Abstract
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer-related death in the Western world. The number of diagnosed cases and the mortality rate are almost equal as the majority of patients present with advanced disease at diagnosis. Between 4 and 10% of pancreatic cancer cases have an apparent hereditary background, known as hereditary pancreatic cancer (HPC) and familial pancreatic cancer (FPC), when the genetic basis is unknown. Surveillance of high-risk individuals (HRI) from these families by imaging aims to detect PDAC at an early stage to improve prognosis. However, the genetic basis is unknown in the majority of HRIs, with only around 10–13% of families carrying known pathogenic germline mutations. The aim of this study was to assess an individual’s genetic cancer risk based on sex and personal and family history of cancer. The Best Linear Unbiased Prediction (BLUP) methodology was used to estimate an individual’s predicted risk of developing cancer during their lifetime. The model uses different demographic factors in order to estimate heritability. A reliable estimation of heritability for pancreatic cancer of 0.27 on the liability scale, and 0.07 at the observed data scale as obtained, which is different from zero, indicating a polygenic inheritance pattern of PDAC. BLUP was able to correctly discriminate PDAC cases from healthy individuals and those with other cancer types. Thus, providing an additional tool to assess PDAC risk HRI with an assumed genetic predisposition in the absence of known pathogenic germline mutations.
Keywords: Pancreatic cancer, Genetic cancer risk, Heritability, High-risk screening
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer death in Europe, behind only lung, colon and breast cancer [1, 2]. The 5-year survival rate is less than 10% [3] as the majority of cases have advanced disease at diagnosis, meaning up to 90% of cases have non curable disease [4]. In recent decades, there have been few advances in treatment, with small innovations in neoadjuvant chemotherapy and chemoradiotherapy showing minimal improvements, and surgical resection remains the only potentially curative treatment option [5]. Therefore, there is a clear need for both improved treatment strategies and early detection techniques to increase survival.
The most important non-modifiable risk factor for the development of pancreatic cancer is genetic predisposition. The risk of developing PDAC increases with the number of affected relatives, and the standard HR (Hazard Ratio) is 32 when 3 relatives are affected [6, 7]. Some cases have a hereditary background, with pathogenic germline mutations in cancer risk genes such as BRCA1/2, PALB2, ATM, CHEK2 and CDKN2A [8–11]. Whereas, Familial Pancreatic Cancer (FPC) is defined as families with at least one pair of affected first-degree relatives, with no known genetic basis. This equates to 4–10% of diagnosed cases of PDAC having a familial or hereditary background [12, 13].
PDAC has a relatively low incidence, with 5.7 cases per 100 000 males and 4.1 cases per 100 000 females. This low incidence, combined with the lack of a reliable markers [14], high costs and the limited sensitivity of imaging tests, makes population wide screening impractical. Healthy first- or second-degree relatives of families with hereditary or familial PDAC are the only high-risk population to be offered an imaging based screening program for early detection [15]. The management of high-risk individuals, both in terms of genetic testing and screening, are performed according to European and international guidelines including The International Consortium for Pancreatic Cancer Screening (CAPS) [16], National Comprehensive Cancer Network (NCCN) [17], American Gastroenterological Association (AGA) [18] and American Society for Gastrointestinal Endoscopy (ASGE) [19]. The aim of screening high-risk individuals (HRIs) is early detection of PDAC during a potentially curable stage, as well the detection of pre-malignant precursor lesions, such as intraductal papillary mucinous neoplasms (IPMN) with high-grade dysplasia. HRIs undergo an annual screening that includes magnetic resonance imaging (MRI) and/or echoendoscopy (EUS), with additional blood based tests for the detection of tumor biomarkers associated with the pancreatic cancer. The Spanish registry of familial pancreatic cancer, PANGENFAM, was established in 2009 with the main objective of characterising the phenotype and genotype of FPC [20]. Within our screening program we have detected and successfully treated 4 malignant lesions (3 early PDAC and one neuroendocrine tumor) and together with two other international registries (Leiden and Marburg), we have critically analyzed the follow-up protocol to make it more efficient and cost-effective [21, 22]. Follow-up of HRIs is effective and improves early detection of the disease. However, the genetic basis of FPC is unknown in the majority of families, thus, all HRIs in the family are included in screening programs, even though approximately 50% will not be carriers of pathogenic germline mutations. Thus, there is a need to identify true HRIs by other techniques, without knowing the specific mutation within the family. The Best Linear Unbiased Prediction (BLUP) is used to study complex traits and allows the prediction of individual genetic risk. Using BLUP, the components of phenotypic variance can be estimated to determine heritability, i.e. whether cases of cancer within the same family are actually due to a heritable factor or whether they are the product of shared environmental factors.
The objective of this study was to estimate the value of the heritability of PDAC, based on the hypothesis that there is an additive genetic component and that an additive genetic component exists. We have previously shown that BLUP can be used to obtain a reliable estimation of heritability for cancer using the Minnesota Breast Cancer data set [23]. BLUP uses a mixed model to simulate individual risk of developing cancer, considering fixed and other variable factors within a population. Three models were used, combining different fixed effects (sex + family + generation) and random effects (individual + family + generation) to estimate the heritability of pancreatic cancer and the individual genetic risk of developing cancer. The main aim was to use this value as a follow-up criterion in FPC families, where the genetic basis of the syndrome is unknown. This analysis could complement the on-going phenotypic, molecular and imaging characterization of the HRIs, to further optimize the screening program, reducing the associated psychological and economic burden.
Materials and methods
PANGENFAM inclusion criteria and data collection
Inclusion criteria: (1) FPC families with ≥ 2 affected first or second degree relatives; (2) Hereditary breast and ovarian cancer (HBOC) families with at least one case of PDAC; (3) Families with ATM mutation and at least one case of PDAC; (4) Familial atypical multiple mole melanoma (FAMMM) families with at least one case of PDAC; (5) Hereditary Non Polyposis Colorectal Cancer (HNPCC) or Lynch Syndrome families with at least one case of PDAC; (6) Peutz Jeghers families; (7) Hereditary Pancreatitis (with pathogenic variants in the genes PRSS1 and SPINK1); and (8) Families with PDAC cases diagnosed at ≤ 50 years of age [20].
