Graphical abstract
Keywords: Homo-oligomerization, NMR Spectroscopy, SAXS, AUC, Native MS
Abstract
Cellular production of tryptophan is metabolically expensive and tightly regulated. The small Bacillus subtilis zinc binding Anti-TRAP protein (AT), which is the product of the yczA/rtpA gene, is upregulated in response to accumulating levels of uncharged tRNATrp through a T-box antitermination mechanism. AT binds to the undecameric axially symmetric ring-shaped protein TRAP (trp RNA Binding Attenuation Protein), thereby preventing it from binding to the trp leader RNA. This reverses the inhibitory effect of TRAP on transcription and translation of the trp operon. AT principally adopts two symmetric oligomeric states, a trimer (AT3) featuring three-fold axial symmetry or a dodecamer (AT12) comprising a tetrahedral assembly of trimers, whereas only the trimeric form binds and inhibits TRAP. We apply native mass spectrometry (nMS) and small-angle x-ray scattering (SAXS), together with analytical ultracentrifugation (AUC) to monitor the pH and concentration-dependent equilibrium between the trimeric and dodecameric structural forms of AT. In addition, we use solution nuclear magnetic resonance (NMR) spectroscopy to determine the solution structure of AT3, while heteronuclear 15N relaxation measurements on both oligomeric forms of AT provide insights into the dynamic properties of binding-active AT3 and binding-inactive AT12, with implications for TRAP binding and inhibition.
Introduction
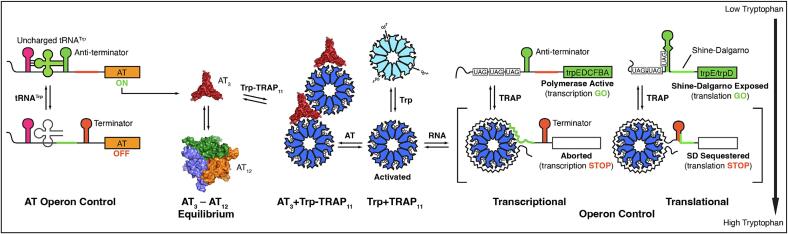
The trp RNA-binding attenuation protein (TRAP) senses intracellular levels of tryptophan in order to regulate transcription (Babitzke et al., 1994, Babitzke and Yanofsky, 1993, Gollnick, 1994, Gollnick et al., 1990) and translation (Du and Babitzke, 1998, Merino et al., 1995, Sarsero et al., 2000, Yakhnin et al., 2004, Yakhnin et al., 2001, Yang et al., 1995) of the genes required for tryptophan biosynthesis (Fig. 1). Transcriptional and translational control by TRAP depends on the ability of tryptophan-bound TRAP to bind to an RNA site with multiple (G/U)AG repeats separated by two or three nonconserved spacer nucleotides (Babitzke et al., 1994). The transcription of the six tryptophan biosynthetic genes clustered in the trpECDFBA operon (Henner et al., 1985) is controlled through an attenuation mechanism by which TRAP binding is thought to remodel competing secondary structural elements in the 5́ leader region of the nascent mRNA: in the absence of tryptophan, RNA binding by TRAP is impaired, and the leader folds into an anti-terminator hairpin that allows transcription of the entire operon. Increased cellular tryptophan binds to TRAP, activating it to bind to its RNA target which overlaps with and prevents formation of the anti-terminator hairpin; instead, a small downstream terminator hairpin forms and transcription of the structural genes is attenuated (Babitzke and Yanofsky, 1993). There is considerable evidence in support of this terminator/anti-terminator model, although TRAP nevertheless induces attenuation in the absence of an intact terminator (McAdams and Gollnick, 2014, Potter et al., 2011).
Fig. 1.
Transcription and translation of the trp operon in B. subtilis is regulated by feedback loops involving the functions of two proteins: the ring-shaped 11-mer TRAP, which senses free Trp, and Anti-TRAP (AT) which is overexpressed in response to accumulation of uncharged tRNATrp. In the absence of AT, Trp-bound TRAP (blue) binds a series of UAG and GAG triplets in the 5′ leader of the trp operon, resulting in transcriptional and translational repression in part by formation of a terminator or sequestering the Shine-Dalgarno (SD) sequence. When expressed, trimeric AT (AT3) can bind to Trp-activated TRAP, preventing RNA binding and blocking its inhibition of Trp biosynthesis. AT exists in equilibrium between trimeric (red) and dodecameric states (blue, orange, green; the red trimer is behind the other three), while only trimeric AT binds and inhibits TRAP.
Translational control of trpE (Du and Babitzke, 1998, Schaak et al., 2003), and perhaps the entire trp operon, is also accomplished through TRAP binding-dependent RNA secondary structure (Yakhnin et al., 2001). When TRAP is inactive and cannot bind to the read-through transcript of the trp operon, the Shine-Dalgarno (SD) sequence of the trpE gene is unstructured and allows ribosome binding. When Trp-activated TRAP binds to the untranslated region of the transcript, the trpE SD sequence becomes sequestered in a hairpin that forms downstream of the TRAP binding site, thereby preventing ribosome binding and translation initiation (Du and Babitzke, 1998). TRAP binding sites are also present in the transcripts of the trpG, trpP, and ycbK genes, but in the translation initiation region, where TRAP binding directly impedes ribosome binding and thereby regulates translation (Babitzke et al., 1994, Du et al., 1997, Lee et al., 1996, Sarsero et al., 2000).
In many bacteria, the extent of charging of tRNATrp is sensed as a regulatory signal in addition to the levels of free tryptophan (Gutiérrez-Preciado et al., 2009, Gutiérrez-Preciado et al., 2007). In studies with a temperature-sensitive mutant of the tryptophanyl-tRNA synthetase in B. subtilis, the accumulation of uncharged tRNATrp leads to over-expression of the trpECDFBA operon (Lee et al., 1996). The operon responsible for this effect was shown to be regulated via the T-box transcription anti-termination mechanism, where uncharged tRNATrp specifically pairs with the leader RNA causing anti-termination and transcriptional read-through (Sarsero et al., 2000). Deletions within the operon identified yczA/rtpA as the responsible gene, and the protein product, anti-TRAP (AT), was shown to inhibit TRAP activity (Sarsero et al., 2000, Sharma and Gollnick, 2014, Valbuzzi and Yanofsky, 2001). Expression of AT was also shown to be regulated at the level of translation by tRNA-dependent ribosome stalling (Chen and Yanofsky, 2004, Chen and Yanofsky, 2003).
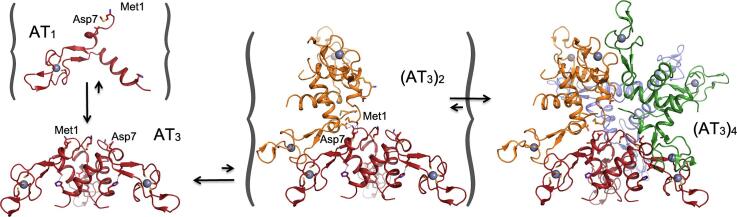
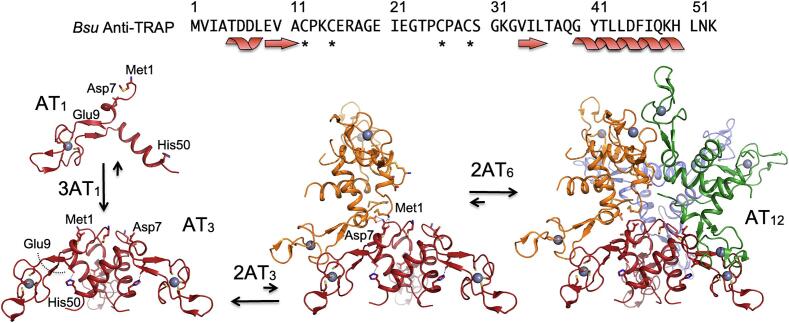
Symmetry mismatch between the oligomeric states of AT and TRAP was evident from early chemical cross-linking, analytical ultracentrifugation, native mass spectrometry and NMR studies (Akashi et al., 2009, Sachleben et al., 2010, Snyder et al., 2004, Valbuzzi and Yanofsky, 2002, Valbuzzi and Yanofsky, 2001). Diffracting crystals of Bsu AT revealed a dodecamer (AT12) arranged as a tetrahedral tetramer of trimers (AT3)4 (Fig. 2) (Shevtsov et al., 2010, Shevtsov et al., 2005, Shevtsov et al., 2004). Each AT protomer forms a short helix (residues 5–8) at the N-terminus, β-strands from residues 9–11 and 34–36 form a short anti-parallel β-sheet, a β-hairpin is formed by strands from residues 20–21 and 24–25, and an α-helix is present at the C-terminus from residues 38–50. Trimers are arranged via a three-helix bundle involving the C-terminal helix, with the zinc binding domains extended outward from the C3 helical axis. The AT dodecamer is formed through complementary inter-trimer ion pair interactions involving an N-terminal amine and a sidechain carboxylate from each trimer, as well as hydrophobic surface burial of the zinc-binding regions (Fig. 2) (Shevtsov et al., 2010, Shevtsov et al., 2005, Shevtsov et al., 2004). Because the stabilizing ion pair requires a protonated N-terminus, which has a pKa near pH 7 (Sereda et al., 1993), assembly of the dodecamer is disfavored above pH ∼ 8 (Sachleben et al., 2010). The B. licheniformis variant of AT (which shares a 76 % sequence identity with B. subtilis AT) crystallized at low pH adopts the same dodecameric structure as the Bsu protein, but at high pH the trimers assemble in an inverted orientation, with their N-termini projecting into solvent (Shevtsov et al., 2010).
Fig. 2.
Oligomeric assembly of Bsu Anti-TRAP (AT). The 53-residue AT protomer (AT1) features a short N-terminal helix leading into a “zinc ribbon” motif and ending in a C-terminal helix. Assembly of AT3 occurs via a helical bundle that buries ∼ 4,200 Å2. In solution AT3 is in an equilibrium with dodecamers (AT12) featuring a tetrahedral arrangement of AT3 stabilized by a network of ion pairs between the amino terminus of Met1 and the sidechain carboxylate of Asp7 of adjacent trimers. A transient AT6 intermediate is hypothesized to arise from dimerization of AT3 via formation of two sets of Met1-Asp7 ion pairs and burial of ∼ 1,100 Å2; dimerization of two AT6 assemblies to form AT12 is favored by formation of ten additional sets of ion pairs and burial of another ∼ 4,600 Å2.
AT binds asymmetrically to Trp-loaded TRAP. Although Bsu TRAP is predominantly undecameric, both undecamers and dodecamers are observed in mass spectra (Babitzke et al., 1994, Bayfield et al., 2012, Chen et al., 2011, McCammon et al., 2004), and crystallographic studies of Bsu AT bound to dodecameric TRAP rings revealed AT3 docked on the RNA binding surface of TRAP12, via protein–protein surfaces of the zinc binding and helical domains that are not accessible in AT12 (Watanabe et al., 2009). Mass spectrometric, solution small angle scattering, and analytical ultracentrifugation experiments (Ihms et al., 2014) are also consistent with a model in which only AT3 can compete with RNA for the TRAP-RNA binding site. These findings suggest that the AT3 ↔ AT12 equilibrium may represent an additional regulatory step.
We found experimentally that preventing formation of the inter-trimer Met1-Asp7 ion pairs favors AT3, even at high protein concentrations (Sachleben et al., 2010). Addition of excess zinc during protein recombinant production of AT in E. coli results in significant accumulation of formylmethionine AT (fAT), presumably due to poisoning of peptide deformylase (Jain et al., 2005, Ragusa et al., 1998); the resulting immature fAT can be chromatographically separated from mature AT under denaturing conditions (Sachleben et al., 2010). Other modification to the N-terminus, such as inclusion of a His-6 tag, similarly prevents AT12 formation (McElroy, 2005). This provides the means to experimentally produce homogenous samples of either AT3 or AT12, to allow characterization of the two oligomers in the absence of confounding effects from their equilibrium.
We have leveraged the ability to selectively produce AT trimers and dodecamers to determine the solution structure of Bsu AT3 and to examine the pH- and concentration-dependent equilibrium between to AT3 and AT12 using small angle x-ray scattering (SAXS), sedimentation velocity analytical ultracentrifugation (AUC), and native mass spectrometry (nMS). In addition, backbone amide 15N NMR relaxation measurements (Barbato et al., 1992, Farrow et al., 1994, Kay et al., 1989) allowed us to study the dynamics of both the trimeric and dodecameric AT forms. These results provide new insights into the structural basis of Anti-TRAP’s homo-oligomeric assembly, and how this process may modulate AT’s function in its regulatory context.
