Abstract
Transcription of the major immediate-early (MIE) genes of cytomegaloviruses (CMV) is driven by a strong promoter-enhancer (MIEPE) complex. Transactivator proteins encoded by these MIE genes are essential for productive infection. Accordingly, the MIEPE is a crucial control point, and its regulation by activators and repressors is pertinent to virus replication. Since the MIEPE contains multiple regulatory elements, it was reasonable to assume that specific sequence motifs are irreplaceable for specifying the cell-type tropism and replication pattern. Recent work on murine CMV infectivity (A. Angulo, M. Messerle, U. H. Koszinowski, and P. Ghazal, J. Virol. 72:8502–8509, 1998) has documented the proposed enhancing function of the enhancer in that its resection or its replacement by a nonregulatory stuffer sequence resulted in a significant reduction of infectivity, even though replication competence was maintained by a basal activity of the spared authentic MIE promoter. Notably, full capacity for productive in vitro infection of fibroblasts was restored in recombinant viruses by the human CMV enhancer. Using two-color in situ hybridization with MIEPE-specific polynucleotide probes, we demonstrated that a murine CMV recombinant in which the complete murine CMV MIEPE is replaced by the paralogous human CMV core promoter and enhancer (recombinant virus mCMVhMIEPE) retained the potential to replicate in vivo in all tissues relevant to CMV disease. Notably, mCMVhMIEPE was also found to replicate in the liver, a site at which transgenic hCMV MIEPE is silenced. We conclude that productive in vivo infection with murine CMV does not strictly depend on a MIEPE type-specific regulation.
The enhancers of the various species-specific strains of cytomegaloviruses (CMV), such as those of human CMV (hCMV) (7), simian CMV (9), rat CMV (42), and murine CMV (mCMV) (11), are regarded as important regulatory elements enforcing transcription of the major immediate-early (MIE) genes that specify transactivator proteins essential for initiating the productive viral cycle in permissive cell types. Thus, the enhancer serves primarily as a genetic amplifier of productive infection. In the specific case of mCMV, the enhancer operates bidirectionally in that it governs the MIE promoter (MIEP) of the ie1-ie3 (hereafter shortened to ie1/3) transcription unit (17), which encodes the principal early gene transactivator IE3 (28) and the cotransactivator IE1 (18, 19), as well as the promoter of gene ie2, which is located upstream of ie1/3, is transcribed in opposite orientation, and encodes the protein murine IE2 (30), for which a critical function has yet to be defined (8). Notably, murine IE2 has no direct counterpart in hCMV, whereas murine IE1 and IE3 are the functional analogs of human IE1 and IE2, respectively. Likewise, the hCMV enhancer controls the hCMV MIEP of the ie1-ie2 transcription unit, and as is the case for murine IE1 and IE3, human IE1 and IE2 are derived from respective mRNAs generated by differential splicing (for reviews, see references 43 and 45). As we have shown recently for mCMV latency and recurrence in the lungs, MIEP activity does not inevitably initiate the productive cycle. Specifically, during latency, focal and stochastic MIEP activity in lung tissue was found to selectively generate spliced ie1 transcripts, while spliced transactivator-specifying ie3 transcripts were missing (21). Although these data indicated a role for posttranscriptional splicing regulation as a second checkpoint in the initiation of productive infection, MIEP activity is unquestionably the first condition for productive primary or recurrent infection. Hence, regulation of MIEP activity by the enhancer and regulation of the enhancer by cellular transcription factors are pertinent to the initiation of the productive cycle. In addition, there is evidence to suggest that viral enhancers may also facilitate viral DNA replication by maintaining an open chromatin structure (33).
CMV enhancers, with the notable exception of the rat CMV enhancer (42), contain multiple regulatory modules consisting of repeat and unique sequence elements (reviewed in reference 27). These regulatory modules, the repeat elements in particular, frequently encompass consensus binding sites for a variety of cellular transcription factors, including, for example, binding sites for NF-κB (within 18-bp repeats), ATF/CREB (within 19-bp repeats), and RAR-RXR family members (reference 2; for an overview see reference 13). Since these transcription factors are involved in signaling pathways, regulation at the enhancer has been implicated in cell-type restricted and differentiation-dependent patterns of hCMV MIE promoter-enhancer (MIEPE)-driven reporter gene expression in transgenic in vivo mouse models during embryogenesis and in mature tissues (5, 6, 20). Likewise, the enhancer is said to serve as a genetic target element for reactivation of latent CMV by extracellular signals (32), such as by tumor necrosis factor alpha effected by NF-κB (37).
While the significance of the enhancer for optimal expression of MIE genes was long predicted from enhanced reporter gene expression in transfection experiments, firm evidence of an amplifying function of the mCMV enhancer in productive viral replication was provided only recently by Angulo et al. (1), who demonstrated severely impaired viral productivity of “enhancerless” mutants of mCMV. Specifically, mCMV mutants with or without a stuffer sequence in place of the enhancer were cloned as bacterial artificial chromosomes (BAC) (29), and compared to parental or revertant mCMV, both mutants were found to replicate with reduced efficacy upon transfection of the respective BAC plasmids in permissive fibroblasts. Notably, viral productivity was fully restored in mutants in which the mCMV enhancer was replaced by the paralogous enhancer of hCMV. This study has thus shown that species specificity of CMVs is not determined by the enhancer and that the hCMV enhancer can substitute for the mCMV enhancer, at least with regard to an optimal infection of permissive fibroblasts in cell culture. The fact that enhancerless mutants were capable of producing some progeny (1) demonstrates that the enhancer is not essential for productive viral replication but performs precisely the function indicated by its name: it enhances! The residual infectivity is likely to be mediated by a basal activity of the authentic mCMV MIEP, located at positions −1 to −48 relative to the ie1/3 transcription start site, which was spared in the enhancerless mutants (1). In addition, promoter activity may be enhanced by downstream sequences in the noncoding exon 1 of the ie1/3 transcription unit that contains regulatory elements, such as SP1 binding sites and CAAT boxes. Accordingly, enhancerless BAC plasmids with an additional deletion of the promoter and downstream sequences did not generate viable virus (1).
Virus replication in diverse cell types in vivo may be subject to enhancer-directed regulation by transcription factors not implicated in the permissive infection of fibroblasts in cell culture. Furthermore, regulation may address the MIEP directly. In this article, we report on the in vivo growth and organ distribution of recombinant virus mCMVhMIEPE, in which the complete MIEPE of mCMV was replaced by the paralogous core promoter and enhancer of hCMV.
(This paper will be part of the Ph.D. thesis of N. K. A. Grzimek at the Faculty of Biology of the Eberhard-Karls-Universität, Tübingen, Germany.)
MATERIALS AND METHODS
Construction of recombinant viruses.
For reasons of biosafety, recombinants containing the hCMV MIEPE were based on an open reading frame (ORF) m152 (38) immune evasion gene deletion mutant (mCMVΔorf152) of mCMV, strain Smith ATCC-VR194. Mutant mCMVΔorf152 and its revertant mCMVorf152-rev were generously provided by U. H. Koszinowski and I. Crnkovic-Mertens, Max von Pettenkofer Institute for Hygiene and Microbiology, Munich, Germany. In vivo replication of mCMVΔorf152 in genetically susceptible BALB/c (H-2d) mice is comparable to that of parental mCMV Smith or mCMVorf152-rev only after an immunoablative treatment by total-body 137Cs γ irradiation with a dose of at least 8 Gy, whereas replication is largely prevented by residual immunity after immunoreductive treatment with doses of up to 6 Gy (not shown). For the sake of brevity, mCMVΔorf152 is herein referred to as mCMV, except when its distinction from parental mCMV Smith is specifically required.
(i) Plasmid constructs for homologous recombination.
Recombinant plasmids were constructed according to established procedures. Enzyme reactions were performed as recommended by the suppliers. Specifically, plasmid pAMB25, containing the sequence from map positions 176,441 to 187,035 of the mCMV Smith strain genome (38) (GenBank accession no. MCU68299 [complete genome]) placed into the BamHI site of pACYC184 (16), was used for construction of the MIEPE deletion plasmid pAMB25ΔMIEPE (Fig. 1A). To construct a plasmid that contains a deletion ranging from nucleotides −5 to −1272, counted relative to the 5′ start site of the ie1/3 transcription unit corresponding to position 182,895 of the complete genome, pAMB25 was digested with MunI and MluI (cleavage positions +209 and −2091, respectively). Surplus deletions of downstream ie1/3 and ie2 sequences were restored by PCR. Specifically, primer Nata-2 (5′-CAATGCATCTTAAGTACCGTCGCAGTCTTCGGTCTG-3′, containing an AflII and a NsiI site) and primer Nata-7 (5′-CCGTCGCTTGTAATATCTGG-3′) were used to amplify a 742-bp fragment of the ie1/3 transcription unit. This fragment was digested with NsiI and MluI. Primer Nata-1 (5′-CAATGCATGCGGCCGCGCAAATTAGGGGATTTCAGTGC-3′, containing a NotI and an NsiI site) and primer Nata-8 (5′-AACAAGAGAGATCAGTCTCG-3′) were used to amplify a 1,433-bp fragment of the ie2 gene that included the complete ie2 promoter. This fragment was digested with MunI and NsiI and was ligated together with the digested ie1/3 PCR fragment into MluI- and MunI-cleaved pAMB25.
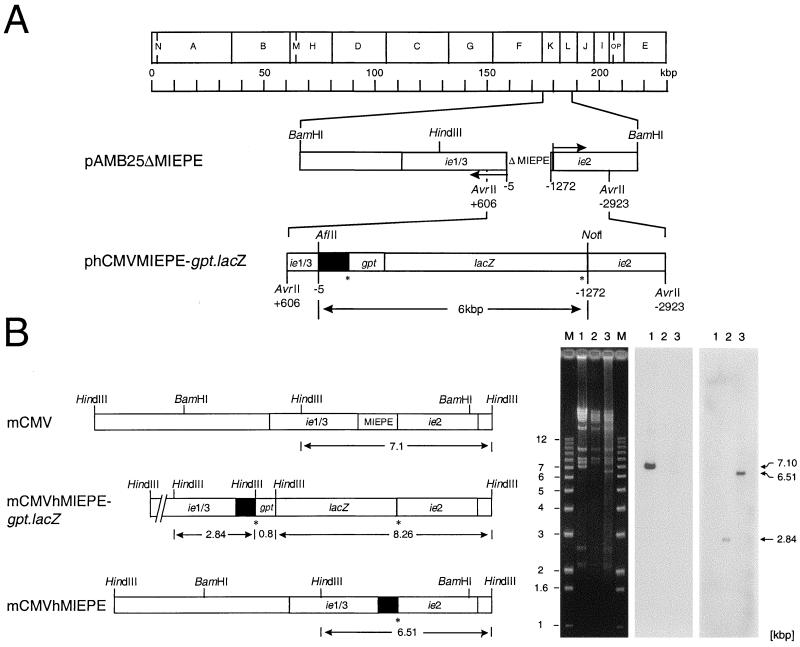
FIG. 1.
