CD68(short)>EYFP, VectorBuilder, cat. no. VB211109–1377wwb) was prepared and packaged by VectorBuilder. Lentiviral transduction was performed on the WTC11-hiPSC line using Lenti-X Accelerator (Takara, cat. no. 631257), according to the manufacturer’s protocol. Selection for stably expressing cells was performed using puromycin and expression of EYFP in target cells was confirmed by fluorescent microscopy (Olympus IX81) and RT-qPCR.
hiPSC culture
All hiPSCs were cultured in mTeSR Plus medium (STEMCELL Technologies, cat. no. 100–0276) on tissue culture plates coated with Matrigel (Corning, cat. no. 354230; diluted 1:100) and passaged every three to four days with 0.5 mM EDTA (Thermo Fisher Scientific, cat. no. 15575) in PBS (Corning, cat. no. 21–040). For the first 24 h after passaging, 5 μM Y-27632 dihydrochloride (Tocris Bioscience, cat. no. 1254), was added to the culture medium. hiPSCs were karyotyped and regularly tested for mycoplasma contamination.
hiPSC-cardiomyocyte differentiation
Cardiomyocytes were differentiated from hiPSCs by adapting the protocol developed by Burridge et al.38 iPSCs were replated at a density of 250,000 cells/cm2 on tissue culture plates coated with Matrigel in mTeSR Plus medium with 5 μM Y-27632 dihydrochloride. 24 h after plating, mTeSR Plus media was refreshed. Following that, for day 0 of the differentiation, cells were switched to CDM3 media (RPMI1640 (Gibco, cat. no. A41923–01), 0.5 mg/mL human Albumin (Sigma, cat. no. A9731–1G), and 213 μg/mL L-ascorbic acid 2-phosphate (Sigma, cat. no. A8960), with 5 μM CHIR 99021 (Tocris, cat. no. 4423). On day 1, medium was changed to CDM3. On day 2, medium was changed to CDM3 supplemented with 2uM Wnt-C59 (Tocris, cat. no. 5148). On day 4, medium was changed to CDM3, and refreshed every other day until day 10, when medium was switched to B27 media (RPMI 1640 + B27 supplement (Thermo Fisher Scientific, cat. no. 17504044). Contracting cells were noted as early as day 7, but typically around day 10.
hiPSC-cardiac fibroblast differentiation
WTC11 hiPSC-derived cardiac fibroblasts were differentiated and maintained according to a previously established protocol.39 In brief, hiPSCs were differentiated into cardiac progenitor cells through modulation of the Wnt signaling pathway, then on day 5 were dissociated and split 1:12 on Matrigel-coated plates in Advanced DMEM (Gibco, cat. no. 12634028) supplemented with 2 μM retinoic acid (Sigma, cat. no. R1514), 5 μM CHIR, and 5 μM Y-27632 dihydrochloride. On day 6 Y-27632 dihydrochloride was removed, and for day 8–12 cells were cultured in Advanced DMEM without any supplementation, differentiating the cells into epicardial cells. On day 12, cells were split and plated at 10,000 cells/cm2 in Fibroblast Growth Medium 3 (Promocell) supplemented with 20 ng/mL FGF2 (Peprotech, cat. no. 100–18B) and 10 μM SB35421 (Selleck Chemicals, cat. no. S1067), which was maintained until day 18 and refreshed every other day. The resulting differentiated iCFs were maintained in Fibroblast Growth Medium 3 and split upon reaching 90% confluency. iCFs were used for engineering tissues between passages 2 and 5.
hiPSC-macrophage differentiation
WTC11 cells expressing CD68-EFYP were replated on tissue culture plates coated with Matrigel (Corning, cat. no. 354230, diluted 1:175) with low agitation to allow for clusters of cells to attach at approximately 8–16 clusters/cm2. Cells were first differentiated into hematopoietic stem and progenitors using a STEMdiff Hematopoietic Kit (Stem Cell Technologies, cat. no. 05310), according to the manufacturer’s instructions, until day 12 of differentiation. At day 12, cells in suspension were removed and the adherent hemogenic endothelial cell fraction was cultured in X-VIVO 15 Serum-free Hematopoietic Cell Medium (Lonza, cat. no. 02–053Q) supplemented with 1:100 GlutaMAX (Invitrogen cat. no. 35050061), 1:100 2-mercaptoethanol, 100 ng/mL recombinant human macrophage colony stimulating factor (M-CSF, Peprotech, cat. no. 300–25), and 25 ng/mL recombinant human interleukin 3 (IL-3, Peprotech, cat. no. 200–03). Hemogenic endothelial cells were maintained for over 8 weeks with weekly media exchange, with cells in the suspension fraction discarded during week 1. Myeloid- and monocytic-specific lineage cells subsequently collected in the suspension fraction were replated onto 6-well ultra-low attachment plastic (Corning) in X-VIVO 15 media containing 100 ng/mL M-CSF for at least 5 days to achieve macrophage differentiation. CD14+ macrophages were positively selected prior to use via CD14 microbeads (Miltenyi, cat. no. 130–050-201), according to the manufacturer’s protocol.
Generation of human engineered cardiac tissues (hECTs)
hECTs were fabricated and cultured as previously described in our recently reported tissue engineering pipeline.37 Briefly, bioreactors were made by casting PDMS in custom-milled molds containing carbon rod electrodes. The resulting PDMS bioreactors were plasma-bonded to glass slides and the electrodes were connected to platinum wires to facilitate electrical stimulation during culture. The complete bioreactor was then autoclaved for sterility prior to use.
To fabricate the tissues, cardiomyocytes and fibroblasts were dissociated using 10X TrypLE Select Enzyme (Fisher, cat. no. A1217702) for 5 min at 37 C. Macrophages were dissociated using 0.5 mM EDTA for 5 min at room temperature. Cells were counted and resuspended at a concentration of 550,000 cells total per tissue in a solution of 3 mg/mL fibrinogen in B27 medium. In hECTs without macrophages (−iM0), tissue composition was 75% iCM and 25% iCF. In hECTs with macrophages (+iM0), the ratio of iCM to iCF was kept consistent, resulting in a tissue composition of 7% iM0, 69.75% iCM, and 23.25% iCF. For each tissue, 12 μL cell suspension was mixed with 3 μL thrombin (2.5 U/mL) in a well of the bioreactor to form the tissue around the pillars. The tissues were placed in a 37°C incubator for 30 min for the hydrogel to crosslink, then B27 media supplemented with 10uM Y-27632 dihydrochloride and 5 mg/mL 6-aminocaproic acid (Sigma-Aldrich, cat. no. A7824) was added to each well containing a tissue. 1–3 h post fabrication, tissues were manually detached from the sides of the well and the underlying glass slide using a 26-gauge needle.