Results
Anti-TRAP (AT) adopts trimeric and dodecameric species in solution
NMR and native MS reveal two states in slow equilibrium
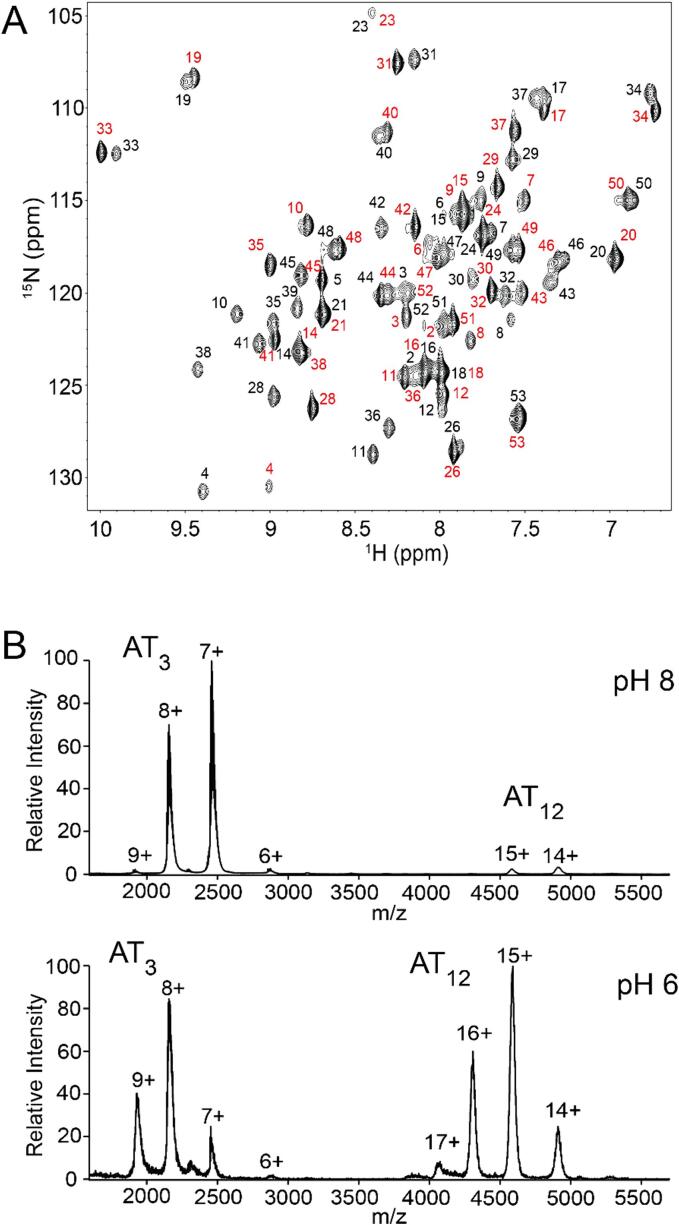
At pH values near 7, purified AT samples at 10-3 M concentrations yield 1H-15N correlation spectra that exhibit doubling of many amide resonances; such doubling arises in cases of chemical heterogeneity or slow conformational exchange between two states. After chromatographically separating mature AT from immature formylated AT (fAT) under denaturing conditions (Sachleben et al., 2010), samples of mature AT produced the same signal doubling (Fig. 3), indicating that it is not due to chemical heterogeneity. Nano electrospray ionization mass spectrometry under soft ionization conditions, at protein concentrations one hundred times lower than NMR (10-5 M), yielded two sets of ions, one corresponding to the mass of AT3, and the other, AT12 (Fig. 3). Adjusting the solution pH to 8 produced a higher proportion of AT3 ions, while at pH 6 similar intensities were observed for both sets of ions, consistent with prior findings from pH-dependent NMR spectra. In addition to affecting the oligomeric state, changing the pH slightly altered the preferred charge state distribution: at pH 8, the 7+ and 14+ charge states were favored for AT3 and AT12, respectively; at pH 6, the 8+ and 15+ charge states dominate the spectrum. Although AT3 and AT12 populations varied with pH, no other oligomeric species (e.g., AT1, AT6) were observed in the nMS data over a wide range of conditions.
Fig. 3.
AT adopts trimeric and dodecameric states in solution. (A) 1H-15N HSQC NMR spectrum of 1 mM AT, pH 7.5 reveals doubling of many signals; residue assignments as indicated (black, AT3; red, AT12). (B) Native MS in 100 mM AmAc shows trimers are favored at pH 8 and 12-mers are favored at pH 6; predicted (Zn-AT)3: 17,144.9 Da; (Zn-AT)12: 68,579.4 Da; observed at pH 6: 17,181.8 ± 63.7 Da and 68,826.5 ± 76.4 Da; observed at pH 8: 17,181.5 ± 55.3 Da and 68,751.5 ± 47.3 Da. Other oligomeric states are not evident. The extra mass can be attributed to salt and/or solvent adducts, typical for the instrument used.
Sedimentation velocity experiments provide evidence for a lowly populated hexamer intermediate
Guided by prior sedimentation velocity experiments at a fixed pH that revealed both AT3 and AT12 in solution (Snyder et al., 2004), we performed analytical ultracentrifugation sedimentation velocity experiments at pH 7, 7.25, 7.5, and 8 and a range of AT concentrations (Fig. 4). For systems with components undergoing rapid exchange on the timescale of the sedimentation experiment, the modeling of boundary phases can provide the sedimentation coefficients for the components as well as the stepwise binding rates (Brautigam, 2011). Sedimentation velocity data at each pH were globally fit across three concentrations in the program SEDPHAT (Zhao et al., 2015), initially to a simple two-state model comprising the two states observed in NMR and MS spectra,
The resulting fits were poor, especially at alkaline conditions, and were better fit with a three-state exchange model,
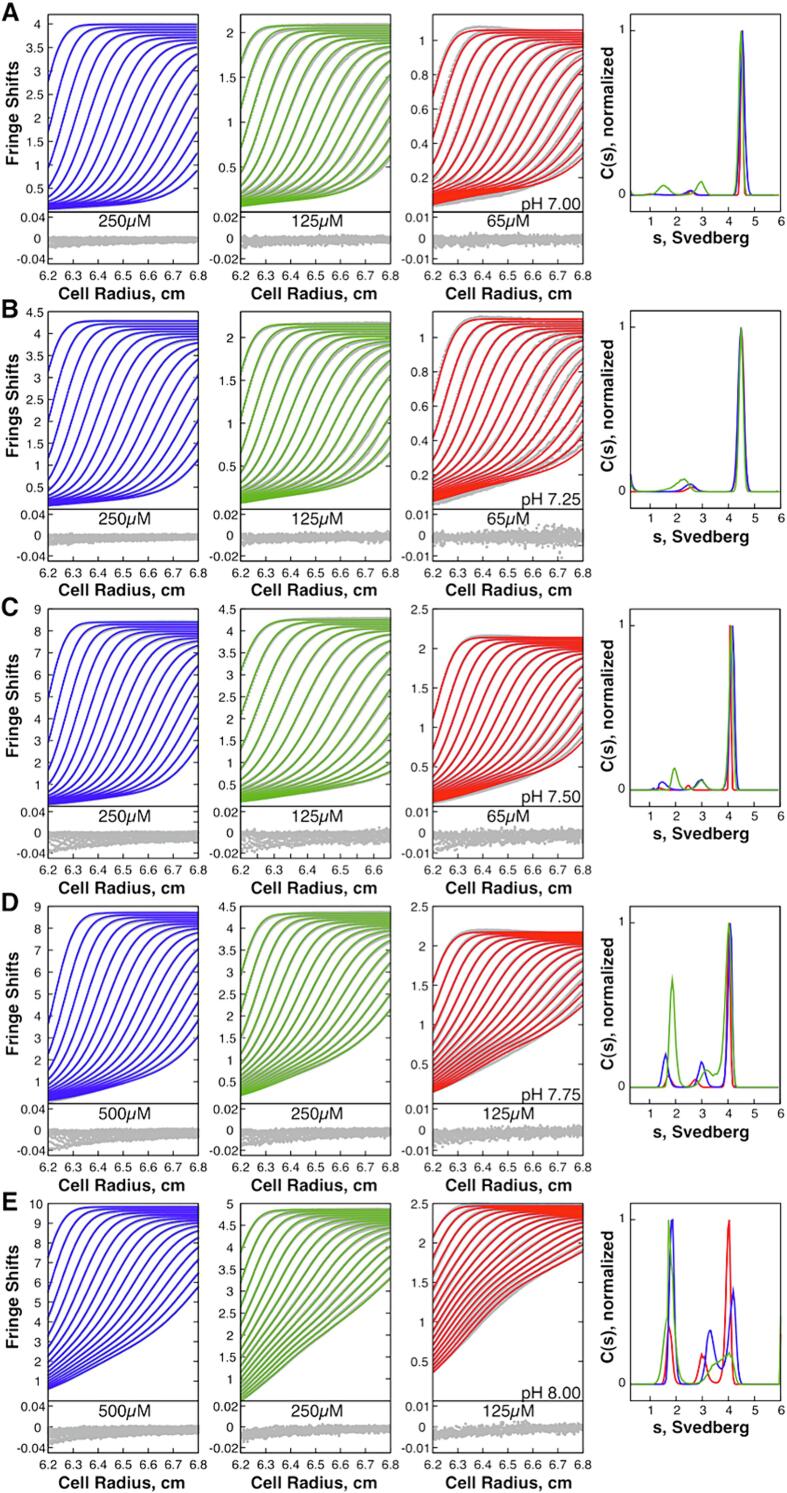
Lamm modeling of the sedimentation data predicts species with an intermediate sedimentation coefficient, which we attribute to a small population of hexamer AT6, despite not observing this species by nMS or NMR (Fig. 3). Fits to experimental interference fringe shifts using this model permitted optimization of two sets of apparent rate constants describing dimerization of AT3 trimers into AT6, and a subsequent dimerization of two AT6 into AT12. From the ratio of the fitted rate constants, we obtained consistent equilibrium dissociation constants for the first step, ranging from 20-90 µM over the experimental pH range. For the second step, the association rate constant dropped over three orders of magnitude over the same pH range, resulting in equilibrium constants from 0.05-50 µM. Thus, at low pH the second step is highly favored compared to the first step, resulting in very low concentrations of the hexameric intermediate AT6. This is generally consistent with the relatively low populations of intermediate species in the fitted c(s) distributions (Fig. 4). The excellent agreement between experimental and modeled data suggests that monomeric AT is not present at a significant population.
Fig. 4.
Sedimentation velocity experiments over a range of pH values can be fit with a three-state association model AT3 ↔ (AT3)2 ↔ AT12. Interference fringe shifts (gray dots) are globally fit across three AT concentrations (first three columns). Fits provided by the mechanistic model are in blue, green, and red lines. Only every tenth scan is shown. Residuals are below the data. Fourth column, sedimentation coefficient distributions with blue, red and green corresponding to the concentrations on the left, at pH 7.00 (A), pH 7.25 (B), pH 7.50 (C), pH 7.75 (D), and pH 8.00 (E).