Construction and verification of recombinant viruses. All map positions are given relative to the 5′ start site (counted as +1) of the ie1/3 transcription unit of mCMV, and illustrations are drawn to scale. (A) Map of plasmid constructs for homologous recombination. The HindIII physical map of the mCMV Smith strain genome is shown at the top. For the construction of the mCMV MIEPE deletion plasmid pAMB25ΔMIEPE, plasmid pAMB25 was digested with MluI and MunI and surplus deletions were restored by PCR, resulting in a final deletion of 1,267 bp (ΔMIEPE). Arrows indicate the orientations of ie1/3 and ie2 transcription. Plasmid phCMVMIEPE-gpt.lacZ was generated by insertion of the hCMV enhancer and core promoter (solid box, representing hCMV nucleotides −14 to −601 relative to the start site of the hCMV ie1-ie2 transcription unit) and of a gpt.lacZ reporter gene cassette flanked by loxP sites (asterisks). (B) MIEPE swap mutants. Maps are shown on the left and the corresponding HindIII cleavage analysis is shown on the right. (Left) Expanded HindIII fragments K and L of the mCMV Smith strain genome are shown on the top, illustrating the location of the MIEPE and flanking ie sequences within the authentic 7.1-kbp L fragment (map corresponding to lane 1). Replacement of the mCMV MIEPE by the hCMV MIEPE (solid box) and insertion of a gpt.lacZ reporter gene cassette flanked by loxP sites (asterisks) created novel HindIII fragments of 0.8, 2.84, and 8.26 kbp in the genome of recombinant virus mCMVhMIEPE-gpt.lacZ (map corresponding to lane 2). For the generation of recombinant virus mCMVhMIEPE, the gpt-lacZ cassette was removed from mCMVhMIEPE-gpt.lacZ via Cre recombinase-mediated loxP-specific recombination, leaving a single loxP site (asterisk) and generating a shortened HindIII L fragment of 6.51 kbp (map corresponding to lane 3). (Right) Purified virion DNA was subjected to cleavage by HindIII, and fragments were analyzed by agarose gel electrophoresis, Southern blot, and hybridization with MIEPE type-specific γ-32P-end-labeled oligonucleotide probes. Lanes: M, indicated size markers; 1, DNA of parental virus mCMVΔorf152; 2, DNA of mCMVhMIEPE-gpt.lacZ; 3, DNA of mCMVhMIEPE. Left panel, ethidium bromide-stained gel; center panel, autoradiograph obtained after hybridization of the Southern blot with the 30-bp probe mE-oligo-P; right panel, autoradiograph obtained after stripping of the same filter followed by hybridization with the 30-bp oligonucleotide probe hE-oligo-P. See Fig. 3 for the map locations of the two probes.
Plasmid phCMVMIEPE-gpt.lacZ (Fig. 1A), which was used for homologous recombination, was constructed as follows. Primers Nata-3 (5′-CAGCGGCCGCCTGCTTCGCGATGTACGGGC-3′, containing a NotI site) and Nata-4 (5′-CACTTAAGGAGAGCTCTGCTTATATAGACC-3′, containing an AflII site) served to amplify a 638-bp fragment from template pcDNA3 (catalog no. V790-20; Invitrogen, Leek, The Netherlands) that included a 587-bp sequence of the hCMV MIEPE encompassing positions −14 to −601 relative to the transcription start site of the ie1-ie2 transcription unit of hCMV. This fragment was digested with NotI and AflII and was inserted into NotI- and AflII-digested pAMB25ΔMIEPE. To facilitate further cloning procedures, phCMVMIEPE was cloned. Specifically, a 2.9-kbp AvrII fragment that included the MIEPE of hCMV flanked by mCMV ie1/3 and ie2 sequences was ligated into the XbaI site of pUC19. For the final construction of phCMVMIEPE-gpt.lacZ, phCMVMIEPE was partially digested with NruI (cleaving at position −629 relative to the 5′ transcription start site of mCMV ie1/3), followed by digestion with NotI and ligation to the 5.3-kbp gpt.lacZ fragment flanked by loxP sites. This marker gene cassette had been generated from plasmid plg1 (kindly provided by M. Messerle, Max von Pettenkofer Institute, Munich, Germany) by XbaI cleavage, filling up of the sticky ends with Klenow DNA polymerase, and NotI cleavage. Throughout, the fidelity of PCR-based cloning steps was verified by sequencing (automated DNA sequencing system, model 4000; LI-COR Inc., Lincoln, Nebr.).
(ii) Generation and purification of recombinant virus mCMVhMIEPE-gpt.lacZ.
Recombinant viruses were generated by homologous recombination in NIH 3T3 fibroblasts (no. CL-163; American Type Culture Collection, Rockville, Md.) transfected with plasmid DNA linearized by ScaI restriction enzyme cleavage and infected with mCMV. Specifically, NIH 3T3 cells were plated in six-well culture plates (catalog no. 3046; Falcon, Meylan Cedex, France) and incubated under standard conditions (37°C, 5% CO2, and humidified atmosphere) in Dulbecco’s modified Eagle’s medium (DMEM) (catalog no. 4196-039; Gibco BRL, Eggenstein, Germany) supplemented with 10% (vol/vol) fetal calf serum, 2 mM l-glutamine, 100 U of penicillin per ml, and 0.1 mg of streptomycin per ml. The next day, an average of 2 × 105 cells per monolayer were transfected with 2 μg of phCMVMIEPE-gpt.lacZ DNA by using 10 μl of the nonliposomal transfection reagent FuGENE 6 (catalog no. 1814443; Boehringer, Mannheim, Germany). One day after transfection, cells were infected under conditions of centrifugal enhancement of infectivity (1,000 × g for 30 min at 20°C) at a multiplicity of 4 PFU* (centrifugal PFU) of sucrose-gradient-purified mCMV (22). Excess virus was washed out, and cultures were refed with fresh DMEM. On day 2 after infection, the culture supernatant was used for centrifugal infection of fetal mouse cells (usually, but incorrectly, referred to as mouse embryo fibroblasts [MEF]) grown to almost confluent monolayers in six-well culture plates. The infection and subsequent cultivation was performed with selection medium MEM-S, which is minimal essential medium (MEM) supplemented as specified previously (40) and containing in addition 12.5 μg of mycophenolic acid (catalog no. 11814-019; Gibco BRL) per ml and 100 μg of xanthine (catalog no. 1.08675.0050; Merck, Darmstadt, Germany) per ml. After 3 days, that is, after plaques became visible, the supernatant was harvested and used for a second round of recombinant virus selection and amplification on MEF. The supernatant thereof was used to infect MEF centrifugally with a low multiplicity of ca. 0.02 PFU, corresponding to 0.4 PFU*. After 24 h, MEF were detached by weak trypsinization, and cells infected with recombinant virus were enriched by cytofluorometric cell sorting using the cell sorter FACSort equipped with a cell concentrator module (Becton Dickinson, San Jose, Calif.). CellQuest software (Becton Dickinson) was used for data acquisition and processing. In brief, cells infected with recombinant virus were identified by the lacZ reporter gene-encoded intracellular β-galactosidase yielding a green 520-nm fluorescence after FACS-Gal vital staining using fluorescein di-β-d-galactopyranoside (FDG) (catalog no. F2756; Sigma-Aldrich, Deisenhofen, Germany) as fluorogenic substrate. The enzyme reaction was performed with 106 cells and 1 mM FDG in a volume of 200 μl according to an established protocol (12). Dead cells were excluded by propidium iodide staining, and the electronic sort window was set on viable cells with high fluorescein fluorescence (FL-1), discarding negative cells as well as cells with low expression. Sorting was performed at a flow rate of ca. 2,000 cells/min and a sort rate of ca. 60 positive cells/min for a total of ca. 5,000 cells sorted. Sorted cells were cocultivated with MEF for 3 days in MEM-S. The supernatant was used to infect MEF at low multiplicity for a second round of cytofluorometric sorting followed by cocultivation. The final supernatant was used for limiting dilution-based plaque purification on MEF monolayers, which was performed by using an overlay of MEM-agarose (0.7% [wt/vol] agarose in MEM with no phenol red) supplemented with Bluo-Gal (300 μg/ml−1) (catalog no. 15519-028; Gibco BRL) for blue vital staining. Plaque-purified recombinant virus was propagated on MEF and concentrated by sedimentation through a sucrose density cushion at 53,000 × g essentially as described previously (22) except that the virus in the culture supernatant was harvested after 3 days instead of after 5 days to minimize contamination by cellular DNA. The infectious titer was determined by a standard 4-day PFU assay on MEF with no centrifugal enhancement and was 2 × 108 PFU per ml for the particular batch used here.
(iii) Generation of recombinant virus mCMVhMIEPE.
The gpt-lacZ cassette was removed from recombinant virus mCMVhMIEPE-gpt-lacZ via loxP-specific recombination by using Cre recombinase (3) introduced by the replication-defective adenovirus vector Cre-Ad (generously provided by W. H. Burns, Medical College of Wisconsin, Milwaukee, Wis.). Specifically, ca. 5 × 106 MEF grown to monolayer in a 10-cm-diameter petri dish were coinfected with mCMVhMIEPE-gpt.lacZ and Cre-Ad at multiplicities of infection of 0.02 PFU and 10 PFU equivalents, respectively. On day 2 after coinfection, that is, when plaques became visible, 20 μl of culture supernatant was used for infection of STO cells (mouse fibroblast cell line) (no. CRL-1503; ATCC), which are resistant to 6-thioguanine. Infected STO cells were cultured in six-well plates in DMEM supplemented with 5% (vol/vol) fetal calf serum and with 20 μg of 6-thioguanine (2-amino-6-methyl-mercaptopurine) (catalog no. A9546; Sigma) per ml for selection against gpt-expressing virus mCMVhMIEPE-gpt.lacZ. On day 4, supernatant of the infected STO cells was used for limiting dilution-based plaque purification on MEF covered by MEM-Bluo-Gal agarose. Unstained “white” plaques were recovered, and recombinant virus mCMVhMIEPE was propagated on MEF and purified by sucrose gradient ultracentrifugation. The titer of the virus batch used in the reported experiments was 1.7 × 108 PFU per ml.