Some high-risk individuals underwent routine genetic testing in the clinic for known familial cancer associated genes, including BRCA2, BRCA1, CDKN2A, MLH1, ATM, PALB2, CHEK2 and SPINK. Of the 71 individuals tested, 21 (30%) were positive for a pathogenic variant, most frequently in the BRCA2 gene (57%). The data for the study are stored in a secure sever in a custom designed database in REDCap (13.4.11, 2024 Vanderbilt University). The database used for this study was downloaded on 5 October 2022 and consisted of 4602 individuals from 125 families. Each family has a 9-digit unique identifier, and each individual has a unique 12-digit identifier. The kinship of each individual was used to construct the correlation matrix based on the offspring relationship and off-kindred individuals were excluded. Information available for each individual on clinical history, sex, age, family identifier and generation were selected to estimate variance components and define heritability. The first phase of the analysis included 3780 individuals, 752 cases of any cancer, of which 213 correspond to PDAC.
Generation of the BLUP mixed model
Data from the families within PANGENFAM were used, assuming a genetic component, although the calculated risk encompassed all cancers with a possible genetic component and also those specifically with PDAC. First, we estimated the heritability of all cancer types with the information PANGENFAM pedigrees. The dependent variable was cancer yes or cancer no. Three models were used, adding sequentially a random effect, model I (individual), II (individual + family) and III (individual + family + generation), to which effects were added, taking as a reference the models described in our previous study that used the same model [23]. Model I was a very simple and biologically implausible model and Model III was more biologically plausible. The components of the 3 models are summarised in Table 1.
Table 1.
Summary of the 3 mixed models, specifying the fixed and random effects in each case
| Model | Fixed effects | Random effects |
|---|---|---|
| 1 → I | Sex + family + generation | Individual |
| 2 → II | Sex + generation | Individual + family |
| 3 → III | Sex | Individual + family + generation |
Subsequently, the dependent variable was refined using just PDAC, yes or no, and applying model III. In this way, the BLUP analysis was used (1) to estimate the heritability of pancreatic cancer and (2) estimate the individual genetic risk of developing pancreatic cancer. The software RStudio [24] was used and several specific packages were employed for further analysis including, “kinship2“ [25], “pedrigreemm” [26], “pedigree” to plot the family trees of each family, the package “MCMCglmm“ [27] for the calculation of the mixed model and the ROCR package [28] to analyse the predictive character of the model obtained.
Statistical methodology for assessing individual risk of developing cancer
For this study, cancer was defined as a phenotypic trait resulting from the additive effect of a large number of genes with a medium-low effect, which supports the hypothesis that this disease has a heritable genetic component. The variable representing the cancer is binary, where a value of 1 is assigned to affected individuals and 0 to unaffected individuals. The usual model to study binary traits is a threshold model. This model assumes a continuous underlying random variable, liability, which when it is over a given threshold, this triggers the expression of one of the binary phenotypes, i.e. cancer or no cancer, and PDAC or no PDAC [29, 30]. The variance for this underlaying normal distribution was set to 1.
For the estimation of risk, the BLUP was calculated using the equations of the Henderson mixed model [31] and the Fisher infinitesimal model [32]. Generalized Linear Mixed Models (glmm) are an extension of the generalized linear models that allow the inclusion of response variables of different distributions, such as binary [27]. The linear mixed model was defined as:
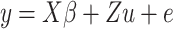 |
1 |
where y is the observed phenotype, β and u are the fixed and random effects respectively, X and Z are matrices and e is the random error. The random effects follow a multivariate normal distribution MVN, u ∼ MV N (0, G) and e ∼ MV B(0, R) where G is the genetic covariance matrix and R the residual. Henderson proposes the following solution to the model:
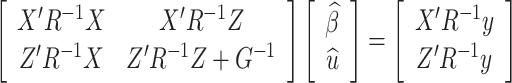 |
2 |
The Fisher model states that genetic inheritance is based on an infinite number of loci with a small additive effect. The phenotypic variance VF is calculated as the sum of the genotypic variance VG and the environmental variance VE. In turn, the genetic variance is the sum of an additive component VA and a non-additive component VNA, related to dominance or epistasis effects. BLUP allows the calculation of the additive part of this heritability that is transmitted between generations. The heritability is calculated using Fisher’s expression:
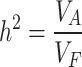 |
3 |
The following formula was used to estimate the heritability from the components of the calculation of model III:
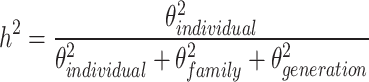 |
4 |
Where 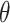 2, is the variance. Denominator of formula (4) is the phenotypic variance decompose on their components, given that we are using a threshold model. Numerator of formula (4) is the additive component of the variance attributable to individual. The consistency of this estimate of h2 was assessed by testing the null hypothesis of heritability, i.e. h2 = 0 using a Bayes factor. The input parameters for the calculation include the kinship matrix, which establishes the relationships between individuals, and a matrix with the values of each individual for each of the effects included in the models. For the calculation of each of these variances necessary for the estimation, Bayesian inference was used because it is a stochastic calculation using a binary variable. The models were run with 5.25 million iterations, with an initial burn-in of 250 000 iterations and sampling every 2500 iterations, resulting in a sample size of 2000. When the model was run for only PDAC as a dependent variable, a model with 6.25 million iterations, with the same burn-in and thinning interval as previous models lead to a sample size of 2400.
2, is the variance. Denominator of formula (4) is the phenotypic variance decompose on their components, given that we are using a threshold model. Numerator of formula (4) is the additive component of the variance attributable to individual. The consistency of this estimate of h2 was assessed by testing the null hypothesis of heritability, i.e. h2 = 0 using a Bayes factor. The input parameters for the calculation include the kinship matrix, which establishes the relationships between individuals, and a matrix with the values of each individual for each of the effects included in the models. For the calculation of each of these variances necessary for the estimation, Bayesian inference was used because it is a stochastic calculation using a binary variable. The models were run with 5.25 million iterations, with an initial burn-in of 250 000 iterations and sampling every 2500 iterations, resulting in a sample size of 2000. When the model was run for only PDAC as a dependent variable, a model with 6.25 million iterations, with the same burn-in and thinning interval as previous models lead to a sample size of 2400.
For the a priori distribution, an inverse-gamma distribution with parameter expansion was used, which is that recommended by the author of the calculation package for small variances as indicated in the manual [33]. The residual variance was set to θ2 = 1. Once the models were calculated, an analysis of the convergence of the Markov chain was carried out using the Heidelberg and Welch test [34] to reject or fail to reject the results obtained in the model calculation.
Estimated genetic value calculation
The estimated genetic value (EGV) was obtained as the solution of the individual random effect, by calculating the mean of the 2000 samples obtained in the calculation of the variance associated with each one. The sampling interval chosen was sufficiently wide to reduce autocorrelation. Furthermore, to identify individuals at high risk for cancer based solely on family pedigree information, the individual risk was calculated as the mean of the values of their parents. This is because each individual inherits half of his additive genetic component from the father and half from the mother [31]. Pearson´s correlation coefficient was used to assess the correlation between family incidence of cancer and median EGV of the family. In order to assess differences in EGVs between groups non –parametric Kruskall Wallis test was used.