SAXS profiles of AT can be reconstructed from those of AT3 and AT12 oligomers
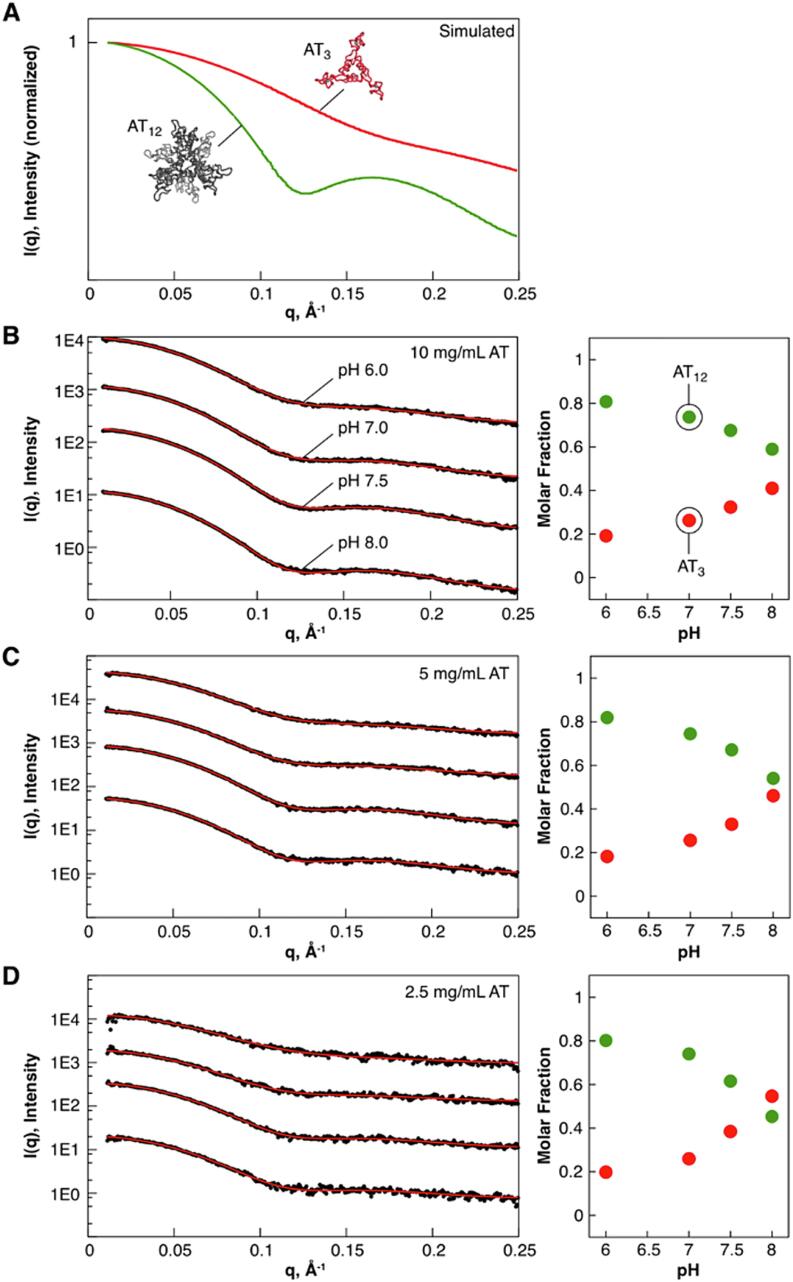
We investigated the pH-dependent structure of AT using small angle x-ray scattering (SAXS). For mixtures, the resulting scattering profiles are the weighted sum of scattering profiles of the individual components (Konarev et al., 2003). Experimental scattering curves were de-convoluted using form factors for trimeric and dodecameric AT (Fig. 5). The predicted scattering profiles for AT3 and AT12 were calculated with CRYSOL (Svergun et al., 1995) from the trimer and dodecamer assemblies in the crystal structure (2BX9) (Shevtsov et al., 2005). Deconvolution of the experimental data from the polydisperse AT solutions was performed using OLIGOMER (Konarev et al., 2003). The resulting fits using these two components describe the experimental data well, with AT12 dominating below pH 7.5. We also tested for the presence of an inverted dodecameric structure such as that observed in crystallographic studies of B. lichenformis AT (PDB 3ld0) (Shevtsov et al., 2010) by including its form factors in fitting the experimental data; this component was found to have zero weight in all of the tested experimental conditions. These data are consistent with a slow exchange between AT3 and AT12 under near-physiological conditions, and a negligible population of intermediates.
Fig. 5.
SAXS profiles of AT can be described as mixtures of AT3 and AT12. A, scattering curves for AT3 and AT12, computed from the dodecameric crystal structure (2bx9). B-D, left, experimental scattering intensities (black) for AT recorded over a range of pHs (6, 7, 7.5, 8) and concentrations (1.77, 0.89 and 0.44 mM), fit to a two-state model with varying fractions of AT3 and AT12. Profiles are offset in the y-axis from each other for visibility. Right, fitted volume fractions as a function of pH for each sampled concentration.
NMR studies of fAT3
Formylated AT is trimeric with high quality NMR spectra
We used immature fAT for detailed investigation of the structure and dynamics of the trimeric TRAP-binding oligomeric form. Since an ion pair between the protonated amine of Met1 and the sidechain carboxylate of Asp7 is proposed to stabilize the dodecameric structure (Sachleben et al., 2010, Shevtsov et al., 2005), the formyl group prevents formation of the ion pair and fAT trimers do not oligomerize to form detectable dodecamers, even at millimolar concentrations at low pH. Resonance assignments and NMR structural data were recorded at 55 °C, to match the conditions used for backbone resonance assignments of Bst TRAP (Kleckner et al., 2012, McElroy et al., 2002, Sachleben et al., 2010).
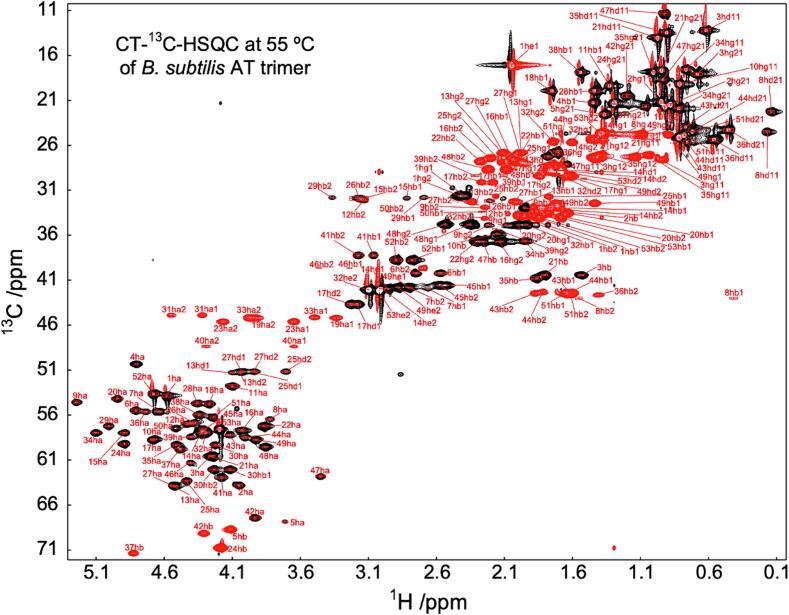
NMR spectra of fAT3 are of exceptionally high quality (e.g., Fig. 6), enabling nearly complete backbone and side chain resonance assignments. Bsu AT comprises 53 amino acids, and zinc-bound fAT3 has a nominal molecular mass of 17.1 kDa. Three-fold symmetry results in spectra reflecting the number of signals of a single chain, while fast tumbling result in narrow NMR resonances and high-quality heteronuclear correlation spectra. Backbone resonances were assigned from standard triple resonance spectra, while sidechain assignments were obtained from a combination of 2D and 3D double- and triple-resonance spectra. This allowed for assignment of every proton in the protein except for the side chain amine and hydroxyl protons.
Fig. 6.
Symmetry and exceptional NMR spectra of fAT3 allowed nearly complete resonance assignments. Aliphatic region of a two-dimensional constant-time 1H–13C correlation spectrum of fAT3. In this spectrum 13C resonances coupled to even (red) and odd (black) numbers of aliphatic carbons have opposite sign. Aliphatic carbon resonance assignments as indicated.
The structure of fAT3 is unperturbed by pH
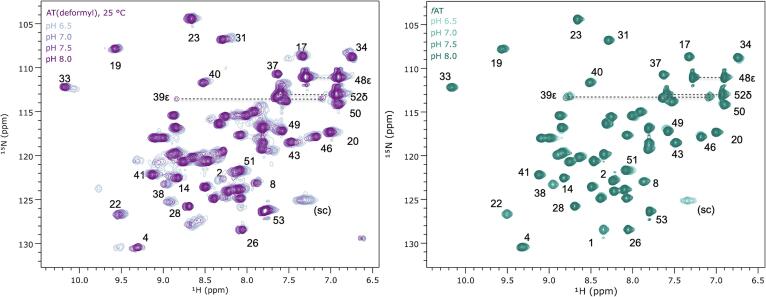
SAXS, nMS and NMR data demonstrate that the trimer-dodecamer equilibrium is strongly affected by pH (Fig. 3, Fig. 4, Fig. 5). This behavior was attributed to a near-neutral pKa for the amine at the N-terminus of the protein (Sereda et al., 1993), and its ion pair with the carboxylate side chain of Asp7 (Sachleben et al., 2010, Shevtsov et al., 2005). There is one other functional group in AT with a predicted pKa near neutrality: His50, which is conserved among AT variants (Shevtsov et al., 2010, Valbuzzi and Yanofsky, 2001). In crystals the side chain imidazole Nε2 of His50, located at the end of the C-terminal helix, is observed to form an intra-chain ion pair with the carboxylate of Glu9, located in the zinc-ribbon motif (Fig. 2). Changes in the protonation state of the imidazole side chain could conceivably alter the geometry between the zinc-binding and helical domains and thus alter trimer packing in the dodecamer, driving changes in oligomeric structure (e.g., such behavior has been engineered into homo-oligomeric assemblies (Boyken et al., 2019)). To examine whether His50 or another titratable group in the protein might induce structural changes that contribute to the pH-dependent oligomerization behavior, we recorded 1H-15N correlation NMR spectra of fAT over a range of pH values (Fig. 7). Since fAT has a blocked N-terminus and does not form stable dodecamers, this allowed distinguishing spectral effects of ionization from those resulting from oligomerization. Correlation spectra of fAT recorded over a pH range that would shift the predominant oligomeric state from AT3 to AT12 in the mature protein result in only minor shifts in the spectra, particularly the amides of Lys49 and Leu51, likely reflecting their proximity to the His50 indole. This is consistent with a dominant role of a near neutral pKa for the N-terminal amide group in stabilizing the dodecamer.
Fig. 7.
Protonation of the Met1 amine is coupled to oligomerization of AT. Left, 1H-15N correlated spectra of AT recorded at 25 °C and pH 6.5, 7.0, 7.5 and 8.0 reveal strong perturbations of several signals, and line narrowing at high pH, which favors trimeric AT. Right, for fAT the N-terminal amine is protected by the formyl group and very minor shift perturbations are observed only for residues flanking His50; this indicates no pH-dependent structural shifts in the AT trimer. Amide assignments transferred from 55 °C via a temperature series. (sc), side chain amide.
Solution structure determination
The solution structure of the TRAP-binding oligomeric state of Bsu AT (AT3) might be inferred from the 2.8 Å crystal structure of the dodecamer (2BX9)(Shevtsov et al., 2005), the 2.1 Å structure of the Bli dodecamer (3LCZ)(Shevtsov et al., 2010), and 3.2 Å structure of the Bsu AT bound to Bst TRAP (2ZP8) (Watanabe et al., 2009), but de novo structure determination was justified by the potential for perturbations from oligomerization and crystal packing. To that end, we determined the solution structure of fAT3 using distance restraints from NOEs, orientation restraints from residual dipolar couplings, dihedral angle restraints from three-bond scalar couplings, three-fold symmetry restraints, and overall shape restraints from SAXS scattering curves.
Because of its homo-oligomeric structure and diverse source of restraints, structure determination proceeded via several steps. First, three-dimensional models of the zinc-bound AT protomer were computed from torsion angle restraints and NOEs that could be unambiguously assigned as being intramolecular. Next, we determined the solution structure of the trimer by replicating the chain and assigning as ambiguous all NOEs for which conservative distance filters prevented their assignment as being intramolecular. Most ambiguous NOEs could be assigned via iterative structure refinement steps, though some NOEs remained ambiguous throughout the refinement process (Table 1).
Table 1.