On day 1 following fabrication, Y-27632 dihydrochloride was removed from the medium, and tissues were manually detached a second time. Medium was changed every day until day 5, when the 6-aminocaproic acid was removed from the medium. From day 5 until the end of culture, tissues were cultured in B27 medium that was refreshed every other day. On day 7, electrical stimulation of the tissues was started using a custom Arduino-based electrical stimulator as previously described 34, which stimulated tissues using a 5 V/cm biphasic pulse (2 ms pulse length) at 2 Hz frequency. Tissues were subjected to a 2-week ramped electrical stimulation regimen, where pacing began at a 2 Hz frequency that increased every 24 h by 1/3 Hz until reaching 6 Hz. After reaching 6 Hz, frequency was reduced back to 2 Hz, which was maintained until the endpoint.
METHOD DETAILS
Experimental design
All experiments involved replication with a minimum of n = 3 individual tissues or samples. Key contractile functional experiments, excluding the β-blocker study, were repeated using multiple batches of tissues, with each batch prepared using independent differentiations of iPSCs. Collection and analysis of videos for evaluation of tissue contractile force and calcium signaling were fully automated. Prior to analysis, tissues were subject to quality selection, and were excluded from analysis if there was no contraction in response to electrical stimulation at a rate of 1 Hz. All other tissues were included in the analysis.
Immunofluorescence
For immunofluorescence imaging of individual cell type monolayers, cells were plated into chambered coverslips designed for imaging (Ibidi, cat. no. 80827). iM0 coverslip chambers were first coated with FBS for 1 h prior to cell attachment, and iCM chambers were coated with Matrigel diluted 1:100 in RPMI1640 for 1 h prior to cell attachment. For human engineered cardiac tissues, the PDMS pillars were gently disconnected from the rest of the bioreactor and the intact tissues were moved to a well-plate for fixation and staining.
All tissues and cell monolayers were fixed with 4% Paraformaldehyde Solution in PBS (Santa Cruz Biotechnology, cat. no. sc-281692) for 20 min at room temperature, permeabilized in 0.25% Triton X-100 (sigma, cat. no. X100–100mL) for 20 min at room temperature, and then blocked with 5% BSA (Sigma, cat. no. A7906–500G) in PBS for 2 h at room temperature. For staining with primary antibody, cells and tissues were incubated with the respective antibodies in 1% BSA in PBS overnight at 4°C, and residual stain was then washed-out using PBS. iCM in monolayer were stained with an Alexa Fluor 488 Phalloidin (1:1000, Fisher, cat. no. A21379), α-actinin (1:100, Sigma, cat. no. A7811), and Cardiac Troponin T (1:400, Abcam, cat. no. ab45932). iCF monolayer was stained with GATA4 (1:100, Santa Cruz Biotechnology, cat. no. sc-25310), TCF21 (1:100, sigma, cat. no. HPA013189), and Vimentin (1:1000, Abcam, cat. no. ab24525). iM0 were stained with CD11b (1:100, ThermoFisher, cat. no. MA5–32793) and CD68 (1:50, Abcam, cat. no. ab955) for initial characterization. iM0 were also stained with Ki-67 (1:500, Abcam, cat. no. ab15580) and CD45 (1:200, Abcam, cat. no. ab8216) for assessment of proliferative capacity. The engineered tissues were stained with α-actinin (1:100, Sigma, cat. no. A7811), Vimentin (1:1000, Abcam, cat. no. ab24525), and CD45 (1:100, Abcam, cat. no. ab8216). Cells and tissues were then incubated with the appropriate secondary antibody in 1% BSA in PBS for 1 h at room temperature, followed by washing in PBS.
Finally, the cell monolayers were mounted using ProLong Glass Antifade Mountant with NucBlue Stain (Fisher, cat. no. P36983). For tissues, nuclei were stained using DAPI (1:1000, BD Biosciences, cat. no. 564907) in PBS for 15 min at room temperature. Tissues were then washed with PBS. Finally, the pillars were gently removed from the tissues using forceps and were embedded in ProLong Diamond Antifade Mountant (Invitrogen, cat. no. P36931) in CoverWell incubation chambers (Grace Bio-labs, cat. no. 645501) sealed with a glass coverslip.
Whole mount z stack images were acquired using either a Nikon Ti Eclipse inverted microscope for spinning-disk confocal microscopy or a Nikon Ti2 inverted microscope with AXR resonant spectral scanning confocal unit. Images were then processed in ImageJ to create z stack maximum projection images. Quantification of %CD45+ from the whole mount images was conducted in ImageJ by thresholding the image and using the ‘analyze particles’ function for both CD45 and DAPI, and normalizing the number of particles for CD45 by the number of nuclei in each image. Alignment of cTNT and Vimentin in the whole mount images was conducted by isolating the stains, thresholding the image, and using the ImageJ Directionality plugin to determine their dispersion (the standard deviation of the orientation of the structures) in the image.
Evaluation of iCM morphology
iCM were plated sparsely with or without iM0 and co-cultured for 1 week, then cells were fixed and stained according to the protocol stated for immunofluorescence above, for α-actinin (1:100, Sigma, cat. no. A7811) and DAPI (1:1000, BD Biosciences, cat. no. 564907), then imaged using a Nikon Ti Eclipse inverted microscope for spinning-disk confocal microscopy. iCM size was determined using ImageJ. Cells where the entire perimeter of the α-actinin structure was clearly visible in the image were identified, the image was thresholded, the area was measured using the analyze particles feature, and then size was determined by dividing area measured by the number of nuclei contained in that area. Circularity was assessed using the same process, but only cells with a single nucleus were considered. Using the ‘analyze particles’ feature, an ellipse was fitted around the area of the cell determined by the α-actinin stain, and circularity was determined by taking the ratio of the major: minor axes.