To assess the predictive ability of this calculation, the area under the Receiver Operating Characteristic (ROC) curve was used to assess the predictive ability of this calculation. Analysis of the ROC curve allows to distinguish between positive and negative cases during prediction, so that an area under the AUC (Area Under Curve) close to 1 indicates that the model has a high predictive ability to identify individuals at risk of developing cancer.
Results
Description of families
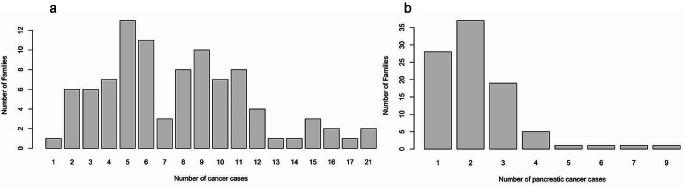
After the cleaning and selection of the families of interest for the study, 96 families consisting of 4578 individuals were used for the final analysis. The remaining families did not meet the criteria for familial or hereditary pancreatic cancer or had data from few individuals available. For each individual, their cancer status was stored, which allowed the calculation of the incidence within the population, obtaining values for females of 0.154 and for males of 0.176. No significant differences were observed according to gender. Regarding the distribution of cancer within the families, each family consisted of 8 to 158 individuals, with between 1 and 22 reported cases of cancer (Fig. 1a) and 1–8 reported cases of PDAC (Fig. 1b). However, only families with 20 or more individuals were considered for the prevalence calculation because a smaller number tends to overestimate the prevalence and 16 families were excluded from this analysis. It is likely that these families were incomplete or that only the family branches and generations most affected by cancer were stored in the study database.
Fig. 1.
(a). Histogram of any cancer prevalence within the families and (b) pancreatic cancer prevalence within the families
Quality control of the data and EGV model
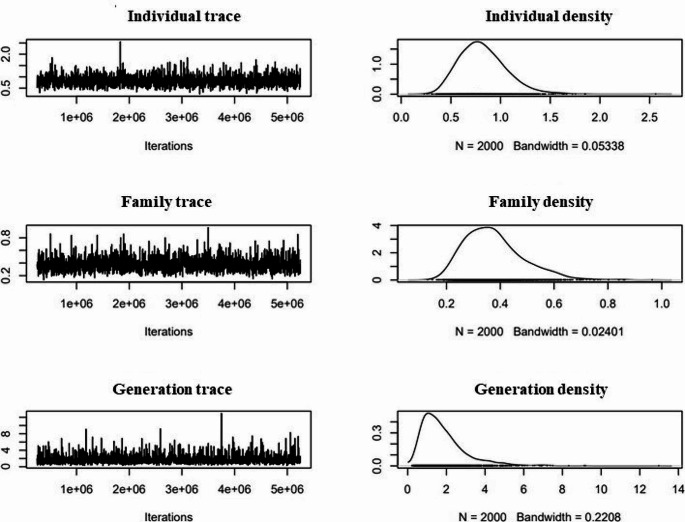
The generation of the genealogical trees was used to verify the quality of the data, as it was a visual way to confirm the correct structure of the data, specifically, ensuring that the kinship matrix was correct, which guaranteed the validity of the model used in the study. Figure 2 shows the trace of model III and the subsequent density obtained for each calculated parameter. The Heidelberg test was applied to each model, which confirmed their convergence. These results validated the model calculation, allowing the analysis to proceed. The high posterior density intervals calculated by MCMCglmm for the variance components in model III were [0.3825–1.265] for the individual, [0.187–0.5932] for the family and [0.3128–4.235] for the generation. From these values, the heritability was estimated, obtaining a heritability value of [0.3128–4.235] for the generation. From these values, the heritability was estimated obtaining a value of 0.44 with a high posterior density interval [0.3030–0.5752] for the PANGENFAM cohort. The value of the interval was not null so the null hypothesis was rejected and allowed us to affirm that there was an additive genetic component in the development of PDAC.
Fig. 2.
MCMC traces of variance (corresponding to VA) and density due to individual, family and generation for model III
Models I and II also converged, however, they were discarded as they were far from the biological reality. When analysing model I, a heritability of 0.99 [0.9843–0.9921] was obtained (Table 2), implying that cancer depends only on its genetic component and that almost all individuals would suffer from the disease. Given that model I attributed too much variability to the genetic component (individual) and this is not biologically likely, new random effects were added to the model until a plausible value was reached. Model II estimated a heritability of 0.39 [0.28–0.52], a value closer to reality. Finally, the generation effect allowed to include a temporal component to the model and to finish adjusting the heritability value to 0.21 [0.1–0.33] in model III (Table 2). The more biologically plausible model III was also run for PDAC only as a more strict assessment of hereditability, with 500,000 and 600,000 iterations, giving a hereditary value of 0.27.
Table 2.