NMR structural statistics for AT ensemble (20 structures):
| NOE-based distance constraints: | |||
|---|---|---|---|
| Total ambiguous (not included in statistics) | 528 | ||
| Total unabiguous | 3396 | ||
| Inter-protomer | 396 | ||
| Intra-residue [i = j] | 611 | ||
| Sequential [| i − j | = 1] | 1007 | ||
| Medium range [1 < | i − j | < 5] | 891 | ||
| Long range [| i − j | > 5] | 887 | ||
| NOE constraints per restrained residue | 64.1 | ||
| Torsion-angle constraints: | 318 | ||
| Total number of restricting constraints | 3714 | ||
| Total number of restricting constraints per restrained residue | 70.1 | ||
| Restricting long-range constraints per restrained residue | 16.7 | ||
| Number of structures used | 20 | ||
| Residual constraint violations | |||
| Distance violations / structure > 0.5 Å | 1 | ||
| RMS of distance violation / constraint | 0.03 Å | ||
| Dihedral angle violations / structure > 10° | 11.3 | ||
| RMS of dihedral angle violation / constraint | 3.42° | ||
| RMSD Values | |||
| Backbone atoms (all residues) | 0.4 Å | ||
| Heavy atoms (all residues) | 0.6 Å | ||
| Structure Quality Factors − overall statistics | Mean | SD | Z-score |
| Procheck G-factor (phi/psi only) | −0.79 | n/a | −2.79 |
| Procheck G-factor (all dihedral angles) | −0.67 | n/a | 3.96 |
| Verify3D | 0.09 | 0.0065 | −5.94 |
| ProsaII (−ve) | 0.40 | 0.0265 | −1.03 |
| MolProbity clash score | 196.35 | 8.4019 | –32.17 |
| Ramachandran Statistics | |||
| Most favored | 74.0 % | ||
| Additionally allowed | 24.2 % | ||
| Generously allowed | 1.8 % | ||
| Disallowed regions | 0.0 % | ||
Summary of conformationally-restricting constraints and structure quality factors from protein structure and validation server (PSVS).
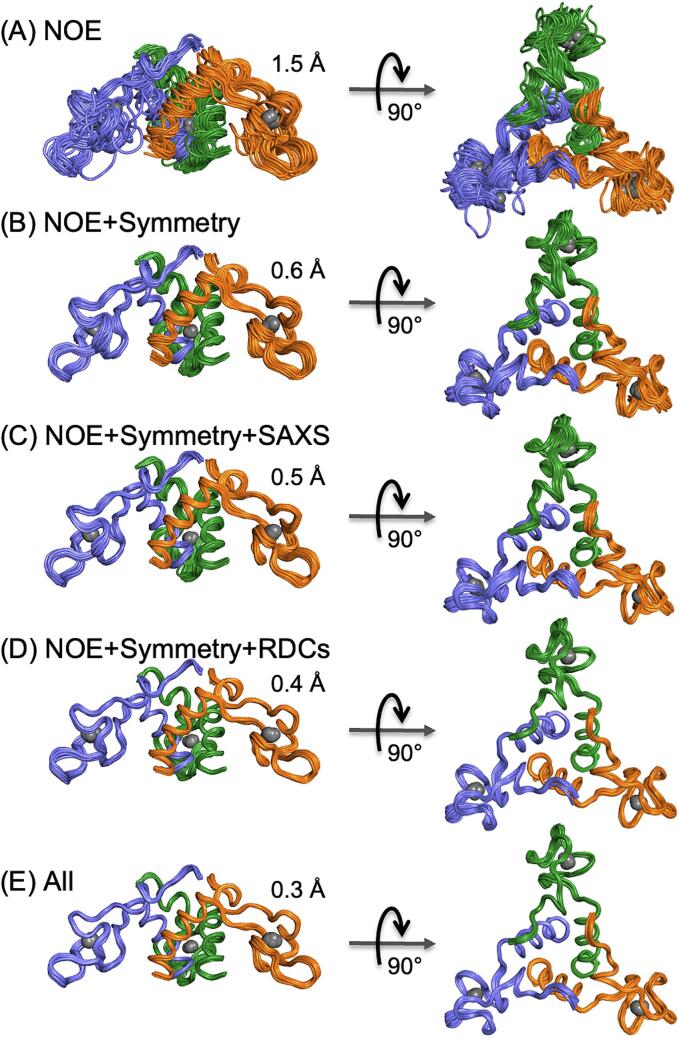
Symmetry, residual dipolar coupling (RDC) and SAXS restraints significantly increased the precision of the NMR structure (Fig. 8). Ensembles computed using only NMR-derived distance and torsion angle restraints yielded poor convergence, with an overall backbone RMSD of 1.5 Å, and per-protomer backbone RMSD of 1.3 Å. RDCs and SAXS data improved the overall precision, but not as much as inclusion of three-fold symmetry restraints, which increased the overall precision by almost an Ångstrom (from 1.5 Å to 0.6 Å).
Fig. 8.
Impact of restraint classes on precision of AT3 NMR ensemble. Each ensemble was computed using NMR-derived torsion angle restraints, and (A) NOEs, (B) NOEs and three-fold symmetry restraints, (C) NOEs, symmetry, special restraints from SAXS data, (D) NOEs, symmetry restraints and residual dipolar couplings, and (E) all restraint classes. Backbone RMSDs as indicated.
Comparison to crystallographic and AlphaFold2 models
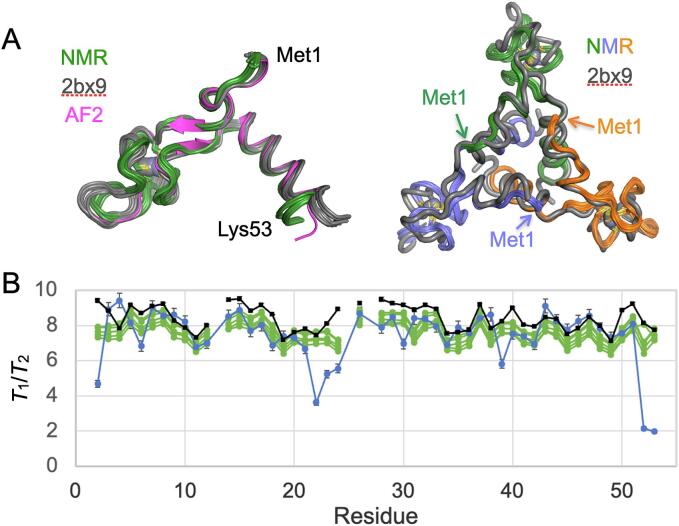
The final NMR ensemble was well defined by the restraints, with a backbone RMSD of 0.3 Å for Cα atoms (Fig. 8, Fig. 9, Table 1), and reveals a high degree of similarity with the trimers in the B. subtilis dodecamer (RMSD ∼ 0.6 Å; Fig. 9). This is similar to that observed between protomers in the 2.8 Å crystal structure of AT12 (∼0.6 Å for backbone atoms in 2BX9), which was determined with 12-fold non-crystallographic symmetry (Shevtsov et al., 2005, Shevtsov et al., 2004), and within the variance between those structures and that predicted by AlphaFold2 (Jumper et al., 2021). We used the longitudinal and transverse NMR relaxation times (T1, T2; the inverse of the relaxation rates R1, R2) of backbone amide 15N nuclei to validate the solution structure (Tjandra et al., 1997). In the absence of internal motions (see below) these values are dominated by reorientation of the amide nitrogen-proton bond vector and reflect the axially symmetric diffusion of the trimer. The experimentally measured T1/T2 ratios for AT3 are in good agreement with those predicted from members of the NMR ensemble, and from the trimer in the AT12 crystal structure (Fig. 9)(Bernadó et al., 2002, García De La Torre et al., 2000, García de la Torre et al., 2000). The largest deviations between the measured and predicted values arise at the N- and C-termini and the zinc binding loops, suggesting both subtle differences between the structure in crystals and in solution, as well as fast and slow conformational averaging in solution that is not possible in the crystal lattice.
Fig. 9.
Validation of the AT3 solution structure. (A) Solution structure of AT3 is congruent with its structure in AT12 crystals and that predicted by AlphaFold2. Left, green, NMR ensemble of 20 conformers for chain A; grey, superposition of the twelve chains in the 2BX9 crystal structure; pink, AlphaFold2 prediction. Right, superposition of the AT3 NMR derived ensemble with chains A-C (blue, orange, green) in the 2BX9 crystal structure. (B) The ratios of the NMR T1 and T2 relaxation times for backbone amide 15N provide an independent validation of the NMR-determined structure. Blue, experimental data; green, predicted ratios based on hydrodynamic modeling of the top five models of the NMR ensemble; black, predicted ratios using chains A-C from the crystal structure. Secondary structure indicated schematically; “*” mark the zinc ligands.
AT dynamics
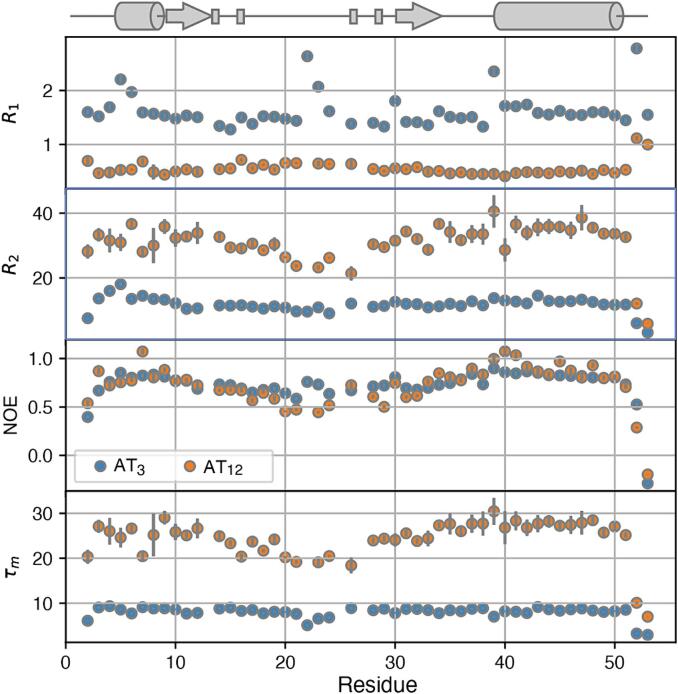
Backbone amide 15N R1, R2, and 1H-15N NOE relaxation rates that are sensitive to reorientation of the amide 1H-15N bond vector (Kay et al., 1989) were measured on both the 68 kDa dodecameric AT12 and 17 kDa trimeric AT3 using conventional two-dimensional heteronuclear correlation spectra (Fig. 10) (Barbato et al., 1992, Farrow et al., 1994). Differences in the R1 and R2 relaxation rates for the two oligomeric states reflected the large difference in their rotational diffusion tensors; overall correlation times τm computed from the 10 % trimmed mean R2/R1 ratios were 8 and 25 ns at 55 °C, respectively (Lee et al., 1997). Hydrodynamic calculations based on the crystal structure of AT12, and the R2/R1 ratios were also consistent with an isotropic diffusion tensor and an overall τm of 25 ns. However, R2/R1 ratios for AT3 were consistent with an axially symmetric diffusion model with an anisotropic diffusion tensor, Diso = 2.00 ± 0.01 x 107 s−1, = 0.82 ± 0.04 (τm = 8.3 ns).
Fig. 10.
Amide 15N relaxation rates for trimeric and dodecameric AT reveal differences in overall correlation and local dynamics. Relaxation rates were measured at 600 MHz, 55 °C. Local correlation times were computed from the R2/R1 ratios. Secondary structures and location of metal ligands are indicated schematically above.
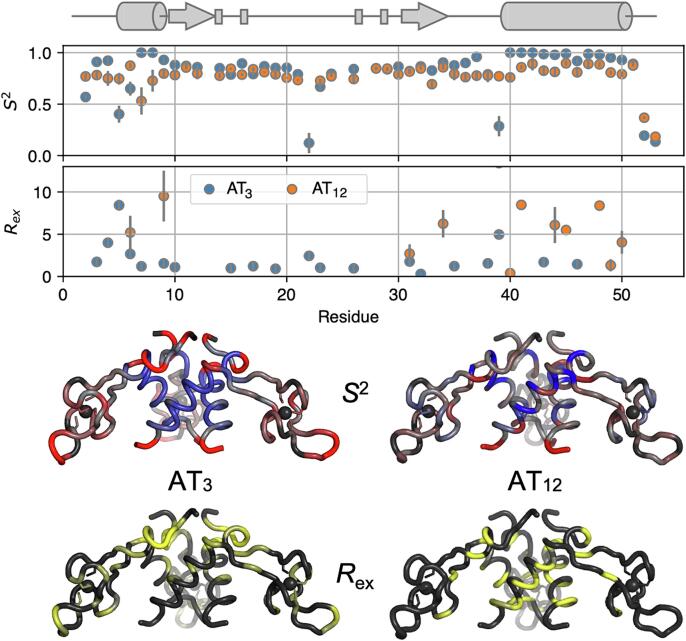
Large differences in the rotational diffusion tensors of AT12 and AT3 are accompanied with differences in internal motions. Amide 15N relaxation rates were analyzed using the extended model-free formalism (Clore et al., 1990, Lipari and Szabo, 1982a, Lipari and Szabo, 1982b) to compare the amplitude internal dynamics of AT3 and AT12 on the ps-ns timescale, as represented by the square of the generalized order parameter, S2 (Fig. 11). While both proteins exhibited generally high-order parameters, AT3 and AT12 were found to exhibit local differences in fast internal motions. High S2 values for AT3 imply rigidity of the C-terminal helical bundle, and larger amplitude fast motions in the zinc binding arm domains and the N-terminal helix. By contrast, the zinc binding domains in AT12 are relatively rigid on the ps-ns time scales reported by S2, likely a reflection of their packing onto adjacent zinc binding domains in the dodecamer (Fig. 2). The two oligomeric forms of AT also exhibit different patterns of the phenomenological exchange broadening term Rex, attributable to dynamics on slower µs-ms time scales (Clore et al., 1990, Mandel et al., 1995). For AT3, the residues with largest Rex terms are located at the N-terminus of the protomers, with smaller Rex terms at the zinc binding domains. For AT 12, almost the inverse is observed, with the largest exchange terms needed to fit data for the helical bundle.