Southern blot analysis of recombinant MIEPE regions.
Virion DNA was prepared from the sucrose-gradient-purified virus stocks of supernatant mCMV and of the recombinants as described previously (22). Purified DNA was then subjected to a HindIII restriction enzyme cleavage performed according to established procedures. Fragments were separated on a 0.7% (wt/vol) agarose gel, stained with ethidium bromide, and visualized by 302-nm UV illumination. After Southern transfer, consecutive hybridizations were performed with fragment-specific, γ-32P-end-labeled oligonucleotide probes mE-oligo-P (5′-AGGTAAGCCAATGGGTTTTTCCCATTACTG-3′) and hE-oligo-P (5′-TACATCTACGTATTAGTCATCGCTATTACC-3′), which are specific for nonhomologous regions of the mCMV and hCMV enhancer, respectively (for a map, see Fig. 3). After the first hybridization, the filter was stripped, radioactivity was washed out, and the same filter was rehybridized with the second probe. Specific bands were visualized by autoradiography.
FIG. 3.
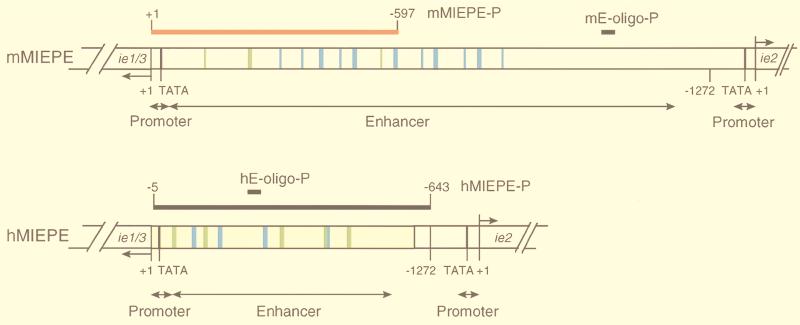
Map of MIEPE type-specific hybridization probes. (Top) Structural organization of the authentic MIEPE of mCMV (mMIEPE), essentially based on data by Dorsch-Häsler et al. (11). Map positions refer to the 5′ start site (counted as +1) of the ie1/3 transcription unit. The map location of oligonucleotide probe mE-oligo-P used for Southern blot hybridization (see Fig. 1B) is indicated. The red bar represents the polynucleotide probe mMIEPE-P used for mCMV MIEPE-specific red staining in the two-color ISH. (Bottom) Structural organization of the chimeric region in recombinant virus mCMVhMIEPE. Sequences of the inserted hCMV core promoter and enhancer (hMIEPE), representing positions −14 to −601 in hCMV, are highlighted by yellow shading. Map positions refer to the 5′ start site (counted as +1) of the mCMV ie1/3 transcription unit. The map location of oligonucleotide probe hE-oligo-P (see Fig. 1B) is indicated. The long black bar represents the polynucleotide probe hMIEPE-P used for hCMV MIEPE-specific black staining in the two-color ISH. The two maps are drawn to scale. Consensus sequences within 18- and 19-bp direct repeats, encompassing NF-κB and CREB/ATF binding sites, are marked by blue and green coloring, respectively.
Determination of genome-to-infectivity ratios.
Viral genomes were quantitated essentially as described previously (references 22 and 44 and with modifications described in reference 21) by serial dilution of purified virion DNA prepared from stocks of known virus infectivity titer, followed by mCMV ie1 gene exon 4-specific PCR, Southern dot blot hybridization, and phosphorimaging. Specifically, the γ-32P-end-labeled oligonucleotide IE1.2135 (4) was used as the hybridization probe to visualize the specific 363-bp amplificate, and plasmid pIE111 (28), which includes the ie1 gene, was titrated as a standard. Infectivity measured in terms of noncentrifugal PFU was related to the number of genomes. For the infection of MEF with the parental mCMV Smith strain, a genome-to-PFU ratio of ca. 500:1 had likewise been determined previously (22).
Infection of immunocompromised mice.
Female BALB/c mice (8 weeks old) were severely immunocompromised by hematoablative total-body γ irradiation with a single dose of 8 Gy delivered by a 137Cs source. In the absence of infection, this dose by itself is 100% lethal within 14 days. At ca. 8 h after the irradiation, recipients were infected intravenously in the tail vein with 104 PFU of sucrose-gradient-purified virus (see above) contained in 0.1 ml of physiological saline. Analyses were usually performed on day 9 after infection, that is, at a prefinal stage of multiple-organ CMV disease (36).
MIEPE-specific two-color in situ hybridization.
The in vivo replication of viruses in host tissues is visualized by in situ hybridization (ISH) detecting the viral DNA that is accumulated in an intranuclear inclusion body during the late phase of the viral replication cycle. mCMV containing the authentic mCMV MIEPE and the recombinant virus mCMVhMIEPE, in which the mCMV MIEPE is replaced by the MIEPE of hCMV, are distinguished by MIEPE-specific polynucleotide probes (see Fig. 3), marked with a red and black label, respectively. Tissue was fixed in 4% (vol/vol) formalin buffered at pH 7.4 and was embedded in paraffin according to established procedures. Deparaffinized 2-μm sections of tissue were subjected to digestion with proteinase K (catalog no. P0390; Sigma), washed, dehydrated, and immersed with hybridization solution consisting of hybridization buffer (HybriBuffer ISH, catalog no. R012.050; Biognostik, Göttingen, Germany) and 1 μg of the respective DNA probes per ml. After denaturation for 10 min at 93°C, hybridization was performed for 16 h at 32°C. The MIEPE-specific staining was performed as specified in more detail below. Sections were counterstained for 5 s with hematoxylin to visualize the nuclei of uninfected cells. Microscopic analysis and documentation were made with a Zeiss research microscope (Axiophot; Carl Zeiss Jena GmbH, Jena, Germany) using oil-immersion optics (plan-Neofluar; Zeiss) for all magnifications.
(i) Single-color ISH specific for the mCMV MIEPE.
A red label is used to visualize viral genomes containing the authentic MIEPE of mCMV. The hybridization probe mMIEPE-P spans positions +1 to −597 (598 bps) within the mCMV MIEPE relative to the 5′ transcription start site of the MIE (ie1/3) transcription unit (see Fig. 3). The probe was synthesized by PCR using plasmid pAMB25 as the template and oligonucleotides Nata-9 (5′-CGGTACCGACGCTGGTCGCGCCTCTTAT-3′) and Nata-10 (5′-TGGTCGCGCCTCTTATACCCACGTAGAA-3′) as forward and reverse primers, respectively. Labeling was achieved by incorporation of fluorescein-conjugated dUTP (fluorescein-12-dUTP, catalog no. 1373242; Boehringer Mannheim). The staining was performed by using alkaline phosphatase-conjugated anti-fluorescein antibody (catalog no. 1426338, Boehringer Mannheim) and new fuchsin as the substrate, yielding a brilliant red color.
(ii) Single-color ISH specific for the hCMV MIEPE.
A black label is used to visualize recombinant mCMV genomes carrying the paralogous MIEPE of hCMV. The hybridization probe hMIEPE-P spans positions −5 to −643 (638 bps) encompassing the inserted hCMV MIEPE relative to the 5′ transcription start site of the mCMV MIE (ie1/3) transcription unit (see Fig. 3). The probe was synthesized by PCR using plasmid pcDNA3 as the template and oligonucleotides Nata-3 and Nata-4 (specified above), respectively, as forward and reverse primers. Labeling was achieved by incorporation of digoxigenin (DIG)-11-dUTP (catalog no. 1093088; Boehringer Mannheim). The staining was performed by using peroxidase-conjugated anti-DIG antibody (catalog no. 1207733; Boehringer Mannheim) and diaminobenzidine tetrahydrochloride (catalog no. D-5637; Sigma) as the substrate. The staining was enhanced by ammonium nickel sulfate hexahydrate (catalog no. 09885; Fluka, Neu-Ulm, Germany), yielding a black precipitate.
(iii) Two-color ISH for the simultaneous detection of mCMV and recombinant mCMVhMIEPE.
Viruses mCMV (red label) and mCMVhMIEPE (black label) were detected simultaneously by two-color ISH in tissues of mice coinfected with both viruses. For hybridization, 1 ml of hybridization buffer contained 1 μg of each probe. The antibodies directed against fluorescein and DIG (see above) were added as a mixture for 1 h. The new fuchsin substrate for red staining was added before the enzyme reaction and enhancement for black staining were performed. Comparable sensitivity of the two MIEPE-specific hybridization probes was verified in tissues infected with either mCMV or mCMVhMIEPE by comparing the number of infected cells detected by the respective probe with the number of infected cells detected in the same tissues by using a previously published mixture of digoxigenized plasmids containing HindIII fragments A, I, and K, thereby representing 51.2 kbp of the mCMV genome (36).
IHC detection of the IE1 protein.
The expression of the mCMV ie1 gene-encoded intranuclear IE1 protein pp89 (19) in mouse tissues was analyzed by immunohistochemical (IHC) staining. Paraffin sections (2 μm) were dewaxed in xylene. Sections were incubated for 15 min at 37°C in trypsin solution (1.25 mg/ml). Endogenous peroxidase was inactivated by an incubation for 30 min at 20°C in 0.5% (vol/vol) hydrogen peroxide in a 1:1 mixture of methanol and phosphate-buffered saline (PBS). Unspecific antibody binding sites were blocked with a 1:10 dilution of normal rabbit serum in PBS for 20 min at 20°C. The sections were then labeled for 60 min at 37°C with the IE1-specific monoclonal antibody CROMA 101 (mouse immunoglobulin isotype G1 [IgG1]). Mouse IgG1 (catalog no. X-0931; Dako, Hamburg, Germany) was used for the isotype control. The staining was performed by the ABC method, by using a biotinylated goat anti-mouse IgG-Fab antibody (catalog no. B0529; Sigma) at a 1:200 dilution in PBS for 30 min, followed by detection with an avidin-biotin-peroxidase complex (Vectastain ABC kit standard PK-4000) and diaminobenzidine tetrahydrochloride as the substrate. The staining was enhanced by ammonium nickel sulfate hexahydrate, yielding a black precipitate. Counterstaining of the sections was performed for 5 s with hematoxylin to visualize uninfected nuclei.
Area morphometry for quantitating multistep virus replication in tissue.
Diapositives (Fujichrome 64T-RTP 135 for color transparencies; Fuji, Tokyo, Japan) of microphotographs taken from stained histological sections were scanned for computing (35-mm film desktop scanner LS-1000; Nikon, Tokyo, Japan). The extension of virus foci in tissue, specifically in the liver, was measured by using the area morphometry utility of the OPTIMAS 6.0 software for image analysis (Optimas Corporation, Bothell, Wash.).