Heritability estimates for different models using all cancer types or PDAC as the dependent variable. Real scale is the observable scale of the trait versus the liability scale which is non-observable. Liability follows a standardized normal distribution that after a certain threshold triggers the illness
| Dependent variable | Heritability | Liability scale | Real scale |
|---|---|---|---|
| All cancer types | Model I | 0.99 [0.98–0.99] | 0.55 [0.41–0.63] |
| Model II | 0.39 [0.28–0.52] | 0.22 [0.15–0.29] | |
| Model III | 0.21 [0.1–0.33] | 0.1 [0.04–0.16] | |
| Only PDAC (Model III) | 500,000 iterations | 0.27 [0.06–0.47] | 0.07 [0.02–0.12] |
| Only PDAC (Model III) | 600,000 iterations | 0.27 [0.07–0.47] | 0.07 [0.02–0.13] |
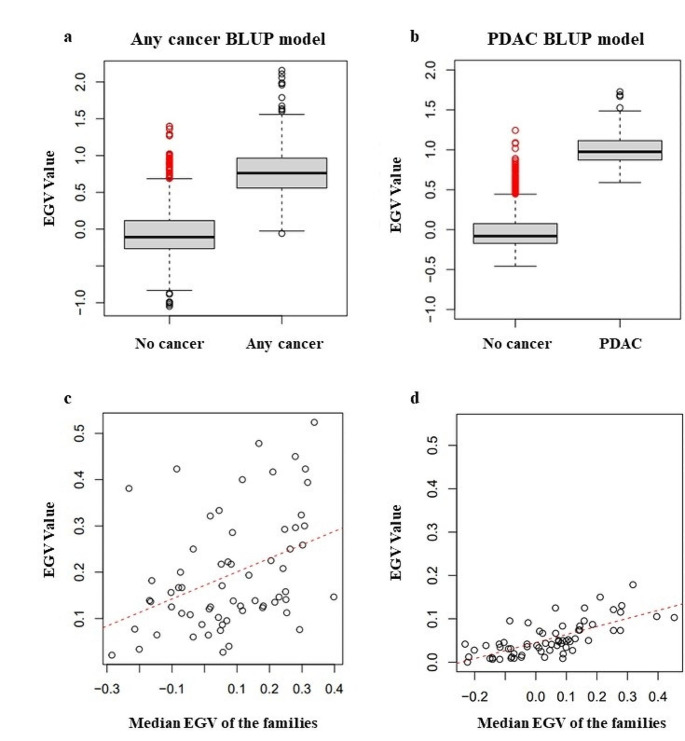
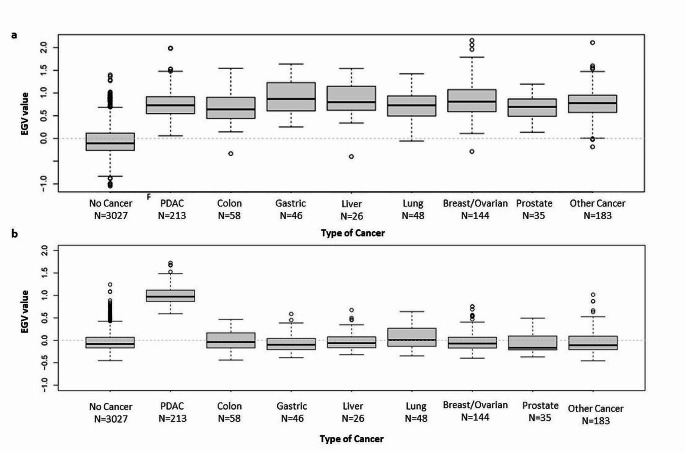
PDAC heritability was estimated only using Model III. The EGV is a measure of the additive genetic risk of developing cancer, so individuals without cancer are expected to have a lower EGV than those who developed the disease. Figure 3a shows that the EGV (with corresponding interquartile range (IQV) using the any cancer BLUP model is significantly higher for individuals with any cancer vs. no cancer (no cancer − 0.108 IQR [-0.264;0.116] vs. any cancer 0.762 IQR [0.558;0.966], p-value < 0.001). Furthermore, the EGV using the PDAC BLUP model was significantly higher for individuals with PDAC versus no PDAC, (no PDAC − 0.081 IQR [-0.172;0.075] vs. PDAC 0.974 IQR [0.870;1.116], p-value < 0.001). Subsequent analyses of EGV was performed using the 2 models, the any cancer BLUP model based on individuals with any cancer vs. no cancer and, the more strict PDAC BLUP model based on individuals with PDAC versus no PDAC. For the screening programme, it is of interest to identify individuals with high EGV who have not yet developed the disease (shown in red in Fig. 3a and b). It is also expected that families with a higher prevalence will have a higher mean EGV, assuming a higher genetic predisposition. Figure 3c and d identifies some relationship, although many families deviate from the trend. This is because the prevalence cannot exceed 0.5 in any case despite having a high EGV, since not all individuals carry the risk mutations.
Fig. 3.
(a) Boxplot of comparison of EGVs according to the any cancer BLUP model and (b) PDAC BLUP model; 0 = no cancer, 1 = any cancer. (c) Correlation between family prevalence and median EGV of the family according to the any cancer BLUP model (Pearson´s correlation coefficient 0.65 [0.5–0.76]; p value < 0.001) and PDAC BLUP model (Pearson´s correlation coefficient 0.61 [0.45–0.73]; p value < 0.001)
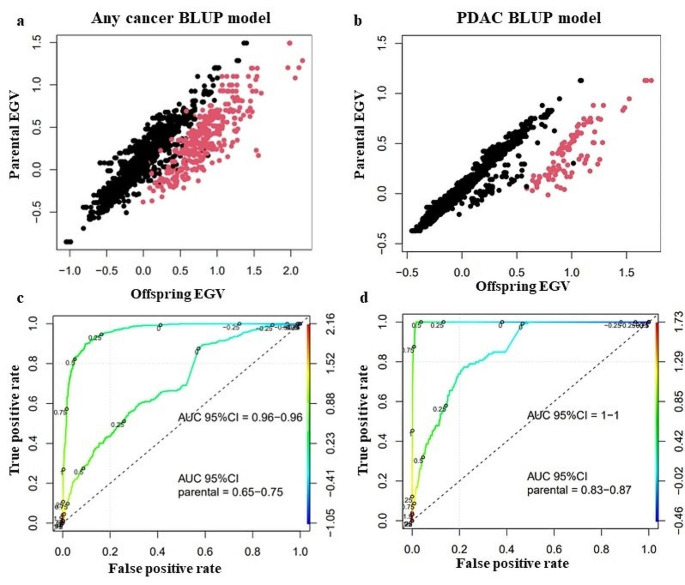
An individual inherits half of the genetic load of each of its parents, and an individual’s EGV can be estimated as the mean of the EGV of the parents and used as a predictive model for new generations. Figure 4a shows the results of the calculation of the mean EGV, showing that individuals with any cancer using the general model had a higher mean EGV than individuals without cancer; (Pearson´s correlation coefficient 0.76 [0.74–0.78] p value < 0.001). Using the more strict model PDAC BLUP model of no PDAC vs. PDAC, individuals with PDAC had a higher mean EGV than individuals without PDAC; (Pearson´s correlation coefficient 0.85 [0.84–0.86] p value < 0.001) (Fig. 4b). In Fig. 4c, the ROC curve allowed the predictive character of the estimate of EGV to be evaluated, as the mean of the parents compared to the cancer status. A reasonably high AUC of [0.65–0.75] was obtained when using the mean of the parents and 0.96 for the any cancer BLUP model. However, an AUC of [0.83–0.87] was obtained with this model (Fig. 4d), indicating that an individual with a high positive EGV has a high genetic predisposition to develop PDAC. Thus this model was used for all subsequent analysis.
Fig. 4.