Fig. 11.
Fitted Model-Free dynamics parameters suggest a stable helical bundle and flexible zinc-binding domains in AT3, but less variable dynamics in AT12. Top, fitted model free order parameters S2 and Rex for AT3 and AT12. Middle, fitted S2 values mapped to the backbone of the AT trimer using a linear color ramp from the mean value (grey) to + (blue) or – (red) two standard deviations from the mean. Bottom, Rex values mapped to the AT3 structure with a color ramp from 0 to ≥ 4/s (yellow).
Discussion
Tryptophan biosynthesis is highly regulated in bacteria (Gutiérrez-Preciado et al., 2007, Merino et al., 2008, Yanofsky, 2007). Within bacteria, TRAP-mediated transcription attenuation and translation repression via RNA binding represents an unusual regulatory mechanism. In the subset of organisms that also encode Anti-TRAP, its expression represents yet an additional regulatory layer by binding to and reversing the activity of an inhibitory protein. In this work we examined the equilibrium between active and inactive oligomeric states of Bsu Anti-TRAP using nMS, SAXS, AUC and NMR spectroscopy. We determined the solution structure of the active AT3 form of the protein and observed differences in its internal dynamics relative to that of the inactive form AT12. These studies thus help to inform our understanding of how AT inhibits RNA binding by Trp-bound TRAP, and of how the AT3 ↔ AT12 equilibrium may provide yet another regulatory layer.
In our studies of Bsu AT, serendipity played a significant role. The fortuitous requirement of a mature amino-methionine terminus for stabilizing the AT dodecamer coincided with experimental zinc supplementation of minimal media for recombinant protein expression, which likely prevented maturation of a sizeable fraction of the protein. The second residue in Bsu AT is a valine, which is often a poor substrate of methionine aminopeptidase (Frottin et al., 2006, Hirel et al., 1989), thus ensuring retention of Met1 in the mature protein. The application of NMR and MS enabled detection of the formylated N-terminus, while denaturing reversed-phase chromatography allowed separation of immature fAT from mature AT, which was not possible with conventional native purification. While the AT3 ↔ AT12 equilibrium is evident in samples of mature AT, the ability to prepare complexes of pure fAT provided an unambiguous means of establishing the role of the N-terminal amine in the equilibrium and enabling independent structural characterization of the TRAP-binding form of the protein (AT3).
We found that Bsu fAT3 (and AT12) yields NMR spectra of exceptional quality, enabling complete resonance assignments and high-resolution structure determination by application of NMR, symmetry and SAXS restraints. Symmetry restraints were found to strongly increase the precision of the structural ensemble, while SAXS and RDC restraints provided modest improvements. We found the solution structure of AT3 to be indistinguishable from that in the 12-mer crystal, or from the AF2 predicted model, indicating little conformational adjustment upon formation of the dodecamer.
While the average structure of the AT trimer in isolation and in the dodecamer are highly similar, they differ in their dynamics. Consistent with the overall size and shape of the oligomers, backbone 15N relaxation measurements revealed an isotropic rotational diffusion tensor for AT12 and axially symmetric diffusion for the smaller AT3. Model-free analysis of internal dynamics revealed flexible zinc ribbon motifs in AT3, suggesting a role for the dynamics of these motifs in engaging Trp-bound TRAP rings. AT12 has comparatively more rigid zinc-binding domains, likely reflecting packing constraints from the other trimers in the tetrahedral arrangement. Modest Rex terms in the N-terminal and zinc-binding domains of AT3 may reflect exchange dynamics on the µs-ms time scale, or potentially transient but unproductive AT3-AT3 association reactions, whereas the pattern of Rex terms is different in AT12, perhaps reflecting transient dissociation to the AT6 intermediate.
We conclude that the Bsu AT3-AT12 equilibrium is dependent on the near-physiological pKa of the Met1 amine, with AT12 favored by a protonated N-terminus and AT3 favored when unprotonated. Prior NMR observations (Sachleben et al., 2010) are corroborated here by nMS, SAXS, and AUC experiments as a function of pH. NMR spectra of fAT, over a pH range that alters the trimer-dodecamer equilibrium for mature AT, only show minor shift perturbations adjacent to His50, supporting the dominant role of the protonated N-terminus in stabilizing the dodecamer (Fig. 7). Native MS spectra (at lower protein concentration) reveal AT3 as the major species at pH 8, while at pH 6 nearly equal intensity is observed for ions arising from AT3 and AT12 species (Fig. 3); this provides some context to a prior nMS study which found exclusively AT12 when sprayed from pH-unadjusted ammonium acetate (Akashi et al., 2009, Watanabe et al., 2009), which can approach a value of 4.75 in the ESI plume (Konermann, 2017). SAXS data for mature AT recorded as a function of pH and protein concentration could be well described by considering varying ratios of AT3 and AT12 (Fig. 5). Experimental curves could be reproduced form a linear combination of the form factors computed from the crystallographic dodecamer and a trimer extracted from the dodecamer. This finding is consistent with the failure to observe other assembly intermediates under equilibrium conditions. Sedimentation velocity (AUC) experiments further corroborate the pH-dependent equilibrium between AT3 and AT12 and provide evidence for a short-lived AT6 intermediate. If we consider AT3 a “monomer” that can tetramerize, this behavior is consistent with biophysical characterization of other “tetramers”, which predominantly follow a monomer–dimer-tetramer oligomerization path (Powers and Powers, 2003). Small surface burial and minimal polar contacts in AT6, result in transient/weak dimerization; large surface burial and many polar contacts favor AT12 upon collision of two AT6 “dimers of trimers”.
Conclusion
The mechanism by which Anti-TRAP (AT) promotes Trp production by inhibiting RNA binding by tryptophan-activated TRAP remains poorly understood (Ihms et al., 2014, Snyder et al., 2004, Valbuzzi and Yanofsky, 2001, Watanabe et al., 2009). The capacity of AT to form stable dodecameric and trimeric structures, wherein only the trimeric form is competent for binding and inhibiting TRAP is suggestive of a regulatory role for that equilibrium. While the complementary biophysical studies reported here provide qualitatively consistent insights into the properties of AT and its mechanism of oligomerization and TRAP inhibition, further work would be required to establish quantitative consistency. Some sources of uncertainty include the effect of molecular tumbling on observed NMR signal intensities, of ionizable buffers and of protein charge on gas phase ionization and gas phase signal detection, and relatively low structural resolution of SAXS and sedimentation velocity methods. Those caveats aside, it is clear that in vitro AT exhibits a slow equilibrium between active AT3 and inactive AT12 states.
This slow association/dissociation behavior of AT could serve a regulatory function by kinetically modulating the response to changes in Trp concentration (Frieden, 2019). In this model (Fig. 1), TRAP is slow to release Trp upon decrease in its cellular concentration and remains active for RNA binding and inhibiting trp expression. Meanwhile, decreasing cellular Trp results in accumulation of uncharged tRNATrp and T-box mediated activation of AT expression. Expressed AT will trimerize and rapidly bind to RNA-free Trp-activated TRAP rings, preventing them from binding new trp RNA leaders. AT3 that do not find TRAP rings could instead dodecamerize, provided the cellular pH is near the pKa, resulting in slowly equilibrating populations of active AT3 and inactive AT12. This scenario would thus generate a pool of fast-acting AT3 and a reservoir of slowly activated AT12. The resulting steady-state equilibrium between AT3, AT12, AT3-Trp-TRAP11, Trp-TRAP11 and Trp-TRAP11-RNA would achieve a steady state of Trp homeostasis.
An alternative intriguing possibility is that the pH-dependent AT3-AT12 equilibrium serves to regulate gene expression in response to changes in intracellular pH. AT is not known to be associated with bacterial response to changes in pH, but sporulation by B. subtillis is a consequence of changes in gene expression patterns and is associated with decreases in cell volume and pH that would likely affect the AT3-AT12 equilibrium. The notion that pH-driven changes in AT oligomeric state could serve as a mechanism for sensing and responding to such environmental changes is an intriguing possibility that merits further exploration (Errington, 1993, Magill et al., 1994).
The properties of the AT3 “triskelion”, with its polydentate configuration of flexible zinc-binding arms, seem uniquely suited for capturing one or multiple TRAP rings, and thereby preventing RNA wrapping. The use of the amino terminus to mediate oligomerization represents a uniquely tuned equilibrium since the pKa and association constants are coupled, such that high protein concentration would shift the pKa to favor the protonated state, and thus the oligomer. It remains unclear why some species of Bacilli evolved to produce AT as an extra layer of regulation (e.g., Bsu, Bli), while others did not (e.g., Bst, Bha). This study highlights the lengths to which organisms have evolved sophisticated mechanisms for metabolic control, and the power of complementary biophysical measurements to describe those properties that enable regulated function.
Materials and methods
Growth and purification of B. subtilis Anti-TRAP
AT was recombinantly expressed from E. coli from an engineered codon-optimized expression vector (Sachleben et al., 2010). B. subtilis Anti-TRAP is a 53 amino acid polypeptide, with the sequence: MVIATDDLEV ACPKCERAGE IEGTPCPACS GKGVILTAQG YTLLDFIQKH LNK. It has a molecular weight of 5649.57 Da and its predicted extinction coefficient at 280 nm is 1490 M−1 cm−1 in its reduced form (Expasy ProtParam). Uniformly N-terminal formylmethionine Anti-TRAP (ƒAT) and deformylated AT were purified as previously described (Sachleben et al., 2010). Briefly, transformed BL21(DE3) cells grown and induced in modified M9 minimal media were lysed and cellular debris pelleted by centrifugation. Isotope labeling was achieved by replacing unlabeled ammonium chloride with 15N ammonium chloride for 15N labeling, by substitution of 13C glucose for 13C/15N double labeling. Heating of the cell lysate supernatant for 15 min at 70 °C and subsequent centrifugation removed most native E. coli proteins. The resulting supernatant was loaded onto Q-sepharose beads (GE healthcare) and eluted via a NaCl gradient. Eluted Anti-TRAP was denatured by addition of 6 M guanidinium and subjected to reverse-phase HPLC purification. AT (i.e., deformyl) and ƒAT elute at sufficiently resolved retention times from a C4 column to be separated and lyophilized, providing > 99 % desired species. Subsequent refolding results in intact ƒAT trimers for ƒAT, and AT3 + AT12 in equilibrium for (deformylated) AT.
Mass spectrometry
The theoretical mass of AT (from ExPASy ProtParam): Monomer (without Zn) = 5,649.57 Da; monomer with Zn (65.38 Da) = 5714.95 Da; trimer with three Zn = 17,144.85 Da; dodecamer with twelve Zn = 68,579.40 Da. AT samples were buffer exchanged twice into 100 mM ammonium acetate at 3 different pH’s (pH 6.8, unadjusted; pH 8, adjusted using ammonium hydroxide; pH 6, adjusted using acetic acid) using size-exclusion spin columns (MicroBioSpin-6; Bio- Rad); the final concentration of AT was 10 μM. Borosilicate glass capillary emitters were pulled in-house with a Sutter Flaming/Brown Micropipette puller P97; samples were loaded and sprayed using nanoESI. Mass spectra were recorded on a Waters Synapt G2S mass spectrometer modified in-house with a SID device located between the trap cell and the drift cell. Data were recorded with capillary voltage, 0.7–1.0 kV; cone voltage, 20 V; extraction cone, 5–8 V; helium gas flow, 120 mL/min; trap gas flow, 2 mL/min; ion mobility nitrogen gas flow, 60 mL/min; source temperature 20 °C; trap wave velocity, 160 m/s; trap wave height, 4 V; ion mobility wave velocity, 300 m/s; ion mobility wave height, 20 V; transfer wave velocity, 65 m/s; transfer wave height, 2 V; time-of-flight analyzer pressure, ∼ 6 × 10−7 mbar; and backing pressure, ∼ 2.5–3 mbar.