Inflammatory cytokine ELISA panel
Quantification of inflammatory cytokine presence in cell culture supernatant was measured using LEGENDplex Human Inflammation Panel 1 kit (Biolegend, cat. no. 740809). Conditioned supernatant was gathered from the tissues on days 7 and 28 during culture. For each timepoint, the supernatant was cultured for 48 h prior to collection. Supernatant was centrifuged at 400 g for 5 min to remove cellular debris and then stored at −20°C for later use. All supernatant samples were thawed for simultaneous assessment and the assay was run as directed by manufacturer’s protocol.
RNA isolation of hECTs
Tissues were snap frozen in liquid nitrogen and stored at −80°C until use. To isolate RNA, snap-frozen tissues were added to 300 μL of RNA lysis buffer with a stainless-steel bead (BioSpec, cat. no. 11079123) and homogenized with a Mini-Beadbeater (Biospec, cat. no. C321001) for two cycles of 10 s intervals, with storage on ice between cycles. A Qiagen RNeasy Micro Kit (Qiagen, cat. no. 74004) was then used according to the manufacturer’s instructions. Total RNA for each sample was quantified using an Agilent 2100 Bioanalyzer with an RNA Pico Chip at Columbia’s Molecular Pathology core. RNA samples with an RNA integrity number (RIN) > 8 were considered acceptable for further analysis.
Bulk RNA sequencing and analysis
An SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (TaKaRa) was used according to the manufacturer’s protocol for cDNA amplification. A library was prepared through Columbia’s Genome Center using a Nextera XT DNA Library Preparation kit (Illumina) with 100–150pg starting material, following the manufacturer’s instructions. Libraries were sequenced to a targeted depth of ~40M 100bp paired-end reads on a NovaSeq 6000. We used RTA (Illumina) for base calling and bcl2fastq2 (version 2.19) for converting BCL to fastq format, coupled with adaptor trimming. We performed a pseudoalignment to a kallisto index created from transcriptomes (Human: GRCh38.p12) using kallisto (0.44.0). Differential expression analysis was performed in R using DESeq2.
Heatmap clustering in R
Gene count data for genes describing CCR2-MERTK+ tissue resident macrophages20,27 were imported into RStudio Software, running R version 4.3.1 with Bioconductor, and hierarchically clustered using the gplots package. Z scores were generated by scaling the data across samples and NaN values were removed from downstream analysis. A heatmap of clustered z scores was generated using the heatmap.2 function.
Single-sample gene set enrichment analysis (ssGSEA)
Gene lists describing macrophages in the adult human heart were obtained from the human cardiac cell atlas.20 Gene lists describing unique monocyte-derived M1, M2a, and M2c macrophage profiles were obtained from Lurier et al.44 Transcript per million (tpm) values for each gene were imported into RStudio Version 2023.06.1 + 524 running R version 4.3.1. Enrichment analyses were performed for each gene list using the ssGSEA2 package, resulting in 10 enrichment scores per experimental sample. GraphPad Prism was used for visualization of the distribution of scores and statistical analysis was performed using two-tailed unpaired t tests.
Gene ontology (GO) analysis
The top significantly impacted biological processes and the top differentially expressed genes associated with those terms were generated using Advaita Bio’s iPathwayGuide by uploading the list of all genes identified in the RNA Sequencing analysis, their Log2(Fold Change), and their adjusted p-value. For GO analysis, top identified biological processes were identified using the smallest common denominator pruning method, which identifies the GO terms that best encapsulate the set of differentially expressed genes in the dataset, consolidating significance of two or more terms into a common parent term, reducing the redundancy of the reported GO terms.
To identify significant GO terms unrelated to immune processes, a comprehensive list of significant GO terms was generated by loading the list of significantly upregulated genes to the Gene Ontology Resource powered by PANTHER. REVIGO (Reduce and Visualize Gene Ontology) was then used to sort through the list. Terms related specifically to calcium signaling were subsequently identified and extracted by searching the list for terms that included “calcium” in the name.
KEGG pathway analysis
Significantly impacted KEGG pathway diagrams and graphs for the expression of associated genes were analyzed and generated using Advaita Bio’s iPathwayGuide. The list of all genes, the Log2(Fold Change), and adjusted p-value generated from the RNA-seq analysis were uploaded into Advaita’s proprietary software for “Impact Analysis” to identify significantly impacted pathways. Their impact analysis uses two types of evidence: the over-representation of differentially expressed genes in a pathway, and the perturbation of that pathway computed by propagating measured changes in expression across the pathway topology, which are obtained from the KEGG database. The p-values generated from these two types of evidence are combined into an overall pathway score by calculating a p-value using Fisher’s method. This p-value was not corrected for multiple comparisons.
Force generation analysis
Assessment of tissue contractile function and force generation capacity was conducted by capturing video of tissue contraction while stimulated at various voltages and frequencies. Tissues were placed in a live-cell chamber (SFX Temp & CO2 Stage Top Incubator, Tokai Hit) for controlled environment set to 37°C and 5% CO2, and imaged using a sCMOS camera (Zyla 4.2, Andor Technology) connected to an inverted microscope (IX-81, Olympus). Brightfield videos were acquired at 20 frames per second for 2400 frames using an Arduino-based electrical stimulator and custom program to stimulate the cardiac tissues from 0 Hz to 4 Hz. Force generation was determined by analyzing pillar deflection using Beatprofiler.66 Briefly, the script identified the location of the pillar heads to determine deflection of the pillar during tissue contraction, and force was calculated by multiplying the displacement by a coefficient determined by the force displacement calibration curve that was empirically determined for the platform’s pillars via mechanical testing. Additionally, analysis of the deflection trace yields many other metrics of contractility, such as total force, passive force, active force, total stress, passive stress, active stress, contraction velocity, and relaxation velocity. Tissues that did not capture at 1 Hz stimulation at the end of the culture period were excluded from further analysis.
Calcium signaling analysis
The use of GCaMP6f fluorescent protein in the iCMs enabled visualization of calcium transients without need for addition of dyes.67 Tissues were imaged with the same set up as described for force generation analysis, with fluorescence visualized using a standard GFP filter set. Tissues were electrically stimulated at 1 Hz and videos were acquired at 20 frames per second to measure calcium flux. Calcium signals were analyzed using Beatprofiler.66 Briefly, the calcium transient within the tissue was assessed by tracking average pixel intensity over time, creating a trace which was then corrected to account for the gradual decay of the baseline fluorescence that occurs due to photobleaching effect.