Average EGV of parents versus calculated EGV of offspring, individuals with cancer are shown in red and individuals without cancer are shown in black, (a) using the any cancer BLUP model and (b) using the PDAC BLUP model. ROC curve using the mean EGV of the parents (c) for the any cancer BLUP model and (d) for the PDAC BLUP model. EGV was used as a cancer predictor, using either the EGV value assigned to the individual or the parental mean of the EGV. Both ROC curves have an AUC above 0.5
The EGV was significantly higher for cases with PDAC compared with individuals without cancer or PDAC; any cancer BLUP model: no cancer/no-PDAC − 0.108 IQR [-0.264;0.116] vs. PDAC/no PDAC 0.729 IQR [0.547;0.918], (p-value < 0.001) (Fig. 5a), and PDAC BLUP model: no PDAC − 0.082 IQR [-0.169;0.069] vs. PDAC 0.974 IQR [0.870;1.117] (p-value < 0.001) (Fig. 5b). The outliers in the “no cancer” group shown in Fig. 5b are those individuals who theoretically should be followed more intensively periodically, as they are at higher risk of developing PDAC. Furthermore, there are some outliers within the group of individuals with breast and ovarian cancer as well as the “other cancer” group, which may also have a hereditary background. The EGV of other tumor types are around or below zero as they most likely related to sporadic cases that are attributed to certain environmental factors, such as smoking in the case of lung cancer.
Fig. 5.
Comparison of the EGV in individuals with different types of cancer and individuals without cancer, including the number of individuals in each group using the (a) Any cancer BLUP model and (b) PDAC BLUP model
EGV of patients on follow-up
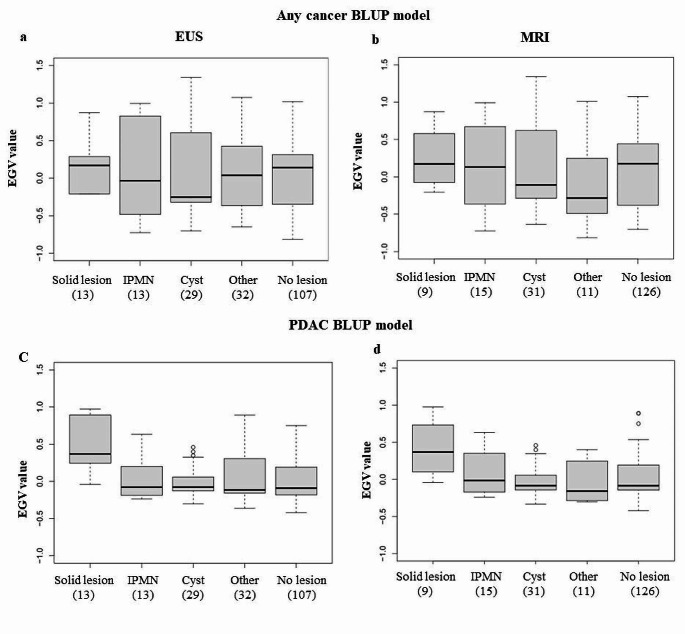
High-risk individuals in follow-up undergo annual MRI and EUS screening, which identifies lesions within the pancreas, as well as outside of the pancreas in the case of MRI. Figure 6 shows the study of the relationship between the EGV of the individuals with the type of pancreatic lesion identified by MRI and EUS and their corresponding age. Figure 6a shows the lesions detected by EUS were and Fig. 6B shows the lesions identified by MRI. Solid lesions had a positive mean EGV (any cancer BLUP model: EUS 0.170 IQR [-0.141;0.285], MRI 0.173 IQR [-0.008;0.532], PDAC BLUP model: EUS 0.369 IQR [0.244;0.792], MRI 0.369 [0.173;0.613]), independently of the screening modality used, as expected. Whereas, pancreatic cysts, IPMN and other lesions (mainly inhomogeneous pancreas parenchyma) had a mean EGV around or lower than zero, similar to the mean EGV in individuals with no pancreatic lesions. Cysts are generally benign or have a low probability of malignant progression. Interestingly, individuals with IPMNs detected by MRI (Fig. 6b) had slightly higher mean EGV (any cancer BLUP model: 0.131 IQR [-0.365;0.673], PDAC BLUP model: -0.014 IQR [-0.171;0.352]), lower than the mean EGV of individuals with solid lesions and slightly higher than the mean EGV for individuals with cysts and no pancreas lesion, although this did not reach statistical significance. This is logical as IPMN are considered as precursor lesions of PDAC, with a variable risk of malignant progression. Interestingly, there were some outliers with high EGVs in the cyst cohort detected by both EUS and MRI and in the no lesion cohort. This is also consistent with the theory that some of the individuals with no solid pancreatic lesions have a potential future diagnosis of PDAC or its precursor lesions. There was no significant difference in age at diagnosis of the different types of pancreatic lesions, although those with IPMN were slightly older compared with those diagnosed with cysts. Furthermore, those with normal pancreatic imaging were slightly older than those with other types of pancreatic lesions.
Fig. 6.
Comparison of EGV in individuals with different types of lesions detected during follow-up as a function of the detection technique used, according to the no cancer-any cancer BLUP model by (a) EUS and (b) MRI and the PDAC BLUP model by (c) EUS and (d) MRI
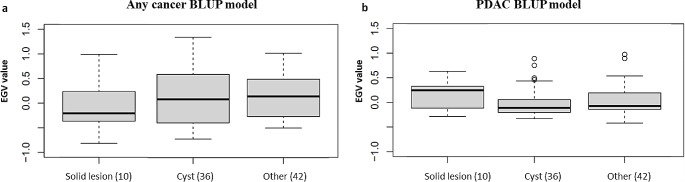
Finally, the relationship of EGV with other types of extra-pancreatic lesions detected during patient follow-up was analysed. Figure 7 shows that individuals with non-pancreatic solid lesions have mean EGV below zero, this is logical as these solid lesions are not expected to be related to familial PDAC risk. The majority of extra-pancreatic lesions were mainly liver and renal cysts, and these individuals had EGV approaching zero. Whereas, the mean EGV for individuals with other extra-pancreatic lesions was slightly higher; any cancer BLUP model: 0.141 IQR [-0.261;0.477] (Fig. 7a), PDAC BLUP model: -0.073 IQR [-0.141;0.192] (Fig. 7b), but did not approach those seen in individuals with solid pancreatic lesions.
Fig. 7.