RESULTS
Generation of recombinant virus mCMVhMIEPE.
The promoter of the ie1/3 transcription unit of mCMV and the upstream enhancer, that is, the complete MIEPE of mCMV, was replaced by the paralogous MIE core promoter and enhancer of hCMV by homologous recombination. To avoid confusion, it must be recalled that the MIEPE of hCMV controls the hCMV transcription unit ie1-ie2 and that the mCMV ie3 gene product IE3 (28) is the main transactivator, corresponding to the hCMV ie2 gene product IE2 (for overviews, see references 43 and 45). For selection by drug resistance and by cytofluorometric cell sorting, recombinant virus mCMVhMIEPE-gpt.lacZ was generated as an intermediate. It should be noted that the promoter of mCMV gene ie2, which is under the control of the same enhancer (30), was spared. Gene ie2 is transcribed in a direction opposite to ie1/3, leading to a 43-kDa protein specified by a spliced 1.75-kb mRNA (30). It has no direct counterpart in hCMV. Although ie2 proved to be nonessential for mCMV replication in vitro and in vivo (8) and was hence used as a site for integration of foreign genes into the mCMV genome (31), a role for the IE2 protein in the biology of mCMV cannot be ruled out. Thus, separating the enhancer from the promoter of ie2 by the 5.3-kbp selection gene cassette might have an unpredictable modulating function in viral pathogenicity. In addition, an influence of the enzymes encoded by gpt and lacZ, namely xanthine-guanine phosphoribosyl transferase and β-d-galactosidase, respectively, is difficult to assess. To avoid all these unnecessary complications, gpt-lacZ was removed by loxP-specific recombination to generate recombinant virus mCMVhMIEPE, in which the hCMV enhancer is flanked upstream of ie1/3 by the hCMV MIE core promoter and upstream of ie2 by the authentic mCMV ie2 promoter (Fig. 1B; resolved to greater detail in Fig. 3). As sketched in the map in Fig. 1B, the HindIII fragment L of mCMV is 7.1 kbp in size. Insertions present in recombinant virus mCMVhMIEPE-gpt.lacZ predict novel HindIII cleavage fragments of 0.8, 2.84, and 8.26 kbp. Replacement of the mCMV MIEPE by the shorter hCMV MIEPE in recombinant virus mCMVhMIEPE predicts an altered HindIII L fragment of 6.51 kbp. The HindIII restriction enzyme cleavage patterns for purified virion DNAs of the three viruses in fact revealed fragments of the predicted sizes in the ethidium bromide-stained agarose gels (Fig. 1B, left panel). Their identity with the predicted fragments of 7.10 kbp (representing mCMV), 2.84 kbp (representing mCMVhMIEPE-gpt.lacZ), and 6.51 kbp (representing mCMVhMIEPE) was confirmed by Southern blotting and hybridization with the enhancer-specific oligonucleotide probes (for map positions, see Fig. 3) mE-oligo-P (center panel) and hE-oligo-P (right panel).
The paralogous MIEPE of hCMV does not alter the in vitro infectivity of mCMV in permissive mouse fibroblasts.
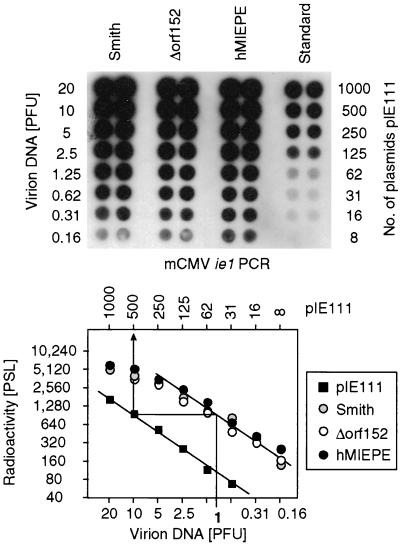
The capacity of recombinant virus mCMVhMIEPE to infect murine fibroblasts in cell culture was assessed in molecular terms by measuring the genome-to-PFU ratio, that is, the number of viral genomes required for initiating a cytolytic, plaque-forming productive infection. Since formation of a plaque over 3 to 5 days in cell culture involves spread of the infection, the genome-to-PFU ratio reflects viral infectivity under multistep growth conditions. For the parental mCMV Smith strain ATCC VR194/1981, a genome-to-PFU ratio of 500:1 has been determined previously (22). That the mutant mCMVhMIEPE is not significantly handicapped with respect to growth in cell culture was already indicated by the comparable virus yields and plaque sizes observed during the preparation of highly titratable stocks of purified virions of mCMVΔorf152 and recombinant mCMVhMIEPE derived therefrom (not shown). For precise quantitation, virion DNA, corresponding to defined infectivity measured as PFU, was isolated and titrated in parallel to defined numbers of plasmids pIE111 encompassing gene ie1 of mCMV that is shared by parental mCMV Smith, mCMVΔorf152, and mCMVhMIEPE. After PCR specific for a 363-bp sequence located within exon 4 of gene ie1, a Southern dot blot in microplate format was hybridized with a γ-32P-end-labeled internal oligonucleotide probe (Fig. 2, top), and quantitation was performed by phosphorimaging (Fig. 2, bottom). In essence, the previously published genome-to-PFU ratio of 500:1 (22) was reproduced here for mCMV Smith, and, remarkably, the same result was obtained for virus mCMVΔorf152 and even for mCMVhMIEPE.
FIG. 2.
Determination of the genome-to-infectivity ratios for prototype mCMV Smith, parental mCMVΔorf152, and MIEPE swap mutant mCMVhMIEPE. Purified virion DNA, corresponding to known infectivity measured as noncentrifugal PFU in fibroblast monolayer cultures, was serially diluted in independent duplicates and was subjected to PCR, amplifying a 363-bp fragment of ie1 gene exon 4. Defined numbers of plasmid pIE111, encompassing gene ie1, were subjected to PCR as a standard. (Top) Autoradiograph of a Southern dot blot obtained after hybridization with a γ-32P-end-labeled internal oligonucleotide probe. (Bottom) Computed phosphorimaging results of the same blot. Log-log plots of radioactivity (means of duplicates) measured as phosphostimulated luminescence (PSL) units (ordinate) versus the DNA dilutions expressed in terms of PFU (abscissa) are shown. The upper rule relates the PFU to the number of pIE111 plasmids in the standard. The calculation from the linear portions of the graphs is demonstrated for 1 PFU corresponding to 500 molecules of pIE111 standard template. Thus, the genome-to-PFU ratio is 500:1 in this example.
Design of MIEPE-specific polynucleotide hybridization probes for two-color in situ detection of recombinant virus replication.
In principle, an in vitro PFU assay performed by plating tissue homogenate onto permissive cell monolayers, such as MEF, could be used to get a relative estimate of in vivo virus replication. However, virus titers are not an absolute measure of virus replication in tissues because the efficacy of detection is unknown and may even differ between organs as a result of virus inactivation during the procedures. In addition, graphs of virus titers do not reveal the cell types infected in tissues, they do not verify the identity of the plaque-forming virus, and they fail to give an impression of the degree of tissue destruction. Specifically, virus titers do not distinguish between the number and the size of infectious foci in tissue. Thus, many small foci and a few large foci can give the same titer. While the number of foci can serve as a measure of the efficacy of virus dissemination to an organ and within an organ, the extension of an infectious focus in a particular tissue more directly reflects the capacity of a virus to grow in the cell types which constitute that tissue. A distinction between these two parameters appeared to us most informative. The detection and quantitation of infected cells in organs by in situ techniques is thus the superior method and was therefore our method of choice.
The difference between the viruses mCMV and mCMVhMIEPE is the nature of the MIEPE. Accordingly, a discrimination between infectious foci caused by these two viruses required MIEPE-specific DNA hybridization probes for two-color ISH detecting viral DNA. In the late phase of productive viral replication, viral DNA is found in the infected cells highly accumulated within an intranuclear inclusion body, that is, at the site of nucleocapsid assembly and DNA packaging. The high concentration of viral genomes in these inclusion bodies contributes to a high sensitivity of detection. The positions of the probes within the aligned hCMV and mCMV MIEPEs are delineated in Fig. 3, with homologous parts within 18- and 19-bp repeats shown in blue and green, respectively. It may be informative to note that our first approach had been to design mixtures of oligonucleotide probes directed selectively against nonhomologous interrepeat regions. However, even though principally successful, the low amount of label provided by an oligonucleotide required a long period of staining and thereby caused some unspecific background in uninfected tissue (not shown). It was a bit of a surprise that the sequence homologies between the two MIEPEs proved not to be critical for the specificity of the chosen polynucleotide hybridization probes mMIEPE-P and hMIEPE-P.