NMR assignments of AT12
AT12 predominates at high protein concentration, pH 7 for the mature, deformylated protein. Two dimensional 1H-15N TROSY-HSQC spectra of AT12 have approximately the number of peaks expected for the protomer, consistent with its molecular symmetry. HNCA, HNCOCA, HNCACB, and HNCO triple resonance spectra recorded at 328 K using water flip-back solvent suppression, sensitivity enhancement, and transverse relaxation optimized spectroscopy (TROSY) coherence selection (Loria et al., 1999, Pervushin et al., 1997, Rance et al., 1999, Salzmann et al., 1999, Salzmann et al., 1998) on uniformly doubly labeled (13C, 15N) AT, 8 mM in 20 mM Tris pH 7.8 (at 298K), 250 mM NaCl, 300 μM ZnCl2, 2 mM TCEP [tris(2-carboxyethyl)phosphine], and 0.02% (wt/vol) NaN3. allowed assignment of every backbone resonance in the spectrum, corresponding to 48 of the 50 non-proline residues (only M1 and E22 were missing).
NMR assignments and structural restraints of AT3
fAT spectra exhibit signals exclusively attributable to trimeric AT. NMR spectra of fAT were recorded on a 3 mM sample in 20 mM d11-Tris pH 7.5, 20 mM β-mercaptoethanol, 1 mM ZnCl2 and 0.02% (wt/vol) NaN3 The NMR spectra of fAT3 also exhibit the number of peaks expected for the monomer, consistent with its molecular symmetry. Assignments of uniformly, doubly labeled (13C, 15N) trimeric AT were obtained using HNCO, HNCACB, CCONH-TOCSY, HCCONH-TOCSY, HCCH-COSY, HCCH-TOCSY, 15N-edited TOCSY, constant-time 13C-edited HSQC, 13C-edited HSQC, constant-time 13C-edited HSQC of the aromatic region, 13C-edited HSQC of the aromatic region, and a 1H–1H plane and 1H–13C plane of the HCCH-COSY of the aromatic region (Cavanagh et al., 2006).
Torsion angle restraints for the backbone φ angles were obtained from the analysis of 3D HNHA spectra (Bax et al., 1994) and additional backbone torsion angle φ, ψ restraints were obtained from chemical shifts using TALOS (Cornilescu et al., 1999). Sidechain χ1 torsion angle restraints were obtained from the HNHB and HACAHB-COSY experiments (Bax et al., 1994). The X-Pro amide bonds for AT3 were determined to be in the trans conformation from analysis of the Cβ and Cγ chemical shifts (Schubert et al., 2002); we did not assign Cγ resonances for AT12 but all amides are modeled as trans in the crystal structure (Shevtsov et al., 2005). One bond 1DHN-N and 1DN-C residual dipolar coupling (RDC) restraints were obtained from IPAP spectra (Ottiger et al., 1998, Permi, 2003), of 13C/15N-labeled AT aligned with 20 mg/ml Pf1 phage (ASLA Biotech). NOE restraints were obtained from a 13C/15N-edited time-shared NOESY (Sattler et al., 1995) in H2O and a 13C-edited NOESY (Cavanagh et al., 2006) in 2H2O.
NOE assignments and initial AT3 structure determination
NOE assignments of the symmetrical homo-trimer were made using the symmetry ambiguous distance restraint (symmetry ADR) method starting with a well-defined monomer (Kovacs et al., 2002, O’Donoghue et al., 2000). The structure of the protomers were first obtained by assigning NOEs presumed to be intramolecular, (i.e., NOEs between zinc-coordinating coordinating cysteines, secondary structural elements inferred from secondary structure predictions based on sequence and chemical shift data), and using these as input for restrained torsion-angle refinement with CNS (Brunger, 2007). The set of 20 lowest energy structures (of 35 total) was then used with a 10 Å cutoff to filter through the remaining NOE data. At this stage, NOEs with less than six ambiguous assignments were used for another round of structure refinement and the cycle was iterated while decreasing the distance cutoff by 2 Å until the cutoff was 6 Å and the monomer structure converged to an RMSD of 2.08 Å for backbone atoms. The trimer structure was then calculated by ambiguously assigning any NOEs that could not be satisfied intramolecularly (i.e., they were defined as being between protomers A, B and C) and a structure calculation was performed utilizing non-crystallographic symmetry restraints in addition to the distance, orientation, and torsion angle restraints. This set of structures was then used with a 6 Å filter to assign the remaining NOEs and sort out the interprotomer contacts.
SAXS and RDC refinement of AT3
To obtain a scattering profile for the trimer, pure ƒAT was purified, refolded and dialyzed against 50 mM sodium-phosphate, pH 8, 100 mM NaCl, 200 µM ZnCl2 and 1 mM TCEP. SAXS profiles at 1, 2, 3.3, 5, and 10 mg/mL ƒAT (monomer) were acquired at the SIBYLS beamline at the Advanced Light Source in Berkeley, California. Data were obtained at a Q spacing range of 0.01 to 0.32 Å-1, and at 0.5, 1, and 6 s exposure times. The scattering contribution from the buffer was removed by subtracting the scattering profile of the dialysis solution. Processing, validation, and analysis of SAXS data was performed with the ATSAS package (Manalastas-Cantos et al., 2021) and custom scripts. For restraints to the AT3 structure, the 6 s exposure profile of the 3.3 mg/mL sample was used, as a small concentration-dependent amount of dodecamer (<3% at 10 mg/mL) was detectable from deconvolution of the higher concentrations.
Isotropic and anisotropic IPAP spectra for RDC data were collected in 50 mM sodium-phosphate pH 6.6, 2 mM DTT at 298 K in the absence and presence of 20 mg/mL Pf1 phage as the alignment medium. The lowest energy initial trimer structure (from above) was used as the starting point for simultaneous refinement against NOE, dihedral, RDC, and SAXS restraints in Xplor-NIH (Schwieters et al., 2018). Agreement between measurements and values calculated from the structure was assessed using the DC server (https://spin.niddk.nih.gov/bax/nmrserver/dc/). Following refinement, the AT3 structure ensemble was validated using protein structure validation server (PSVS) (Bhattacharya et al., 2007) and Procheck-NMR (Laskowski et al., 1996). Final refinement and validation statistics are provided in Table 1.
NMR relaxation data
The 15N relaxation data were recorded on a Bruker DRX 600 MHz spectrometer at 328 K using water flip-back and 3–9-19 WATERGATE solvent suppression, sensitivity improvement, and gradient coherence selection (Cavanagh et al., 2006). All relaxation data were recorded with a spectral width of 8389.26 Hz sampled over 1024 complex points in ω2 (1H) and a spectral width of 1763.75 Hz sampled over 64 complex points in ω1 (15N), with 16 scans per transient except for the NOE which was collected with 64 scans per transient, and with a recovery delay between transients of 3 s. The recovery delays for R1 measurements were (100, 200, 400, 800, 1200, 1600, 2000, 2400 ms for AT12 and 10, 100, 250, 400, 550, 700, 850, 1250, and 2000 ms for AT3); relaxation delays for R2 measurements were (0, 9, 16.2, 23.4, 30.6, 37.8, 41.4, 45, 52.2, and 59.4 ms for AT12 and 0, 17, 33.9, 50.9, 67.8, 84.8, 101.8, 135.7, 169.6 ms for AT3). The NOE values were determined as the ratio of the peak intensities from two interleaved spectra recorded with and without a 3 s 1H presaturation period consisting of a series of 120° pulses separated by 5 ms delays. Relaxation rates were obtained by non-linear least-squares fitting measured peak heights with mono-exponential functions; uncertainties in peak heights were estimated from the standard deviation of the noise.
Rotational diffusion
Overall rotational correlation times, τm, and rotational diffusion tensors were determined experimentally from R2/R1 ratios (Lee et al., 1997) utilizing only R2/R1 ratios for residues with relaxation rates within one standard deviation of the 10 % trimmed mean value. These calculations for AT12 showed no statistical significance for fitting the NMR data to an axially symmetric or fully anisotropic diffusion tensor over an isotropic model. Therefore, an isotropic diffusion tensor with a τm of 25 ns was used for the model-free calculations of the dodecamer. This was supported by calculations of the hydrodynamic parameters by HYDRONMR 7.C (Bernadó et al., 2002, García De La Torre et al., 2000, García de la Torre et al., 2000) based on the dodecameric structure 2BX9 that also predict nearly isotropic tumbling for AT12; the crystallographic coordinates were protonated using PDB2PQR (Jurrus et al., 2018). The calculations based on the R2/R1 ratios of AT3, however, indicated a statistically significant improvement for the axially symmetric model over the isotropic model. This was similarly supported by hydrodynamic calculations that yielded an anisotropic diffusion tensor with two similar and one unique axis. Therefore, an axially symmetric diffusion tensor from the R2/R1 ratios was used for the model-free calculations of AT3. Validation of the resulting structures from the R2/R1 ratios was performed with HYDRONMR 7.C, using a solvent viscosity of 0.005P, T = 328 K, 1.02 Å N-H bond distance, CSA of −160 ppm and 14.09 T field. Diffusion tensors were computed for the symmetric trimers and T1/T2 ratios were analyzed for the first chain (Fig. 9).
Model-free analysis
Model-free analysis for both AT3 and AT12 (Clore et al., 1990) was performed using the program TENSOR v. 2 (Dosset et al., 2000), which was recompiled to allow for input of up to 1000 residues and a CSA of −172 ppm (Lee and Wand, 1999). Five motional models with up to three parameters describing the internal motions were used: (1) S2 (2) S2 and te (3) S2 and Rex (4) S2, te, and Rex (5) S2, S2f, and te. Standard errors in the model-free parameters were obtained from Monte Carlo simulations using 1000 samples. Model fitting was performed using TENSOR v. 2 via a modified step-up hypothesis testing method (Mandel et al., 1995).
Observation of the pH-dependent AT3-AT12 equilibrium via SAXS
The program CRYSOL (Svergun et al., 1995) was used to generate predicted scattering profiles AT3 structure, the B. subtilis AT dodecamer retrieved from the RCSB (3BX9), and the B. licheniformis high-pH “inverted” dodecameric isoform (3LD0). Given the structural similarity of the AT trimer component between B. subtilis and B. licheniformis at nominal SAXS resolution (10–20 Å), the B. licheniformis dodecamer coordinates were used without modification. All three profiles were normalized to the square of the input structure’s molecular weight. The solvent density and contrast of the hydration shell were determined by fitting the scattering of the AT trimer structure against an empirical scattering profile of 5 mg/mL fAT in CRYSOL. The empirical scattering data from mature AT at 16 different pH and concentration conditions was fit with the program OLIGOMER (Konarev et al., 2003), using the three predicted scattering profiles. An additional systematic term was also used to correct for baseline offset arising from incomplete buffer subtraction/mismatch.
Analytical ultracentrifugation
AT stock solutions were split and dialyzed against sample buffers at pH 7, 7.25, 7.5, and 8, consisting of 50 mM sodium phosphate, 100 mM NaCl, 0.5 mM tryptophan, and 0.02 % (w/v) NaN3. Sedimentation samples were subsequently made from the concentrated stock solutions with the corresponding dialysis buffer and allowed to equilibrate overnight. To account for the decreasing self-affinity at increasing pH, samples at pH 7–7.25 were made to 250, 125, and 65 µM AT monomer concentration, and at pH 7.5–8, to 500, 250, and 125 µM. Dilutions were performed on a gravimetric balance accurate to 0.2 mg, with an assumed solution density of 1.0 g/ml. Sedimentation velocity data were obtained at 25 °C using a Beckman Coulter ProteomeLab XL-I ultracentrifuge equipped with an 8-position rotor and double-sector cells with sapphire windows. Matched dialysis buffer was present in the reference sector. Data were recorded using interference optics with a scan interval of one minute and 200 scans per cell at a rotor speed of 50,000 rpm. The position of the meniscus and cell bottom were determined for each cell by fitting a sedimentation coefficient distribution and uniform frictional value f/f0 in SEDFIT (Schuck, 2013) at a sampling interval of 10 to 20 points per Svedberg. Time and radial invariant noise were removed from each interference scan. The buffer density ρ and viscosity η under these conditions were calculated by SEDNTRP (Philo, 2023) to be 1.01260 g/mol and 1.0520 cP, respectively.