Comparison of EGV in individuals with different types of extra-pancreatic lesions detected on MRI and EUS. (a) No cancer-any cancer model (b) No pancreatic cancer-pancreatic cancer model
Discussion
The heritability estimated by BLUP calculation in this study of families with familial pancreatic cancer differs from 0, confirming the hypothesis of polygeny in the inheritance pattern of this disease. EGV allow distinguishing between individuals with cancer and those without the disease, which makes it a useful tool to estimate the cancer risk of an individual and to include them in a high-risk screening programme. In this case, the heritability was estimated at 0.44, a value similar to that obtained in our previous study [23], where a heritability of breast cancer was estimated at [0.017–0.396]. In theory, individuals with a negative EGV are considered to be free of genetic risk for cancer. However, this does not rule out the possibility that they may develop the disease due to the action of other factors such as environmental factors. On the other hand, individuals with a positive EGV have a hereditary basis that predisposes them to the disease to a greater or lesser extent, depending on the value.
The estimated heritability for other complex diseases is 52% for Amyotrophic Lateral Sclerosis [35], 17–21% for schizophrenia [36] and a h2 = 0.67–0.91 for the form of the hippocampal subregion that predisposes to Alzheimer’s disease [37]. In comparison, the heritability value of the pancreas is moderate, with less than half of the variation in disease susceptibility attributable to genetic factors. However, pedigree-based risk estimation is not a widespread procedure and predictive models with a genetic basis and specific mutations are often chosen. As an example, the Gail model is used in breast cancer prediction [38], as shown in the study by Johansson et al. [39] where an AUC of 61.8% is obtained using this logistic regression model incorporating 10 genetic variants. Another example is the prediction model of Wu et al. [40], which obtained an AUC of 80% for the prostate cancer using a similar methodology. Also noteworthy is the study by Lee et al. [41], which uses the predictive model BOADICEA (Breast and Ovarian Analysis of Disease Prevalence and Carrier Estimation Algorithm) which combines high-risk mutations with other associated environmental factors, and shows that the best possible prediction of breast cancer is achieved by combining both genetic and environmental factors. Comparing these values to those obtained using the BLUP calculation, where an AUC of 95% was achieved for the individual EGV estimated by the model and an AUC of 65–75% when the prediction was performed using the mean of EGV of the parents, the latter is at the same predictive level as the previous models, being equally powerful despite not considering specific mutations in the calculation.
In the case of PDAC, current research is focused next generation genomic analysis to identify variants associated with the occurrence of the disease. Several Polygenic Risk Scores (PRS) models for PDAC have been developed, including 4 models that include 5, 30, 33 and 22 SNPs [42–45], respectively, that predispose to the development of PDAC. However, these studies are costly as sequencing is still an expensive technique. Moreover, the clinical application of these techniques has not yet been achieved and there are doubts about their efficacy [46, 47]. Therefore, it is important to highlight the relevance of studies with models such as BLUP, as they only require a computer and a database to identify individuals with a potential risk of developing PDAC. These models can be incorporated in hospitals at no great additional cost. In addition, they allow the study of a large number of patients and, as they are specific to each individual, they provide a more personalised, faster and more effective follow-up.
The mean EGV for individuals with solid extra-pancreatic lesions was much lower than the in individuals with solid pancreatic lesions. Thus, this supports the notion that the BLUP model can predict PDAC risk, as it does not assign a high EGV in the presence of non-PDAC solid lesions in high risk individuals, although a small proportion of these lesions may progress to PDAC. The reported surgical intervention rates due to suspicious lesions in this population type is around 5% [48–50], and specifically 2.6% of individuals from our registry (REFERENCE). To achieve a high-degree of reliability, the BLUP model requires good quality multi generation pedigree information and a large of number of pedigrees with accurate information of cancer diagnosis. The calculation of the EGV can be made more accurate by adding more effects to the model, such as clinical, sociological, or demographic characteristics. However, obtaining these data for the entire population is more difficult. Nevertheless, it would be possible to add the associated pathogenic variants as they are discovered. Another way to improve the BLUP model would be to incorporate genetic information in the kinship matrix, in the form of coefficients to estimate genetic values. The genetic values could be estimated by combining phenotypic, pedigree and genomic information at the same time [51].
At present, early diagnosis and the development of new, truly effective therapeutic strategies for the treatment of pancreatic cancer remain essential. Widespread screening at the population level is not feasible due to the low prevalence of this type of cancer and the invasive techniques used to detect pancreatic lesions. It is only justified in defined high-risk populations, such as those with hereditary or familial pancreatic cancer. Therefore, new specific, sensitive and minimally invasive approaches are needed, and the calculation of EGV for HRI can serve as a starting point to predict the risk of this deadly disease. However, in order for algorithms such as these to be accurate and useful assessments for the management of high-risk individuals, we must ensure that input data is of high-quality, including pedigree information, full medical history and epidemiological data. Importantly, there are several international consortiums that provide guidelines and recommendations for high-risk screening [16–19], as well as templates for standardized reporting and interpretation of the EUS [52] and MRI [53] imaging tests.
Conclusions
The BLUP model offers a valuable tool for the study of hereditary cancer, allowing to estimate the degree of heritability of the cancer and to calculate the individual genetic risk in the members of a family with a history, as well as an approximation to the genetic risk in future generations. In addition, this tool can be used to identify the likely carriers of pathogenic variants, that should be prioritized for next-generation sequencing analysis. As a next step in the validation of the model, it is suggested to apply it to a larger population, standardising the data of the families involved.
Acknowledgements
We would like to acknowledge the support of the following people Celia Calcedo-Arnaíz, Victoria López Gómez, Carmen Guillen Ponce, María Teresa Salazar López, Andrea Santos Gil, Maria del Carmen Perez Ruiz, Manuela Hernando, Maria Jesus Casado Cespedosa and Clara Pacheco Torres.
Author contributions
Conceptualization: Julie Earl and Jose Carlos Martínez Ávila. Data curation, All authors. Methodology: Julie Earl, Jose Carlos Martínez Ávila and Cristina-Marianini-Rios. Writing and review of the manuscript: All authors.
Funding
This study was funded by the Instituto de Salud Carlos III (Plan Estatal de I + D + i 2013–2016): ISCIII (PI09/02221, PI12/01635, PI15/02101 and PI18/0135) and co-financed by the European Development Regional Fund ‘‘A way to achieve Europe’’ (ERDF), the Biomedical Research Network in Cancer: CIBERONC (CB16/12/00446), Red Temática de investigación cooperativa en cáncer: RTICC (RD12/0036/0073), La Asociación Española contra el Cáncer: AECC (Grupos Coordinados Estables 2016), Fundación Mutua Madrileña (FMM) / XVI Convocatoria de Ayudas a la Investigación en Salud and Asociación Cáncer de Páncreas (ACanPan); Asociación Española de Pancreatología (AESPANC) / IV Becas de Investigación Carmen Delgado/Miguel Pérez-Mateo. The funding sources of this study did not play any role in the study design, the collection, analysis and interpretation of the data, in the writing of the manuscript and in the decision to submit the paper for publication.