Isoproterenol treatment
After 4 weeks of culture, cardiac tissues were serially treated with increasing concentrations of isoproterenol (Sigma, cat. no. I6504–500MG), from 1E-13 M to 1E-5 M, added in 1 μL volumes to each well containing a tissue. After the addition of isoproterenol to the tissues, the tissues were incubated in a live cell chamber at 37°C and 5% CO2 for 5 min, then imaged to assess changes in calcium handling using the encoded calcium-responsive fluorescent protein in the cardiomyocytes. Following imaging, the process was repeated with a dosage of isoproterenol one order of magnitude higher than the previous, until the concentration of 10 μM was reached. Calcium signaling analysis was conducted as described above.
Forskolin treatment
Following 4 weeks of culture, cardiac tissues were treated with 10 μM Forskolin (Tocris, cat. no. 1099). Live imaging of calcium handling of the tissues was visualized using the encoded GCaMP6f calcium-responsive fluorescent protein both prior to and following 30 min post treatment. Calcium signaling analysis was conducted as described above.
Catecholamine quantification
Measurement of the catecholamines Adrenaline (epinephrine) and Norepinephrine from cell and hECT supernatant was quantified using an Adrenaline ELISA kit (Abcam, cat. ab287788) and Norepinephrine ELISA kit (Abcam, cat. ab287789) respectively. For tissues, conditioned supernatants from +iM0 and −iM0 hECTs were collected after 48 h of culture. For individual cell populations, iM0, iCM, and iCF were plated at similar cell densities (100,000 cells/well) and maintained in B27 media. Conditioned supernatant was collected from all groups after 72 h of culture. All collected supernatants alongside fresh B27 media controls were assayed according to manufacturer’s protocol for both the Adrenaline and Norepinephrine ELISAs.
Beta-blocker treatment
Following 4 weeks of culture, cardiac tissues were treated with 20 μM Bisoprolol hemifumarate (Medchemexpress, cat. no. HY-B0076), a β1-adrenergic receptor blocker. Force generation analysis was conducted as described above both prior to and following 1 h of treatment to determine the effect on tissue contractile function.
RT-qPCR
mRNA from iPSCs and iCFs was isolated from cells using the RNeasy Mini kit (Qiagen, cat. no. 74106) according to the manufacturer’s instructions. mRNA from iM0 was isolated from cells using the RNeasy Micro kit (Qiagen, cat. No. 74004). cDNA samples were obtained from mRNA using the High-Capacity cDNA Reverse Transcription kit (Applied biosystems, cat. no. 4368813) according to the manufacturer’s instructions. Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) was performed with the Fast SYBR Green Master Mix (Applied Biosystems, cat. no. 4385612) according to manufacturer’s instructions in a StepOnePlus Real-Time PCR System (Applied Biosystems) thermocycler using 40 cycles of two-step PCR protocol of 95°C melting for 3 s and 60°C annealing and extension for 30 s, prior to an initial hot start at 95°C for 20 s. Targets and oligo sequences used are listed in Table S1.
Collagen secretion
Supernatant of iPSCs and iCFs with and without 10 ng/mL TGF-β1 for 48h treatment was collected, and the collagen was concentrated using a concentrating solution (Chondrex, cat. no. 90626), according to the manufacturer’s instructions. Media only samples (FGM and mTeasR, respectively) served as background control. Secreted collagen was determined using Sirius Red Total Collagen Detection Kit (Chondrex, cat. no. 9062), according to manufacturer’s instructions, and absorbance was measured at 530 nm in a spectrophotometer. Total secreted collagen concentrations were normalized to the total protein content, using the Pierce BCA protein assay (ThermoFisher, cat. no. 23225).
iM0 surface marker validation
iM0 were detached from ultra-low attachment culture dishes by incubation with TrypLE (ThermoFisher, cat. no. 12605010) for 3 min. For flow cytometry characterization of surface marker expression, cells were centrifuged, resuspended in FACS buffer (2% FBS, 0.5 mM EDTA), and blocked in FcR blocking solution (Miltenyi, cat. no. 130-059-901) for 15 min at 4°C. Cells were subsequently resuspended in FACs buffer containing AlexaFluor 700-conjugated anti-human CD45 (BioLegend, cat. no. 304024, 1:50 dilution), Brilliant Violet 420-conjugated anti-human CD11b (BioLegend, cat. no. 301324, 1:50 dilution), Brilliant Violet 605-conjugated anti-human CD14 (BioLegend, cat. no. 301834, 1:50 dilution), PE/Cyanine7-conjugated anti-human HLA-DR (BioLegend, cat. no. 307615, 1:50 dilution), and APC-conjugated anti-human CD172/SIRPa (BioLegend, cat. no. 372105, 1:50 dilution). Cells were then stained with propidium iodide (ThermoFisher), and washed 2x with FACS buffer. Data were acquired on a Novocyte Penteon flow cytometer at the Columbia Stem Cell Initiative Flow Cytometry core facility. Data analyses were performed using FlowJo software (BD BioSciences).
Phagocytosis assay
iM0 were replated onto FBS-coated wells at a density of 100,000 cells/well of a 96-well plate and incubated overnight in X-VIVO 15 media containing 100 ng/mL M-CSF. The cells were subsequently incubated with fresh media containing 20 μL/mL fluorescent red Latex beads (Sigma, cat. no. L3030) for 2 h at 37°C. Post-incubation, cells were detached using trypsin, centrifuged, washed 2X with FACS buffer, and stained with CD45 and CD14 antibodies for flow cytometry analysis (as described above).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using GraphPad Prism 10.2.1. Single comparisons between two groups were analyzed using a two-tailed, unpaired Student’s t test assuming a Gaussian distribution to determine statistical significance. Outliers were identified using the ROUT method with Q = 1% and excluded from analysis. When comparing multiple factors, unless otherwise indicated, a two-way ANOVA corrected for multiple comparisons using Tukey’s postdoc analysis was conducted. For all analyses, p < 0.05 was considered statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). All data are represented as mean ± SEM.
Supplementary Material
Highlights.
