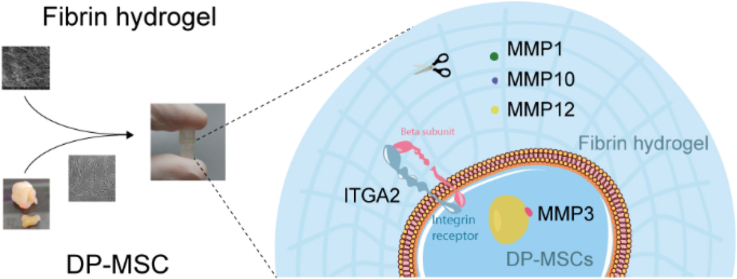
Abstract
Regenerative endodontic procedures (REP) aim at reestablishing tooth vitality by replacing the irreversibly damaged dental pulp removed by the dental practitioner with a new functional one. The current treatment of advanced caries relies on the replacement of the inflamed or necrosed dental pulp with an inert filling material. This leads to a functional but non-vital tooth, which lacks the ability to sense dental tissue damage, and to protect from further bacterial attack. Therapeutic strategies inspired by tissue engineering called REP propose to regenerate a fully functional dental pulp directly in the canal space. Promising results were obtained using dental pulp mesenchymal stem cells (DP-MSCs) in combination with bio-inspired artificial and temporary 3D hydrogels made of extracellular matrix molecules such as collagen and fibrin biomacromolecules. However, the uncontrolled mechanisms of DP regeneration from DP-MSCs in 3D biomacromolecules fail to regenerate a fully functional DP and can induce fibrotic scarring or mineralized tissue formation to a non-negligible extent. The lack of knowledge regarding the early molecular mechanisms initiated by DP-MSCs seeded in ECM-made hydrogels is a scientific lock for REP. In this study, we investigated the early DP-MSC–response in a 3D fibrin hydrogel. DP-MSCs isolated from human third molars were cultured for 24 h in the fibrin hydrogel. The differential transcript levels of extracellular and cell surface genes were screened with 84-gene PCR array. Out of the 84 genes screened, 9 were found to be overexpressed, including those coding for the integrin alpha 2 subunit, the collagenase MMP1 and stromelysins MMP3, MMP10 and MMP12. Over-expression of ITGA2 was confirmed by RT-qPCR. The expression of alpha 2 integrin subunit protein was assessed over time by immunoblot and immunofluorescence staining. The increase in the transcript level of MMP1, MMP3, MM10 and MMP12 was confirmed by RT-qPCR. The overexpression of MMP1 and 3 at the protein level was assessed by immunoblot. MMP3 expression by DP-MSCs was observed by immunofluorescence staining. This work demonstrates overexpression of ITGA2 and of MMP1, 3, 10 and 12 by DP-MSCs cultured in a fibrin hydrogel. The main preliminary extracellular and cell surface response of the DP-MSCs to fibrin hydrogel seems to rely on a ITGA2/MMP3 axis. Further investigations are needed to precisely decipher the role of this axis in dental pulp tissue building. Nevertheless, this work identifies extracellular and cell surface molecules that could be potential checkpoints to be targeted to guide proper dental pulp tissue regeneration.
Keywords: Regenerative endodontic procedures, Fibrin, Dental pulp, Metalloproteases, Integrins
Graphical abstract
DP-MSCs cultured in fibrin hydrogel express ITGA2 and the metalloproteases MMP1, MMP3, MMP10 and MMP12.
1. Introduction
Regenerative endodontic procedures (REP) have been proposed to regenerate tooth vitality in case of endodontic treatment of irreversible pulpitis [[1], [2], [3]]. Various synthetic, natural or bio-inspired scaffolds have been tested to promote dental pulp (DP) regeneration [[4], [5], [6], [7], [8], [9]]. However, the poorly understood and uncontrolled mechanisms of regeneration could induce the formation of scar fibrous or mineralized dysfunctional tissues [[9], [10], [11], [12], [13], [14]]. Proper tissue regeneration requires finely tuned degradation of the temporary or damaged tissue and neo-synthesis of a scar-free functional tissue [15]. This process involves a battery of cell surface receptors and proteases, such as integrins and metalloproteases (MMPs) to sense the environment and orchestrate ECM remodeling [16,17].