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Cristina-Marianini-Rios and María E Castillo Sanchez contributed equally to this work.
Contributor Information
Jose Carlos Martínez Ávila, Email: jc.martinez.avila@upm.es.
Julie Earl, Email: julie.earl@live.co.uk.
References
- 1.Huang J, Lok V, Ngai CH, et al. Worldwide Burden of, risk factors for, and trends in Pancreatic Cancer. Gastroenterology. 2021;160:744–754. doi: 10.1053/j.gastro.2020.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Carioli G, Malvezzi M, Bertuccio P, et al. European cancer mortality predictions for the year 2021 with focus on pancreatic and female lung cancer. Ann Oncol. 2021;32:478–487. doi: 10.1016/J.ANNONC.2021.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Pourshams A, Sepanlou SG, Ikuta KS, et al. The global, regional, and national burden of pancreatic cancer and its attributable risk factors in 195 countries and territories, 1990–2017: a systematic analysis for the global burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2019;4:934–947. doi: 10.1016/S2468-1253(19)30347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/CAAC.21654. [DOI] [PubMed] [Google Scholar]
- 5.Kolbeinsson HM, Chandana S, Wright GP, Chung M (2023) Pancreatic Cancer: a review of current treatment and Novel therapies. J Investig Surg 36 [DOI] [PubMed]
- 6.Klein AP. Prospective risk of pancreatic Cancer in familial pancreatic Cancer kindreds. Cancer Res. 2004;64:2634–2638. doi: 10.1158/0008-5472.CAN-03-3823. [DOI] [PubMed] [Google Scholar]
- 7.Porter N, Laheru D, Lau B, et al. Risk of pancreatic Cancer in the long-term prospective Follow-Up of familial pancreatic Cancer kindreds. JNCI J Natl Cancer Inst. 2022;114:1681–1688. doi: 10.1093/jnci/djac167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaffee KG, Oberg AL, McWilliams RR, et al. Prevalence of germline mutations in Cancer genes among pancreatic Cancer patients with positive family history. Genet Med. 2018;20:119–127. doi: 10.1038/gim.2017.85.PREVALENCE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhen DB, Rabe KG, Gallinger S, et al. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic Cancer (FPC): a PACGENE study. Genet Med. 2015;17:569–577. doi: 10.1038/gim.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts NJ, Norris AL, Petersen GM, et al. Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discov. 2016;6:166–175. doi: 10.1158/2159-8290.CD-15-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Earl J, Galindo-Pumariño C, Encinas J et al (2020) A comprehensive analysis of candidate genes in familial pancreatic cancer families reveals a high frequency of potentially pathogenic germline variants. 10.1016/j.ebiom.2020.102675. EBioMedicine 53: [DOI] [PMC free article] [PubMed]
- 12.Matsubayashi H, Takaori K, Morizane C, et al. Familial pancreatic cancer: Concept, management and issues. World J Gastroenterol. 2017;23:935–948. doi: 10.3748/wjg.v23.i6.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen GM. Familial pancreatic Cancer. Semin Oncol. 2016;43:548–553. doi: 10.1053/j.seminoncol.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19 – 9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: an evidence based appraisal. J Gastrointest Oncol. 2012;3:105–119. doi: 10.3978/j.issn.2078-6891.2011.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brand RE, Lerch MM, Rubinstein WS et al (2007) Advances in counselling and surveillance of patients at risk for pancreatic cancer. Gut 56 [DOI] [PMC free article] [PubMed]
- 16.Goggins M, Overbeek KA, Brand R, et al. Management of patients with increased risk for familial pancreatic cancer: updated recommendations from the International Cancer of the pancreas Screening (CAPS) Consortium. Gut. 2020;69:7–17. doi: 10.1136/gutjnl-2019-319352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daly MB, Pilarski R, Yurgelun MB, et al. NCCN guidelines insights: Genetic/Familial High-Risk Assessment: breast, ovarian, and pancreatic, Version 1.2020. J Natl Compr Canc Netw. 2020;18:380–391. doi: 10.6004/jnccn.2020.0017. [DOI] [PubMed] [Google Scholar]
- 18.Aslanian HR, Lee JH, Canto MI. AGA clinical practice update on Pancreas Cancer Screening in High-Risk individuals. Expert Rev Gastroenterol. 2020;159:358–362. doi: 10.1053/j.gastro.2020.03.088. [DOI] [PubMed] [Google Scholar]
- 19.Sawhney MS, Calderwood AH, Thosani NC, et al. ASGE guideline on screening for pancreatic cancer in individuals with genetic susceptibility: summary and recommendations. Gastrointest Endosc. 2022;95:817–826. doi: 10.1016/J.GIE.2021.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Mocci E, Guillen-Ponce C, Earl J, et al. PanGen-Fam: Spanish registry of hereditary pancreatic cancer. Eur J Cancer. 2015;51:1911–1917. doi: 10.1016/j.ejca.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Vasen H, Ibrahim I, Robbers K, et al. Benefit of surveillance for pancreatic cancer in high-risk individuals: outcome of long-term prospective follow-up studies from three European expert centers. J Clin Oncol. 2016;34:2010–2019. doi: 10.1200/JCO.2015.64.0730. [DOI] [PubMed] [Google Scholar]
- 22.Bartsch DK, Slater EP, Carrato A, et al. Refinement of screening for familial pancreatic cancer. Gut. 2016;65:1314. doi: 10.1136/gutjnl-2015-311098. [DOI] [PubMed] [Google Scholar]
- 23.Martínez-Ávila J, Guillén-Ponce C, Earl J, García-Cortés L. Hereditary Lifetime Cancer Risk Assessment modeling: a case study in breast Cancer. Int J Mol Genet Gene Ther. 2017 doi: 10.16966/2471-4968.106. [DOI] [Google Scholar]
- 24.R Core Team (2022) (2020) R: A language and environment for statistical computing. R Found. Stat. Comput. Vienna, Austria
- 25.Sinnwell JP, Therneau TM, Schaid DJ. The kinship2 R Package for Pedigree Data. Hum Hered. 2014;78:91–93. doi: 10.1159/000363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vazquez AI, Bates DM, Rosa GJM, et al. Technical note: an R package for fitting generalized linear mixed models in animal breeding. J Anim Sci. 2010;88:497–504. doi: 10.2527/jas.2009-1952. [DOI] [PubMed] [Google Scholar]