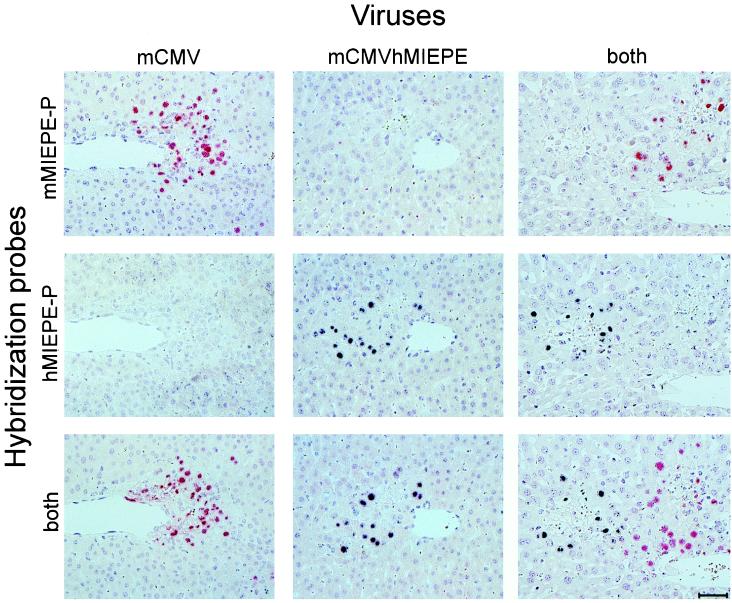
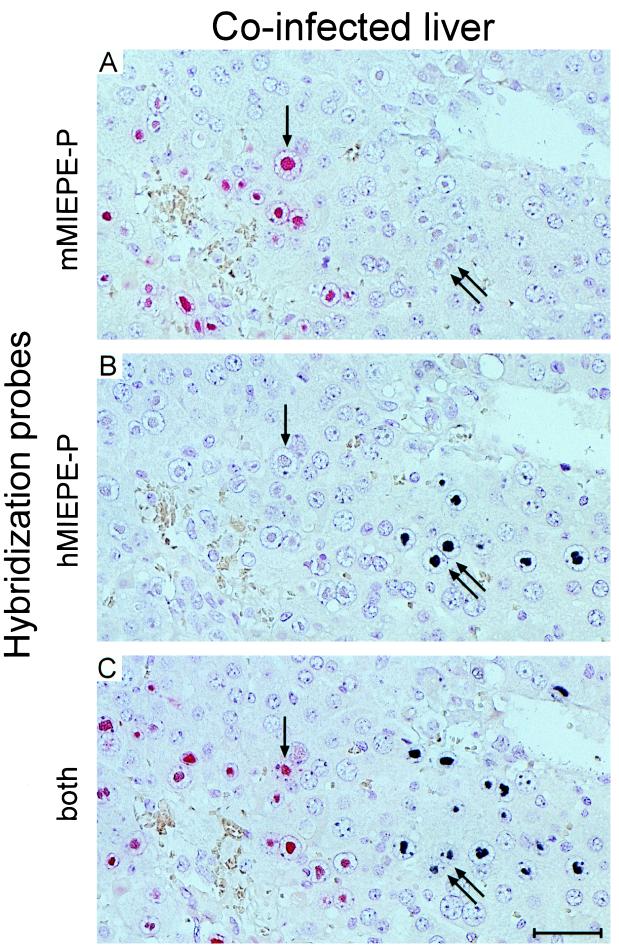
For testing the specificity of the ISH probes, BALB/c mice were immunodepleted by γ irradiation with a dose of 8 Gy and were infected intravenously with either 104 PFU of mCMV or 104 PFU of mCMVhMIEPE or with a mixture (5 × 103 PFU plus 5 × 103 PFU) of both viruses. At the time of clinically manifested severe CMV disease, that is, usually on day 9 after infection, liver sections were stained for single-color ISH with either probe mMIEPE-P (red label) specific for mCMV or probe hMIEPE-P (black label) specific for recombinant virus mCMVhMIEPE or were stained for two-color ISH with a mixture of both probes. The results obtained with all possible combinations of viruses and hybridization probes are shown in Fig. 4. The analysis of individual foci was made possible by serial, neighboring sections, and in all instances a central vein was chosen as a landmark in order to facilitate recognition of individual foci. The hybridizations document the exquisite specificity of the chosen probes. Thus, with probe mMIEPE-P, mCMV-derived foci appear red and mCMVhMIEPE-derived foci remain unstained (Fig. 4, top). In like manner, with probe hMIEPE-P, mCMV-derived foci remain unstained while mCMVhMIEPE-derived foci appear black (Fig. 4, center). Finally, by using a mixture of the two probes, mCMV-derived and mCMVhMIEPE-derived foci are distinguished by red and black staining, respectively (Fig. 4, bottom).
FIG. 4.
Two-color MIEPE type-specific ISH in liver tissue sections. After immunoablative treatment, groups of BALB/c mice were infected intravenously with either mCMV or recombinant virus mCMVhMIEPE or were coinfected with both viruses. Histological analysis was performed on day 9 after infection. Three serial sections, sharing a central vein as a landmark, were selected for hybridization. The first section of each series was hybridized with a polynucleotide probe specific for the MIEPE of mCMV (mMIEPE-P; red staining), the second was hybridized with a polynucleotide probe specific for the MIEPE of hCMV (hMIEPE-P; black staining), and the third was hybridized with a mixture of both probes. See Fig. 3 for the map locations of the two probes. Results are documented for all nine possible combinations. Counterstaining was performed with hematoxylin. The bar in the bottom right panel represents 50 μm.
Recombinant virus mCMVhMIEPE and parental virus mCMV replicate independently.
The presence of separate red and black foci in liver parenchyma (Fig. 4) already implied that mCMVhMIEPE does not depend on parental mCMV as a helper virus for replication in hepatocytes. This is even more elegantly revealed by an in situ single cell analysis as documented by serial sections tracking individual hepatocytes in a liver coinfected with mCMV and mCMVhMIEPE (Fig. 5). Probe mMIEPE-P stains mCMV-derived intranuclear inclusion bodies red. Out of many mCMV-infected cells present in the red focus, a single arrow highlights a hepatocyte with a prominent red-stained intranuclear inclusion body (Fig. 5A). A twin arrow points to a cell couple with clearly visible but only hematoxylin-stained intranuclear inclusion bodies, indicating the presence of a second focus apparently not caused by parental mCMV. The immediate neighbor in the series of sections, that is, only 2 μm from the first, was hybridized with probe hMIEPE-P (Fig. 5B). The twin arrow points to the very same cell couple seen before, but now black staining of their intranuclear inclusion bodies identifies them as hepatocytes being infected by mCMVhMIEPE, whereas the inclusion body of the hepatocyte in the left focus is no longer specifically stained. The third section in the series, 4 μm from the first, was hybridized with both probes (Fig. 5C). Although the mCMVhMIEPE-infected cell couple begins to disappear from sight, we can still recognize parts of the black-stained intranuclear inclusion bodies. Likewise, the mCMV-infected cell in the left focus can still be identified. In addition to the cells highlighted by the arrows, one can easily find many further examples of hepatocytes infected in a mutually exclusive manner by either mCMV or mCMVhMIEPE, some emerging and some disappearing in the section series.
FIG. 5.
Replication of mCMV and mCMVhMIEPE in mouse liver parenchyma after coinfection. Mutually exclusive, independent infection of hepatocytes by the two viruses is documented by a MIEPE type-specific, single-cell two-color ISH analysis. Three serial, directly neighboring 2-μm sections were hybridized with probe mMIEPE-P, staining mCMV DNA in red (A); with probe hMIEPE-P, staining mCMVhMIEPE DNA in black (B); and with both probes (C). A central vein in the upper right corner serves as a landmark. The single arrow tracks an mCMV-infected hepatocyte through the section series. Note the particularly prominent intranuclear inclusion body stained by mMIEPE-P but not by hMIEPE-P. Likewise, the twin arrows mark a cell couple that is infected by the MIEPE swap recombinant virus mCMVhMIEPE, as we can conclude from the black staining with hMIEPE-P. Counterstaining was performed with hematoxylin. The bar in panel C represents 50 μm.
In conclusion, an mCMV recombinant virus containing the enhancer and core promoter of hCMV can independently replicate in vivo in murine hepatocytes and generates foci of infection in mouse liver parenchyma spatially separated from the foci that are generated by parental mCMV.
The core promoter and enhancer swap does not impair the multistep growth of mCMV in mouse liver parenchyma.
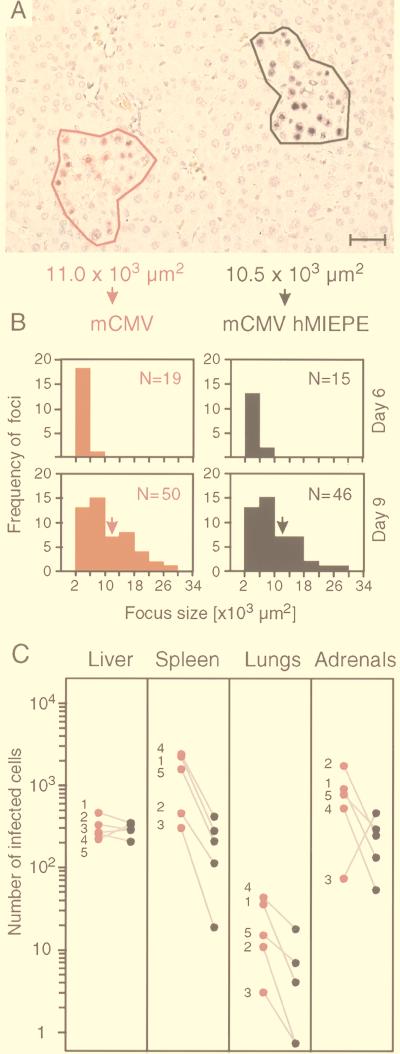
So far, the data have documented that mCMV is principally capable of infecting mouse hepatocytes in vivo after replacement of its complete autologous MIEPE by the paralogous MIE core promoter and enhancer of hCMV. We next asked whether a careful quantitative analysis would reveal any attenuation of recombinant virus mCMVhMIEPE with respect to infection of hepatocytes. It is instructive to recall the often-documented and generally accepted experience that virus titers in tissues may differ markedly between individual mice, even when mice are inbred and age matched and when all experimental parameters are kept bona fide constant. Specifically, after hematoablative treatment by γ irradiation, titer variances by a factor of 10 to 100 were not unusual for mCMV in organs, in particular in the liver (40). The phenomenon has never been investigated in detail, but since the genotoxic effect of radiation is by its nature a stochastic event, differences in residual immune functions are likely to be one significant cause of variance. Such variances clearly complicate the approach of comparing the in vivo infectivity of two viruses by comparing groups of mice infected with either virus, and conclusions then always depend on a statistical evaluation. However, there now exists a much better way: since we can distinguish mCMV and recombinant mCMVhMIEPE in tissue by two-color ISH, quantitative analysis can be made after coinfection of an animal so that all parameters imposed by the individual recipient are absolutely the same for the two viruses being compared.
A focus of infection, as it is seen in a liver tissue section, always represents a section through a three-dimensional “virus plaque” with a necrotic center and infected cells at its margin. During the screening of many sections, we never found a mixed focus. This indicates that each focus is caused by the progeny of a single PFU of either mCMV or mCMVhMIEPE. Accordingly, counting the foci in defined areas of tissue sections can be viewed as a two-color in vivo plaque assay. In addition to counting the number of foci, the spread of the viruses from hepatocyte to hepatocyte can be quantitated by measuring the size of foci. To do so, stained cells at the periphery of a focus were connected by a virtual line, and the area enclosed by the resulting polygon was determined by using area morphometry software. The procedure is illustrated for two foci, which have an almost equal, intermediate size on day 9 after coinfection (Fig. 6A). It is understood that the area covered by a focus in a 2-μm section is not representative of the total size of a three-dimensional focus. Despite this topological problem, measuring the areas of a large number of two-dimensional focus segments gives us a reliable measure for comparing the in vivo growth of different viruses. The results of such analyses, performed on days 6 and 9 after coinfection, are shown in Fig. 6B for representative total section areas of 30 mm2 compiled from three livers per time point. It is apparent that mCMV and mCMVhMIEPE were replicating in the liver with no significant difference in terms of focus number, focus size, and focus growth kinetics. Specifically, for both viruses, foci defined by more than four infected hepatocytes became detectable on day 6, and the number and size of foci increased similarly with time. In conclusion, recombinant virus mCMVhMIEPE is not significantly attenuated with respect to replication in mouse liver parenchyma.