Determination of kinetic parameters for the AT3 ↔ AT6 ↔ AT12 model in SEDPHAT (Zhao et al., 2015) was performed by globally fitting 200 interference scans from each set of AT concentrations at one pH (600 scans total). In addition to model kinetic parameters, the fringe shift coefficient, apparent sedimentation coefficients for AT3, AT6, and AT12, as well as the molar mass for the trimer were also optimized. pH 8 data were fit first, using the HYDROPRO (García De La Torre et al., 2000, García de la Torre et al., 2000) -predicted sedimentation coefficients of 2.1 S and 4.1 S at 25 °C for AT3 and AT12, respectively, as starting sedimentation coefficients. The AT6 starting sedimentation coefficient was set to 3 S. Initial parameters were 105 M−1 for k1, 10-3 s−1 for k-1, 106 M−1 for k2, and 10-4 s−1 for k-2, as these provided reasonable predicted interference curves. The apparent molar weight of AT in the experiment was also optimized, using the expected mass of the trimer as a starting value. A reasonable starting point for the molar absorptivity was determined by manual iterative evaluations of the model using different values until a fringe shift of appropriate magnitude was achieved. (Uncertainty analysis in the kinetic parameters was attempted through Monte-Carlo estimation of errors, but after 1000 evaluations no variance in the fitted parameters was detected.).
Interfaces
Solvent accessible surface (SAS) area burial between protomers was computed using PyMOL 2.5.2 (Schrödinger, LLC) using a dot density of 3. Burial was computed from difference between the SAS of components and their complex, using PyMOL get_area. SAS burial for folding of the protomer was estimated from multiple conformations using PROSA (Estrada et al., 2009). Similar interface and surface areas were obtained using PISA (Krissinel and Henrick, 2007).
Coordinates of fAT3 have been deposited to the RCSB, accession 9CCH, and NMR data, including assignments and restraints obtained 328 K deposited to the BMRB, accession 16492. Raw data, including NMR spectra, SAXS curves and sedimentation velocity data are being deposited to BMRBig, accession (bmrbig100).
CRediT authorship contribution statement
Craig A. McElroy: Writing – original draft, Visualization, Validation, Software, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Elihu C. Ihms: . Deepak Kumar Yadav: Writing – original draft, Visualization, Validation, Investigation, Formal analysis, Data curation. Melody L. Holmquist: Writing – original draft, Visualization, Validation, Methodology, Formal analysis, Data curation. Vibhuti Wadwha: Investigation, Formal analysis, Data curation. Vicki H. Wysocki: Writing – review & editing, Supervision, Resources, Methodology, Funding acquisition, Data curation. Paul Gollnick: Supervision, Resources, Project administration, Investigation, Funding acquisition, Conceptualization. Mark P. Foster: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Mark P Foster reports financial support was provided by National Institutes of Health. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by grants from the NIH to MPF, PG and VW (GM077234, GM062750, GM120923, P41 GM 128577). SAXS data were collected at the SIBYLS beamline at the Advanced Light Source (ALS), a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the Department of Energy, Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies (IDAT) program, supported by DOE Office of Biological and Environmental Research. Additional support comes from the National Institute of Health project ALS-ENABLE (P30 GM124169) and a High-End Instrumentation Grant S10OD018483. This work made use of NMRBOX, which is supported by NIH grant P41 GM111135. CAM was supported in part by NIH T32 GM008512, and MH was supported in part by NIH T32 GM141955. Charles Cottrell (deceased) and Chunhua Yuan (CCIC) provided support for data collection experiments, Charles D. Schwieters for help with structure calculations, and the members of the Foster laboratory for helpful discussions.
Data availability
Data will be made available on request.
References
- Akashi S., Watanabe M., Heddle J.G., Unzai S., Park S.-Y., Tame J.R.H. RNA and Protein Complexes of trp RNA-Binding Attenuation Protein Characterized by Mass Spectrometry. Anal. Chem. 2009;81:2218–2226. doi: 10.1021/ac802354j. [DOI] [PubMed] [Google Scholar]
- Babitzke P., Stults J.T., Shire S.J., Yanofsky C. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J. Biol. Chem. 1994;269:16597–16604. [PubMed] [Google Scholar]
- Babitzke P., Yanofsky C. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc. Natl. Acad. Sci. U. S. A. 1993;90:133–137. doi: 10.1073/pnas.90.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbato G., Ikura M., Kay L.E., Pastor R.W., Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- Bax A., Vuister G.W., Grzesiek S., Delaglio F., Wang A.C., Tschudin R., Zhu G. Measurement of homo- and heteronuclear J couplings from quantitative J correlation. Methods Enzym. 1994;239:79–105. doi: 10.1016/s0076-6879(94)39004-5. [DOI] [PubMed] [Google Scholar]
- Bayfield O.W., Chen C.-S., Patterson A.R., Luan W., Smits C., Gollnick P., Antson A.A. Trp RNA-Binding Attenuation Protein: Modifying Symmetry and Stability of a Circular Oligomer. PLoS ONE. 2012;7:e44309. doi: 10.1371/journal.pone.0044309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernadó P., García de la Torre J., Pons M. Interpretation of 15N NMR relaxation data of globular proteins using hydrodynamic calculations with HYDRONMR. J. Biomol. NMR. 2002;23:139–150. doi: 10.1023/a:1016359412284. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A., Tejero R., Montelione G.T. Evaluating protein structures determined by structural genomics consortia. Proteins Struct. Funct. Bioinforma. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- Boyken S.E., Benhaim M.A., Busch F., Jia M., Bick M.J., Choi H., Klima J.C., Chen Z., Walkey C., Mileant A., Sahasrabuddhe A., Wei K.Y., Hodge E.A., Byron S., Quijano-Rubio A., Sankaran B., King N.P., Lippincott-Schwartz J., Wysocki V.H., Lee K.K., Baker D. De novo design of tunable, pH-driven conformational changes. Science. 2019;364:658–664. doi: 10.1126/science.aav7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam C.A. Using Lamm-Equation modeling of sedimentation velocity data to determine the kinetic and thermodynamic properties of macromolecular interactions. Methods, Modern Analytical Ultracentrifugation. 2011;54:4–15. doi: 10.1016/j.ymeth.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Cavanagh J., Fairbrother W.J., Iii A.G.P., Skelton N.J., Rance M. Second ed. Academic Press; Principles and Practice: 2006. Protein NMR Spectroscopy. [Google Scholar]
- Chen C.-S., Smits C., Dodson G.G., Shevtsov M.B., Merlino N., Gollnick P., Antson A.A. How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit TRAP Suggests a General Mechanism. PLOS ONE. 2011;6:e25296. doi: 10.1371/journal.pone.0025296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Yanofsky C. Tandem transcription and translation regulatory sensing of uncharged tryptophan tRNA. Science. 2003;301:211–213. doi: 10.1126/science.1084902. [DOI] [PubMed] [Google Scholar]
- Chen G., Yanofsky C. Features of a leader peptide coding region that regulate translation initiation for the anti-TRAP protein of B. subtilis. Mol. Cell. 2004;13:703–711. doi: 10.1016/s1097-2765(04)00085-1. [DOI] [PubMed] [Google Scholar]
- Clore G.M., Szabo A., Bax A., Kay L.E., Driscoll P.C., Gronenborn A.M. Deviations from the simple two-parameter model-free approach to the interpretation of nitrogen-15 nuclear magnetic relaxation of proteins. J. Am. Chem. Soc. 1990;112:4989–4991. doi: 10.1021/ja00168a070. [DOI] [Google Scholar]
- Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Dosset P., Hus J.C., Blackledge M., Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- Du H., Babitzke P. trp RNA-binding attenuation protein-mediated long distance RNA refolding regulates translation of trpE in Bacillus subtilis. J. Biol. Chem. 1998;273:20494–20503. doi: 10.1074/jbc.273.32.20494. [DOI] [PubMed] [Google Scholar]
- Du H., Tarpey R., Babitzke P. The trp RNA-binding attenuation protein regulates TrpG synthesis by binding to the trpG ribosome binding site of Bacillus subtilis. J. Bacteriol. 1997;179:2582–2586. doi: 10.1128/jb.179.8.2582-2586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada J., Bernadó P., Blackledge M., Sancho J. ProtSA: a web application for calculating sequence specific protein solvent accessibilities in the unfolded ensemble. BMC Bioinformat. 2009;10:104. doi: 10.1186/1471-2105-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow N.A., Muhandiram R., Singer A.U., Pascal S.M., Kay C.M., Gish G., Shoelson S.E., Pawson T., Forman-Kay J.D., Kay L.E. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Frieden C. Protein oligomerization as a metabolic control mechanism: Application to apoE. Protein Sci. 2019;28:837–842. doi: 10.1002/pro.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frottin F., Martinez A., Peynot P., Mitra S., Holz R.C., Giglione C., Meinnel T. The proteomics of N-terminal methionine cleavage. Mol. Cell. Proteomics MCP. 2006;5:2336–2349. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- García de la Torre J., Huertas M.L., Carrasco B. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. San Diego Calif. 2000;1997(147):138–146. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
- García De La Torre J., Huertas M.L., Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollnick P. Regulation of the Bacillus subtilis trp operon by an RNA-binding protein. Mol. Microbiol. 1994;11:991–997. doi: 10.1111/j.1365-2958.1994.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Gollnick P., Ishino S., Kuroda M.I., Henner D.J., Yanofsky C. The mtr locus is a two-gene operon required for transcription attenuation in the trp operon of Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 1990;87:8726–8730. doi: 10.1073/pnas.87.22.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Preciado A., Yanofsky C., Merino E. Comparison of tryptophan biosynthetic operon regulation in different Gram-positive bacterial species. Trends Genet. 2007;23:422–427. doi: 10.1016/j.tig.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Preciado A., Henkin T.M., Grundy F.J., Yanofsky C., Merino E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol. Mol. Biol. Rev. MMBR. 2009;73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner D.J., Band L., Shimotsu H. Nucleotide sequence of the Bacillus subtilis tryptophan operon. Gene. 1985;34:169–177. doi: 10.1016/0378-1119(85)90125-8. [DOI] [PubMed] [Google Scholar]
- Hirel P.H., Schmitter M.J., Dessen P., Fayat G., Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihms E.C., Zhou M., Zhang Y., Kleckner I.R., McElroy C.A., Wysocki V.H., Gollnick P., Foster M.P. Gene regulation by substoichiometric heterocomplex formation of undecameric TRAP and trimeric anti-TRAP. Proc. Natl. Acad. Sci. 2014;111:3442–3447. doi: 10.1073/pnas.1315281111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Hao B., Liu R., Chan M.K. Structures of E. coli Peptide Deformylase Bound to Formate: Insight into the Preference for Fe2+ over Zn2+ as the Active Site Metal. J. Am. Chem. Soc. 2005;127:4558–4559. doi: 10.1021/ja0503074. [DOI] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurrus E., Engel D., Star K., Monson K., Brandi J., Felberg L.E., Brookes D.H., Wilson L., Chen J., Liles K., Chun M., Li P., Gohara D.W., Dolinsky T., Konecny R., Koes D.R., Nielsen J.E., Head-Gordon T., Geng W., Krasny R., Wei G.-W., Holst M.J., McCammon J.A., Baker N.A. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018;27:112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay L.E., Torchia D.A., Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- Kleckner I.R., Gollnick P., Foster M.P. Mechanisms of allosteric gene regulation by NMR quantification of microsecond-millisecond protein dynamics. J. Mol. Biol. 2012;415:372–381. doi: 10.1016/j.jmb.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. doi: 10.1107/S0021889803012779. [DOI] [Google Scholar]