- 27.Hadfield JD. MCMC methods for Multi-response Generalized Linear mixed models: the MCMCglmm R Package. J Stat Softw. 2010;33:1–22. doi: 10.18637/JSS.V033.I02. [DOI] [Google Scholar]
- 28.Sing T, Sander O, Beerenwinkel N, Lengauer T (2005) ROCR: visualizing classifier performance in R. Bioinformatics 21. 10.1093/bioinformatics/bti623 [DOI] [PubMed]
- 29.de Villemereuil P, Schielzeth H, Nakagawa S, Morrissey M (2016) General methods for evolutionary quantitative genetic inference from generalized mixed models. Genetics 204. 10.1534/genetics.115.186536 [DOI] [PMC free article] [PubMed]
- 30.De Villemereuil P, Gimenez O, Doligez B (2013) Comparing parent-offspring regression with frequentist and bayesian animal models to estimate heritability in wild populations: a simulation study for Gaussian and binary traits. Methods Ecol Evol 4. 10.1111/2041-210X.12011
- 31.Henderson CR (1975) Best Linear unbiased estimation and prediction under a selection model. Biometrics 31. 10.2307/2529430 [PubMed]
- 32.Fisher RA (1919) XV.—The correlation between relatives on the supposition of mendelian inheritance. Trans R Soc Edinb 52. 10.1017/S0080456800012163
- 33.de Villemereuil P (2018) Quantitative genetic methods depending on the nature of the phenotypic trait. Ann N Y Acad Sci 1422 [DOI] [PubMed]
- 34.Heidelberger P, Welch PD (1981) A spectral method for confidence interval generation and run length control in simulations. Commun ACM 24. 10.1145/358598.358630
- 35.Zhang S, Cooper-Knock J, Weimer AK, et al. Genome-wide identification of the genetic basis of amyotrophic lateral sclerosis. Neuron. 2022;110:992–1008e11. doi: 10.1016/j.neuron.2021.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sada-Fuente E, Aranda S, Papiol S et al (2023) Common genetic variants contribute to heritability of age at onset of schizophrenia. Transl Psychiatry 13. 10.1038/s41398-023-02508-0 [DOI] [PMC free article] [PubMed]
- 37.Whelan CD, Hibar DP, Van Velzen LS, et al. Heritability and reliability of automatically segmented human hippocampal formation subregions. NeuroImage. 2016;128:125–137. doi: 10.1016/J.NEUROIMAGE.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gail MH, Brinton LA, Byar DP, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–1886. doi: 10.1093/JNCI/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 39.Johansson M, Holmström B, Hinchliffe SR, et al. Combining 33 genetic variants with prostate-specific antigen for prediction of prostate cancer: longitudinal study. Int J Cancer. 2012;130:129–137. doi: 10.1002/ijc.25986. [DOI] [PubMed] [Google Scholar]
- 40.Wu X, Lin J, Grossman HB, et al. Projecting individualized probabilities of developing bladder cancer in white individuals. J Clin Oncol. 2007;25:4974–4981. doi: 10.1200/JCO.2007.10.7557. [DOI] [PubMed] [Google Scholar]
- 41.Lee A, Mavaddat N, Wilcox AN, et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet Med. 2019;21:1708–1718. doi: 10.1038/S41436-018-0406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia G, Lu Y, Wen W, et al. Evaluating the utility of polygenic risk scores in identifying high-risk individuals for eight common cancers. JNCI cancer Spectr. 2020;4:pkaa021. doi: 10.1093/jncics/pkaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakatochi M, Lin Y, Ito H et al (2018) Prediction model for pancreatic cancer risk in the general Japanese population. PLoS ONE 13. 10.1371/journal.pone.0203386 [DOI] [PMC free article] [PubMed]
- 44.Galeotti AA, Gentiluomo M, Rizzato C, et al. Polygenic and multifactorial scores for pancreatic ductal adenocarcinoma risk prediction. J Med Genet. 2020 doi: 10.1136/jmedgenet-2020-106961. [DOI] [PubMed] [Google Scholar]
- 45.Molina-Montes E, Coscia C, Gómez-Rubio P, et al. Deciphering the complex interplay between pancreatic cancer, diabetes mellitus subtypes and obesity/BMI through causal inference and mediation analyses. Gut. 2021;70:319–329. doi: 10.1136/GUTJNL-2019-319990. [DOI] [PubMed] [Google Scholar]
- 46.Koch S, Schmidtke J, Krawczak M, Caliebe A. Clinical utility of polygenic risk scores: a critical 2023 appraisal. J Community Genet. 2023;14:471–487. doi: 10.1007/s12687-023-00645-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klein RJ, Gümüş ZH. Are polygenic risk scores ready for the cancer clinic?—a perspective. Transl Lung Cancer Res. 2022;11:910–919. doi: 10.21037/TLCR-21-698/COIF). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paiella S, Capurso G, Cavestro GM, et al. Results of First-Round of Surveillance in individuals at high-risk of pancreatic Cancer from the AISP (Italian Association for the study of the Pancreas) Registry. Am J Gastroenterol. 2019;114:665–670. doi: 10.1038/S41395-018-0414-Z. [DOI] [PubMed] [Google Scholar]
- 49.Overbeek KA, Goggins MG, Dbouk M, et al. Timeline of Development of Pancreatic Cancer and implications for successful early detection in high-risk individuals. Gastroenterology. 2022;162:772–785e4. doi: 10.1053/j.gastro.2021.10.014. [DOI] [PubMed] [Google Scholar]
- 50.Canto MI, Hruban RH, Fishman EK, et al. Frequent detection of pancreatic lesions in Asymptomatic High-Risk individuals. Gastroenterology. 2012;142:796–804. doi: 10.1053/J.GASTRO.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forni S, Aguilar I, Misztal I (2011) Different genomic relationship matrices for single-step analysis using phenotypic, pedigree and genomic information. Genet Sel Evol 43. 10.1186/1297-9686-43-1 [DOI] [PMC free article] [PubMed]
- 52.Gonda TA, Farrell J, Wallace M, et al. Standardization of EUS imaging and reporting in high-risk individuals of pancreatic adenocarcinoma: consensus statement of the pancreatic Cancer Early Detection Consortium. Gastrointest Endosc. 2022;95:723–732e7. doi: 10.1016/J.GIE.2021.10.025. [DOI] [PubMed] [Google Scholar]
- 53.Huang C, Simeone DM, Luk L, et al. Standardization of MRI screening and reporting in individuals with elevated risk of pancreatic ductal adenocarcinoma: Consensus Statement of the PRECEDE Consortium. AJR Am J Roentgenol. 2022;219:903–914. doi: 10.2214/AJR.22.27859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.