Engineered cardiac tissues can maintain an iPSC-macrophage (iM0) population for 28 days
iM0s in engineered cardiac tissues take on a tissue-resident-like phenotype
iM0s alter cardiac tissue function, increasing contractile force production
Observed functional changes may be due to iM0 stimulation of the β-adrenergic pathway in iCMs
ACKNOWLEDGMENTS
The authors gratefully acknowledge the funding support of our research by the NIH (P41 EB027062 and R01 HL076485) and NSF (NSF1647837). The authors also acknowledge that these studies used the resources of the Herbert Irving Comprehensive Cancer Center Confocal and Specialized Microscopy Core and the Columbia Genomics and High Throughput Screening Shared Resource, funded in part through NIH/NCI Cancer Center Support Grant P30CA013696. We would also like to thank the staff of the Columbia Stem Cell Initiative Flow Cytometry Core Facility, under the leadership of Michael Kissner, at the Columbia University Irving Medical Center for their contributions to the work presented in this manuscript. All schematics were created using BioRender.com, and chord graphs and KEGG diagrams were generated from Advaita Bio’s iPathwayGuide.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114302.
REFERENCES
- 1.Zhuang RZ, Lock R, Liu B, and Vunjak-Novakovic G (2022). Opportunities and challenges in cardiac tissue engineering from an analysis of two decades of advances. Nat. Biomed. Eng. 6, 327–338. 10.1038/s41551-022-00885-3. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y, Rafatian N, Feric NT, Cox BJ, Aschar-Sobbi R, Wang EY, Aggarwal P, Zhang B, Conant G, Ronaldson-Bouchard K, et al. (2019). A Platform for Generation of Chamber-Specific Cardiac Tissues and Disease Modeling. Cell 176, 913–927.e18. 10.1016/j.cell.2018.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldfracht I, Protze S, Shiti A, Setter N, Gruber A, Shaheen N, Nartiss Y, Keller G, and Gepstein L (2020). Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat. Commun. 11, 75. 10.1038/s41467-019-13868-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giacomelli E, Bellin M, Sala L, van Meer BJ, Tertoolen LGJ, Orlova VV, and Mummery CL (2017). Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development 144, 1008–1017. 10.1242/dev.143438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noor N, Shapira A, Edri R, Gal I, Wertheim L, and Dvir T (2019). 3D Printing of Personalized Thick and Perfusable Cardiac Patches and Hearts. Adv. Sci. 6, 1900344. 10.1002/advs.201900344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braam SR, Tertoolen L, van de Stolpe A, Meyer T, Passier R, and Mummery CL (2010). Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. 4, 107–116. 10.1016/j.scr.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Gintant G, Burridge P, Gepstein L, Harding S, Herron T, Hong C, Jalife J, and Wu JC (2019). Use of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes in Preclinical Cancer Drug Cardiotoxicity Testing: A Scientific Statement From the American Heart Association. Circ. Res. 125, e75–e92. 10.1161/RES.0000000000000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang BZ, Nash TR, Zhang X, Rao J, Abriola L, Kim Y, Zakharov S, Kim M, Luo LJ, Morsink M, et al. (2023). Engineered cardiac tissue model of restrictive cardiomyopathy for drug discovery. Cell Rep. Med. 4, 100976. 10.1016/j.xcrm.2023.100976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Passier R, Orlova V, and Mummery C (2016). Complex Tissue and Disease Modeling using hiPSCs. Cell Stem Cell 18, 309–321. 10.1016/j.stem.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, and Yamanaka S (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Robertson C, Tran DD, and George SC (2013). Concise Review: Maturation Phases of Human Pluripotent Stem Cell-Derived Cardiomyocytes. STEM CELLS 31, 829–837. 10.1002/stem.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koivumäki JT, Naumenko N, Tuomainen T, Takalo J, Oksanen M, Puttonen KA, Lehtonen Š, Kuusisto J, Laakso M, Koistinaho J, and Tavi P (2018). Structural Immaturity of Human iPSC-Derived Cardiomyocytes: In Silico Investigation of Effects on Function and Disease Modeling. Front. Physiol. 9, 80. 10.3389/fphys.2018.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ah F, M R-L, Cm H, M K, D T, A S, Aq T, Hh P, C F, Pd G, et al. (2016). Three-Dimensional Adult Cardiac Extracellular Matrix Promotes Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Tissue Eng. Part A, 22. 10.1089/ten.TEA.2016.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majid QA, Fricker ATR, Gregory DA, Davidenko N, Hernandez Cruz O, Jabbour RJ, Owen TJ, Basnett P, Lukasiewicz B, Stevens M, et al. (2020). Natural Biomaterials for Cardiac Tissue Engineering: A Highly Biocompatible Solution. Front. Cardiovasc. Med. 7, 554597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kharaziha M, Nikkhah M, Shin S-R, Annabi N, Masoumi N, Gaharwar AK, Camci-Unal G, and Khademhosseini A (2013). PGS:Gelatin Nanofibrous Scaffolds with Tunable Mechanical and Structural Properties for Engineering Cardiac Tissues. Biomaterials 34, 6355–6366. 10.1016/j.biomaterials.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacQueen LA, Sheehy SP, Chantre CO, Zimmerman JF, Pasqualini FS, Liu X, Goss JA, Campbell PH, Gonzalez GM, Park S-J, et al. (2018). A tissue-engineered scale model of the heart ventricle. Nat. Biomed. Eng. 2, 930–941. 10.1038/s41551-018-0271-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feiner R, Engel L, Fleischer S, Malki M, Gal I, Shapira A, Shacham-Diamand Y, and Dvir T (2016). Engineered hybrid cardiac patches with multifunctional electronics for online monitoring and regulation of tissue function. Nat. Mater. 15, 679–685. 10.1038/nmat4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidorov VY, Samson PC, Sidorova TN, Davidson JM, Lim CC, and Wikswo JP (2017). I-Wire Heart-on-a-Chip I: Three-dimensional cardiac tissue constructs for physiology and pharmacology. Acta Biomater. 48, 68–78. 10.1016/j.actbio.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Neste CC, Wiley KA, Chang SW, Borrello J, Turnbull IC, and Costa KD (2023). Designing a Bioreactor to Improve Data Acquisition and Model Throughput of Engineered Cardiac Tissues. J. Vis. Exp. 2, e64368. 10.3791/64368. [DOI] [PubMed] [Google Scholar]