FIG. 6.
Quantitative analysis of virus growth in coinfected tissues. (A) Illustration of area morphometric analysis. MIEPE type-specific two-color ISH with hybridization probes mMIEPE-P (red staining) and hMIEPE-P (black staining) was employed to distinguish infectious foci caused by the viruses mCMV and mCMVhMIEPE, respectively, in liver parenchyma on day 9 after coinfection. Foci were circumscribed by a virtual line, and the area enclosed by the resulting polygon was calculated by using area morphometry software. The bar represents 40 μm. (B) Kinetics of virus growth in liver parenchyma. Results of area morphometric analyses performed on days 6 and 9 after coinfection are documented for representative 30-mm2 areas of tissue compiled from three livers per time point. Histograms represent the number of foci (ordinate) per classified focus size (abscissa) with size classes of 4,000 μm2. A threshold was set at 2,000 μm2, which represents ca. four hepatocytes. N, total number of foci within a 30-mm2 area of tissue. Arrows point to the size class of the particular foci shown in panel A. (C) Comparative quantitation of virus replication in the liver and at extrahepatic sites. The numbers of cells infected with mCMV (red) and mCMVhMIEPE (black) were determined on day 9 after coinfection for representative 10-mm2 areas of tissue sections derived from the organs indicated. In the case of adrenal glands, infected cells were counted for 2-mm2 areas, and the number was extrapolated to 10 mm2, whereas infected cells were counted for 10-mm2 areas in the case of the other organs. Shown are linked data pairs for mouse 1 through mouse 5 in order to document the variance between individual mice.
Evidence for attenuation of recombinant virus mCMVhMIEPE at extrahepatic sites.
We next asked whether the results obtained for liver parenchyma, that is, for hepatocytes as its principal cellular constituent, would apply also to tissues composed of different cell types. The analysis was made by two-color ISH on day 9 after coinfection. The red- and black-stained cells, representing the replication of mCMV and mCMVhMIEPE, respectively, were counted for a representative area of tissue, and the results are presented as data pairs for five individual recipients (Fig. 6C). The analysis for the liver is included as a reference. Again, both viruses were found to have replicated with similar efficacy in the liver. There may be a tiny advantage for parental mCMV, as also seen in Fig. 6B, but in individual cases, such as in mouse 5, replication of mCMVhMIEPE predominates. By contrast, with no exception, there was an unequivocal replication advantage for parental mCMV in the spleen and the lungs of the same individual mice. Parental mCMV also predominated in the adrenal glands, with the exception of mouse 3, even though parental mCMV predominated in spleen and lungs of mouse 3. It must be noted, however, that colonization of adrenal glands by virus can be a random, clonal event. We even saw a case in which one of the paired adrenal glands was occupied by parental mCMV while its contralateral twin was occupied by mCMVhMIEPE (not shown). The molecular basis of the attenuation of mCMVhMIEPE is not yet known, and we wish to emphasize that we do not presently state that the foreign enhancer is responsible for a reduced replication in cell types other than hepatocytes. Our current impression is that mCMVhMIEPE has difficulty colonizing extrahepatic tissues although it is effectual in replicating within different tissues. One may speculate that mCMVhMIEPE is less efficient in breaking through endothelial barriers.
Recombinant virus mCMVhMIEPE is principally capable of infecting relevant target organs of CMV disease.
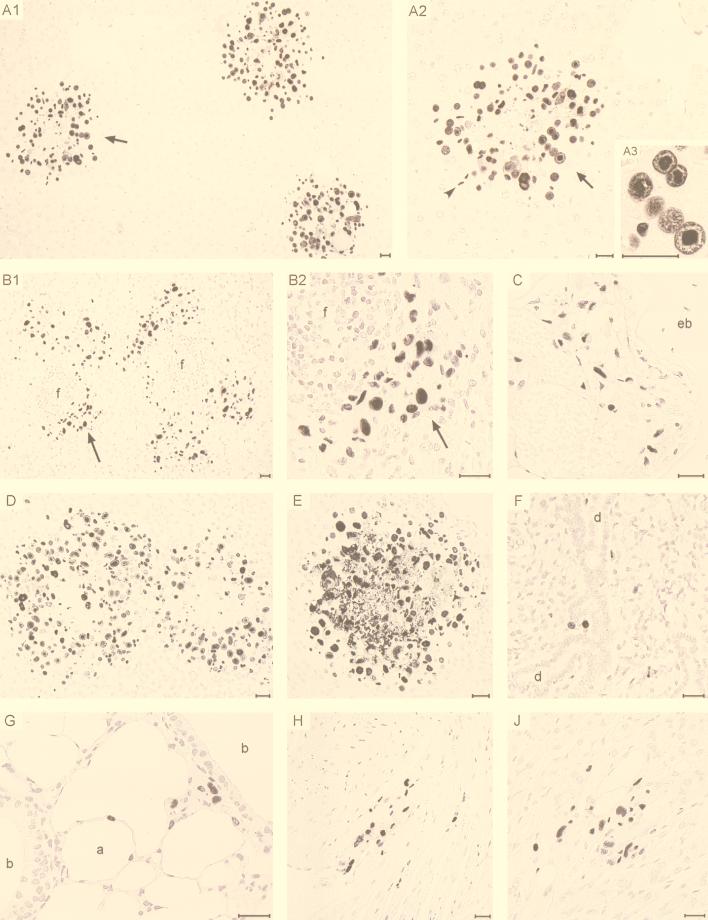
Finally, we asked whether the paralogous MIEPE has an influence on organ tropism of mCMV in a qualitative sense. As we have shown previously, mCMV is a polytropic virus that productively infects many different cell types and thereby causes fatal multiple-organ histopathology, provided that controlling CD8 T cells are absent (36). This condition is fulfilled after the immunoablative treatment used in this study. The MIEPE governs the transcription of the ie1/3 transcription unit. One of the resultant proteins is IE1 (pp89), which is produced in large amounts, is located in the nucleus throughout the replicative cycle, and condenses during the late phase of the cycle in the intranuclear inclusion body. Thus, labeling of IE1 by IHC staining detects all infected cells and, unlike ISH, not just those that have reached the late phase. IE1-specific IHC detection performed with nickel-enhanced black staining is therefore the most sensitive method for documenting the full extent of infection in tissues. Mice were infected intravenously with recombinant virus mCMVhMIEPE, and replication in various organs was studied on day 9 (Fig. 7). Again, livers were found to be heavily infected (see panel A1 for overview and panels A2 and A3 for details). The focus resolved to greater detail in Fig. 7A2 nicely demonstrates a plaque-like character with a necrotic center of already lysed hepatocytes and a margin of more recently infected hepatocytes. Few infected endothelial cells can easily be distinguished from hepatocytes by much smaller size and by an elongated nucleus (panel A2, arrowhead). Infected Kupffer cells are rarely found after 8 Gy immunoablation. Note the prominent inclusion bodies in hepatocyte nuclei highlighted in Fig. 7A3. In addition, mCMVhMIEPE was found to replicate in perifollicular stromal cells of the spleen (panels B1 and B2) as well as in reticular stromal cells of the bone marrow (panel C). Among glandular tissues, adrenal gland medulla (panel D) and cortex (panel E) as well as salivary glands (panel F; shown are acini of the submandibular gland) proved to be target sites for the replication of mCMVhMIEPE. While only single glandular epithelial cells are infected in the salivary glands with no signs of a cytopathic effect, which is a known characteristic of persistent salivary gland infection (15), foci in adrenal cortex and medulla are in a highly advanced stage and are thus associated with extended tissue necrosis. This finding demonstrates that mCMVhMIEPE spreads very efficiently within adrenal gland cortical and medullary tissues, provided that the colonization of the organ was successful. Further target sites include the lungs (panel G), the heart muscle (panel H), and the digestive tract (panel J; shown is the gastric mucosa). In conclusion, recombinant virus mCMVhMIEPE infects all relevant target organs known before from the tissue distribution of the CMV Smith strain (36) and can cause the multiple-organ histopathology that is characteristic of full-blown CMV disease.
FIG. 7.
Tissue distribution and cell-type tropism of MIEPE swap mutant mCMVhMIEPE. Immunohistological analysis specific for the IE1 protein was performed on day 9 after infection with mCMVhMIEPE. (A1 to A3) Infection of liver parenchyma. A1, overview demonstrating advanced foci of infection; A2, enlargement of the portion of A1 indicated by the arrow, demonstrating the infection’s plaque-like character with a necrotic center. Infected cells were mostly hepatocytes, but endothelial cells (arrowhead) and a few Kupffer cells (not visible) were also present. A3, detail showing condensation of the IE1 protein in the intranuclear inclusion body of infected hepatocytes. (B1 to B2) Infection of perifollicular stromal cells in the spleen. B1, overview; B2, detail of the portion of B1 indicated by the arrow. f, remnants of follicles. (C) Infection of bone marrow stromal cells in the epiphysial region of a femur. eb, epiphysial bone. (D) Infection of adrenal medulla. Shown is a pair of huge, plaque-like foci in the medullary parenchyma. (E) Infection of adrenal cortex. Shown is an advanced focus extending from the zona glomerulosa deeply into the zona fasciculata. (F) Few infected glandular epithelial cells were present in the acini of the submandibular gland. d, duct system of salivary gland. (G) Infection of the lungs. Visible are infected cells in the alveolar septa and in the peribronchiolar connective tissue. Other sections show also infected capillary endothelial cells. a, alveolus; b, bronchioli. (H) Infection of cardiac muscle cells. (J) Infection of gastric mucosa. Counterstaining was performed with hematoxylin. Bars represent 25 μm.
DISCUSSION
Previous work by Angulo et al. (1) has demonstrated that the MIE enhancer of hCMV can functionally replace the MIE enhancer of mCMV in mediating efficient productive infection of fibroblasts in cell culture by the respective recombinant virus. In that study, the authentic promoter of mCMV, including the TATA box, was spared. The data presented in this article fully confirm the previous results by an independent approach and extend them by providing an mCMV recombinant virus, namely mCMVhMIEPE, in which the promoter and enhancer of mCMV (that is, the complete MIEPE of mCMV) are replaced by the paralogous core promoter and enhancer of hCMV. For the sake of precision it should be mentioned that the enhancer or promoter-enhancer swap mutants are all based on deletion mutants of mCMV Smith. Mutants hMCMV-ES1 and -ES2 contain a deletion of an array of genes within HindIII fragment E′ (1), namely, ORFs m151 to m158, whereas mCMVhMIEPE contains a deletion of ORF m152 only.