- Konermann L. Addressing a Common Misconception: Ammonium Acetate as Neutral pH “Buffer” for Native Electrospray Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2017;28:1827–1835. doi: 10.1007/s13361-017-1739-3. [DOI] [PubMed] [Google Scholar]
- Kovacs H., O’Ddonoghue S.I., Hoppe H.-J., Comfort D., Reid K.B.M., Campbell Lain D., Nilges M. Solution structure of the coiled-coil trimerization domain from lung surfactant protein D. J. Biomol. NMR. 2002;24:89–102. doi: 10.1023/a:1020980006628. [DOI] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Lee L.K., Rance M., Chazin W.J., Palmer A.G. Rotational diffusion anisotropy of proteins from simultaneous analysis of 15N and 13C alpha nuclear spin relaxation. J. Biomol. NMR. 1997;9:287–298. doi: 10.1023/a:1018631009583. [DOI] [PubMed] [Google Scholar]
- Lee A.I., Sarsero J.P., Yanofsky C. A temperature-sensitive trpS mutation interferes with trp RNA-binding attenuation protein (TRAP) regulation of trp gene expression in Bacillus subtilis. J. Bacteriol. 1996;178:6518–6524. doi: 10.1128/jb.178.22.6518-6524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.L., Wand A.J. Assessing potential bias in the determination of rotational correlation times of proteins by NMR relaxation. J. Biomol. NMR. 1999;13:101–112. doi: 10.1023/a:1008304220445. [DOI] [PubMed] [Google Scholar]
- Lipari G., Szabo A. A Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- Lipari G., Szabo A. A model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc. 1982;104:4559–4570. [Google Scholar]
- Loria J.P., Rance M., Palmer A.G. Transverse-relaxation-optimized (TROSY) gradient-enhanced triple-resonance NMR spectroscopy. J. Magn. Reson. San Diego Calif. 1999;1997(141):180–184. doi: 10.1006/jmre.1999.1891. [DOI] [PubMed] [Google Scholar]
- Magill N.G., Cowan A.E., Koppel D.E., Setlow P. The internal pH of the forespore compartment of Bacillus megaterium decreases by about 1 pH unit during sporulation. J. Bacteriol. 1994;176:2252–2258. doi: 10.1128/jb.176.8.2252-2258.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manalastas-Cantos K., Konarev P.V., Hajizadeh N.R., Kikhney A.G., Petoukhov M.V., Molodenskiy D.S., Panjkovich A., Mertens H.D.T., Gruzinov A., Borges C., Jeffries C.M., Svergun D.I., Franke D. ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr. 2021;54:343–355. doi: 10.1107/S1600576720013412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel A.M., Akke M., Palmer A.G., 3rd Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- McAdams N.M., Gollnick P. The Bacillus subtilis TRAP Protein Can Induce Transcription Termination in the Leader Region of the Tryptophan Biosynthetic (trp) Operon Independent of the trp Attenuator RNA. PLoS ONE. 2014;9:e88097. doi: 10.1371/journal.pone.0088097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCammon M.G., Hernández H., Sobott F., Robinson C.V. Tandem Mass Spectrometry Defines the Stoichiometry and Quaternary Structural Arrangement of Tryptophan Molecules in the Multiprotein Complex TRAP. J. Am. Chem. Soc. 2004;126:5950–5951. doi: 10.1021/ja0317170. [DOI] [PubMed] [Google Scholar]
- McElroy C.A. The Ohio State University; 2005. Dynamics and function: mechanistic studies of the gene regulatory proteins TRAP and anti-TRAP. [Google Scholar]
- McElroy C., Manfredo A., Wendt A., Gollnick P., Foster M. TROSY-NMR Studies of the 91kDa TRAP Protein Reveal Allosteric Control of a Gene Regulatory Protein by Ligand-altered Flexibility. J. Mol. Biol. 2002;323:463–473. doi: 10.1016/S0022-2836(02)00940-3. [DOI] [PubMed] [Google Scholar]
- Merino E., Babitzke P., Yanofsky C. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J. Bacteriol. 1995;177:6362–6370. doi: 10.1128/jb.177.22.6362-6370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino E., Jensen R.A., Yanofsky C. Evolution of bacterial trp operons and their regulation. Curr. Opin. Microbiol. 2008;11:78–86. doi: 10.1016/j.mib.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue S.I., Chang X., Abseher R., Nilges M., Led J.J. Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure. J Biomol NMR. 2000;16:93–108. doi: 10.1023/a:1008323819099. [DOI] [PubMed] [Google Scholar]
- Ottiger M., Delaglio F., Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- Permi P. Measurement of residual dipolar couplings from 1Halpha to 13Calpha and 15N using a simple HNCA-based experiment. J Biomol NMR. 2003;27:341–349. doi: 10.1023/a:1025866606252. [DOI] [PubMed] [Google Scholar]
- Pervushin K., Riek R., Wider G., Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo, J.S., 2023. SEDNTERP: a calculation and database utility to aid interpretation of analytical ultracentrifugation and light scattering data. Eur. Biophys. J. EBJ. https://doi.org/10.1007/s00249-023-01629-0. [DOI] [PubMed]
- Potter K.D., Merlino N.M., Jacobs T., Gollnick P. TRAP binding to the Bacillus subtilis trp leader region RNA causes efficient transcription termination at a weak intrinsic terminator. Nucleic Acids Res. 2011;39:2092–2102. doi: 10.1093/nar/gkq965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers E.T., Powers D.L. A Perspective on Mechanisms of Protein Tetramer Formation. Biophys. J. 2003;85:3587–3599. doi: 10.1016/S0006-3495(03)74777-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa S., Blanquet S., Meinnel T. Control of peptide deformylase activity by metal cations 11Edited by A. R. Fersht. J. Mol. Biol. 1998;280:515–523. doi: 10.1006/jmbi.1998.1883. [DOI] [PubMed] [Google Scholar]
- Rance M., Loria J.P., Palmer AG3rd, null Sensitivity improvement of transverse relaxation-optimized spectroscopy. J. Magn. Reson. San Diego Calif. 1999;1997(136):92–101. doi: 10.1006/jmre.1998.1626. [DOI] [PubMed] [Google Scholar]
- Sachleben J.R., McElroy C.A., Gollnick P., Foster M.P. Mechanism for pH-dependent gene regulation by amino-terminus-mediated homooligomerization of Bacillus subtilis anti-trp RNA-binding attenuation protein. Proc. Natl. Acad. Sci. u. s. a. 2010;107:15385–15390. doi: 10.1073/pnas.1004981107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann M., Pervushin K., Wider G., Senn H., Wuthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci U A. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann M., Wider G., Pervushin K., Wuthrich K. Improved sensitivity and coherence selection for [15N,1H]-TROSY elements in triple resonance experiments. J Biomol NMR. 1999;15:181–184. doi: 10.1023/a:1008358030477. [DOI] [PubMed] [Google Scholar]
- Sarsero J.P., Merino E., Yanofsky C. A Bacillus subtilis operon containing genes of unknown function senses tRNATrp charging and regulates expression of the genes of tryptophan biosynthesis. Proc Natl Acad Sci U A. 2000;97:2656–2661. doi: 10.1073/pnas.050578997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M., Maurer M., Schleucher J., Griesinger C. A simultaneous (15)N, (1)H- and (13)C, (1)H-HSQC with sensitivity enhancement and a heteronuclear gradient echo. J. Biomol. NMR. 1995;5:97–102. doi: 10.1007/BF00227475. [DOI] [PubMed] [Google Scholar]
- Schaak J.E., Yakhnin H., Bevilacqua P.C., Babitzke P. A Mg2+-dependent RNA tertiary structure forms in the Bacillus subtilis trp operon leader transcript and appears to interfere with trpE translation control by inhibiting TRAP binding. J. Mol. Biol. 2003;332:555–574. doi: 10.1016/s0022-2836(03)00969-0. [DOI] [PubMed] [Google Scholar]
- Schubert M., Labudde D., Oschkinat H., Schmieder P. A software tool for the prediction of Xaa-Pro peptide bond conformations in proteins based on 13C chemical shift statistics. J. Biomol. NMR. 2002;24:149–154. doi: 10.1023/a:1020997118364. [DOI] [PubMed] [Google Scholar]
- Schuck P. Analytical Ultracentrifugation as a Tool for Studying Protein Interactions. Biophys. Rev. 2013;5:159–171. doi: 10.1007/s12551-013-0106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters C.D., Bermejo G.A., Clore G.M. Xplor-NIH for molecular structure determination from NMR and other data sources. Protein Sci. 2018;27:26–40. doi: 10.1002/pro.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sereda T.J., Mant C.T., Quinn A.M., Hodges R.S. Effect of the α-amino group on peptide retention behaviour in reversed-phase chromatography Determination of the pKa values of the α-amino group of 19 different N-terminal amino acid residues. J. Chromatogr. A. 1993;646:17–30. doi: 10.1016/S0021-9673(99)87003-4. [DOI] [PubMed] [Google Scholar]
- Sharma S., Gollnick P. Modulating TRAP-mediated transcription termination by AT during transcription of the leader region of the Bacillus subtilis trp operon. Nucleic Acids Res. 2014;42:5543–5555. doi: 10.1093/nar/gku211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov M.B., Chen Y., Gollnick P., Antson A.A. Anti-TRAP protein from Bacillus subtilis: crystallization and internal symmetry. Acta Crystallogr Biol Crystallogr. 2004;60:1311–1314. doi: 10.1107/S0907444904011199. [DOI] [PubMed] [Google Scholar]
- Shevtsov M.B., Chen Y., Gollnick P., Antson A.A. Crystal structure of Bacillus subtilis anti-TRAP protein, an antagonist of TRAP/RNA interaction. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17600–17605. doi: 10.1073/pnas.0508728102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov M.B., Chen Y., Isupov M.N., Leech A., Gollnick P., Antson A.A. Bacillus licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers. J. Struct. Biol. 2010;170:127–133. doi: 10.1016/j.jsb.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder D., Lary J., Chen Y., Gollnick P., Cole J.L. Interaction of the trp RNA-binding Attenuation Protein (TRAP) with Anti-TRAP. J. Mol. Biol. 2004;338:669–682. doi: 10.1016/j.jmb.2004.03.030. [DOI] [PubMed] [Google Scholar]
- Svergun D., Barberato C., Koch M.H.J. CRYSOL – a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J. Appl. Crystallogr. 1995;28:768–773. doi: 10.1107/S0021889895007047. [DOI] [Google Scholar]
- Tjandra N., Garrett D.S., Gronenborn A.M., Bax A., Clore G.M. Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy. Nat. Struct. Biol. 1997;4:443–449. doi: 10.1038/nsb0697-443. [DOI] [PubMed] [Google Scholar]
- Valbuzzi A., Yanofsky C. Inhibition of the B. subtilis regulatory protein TRAP by the TRAP- inhibitory protein. At. Science. 2001;293:2057–2059. doi: 10.1126/science.1062187. [DOI] [PubMed] [Google Scholar]
- Valbuzzi A., Yanofsky C. Zinc is required for assembly and function of the anti-trp RNA-binding attenuation protein. At. J. Biol. Chem. 2002;277:48574–48578. doi: 10.1074/jbc.M208980200. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Heddle J.G., Kikuchi K., Unzai S., Akashi S., Park S.-Y., Tame J.R.H. The nature of the TRAP-Anti-TRAP complex. Proc. Natl. Acad. Sci. u. s. a. 2009;106:2176–2181. doi: 10.1073/pnas.0801032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin H., Babiarz J.E., Yakhnin A.V., Babitzke P. Expression of the Bacillus subtilis trpEDCFBA Operon Is Influenced by Translational Coupling and Rho Termination Factor. J. Bacteriol. 2001;183:5918–5926. doi: 10.1128/JB.183.20.5918-5926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin H., Zhang H., Yakhnin A.V., Babitzke P. The trp RNA-binding attenuation protein of Bacillus subtilis regulates translation of the tryptophan transport gene trpP (yhaG) by blocking ribosome binding. J. Bacteriol. 2004;186:278–286. doi: 10.1128/JB.186.2.278-286.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., de Saizieu A., van Loon A.P., Gollnick P. Translation of trpG in Bacillus subtilis is regulated by the trp RNA- binding attenuation protein (TRAP) J Bacteriol. 1995;177:4272–4278. doi: 10.1128/jb.177.15.4272-4278.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria. RNA. 2007;13:1141–1154. doi: 10.1261/rna.620507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Piszczek G., Schuck P. SEDPHAT – A platform for global ITC analysis and global multi-method analysis of molecular interactions. Methods, Biocalorimetry. 2015;76:137–148. doi: 10.1016/j.ymeth.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.