- 20.Litviňková M, Talavera-López C, Maatz H, Reichart D, Worth CL, Lindberg EL, Kanda M, Polanski K, Heinig M, Lee M, et al. (2020). Cells of the adult human heart. Nature 588, 466–472. 10.1038/s41586-020-2797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suku M, Forrester L, Biggs M, and Monaghan MG (2022). Resident Macrophages and Their Potential in Cardiac Tissue Engineering. Tissue Eng. Part B Rev. 28, 579–591. 10.1089/ten.teb.2021.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masumoto H, Nakane T, Tinney JP, Yuan F, Ye F, Kowalski WJ, Minakata K, Sakata R, Yamashita JK, and Keller BB (2016). The myocardial regenerative potential of three-dimensional engineered cardiac tissues composed of multiple human iPS cell-derived cardiovascular cell lineages. Sci. Rep. 6, 29933. 10.1038/srep29933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giacomelli E, Meraviglia V, Campostrini G, Cochrane A, Cao X, van Helden RWJ, Krotenberg Garcia A, Mircea M, Kostidis S, Davis RP, et al. (2020). Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease. Cell Stem Cell 26, 862–879.e11. 10.1016/j.stem.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinto AR, Ilinykh A, Ivey MJ, Kuwabara JT, D’Antoni ML, Debuque R, Chandran A, Wang L, Arora K, Rosenthal NA, and Tallquist MD (2016). Revisiting Cardiac Cellular Composition. Circ. Res. 118, 400–409. 10.1161/CIRCRESAHA.115.307778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heidt T, Courties G, Dutta P, Sager HB, Sebas M, Iwamoto Y, Sun Y, Da Silva N, Panizzi P, van der Laan AM, et al. (2014). Differential contribution of monocytes to heart macrophages in steady-state and after myocardial infarction. Circ. Res. 115, 284–295. 10.1161/CIRCRESAHA.115.303567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sansonetti M, Waleczek FJG, Jung M, Thum T, and Perbellini F (2020). Resident cardiac macrophages: crucial modulators of cardiac (patho)physiology. Basic Res. Cardiol. 115, 77. 10.1007/s00395-020-00836-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bajpai G, Schneider C, Wong N, Bredemeyer A, Hulsmans M, Nahrendorf M, Epelman S, Kreisel D, Liu Y, Itoh A, et al. (2018). The Human Heart Contains Distinct Macrophage Subsets with Divergent Origins and Functions. Nat. Med. 24, 1234–1245. 10.1038/s41591-018-0059-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Epelman S, Lavine KJ, and Randolph GJ (2014). Origin and Functions of Tissue Macrophages. Immunity 41, 21–35. 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies LC, Jenkins SJ, Allen JE, and Taylor PR (2013). Tissue-resident macrophages. Nat. Immunol. 14, 986–995. 10.1038/ni.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon S, and Plüddemann A (2017). Tissue macrophages: heterogeneity and functions. BMC Biol. 15, 53. 10.1186/s12915-017-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hulsmans M, Clauss S, Xiao L, Aguirre AD, King KR, Hanley A, Hucker WJ, Wu€lfers EM, Seemann G, Courties G, et al. (2017). Macrophages Facilitate Electrical Conduction in the Heart. Cell 169, 510–522.e20. 10.1016/j.cell.2017.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaman R, Hamidzada H, Kantores C, Wong A, Dick SA, Wang Y, Momen A, Aronoff L, Lin J, Razani B, et al. (2021). Selective loss of resident macrophage-derived insulin-like growth factor-1 abolishes adaptive cardiac growth to stress. Immunity 54, 2057–2071.e6. 10.1016/j.immuni.2021.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Dick SA, Macklin JA, Nejat S, Momen A, Clemente-Casares X, Althagafi MG, Chen J, Kantores C, Hosseinzadeh S, Aronoff L, et al. (2019). Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat. Immunol. 20, 29–39. 10.1038/s41590-018-0272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao X, Yakala GK, van den Hil FE, Cochrane A, Mummery CL, and Orlova VV (2019). Differentiation and Functional Comparison of Monocytes and Macrophages from hiPSCs with Peripheral Blood Derivatives. Stem Cell Rep. 12, 1282–1297. 10.1016/j.stemcr.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Armitage LH, Stimpson SE, Santostefano KE, Sui L, Ogundare S, Newby BN, Castro-Gutierrez R, Huber MK, Taylor JP, Sharma P, et al. (2021). Use of Induced Pluripotent Stem Cells to Build Isogenic Systems and Investigate Type 1 Diabetes. Front. Endocrinol. 12, 737276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchrieser J, James W, and Moore MD (2017). Human Induced Pluripotent Stem Cell-Derived Macrophages Share Ontogeny with MYB-Independent Tissue-Resident Macrophages. Stem Cell Rep. 8, 334–345. 10.1016/j.stemcr.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamargo MA, Nash TR, Fleischer S, Kim Y, Vila OF, Yeager K, Summers M, Zhao Y, Lock R, Chavez M, et al. (2021). milliPillar: A Platform for the Generation and Real-Time Assessment of Human Engineered Cardiac Tissues. ACS Biomater. Sci. Eng. 7, 5215–5229. 10.1021/acsbiomaterials.1c01006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, et al. (2014). Chemically defined generation of human cardiomyocytes. Nat. Methods 11, 855–860. 