Our data contribute the following novel information. (i) Replacement of the mCMV MIEP by the hCMV MIE core promoter in addition to replacement of the enhancer does not affect the infectivity of mCMV in permissive murine fibroblasts in cell culture. (ii) The paralogous hCMV MIEPE in recombinant mCMVhMIEPE provides full support for productive mCMV replication in the mouse liver. (iii) The qualitative distribution of mCMV replication in differentiated tissues of adult mice is not determined by type-specific regulation operating at MIE enhancer or core promoter sequences.
Replacement of the MIEP.
Recombinant viruses mCMVhMIEPE (this report) and hMCMV-ES1 and -ES2 (1) differ in that the ES strains still contain the complete mCMV MIEP represented by the sequence from positions −1 to −48 upstream of the 5′ start site of the mCMV ie1/3 transcription unit, whereas in mCMVhMIEPE, the mCMV MIEP is deleted in total and is replaced by the hCMV MIE core promoter starting with position −14 (relative to the hCMV ie1-ie2 transcription start site) and including the TATA box. Thus, the hCMV sequence 5′-GTTTAGTGAACCG-3′ (positions −13 to −1) is not present in recombinant virus mCMVhMIEPE. This fact has an important implication. Previous work has shown that this sequence in the MIEP of hCMV contains a cis-acting, position-dependent repressor element (10, 23, 34) that responds to autoregulation by the human IE2 protein. Notably, this element is not present in the mCMV MIEP. Accordingly, human IE2 cannot repress the mCMV MIEPE (14). To our knowledge, it is not known whether murine IE3 can substitute for human IE2 in addressing this repressor. However, since the sequence is knocked out in recombinant virus mCMVhMIEPE, this type of autoregulation is definitely not operative in cells infected with recombinant virus mCMVhMIEPE. Yet, murine IE3 does exert an autoregulatory repression on the mCMV MIEPE (28), although the target sequence has yet to be defined. Thus, mCMV is subject to autoregulatory repression of its MIEPE by IE3, while mCMVhMIEPE should be protected, and this difference should have led to a difference in the replication efficacies. Contrary to existing theory, however, mCMV and mCMVhMIEPE were found to replicate with the same efficacy in vitro, namely, in permissive fibroblasts, as well as in vivo, namely, in hepatocytes. We must hence assume that autoregulatory repression of MIE gene expression by IE3 is operative in both viruses, possibly by targeting a consensus motif in the enhancers, or we must propose that replication efficacy is not influenced by absence or presence of IE3-mediated repression.
In conclusion, efficient growth of the “core promoter plus enhancer swap” and “promoter-repressor element knock-out” mutant mCMVhMIEPE shows that the hCMV core promoter (upstream of position −13) and enhancer are sufficient for virus replication, whereas the repressor module of the promoter is dispensable, at least in fibroblasts in vitro and in hepatocytes in vivo.
Growth of mCMVhMIEPE in mouse liver parenchyma.
That the hCMV enhancer can substitute for the mCMV enhancer for efficient replication in permissive, proliferating fibroblasts in cell culture was known previously (1) and has been confirmed in this study. It was open to question, however, whether this would apply in vivo to different cell types of different differentiation stages and in tissues in which cells are contact inhibited. Thus, regulation by cellular transcription factors, either activating or silencing the enhancer, may differ between different tissues and, in particular, may differ from the regulation in cultured fibroblasts. That this is not just a theoretical argument has been shown previously by meticulous studies on the tissue-specific activity of the hCMV MIEPE in transgenic mice expressing lacZ as reporter transgene under the control of the hCMV MIEPE during embryonal development and in mature tissues (5, 6, 20). It became very clear that the hCMV MIEPE is not pan-active in all cell types and during all developmental stages but is highly regulated, resulting in specific expression patterns of the transgene. It was hence concluded that the hCMV MIEPE is a critical control point for determining viral tropism in vivo. In adult mice, the reporter transgene was found to be expressed in many organs and in many cell types therein, including most known target tissues of hCMV replication during CMV disease in the human host (for an extensive list, see reference 6). Notable exceptions, surprisingly, included the liver in adult mice (6), even though CMV hepatitis is a common manifestation of CMV disease, with hepatocytes being undoubtedly the principal target cells of hCMV (35) and mCMV (reference 36 and this report) replication in the liver. Some hepatic expression of the transgene has been reported for embryofetal and neonatal liver (5, 20), suggesting that the activity of the hCMV MIEPE in the liver is dependent upon the developmental stage. However, the fetal liver is a site of extramedullary hematopoiesis, and the possibility that the reporter transgene was expressed in hematopoietic fetal liver cells instead of in hepatocytes was not ruled out in these studies. It should be noted, as can clearly be seen in the documented liver tissue sections, that infiltrates of hematopoietic origin were completely absent in the liver after the effective hematoablative treatment used in our model.
Silencing of the hCMV MIEPE in the murine liver is a recognized phenomenon and is actually causing problems in approaches to experimental in vivo gene therapy employing the hCMV MIEPE for expression of a transgene. Recent work has indicated that this enhancer silencing is associated with the absence of transcription factor NF-κB in adult mouse hepatocytes in vivo and that stimuli inducing NF-κB can reactivate a previously silenced hCMV MIEPE (25). In this context, it is notable that the mCMV MIEPE contains more NF-κB binding sites than does the hCMV MIEPE, and this may confer to mCMV a higher resistance to NF-κB deficiency.
In view of these data, it was of interest to see whether the hCMV MIEPE promotes the replication of recombinant virus mCMVhMIEPE in the mouse liver parenchyma. The answer given unequivocally by our experiments is yes, it does, and hepatocytes are identified by the histology as the most prominent target cell type in the liver. The formation of extended, plaque-like lesions in the liver parenchyma reflects lytic infection of hepatocytes. Infected endothelial cells were also visible in most sections, while infected Kupffer cells were rarely seen but did exist. There was no significant difference between mCMV and mCMVhMIEPE with respect to focus number and focus growth in the liver. It is thus likely that cis- or trans-regulating elements of mCMV specified outside of the MIEPE, and therefore not present in the transgene systems discussed above, are responsible for the activity of the MIEPE. Previous work by Sambucetti et al. has indicated that the IE1 protein of hCMV can mediate an activation of the enhancer by induction of NF-κB (41), and Liu and Stinski have shown that tegument proteins of hCMV can enhance the activity of certain promoters (24). Similar mechanisms might apply to mCMV. We have not yet addressed the question of whether the infection or the genotoxic stress associated with the hematoablative treatment lead to an upregulation of NF-κB in the liver, but this is obviously an issue for future experiments and may help to explain the difference from the transgene expression models.
Implication of the MIEPE in tissue tropism of CMV.
Disease caused by mCMV in the immunocompromised host is typically a fatal multiple-organ CMV disease characterized by viral hepatitis (36), splenitis (36), adrenalitis (36, 39), interstitial pneumonia (40), moderate carditis (36), gastritis-enterocolitis (36), and bone marrow aplasia caused by infection of bone marrow stromal cells (26). The salivary glands are not a site of histopathology, but few highly productive glandular epithelial cells of serous as well as of mucous acini account for long-persisting infection (15) relevant to CMV transmission. It was unclear whether this typical pattern of tissue tropism and histopathology could be reproduced with recombinant virus mCMVhMIEPE. At most of these sites, transgenic hCMV MIEPE is active, with the notable exceptions of liver (discussed above), lungs, and most parts of the gastrointestinal tract (6). Data documented herein have shown that recombinant virus mCMVhMIEPE replicates at all these tissue sites, including those at which transgenic hCMV MIEPE is silenced. Since hepatitis, pneumonitis, and gastritis-enterocolitis are among the most prominent manifestations of clinical CMV disease, results obtained with mCMVhMIEPE reflect CMV pathogenesis more closely than was predicted by the transgene models. Altogether, the paralogous MIEPE does not alter the tissue tropism of mCMV in a qualitative sense. The observed lower rate of replication of mCMVhMIEPE at the analyzed extrahepatic sites awaits further analysis.
Conclusion.
Collectively, our data have shown that the complete MIEPE of mCMV can be replaced by the paralogous enhancer plus core promoter of hCMV without a significant influence on the efficacy of virus replication in fetal fibroblasts in vitro and, notably, in hepatocytes in vivo. Furthermore, at least in a qualitative sense, tissue tropism is not altered in the immunodepleted host. Thus, productive infection of permissive cell types does not strictly depend on MIEPE type-specific sequence elements. This is not a trivial finding in view of the fact that during evolutionary adaptation to the respective host species, mCMV MIEPE and hCMV MIEPE have evolved quite different arrangements and numbers of regulatory modules. For example, mCMV MIEPE is longer than hCMV MIEPE and contains more NF-κB but fewer CREB/ATF binding sites, and further differences could be listed. Our data do not say that regulation at the MIEPE is not involved in replication efficacy and cell type tropism but may rather indicate that these parameters of productive infection are largely addressed by the conserved common elements. What then could have been the selective pressure for the evolution of the differences? Most likely, the fate of the virus species is not decided by its replication in the immunocompromised host but rather by its smartness in dealing with the hostile immune system, by establishing latency, and by awakening for recurrence and horizontal transmision. The role of the MIEPE in these aspects of CMV biology can be addressed in vivo with the mCMVhCMV MIEPE swap mutants now available.
ACKNOWLEDGMENTS
We thank Ulrich H. Koszinowski, Max von Pettenkofer Institute, Munich, Germany, for the permission to use recombinant virus mCMVΔorf152, and William H. Burns, Medical College of Wisconsin, Milwaukee, Wis., for the permission to use adenovirus vector Cre-Ad. Stipan Jonjic, Medical Faculty of Rijeka, Croatia, helped by supplying monoclonal antibody CROMA 101. Maria Rapp, now at the Institute for Anatomy, Johannes Gutenberg University, Mainz, Germany, made contributions in earlier stages of the project. Irena Crnkovic-Mertens, Martin Messerle, and Armin Saalmüller helped us by offering advice.