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Tian L, Shen M, Tu C, Wu H, Gu M, Paik DT, and Wu JC (2019). Generation of Quiescent Cardiac Fibroblasts From Human Induced Pluripotent Stem Cells for In Vitro Modeling of Cardiac Fibrosis. Circ. Res. 125, 552–566. 10.1161/CIRCRESAHA.119.315491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou P, and Pu WT (2016). Recounting Cardiac Cellular Composition. Circ. Res. 118, 368–370. 10.1161/CIRCRESAHA.116.308139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiburcy M, Hudson JE, Balfanz P, Schlick S, Meyer T, Chang Liao M-L, Levent E, Raad F, Zeidler S, Wingender E, et al. (2017). Defined Engineered Human Myocardium With Advanced Maturation for Applications in Heart Failure Modeling and Repair. Circulation 135, 1832–1847. 10.1161/CIRCULATIONAHA.116.024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liau B, Jackman CP, Li Y, and Bursac N (2017). Developmental stage-dependent effects of cardiac fibroblasts on function of stem cell-derived engineered cardiac tissues. Sci. Rep. 7, 42290. 10.1038/srep42290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, and Vunjak-Novakovic G (2018). Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239–243. 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lurier EB, Dalton D, Dampier W, Raman P, Nassiri S, Ferraro NM, Rajagopalan R, Sarmady M, and Spiller KL (2017). Transcriptome analysis of IL-10-stimulated (M2c) macrophages by next-generation sequencing. Immunobiology 222, 847–856. 10.1016/j.imbio.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou Y, Yao A, Zhu W, Kudoh S, Hiroi Y, Shimoyama M, Uozumi H, Kohmoto O, Takahashi T, Shibasaki F, et al. (2001). Isoproterenol activates extracellular signal-regulated protein kinases in cardiomyocytes through calcineurin. Circulation 104, 102–108. 10.1161/hc2601.090987. [DOI] [PubMed] [Google Scholar]
- 46.Shen J.x. (2006). Isoprenaline enhances local Ca2+ release in cardiac myocytes. Acta Pharmacol. Sin. 27, 927–932. 10.1111/j.1745-7254.2006.00383.x. [DOI] [PubMed] [Google Scholar]
- 47.Lymperopoulos A, Rengo G, and Koch WJ (2013). Adrenergic nervous system in heart failure: pathophysiology and therapy. Circ. Res. 113, 739–753. 10.1161/CIRCRESAHA.113.300308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park DS, Kozaki T, Tiwari SK, Moreira M, Khalilnezhad A, Torta F, Olivié N, Thiam CH, Liani O, Silvin A, et al. (2023). iPS-cell-derived microglia promote brain organoid maturation via cholesterol transfer. Nature 623, 397–405. 10.1038/s41586-023-06713-1. [DOI] [PubMed] [Google Scholar]
- 49.Múnera JO, Kechele DO, Bouffi C, Qu N, Jing R, Maity P, Enriquez JR, Han L, Campbell I, Mahe MM, et al. (2023). Development of functional resident macrophages in human pluripotent stem cell-derived colonic organoids and human fetal colon. Cell Stem Cell 30, 1434–1451.e9. 10.1016/j.stem.2023.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen T, and Vunjak-Novakovic G (2019). Human Tissue-Engineered Model of Myocardial Ischemia-Reperfusion Injury. Tissue Eng. Part A 25, 711–724. 10.1089/ten.TEA.2018.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma A, Marceau C, Hamaguchi R, Burridge PW, Rajarajan K, Churko JM, Wu H, Sallam KI, Matsa E, Sturzu AC, et al. (2014). Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes as an In Vitro Model for Coxsackievirus B3-Induced Myocarditis and Antiviral Drug Screening Platform. Circ. Res. 115, 556–566. 10.1161/CIRCRESAHA.115.303810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tavakol DN, Nash TR, Kim Y, He S, Fleischer S, Graney PL, Brown JA, Liberman M, Tamargo M, Harken A, et al. (2023). Modeling and countering the effects of cosmic radiation using bioengineered human tissues. Biomaterials 301, 122267. 10.1016/j.biomaterials.2023.122267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takata K, Kozaki T, Lee CZW, Thion MS, Otsuka M, Lim S, Utami KH, Fidan K, Park DS, Malleret B, et al. (2017). Induced-Pluripotent-Stem-Cell-Derived Primitive Macrophages Provide a Platform for Modeling Tissue-Resident Macrophage Differentiation and Function. Immunity 47, 183–198.e6. 10.1016/j.immuni.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 54.Chan Y-C, Ting S, Lee Y-K, Ng K-M, Zhang J, Chen Z, Siu C-W, Oh SKW, and Tse H-F (2013). Electrical stimulation promotes maturation of cardiomyocytes derived from human embryonic stem cells. J. Cardiovasc. Transl. Res. 6, 989–999. 10.1007/s12265-013-9510-z. [DOI] [PubMed] [Google Scholar]
- 55.Kong Y, Liu F, Ma B, Duan J, Yuan W, Sang Y, Han L, Wang S, and Liu H (2021). Wireless Localized Electrical Stimulation Generated by an Ultrasound-Driven Piezoelectric Discharge Regulates Proinflammatory Macrophage Polarization. Adv. Sci. 8, 2100962. 10.1002/advs.202100962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li C, Levin M, and Kaplan DL (2016). Bioelectric modulation of macrophage polarization. Sci. Rep. 6, 21044. 10.1038/srep21044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoare JI, Rajnicek AM, McCaig CD, Barker RN, and Wilson HM (2016). Electric fields are novel determinants of human macrophage functions. J. Leukoc. Biol. 99, 1141–1151. 10.1189/jlb.3A0815-390R. [DOI] [PubMed] [Google Scholar]
- 58.Chramiec A, Teles D, Yeager K, Marturano-Kruik A, Pak J, Chen T, Hao L, Wang M, Lock R, Tavakol DN, et al. (2020). Integrated human organ-on-a-chip model for predictive studies of anti-tumor drug efficacy and cardiac safety. Lab Chip 20, 4357–4372. 10.1039/D0LC00424C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ronaldson-Bouchard K, Teles D, Yeager K, Tavakol DN, Zhao Y, Chramiec A, Tagore S, Summers M, Stylianos S, Tamargo M, et al. (2022). A multi-organ chip with matured tissue niches linked by vascular flow. Nat. Biomed. Eng. 6, 351–371. 10.1038/s41551-022-00882-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sucharov CC, Mariner PD, Nunley KR, Long C, Leinwand L, and Bristow MR (2006). A beta1-adrenergic receptor CaM kinase II-dependent pathway mediates cardiac myocyte fetal gene induction. Am. J. Physiol. Heart Circ. Physiol. 291, H1299–H1308. 10.1152/ajpheart.00017.2006. [DOI] [PubMed] [Google Scholar]
- 61.Flierl MA, Rittirsch D, Huber-Lang M, Sarma JV, and Ward PA (2008). Catecholamines—Crafty Weapons in the Inflammatory Arsenal of Immune/Inflammatory Cells or Opening Pandora’s Box? Mol. Med. 14, 195–204. 10.2119/2007-00105.Flierl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nguyen KD, Qiu Y, Cui X, Goh YPS, Mwangi J, David T, Mukundan L, Brombacher F, Locksley RM, and Chawla A (2011). Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature 480, 104–108. 10.1038/nature10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Freire BM, de Melo FM, and Basso AS (2022). Adrenergic signaling regulation of macrophage function: do we understand it yet? Immunother. Adv. 2, ltac010. 10.1093/immadv/ltac010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown SW, Meyers RT, Brennan KM, Rumble JM, Narasimhachari N, Perozzi EF, Ryan JJ, Stewart JK, and Fischer-Stenger K (2003). Catecholamines in a macrophage cell line. J. Neuroimmunol. 135, 47–55. 10.1016/S0165-5728(02)00435-6. [DOI] [PubMed] [Google Scholar]
- 65.Huebsch N, Loskill P, Deveshwar N, Spencer CI, Judge LM, Mandegar MA, Fox CB, Mohamed TMA, Ma Z, Mathur A, et al. (2016). Miniaturized iPS-Cell-Derived Cardiac Muscles for Physiologically Relevant Drug Response Analyses. Sci. Rep. 6, 24726. 10.1038/srep24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim Y, Wang K, Lock RI, Nash TR, Fleischer S, Wang BZ, Fine BM, and Vunjak-Novakovic G (2024). BeatProfiler: Multimodal In Vitro Analysis of Cardiac Function Enables Machine Learning Classification of Diseases and Drugs . IEEE Open J. Eng. Med. Biol. 5, 238–249. 10.1109/OJEMB.2024.3377461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk RNA-sequencing data have been deposited at Gene Expression Omnibus (GEO) (GEO: GSE266797) and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Alexa Fluor 488 Phalloidin | ThermoFisher | Cat# A12379 |
| Cardiac Troponin T | Abcam | Cat# ab45932; RRID: AB_956386 |
| TCF21 | Sigma Aldrich | Cat# HPA013189; RRID: AB_10601215 |
| Vimentin | Abcam | Cat# ab24525; RRID: AB_778824 |
| CD11b | ThermoFisher | Cat# MA5-32793; RRID: AB_2810069 |
| CD68 | Abcam | Cat# ab955; RRID: AB_307338 |
| Alpha actinin | Sigma | Cat# A7811; RRID: AB_476766 |
| CD45 | Abcam | Cat# ab8216; RRID: AB_306361 |
| DAPI | BD Biosciences | Cat# 564907; RRID: AB_2869624 |
| AlexaFluor 700-conjugated anti-human CD45 | BioLegend | Cat# 304024; RRID: AB_493761 |
| Brilliant Violet 420-conjugated anti-human CD11b | BioLegend | Cat# 301324; RRID: AB_11219589 |
| Brilliant Violet 605-conjugated anti-human CD14 | BioLegend | Cat# 301834; RRID: AB_2563798 |
| PE/Cyanine7-conjugated anti-human HLA-DR | BioLegend | Cat# 307615; RRID: AB_493589 |
| APC-conjugated anti-human CD172/SIRPa | BioLegend | Cat# 372105; RRID: AB_2650862 |
| Ki-67 | Abcam | Cat# ab15580; RRID: AB_443209 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| (pLV(Exp)-Neo-CD68(short)>EYFP | VectorBuilder | Cat# VB211109-1377wwb |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Lenti-X Accelerator | Takara | Cat# 631257 |
| Isoproterenol | Sigma | Cat# I6504-500mg |
| Forskolin | Tocris | Cat# 1099 |
| Bisoprolol Hemifumarate | MedChemExpress | Cat# HY-B0076 |
| Y-27632 dihydrochloride | Tocris | Cat# 1254 |
| CHIR 99021 | Tocris | Cat# 4423 |
| Wnt-C59 | Tocris | Cat# 5148 |
| 6-aminocaproic acid | Sigma-Aldrich | Cat# A7824 |
| Fibrinogen | Sigma-Aldrich | Cat# F3879-100mg |
| Thrombin | Sigma-Aldrich | Cat# T6884 |
| PDMS/Sylgard 184 | Dow | Cat# 1024001 |
| Human Albumin | Sigma-Aldrich | Cat# A9731-1G |
| L-ascorbic acid 2-phosphate | Sigma-Aldrich | Cat# A8960 |
| Retinoic acid | Sigma-Aldrich | Cat# R1514 |
| FGF2 | Peprotech | Cat# 100-18B |
| SB35421 | Selleck Chemicals | Cat# S1067 |
| GlutaMAX | Invitrogen | Cat# 35050061 |
| Recombinant human macrophage colony stimulating factor | Peprotech | Cat# 300-25 |
| Human interleukin 3 | Peprotech | Cat# 200-03 |
|
| ||
| Critical commercial assays | ||
|
| ||
| STEMdiff Hematopoietic Kit | Stem Cell Technologies | Cat# 05310 |
| CD14 Microbeads, human | Miltenyi Biotec | Cat# 130-050-201 |
| LEGENDplex Human Inflammation Panel 1 | Biolegend | Cat# 740809 |
| RNeasy Micro Kit | Qiagen | Cat# 74004 |
| High-Capacity cDNA Reverse Transcription kit | Applied Biosystems | Cat# 4368813 |
| Sirius red total collagen detection kit | Chondrex | Cat# 9062 |
| Fluorescent red Latex beads | Sigma | Cat# L3030 |
| Adrenaline ELISA kit | Abcam | Cat# ab287788 |
| Norepinephrine ELISA kit | Abcam | Cat# ab287789 |
|
| ||
| Deposited data | ||
|
| ||
| Bulk RNA-sequencing | This paper | GEO: GSE266797 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| WTC11 | Material Transfer Agreements from Bruce Conklin, Gladstone Institute | Huebsch et al.65 |
| WTC11-GCAMP6f | Material Transfer Agreements from Bruce Conklin, Gladstone Institute | Huebsch et al.65 |
| WTC11-CD68-EYFP | Generated in this study | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S1 for list of RNA sequences used for RT-qPCR. | N/A | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| R Studio (version 4.3.1) | Posit | https://www.rstudio.com/ |
| GraphPad Prism 10.2.1 | GraphPad | https://www.graphpad.com/ |
| iPathwayGuide | Advaita Bioinformatics | https://advaitabio.com/ |
| Gene Ontology Resource | Global Core Biodata Resource | https://geneontology.org/ |
| REVIGO | REVIGO | http://revigo.irb.hr/ |