REFERENCES
- 1.Angulo A, Messerle M, Koszinowski U H, Ghazal P. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J Virol. 1998;72:8502–8509. doi: 10.1128/jvi.72.11.8502-8509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angulo A, Suto C, Heyman R A, Ghazal P. Characterization of the sequences of the human cytomegalovirus enhancer that mediate differential regulation by natural and synthetic retinoids. Mol Endocrinol. 1996;10:781–793. doi: 10.1210/mend.10.7.8813719. [DOI] [PubMed] [Google Scholar]
- 3.Anton M, Graham F L. Site-specific recombination mediated by an adenovirus vector expressing the Cre recombinase protein: a molecular switch for control of gene expression. J Virol. 1995;69:4600–4606. doi: 10.1128/jvi.69.8.4600-4606.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balthesen M, Messerle M, Reddehase M J. Lungs are a major organ site of cytomegalovirus latency and recurrence. J Virol. 1993;67:5360–5366. doi: 10.1128/jvi.67.9.5360-5366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baskar J F, Smith P P, Ciment G S, Hoffmann S, Tucker C, Tenney D J, Colberg-Poley A M, Nelson J A, Ghazal P. Developmental analysis of the cytomegalovirus enhancer in transgenic animals. J Virol. 1996;70:3215–3226. doi: 10.1128/jvi.70.5.3215-3226.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baskar J F, Smith P P, Nilaver G, Jupp R A, Hoffmann S, Peffer N J, Tenney D J, Colberg-Poley A M, Ghazal P, Nelson J A. The enhancer domain of the human cytomegalovirus major immediate-early promoter determines cell type-specific expression in transgenic mice. J Virol. 1996;70:3207–3214. doi: 10.1128/jvi.70.5.3207-3214.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boshart M, Weber F, Jahn G, Dorsch-Häsler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 8.Cardin R D, Abenes G B, Stoddard C A, Mocarski E S. Murine cytomegalovirus IE2, an activator of gene expression, is dispensable for growth and latency in mice. Virology. 1995;209:236–241. doi: 10.1006/viro.1995.1249. [DOI] [PubMed] [Google Scholar]
- 9.Chang Y N, Crawford S, Stall J, Rawlins D R, Jeang K T, Hayward G S. The palindromic series I repeats in the simian cytomegalovirus major immediate-early promoter behave as both strong basal enhancers and cyclic AMP response elements. J Virol. 1990;64:264–277. doi: 10.1128/jvi.64.1.264-277.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherrington J M, Khoury E L, Mocarski E S. Human cytomegalovirus ie2 negatively regulates α gene expression via a short target sequence near the transcription start site. J Virol. 1991;65:887–896. doi: 10.1128/jvi.65.2.887-896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorsch-Häsler K, Keil G M, Weber F, Jasin M, Schaffner W, Koszinowski U H. A long and complex enhancer activates transcription of the gene coding for the highly abundant immediate early mRNA in murine cytomegalovirus. Proc Natl Acad Sci USA. 1985;82:8325–8329. doi: 10.1073/pnas.82.24.8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiering S N, Roederer M, Nolan G P, Micklem D R, Parks D R, Herzenberg L A. Improved FACS-Gal: flow cytometric analysis and sorting of viable eukaryotic cells expressing reporter gene constructs. Cytometry. 1991;12:291–301. doi: 10.1002/cyto.990120402. [DOI] [PubMed] [Google Scholar]
- 13.Ghazal P, Nelson J A. Transcription factors and viral regulatory proteins as potential mediators of human cytomegalovirus pathogenesis. In: Becker Y, Darai G, Huang E S, editors. Molecular aspects of human cytomegalovirus diseases. Heidelberg, Germany: Springer-Verlag Publishers; 1993. pp. 360–383. [Google Scholar]
- 14.Hermiston T W, Malone C L, Stinski M F. Human cytomegalovirus immediate-early 2 protein region involved in negative regulation of the major immediate-early promoter. J Virol. 1990;64:3532–3536. doi: 10.1128/jvi.64.7.3532-3536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonjic S, Mutter W, Weiland F, Reddehase M J, Koszinowski U H. Site-restricted persistent cytomegalovirus infection after selective long-term depletion of CD4+ T lymphocytes. J Exp Med. 1989;169:1199–1212. doi: 10.1084/jem.169.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keil G M, Ebeling-Keil A, Koszinowski U H. Temporal regulation of murine cytomegalovirus transcription and mapping of viral RNA synthesized at immediate early times after infection. J Virol. 1984;50:784–795. doi: 10.1128/jvi.50.3.784-795.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keil G M, Ebeling-Keil A, Koszinowski U H. Immediate-early genes of murine cytomegalovirus: location, transcripts, and translation products. J Virol. 1987;61:526–533. doi: 10.1128/jvi.61.2.526-533.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keil G M, Ebeling-Keil A, Koszinowski U H. Sequence and structural organization of murine cytomegalovirus immediate-early gene 1. J Virol. 1987;61:1901–1908. doi: 10.1128/jvi.61.6.1901-1908.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keil G M, Fibi M R, Koszinowski U H. Characterization of the major immediate-early polypeptides encoded by murine cytomegalovirus. J Virol. 1985;54:422–428. doi: 10.1128/jvi.54.2.422-428.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koedood M, Fichtel A, Meier P, Mitchell P J. Human cytomegalovirus (HCMV) immediate-early enhancer/promoter specificity during embryogenesis defines target tissues of congenital HCMV infection. J Virol. 1995;69:2194–2207. doi: 10.1128/jvi.69.4.2194-2207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurz S K, Rapp M, Steffens H-P, Grzimek N K A, Schmalz S, Reddehase M J. Focal transcriptional activity of murine cytomegalovirus during latency in the lungs. J Virol. 1999;73:482–494. doi: 10.1128/jvi.73.1.482-494.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurz S, Steffens H-P, Mayer A, Harris J R, Reddehase M J. Latency versus persistence or intermittent recurrences: evidence for a latent state of murine cytomegalovirus in the lungs. J Virol. 1997;71:2980–2987. doi: 10.1128/jvi.71.4.2980-2987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B, Hermiston T W, Stinski M F. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J Virol. 1991;65:897–903. doi: 10.1128/jvi.65.2.897-903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu B, Stinski M F. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Löser P, Jennings G S, Strauss M, Sandig V. Reactivation of the previously silenced cytomegalovirus major immediate-early promoter in the mouse liver: involvement of NFκB. J Virol. 1998;72:180–190. doi: 10.1128/jvi.72.1.180-190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer A, Podlech J, Kurz S, Steffens H-P, Maiberger S, Thalmeier K, Angele P, Dreher L, Reddehase M J. Bone marrow failure by cytomegalovirus is associated with an in vivo deficiency in the expression of essential stromal hemopoietin genes. J Virol. 1997;71:4589–4598. doi: 10.1128/jvi.71.6.4589-4598.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meier J L, Stinski M F. Regulation of human cytomegalovirus immediate-early gene expression. Intervirology. 1996;39:331–342. doi: 10.1159/000150504. [DOI] [PubMed] [Google Scholar]
- 28.Messerle M, Bühler B, Keil G M, Koszinowski U H. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J Virol. 1992;66:27–36. doi: 10.1128/jvi.66.1.27-36.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski U H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messerle M, Keil G M, Koszinowski U H. Structure and expression of murine cytomegalovirus immediate-early gene 2. J Virol. 1991;65:1638–1643. doi: 10.1128/jvi.65.3.1638-1643.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mocarski E S, Abenes G B, Manning W C, Sambucetti L C, Cherrington J M. Molecular genetic analysis of cytomegalovirus gene regulation in growth, persistence and latency. Curr Top Microbiol Immunol. 1990;154:47–74. doi: 10.1007/978-3-642-74980-3_3. [DOI] [PubMed] [Google Scholar]
- 32.Nelson J A, Gnann J W, Jr, Ghazal P. Regulation and tissue-specific expression of human cytomegalovirus. Curr Top Microbiol Immunol. 1990;154:75–100. doi: 10.1007/978-3-642-74980-3_4. [DOI] [PubMed] [Google Scholar]
- 33.Nelson J A, Groudine M. Transcriptional regulation of the human cytomegalovirus major immediate-early gene is associated with induction of DNase I-hypersensitive sites. Mol Cell Biol. 1986;6:452–461. doi: 10.1128/mcb.6.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pizzorno M C, Hayward G S. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J Virol. 1990;64:6154–6165. doi: 10.1128/jvi.64.12.6154-6165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plachter B, Sinzger C, Jahn G. Cell types involved in replication and distribution of human cytomegalovirus. Adv Virus Res. 1996;46:195–261. doi: 10.1016/s0065-3527(08)60073-1. [DOI] [PubMed] [Google Scholar]
- 36.Podlech J, Holtappels R, Wirtz N, Steffens H-P, Reddehase M J. Reconstitution of CD8 T cells is essential for the prevention of multiple-organ cytomegalovirus histopathology after bone marrow transplantation. J Gen Virol. 1998;79:2099–2104. doi: 10.1099/0022-1317-79-9-2099. [DOI] [PubMed] [Google Scholar]
- 37.Prösch S, Staak K, Stein J, Liebenthal C, Stamminger T, Volk H-D, Krüger D H. Stimulation of the human cytomegalovirus IE enhancer/promoter in HL-60 cells by TNFα is mediated via induction of NF-κB. Virology. 1995;208:197–206. doi: 10.1006/viro.1995.1143. [DOI] [PubMed] [Google Scholar]
- 38.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reddehase M J, Jonjic S, Weiland F, Mutter W, Koszinowski U H. Adoptive immunotherapy of murine cytomegalovirus adrenalitis in the immunocompromised host: CD4-helper-independent antiviral function of CD8-positive memory T lymphocytes derived from latently infected donors. J Virol. 1988;62:1061–1065. doi: 10.1128/jvi.62.3.1061-1065.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddehase M J, Weiland F, Münch K, Jonjic S, Lüske A, Koszinowski U H. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J Virol. 1985;55:264–273. doi: 10.1128/jvi.55.2.264-273.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambucetti L C, Cherrington J M, Wilkinson G W, Mocarski E S. NF-kappaB activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 1989;8:4251–4258. doi: 10.1002/j.1460-2075.1989.tb08610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandford G R, Burns W H. Rat cytomegalovirus has a unique immediate early gene enhancer. Virology. 1996;222:310–317. doi: 10.1006/viro.1996.0428. [DOI] [PubMed] [Google Scholar]
- 43.Spector D H. Activation and regulation of human cytomegalovirus early genes. Intervirology. 1996;39:361–377. doi: 10.1159/000150507. [DOI] [PubMed] [Google Scholar]
- 44.Steffens H-P, Kurz S, Holtappels R, Reddehase M J. Preemptive CD8 T-cell immunotherapy of acute cytomegalovirus infection prevents lethal disease, limits the burden of latent viral genomes, and reduces the risk of virus recurrence. J Virol. 1998;72:1797–1804. doi: 10.1128/jvi.72.3.1797-1804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stenberg R M. The human cytomegalovirus major immediate-early gene. Intervirology. 1996;39:343–349. doi: 10.1159/000150505. [DOI] [PubMed] [Google Scholar]