The use of dental pulp mesenchymal stem cells (DP-MSCs) for pulp regeneration gave promising results [5,18,19]. Their origin and their easy isolation from human third molars extracted for orthodontic reasons make them the best candidates to regenerate the DP [20]. The immunomodulatory properties of DP-MSCs lead to the inhibition of T-cell response and to the modulation of the M1/M2 ratio to the benefit of an anti-inflammatory response required for ECM building during tissue regeneration [21]. DP-MSCs show a fibroblastic shape in cell culture and an immunophenotypic profile of mesenchymal cells by the expression of stemness/progenitor markers including CD271, Stro-1, CD146, and/or MSCA-1 [22]. These cells display a trilineage differentiation capacity (osteoblasts, adipocytes and chondrocytes) which confirmed their stemness property [23]. DP-MSCs express metalloproteases such as MMP2, MMP9, MMP7, MMP23 and MMP25 [[24], [25], [26]]. However, the mechanisms initiated by DP-MSC in a fibrin hydrogel need to be further investigated to control the regeneration process and minimize scar or mineralized tissue formation.
In physiological conditions, fibrin plays a crucial role in hemostasis and wound healing by forming a fibrous network [27]. The success of tooth revitalization based on induced peri-apical bleeding and blood clot formation into the root canal strongly suggests that fibrin-based biomaterials could be used as 3D scaffolds in dental pulp regeneration procedures [1,28,29]. These experiments have shown that a fibrin meshwork can form a natural hydrogel that constitutes a cell scaffold biologically adapted to DP regeneration. The fibrin scaffold is cytocompatible, physiologically degradable and is replaced with a new cell-derived extracellular matrix (ECM) within few days [30]. The cryptic peptides released following fibrin degradation support tissue remodeling by promoting angiogenesis and chemotaxis [31,32]. Moreover, mechanical properties of the fibrin hydrogel can be fine-tuned by modifying fibrinogen concentration and ionic strength [30]. Several studies demonstrated that fibrin hydrogels appear to be particularly well suited to dental pulp regeneration as they are propitious for DP-MSC viability and for the formation of a collagen-rich pulp-like tissue in vitro and in vivo [7,28,33,34]. Fibrin therefore appears to be a biomacromolecule of choice to design of hydrated 3D scaffolds for tissue regeneration. However, very little is known about DP-MSC behavior into the fibrin scaffold, and in particular how they interact with the scaffold and how they degrade and replace it with a new extracellular matrix during the regenerative process. We postulated that the recognition of matrix environment by the cells and the resulting cell adhesion process may be early events important to orientate DP-MSC phenotype towards acquisition by the cells of the ability to produce a DP-like ECM.
Human cells interact with fibrin mainly through an integrin-driven mechanism mediated by Arg-Gly-Asp (RGD) sequences. The integrins involved differ according to the cell type: platelets bind fibrin through integrins αIIbβ3 [35], leukocytes through αMβ2/Mac-1 [36] and fibroblasts through αVβ3 [28,37]. These interactions induce blood clot formation and lead to ECM remodeling and inflammatory response. Fibrin has also binding sites for other ECM proteins, such as fibronectin and vitronectin, that could regulate cell adhesion and response through multiple mechanisms involving cell-surface receptor clustering and cross-talks [[38], [39], [40]]. Fibrin degradation occurs in vivo by the plasmin-centered fibrinolytic cascade originating mainly from endothelial cells [17,41,42]. Activated plasmin forms the center of a proteolytic hub constituted by several MMPs (MMP1, MMP2, MMP3, MMP7, MMP8, MMP9, MMP12, MMP13, MMP14). This protease hub leads to fibrin cleavage by some MMPs (MMP3, MMP8, MMP9, MMP12, MMP13, MMMP14) [[43], [44], [45]] and to the activation of many cytokines, chemokines and growth factors to promote tissue remodeling. To date, DP-MSC response in fibrin hydrogels including integrin and MMP expression remain unknown and should be investigated to optimize the use of fibrin as a biomaterial by controlling fibrin degradation and ECM replacement.
This study investigates the early response of DP-MSCs seeded in a fibrin hydrogel by focusing on the extracellular and cell surface molecules that could initiate the regeneration process to identify molecular actors that might be targeted to rationally control the regeneration process and drive proper dental pulp tissue formation in different pathological contexts.
2. Materials and methods
2.1. DP-MSCs collection
Healthy impacted human third molars were collected from donors aged 11–17 years with informed consent of the donors and their parents, in accordance with the recommendations of the World Medical Association's Declaration of Helsinki and following a protocol locally and nationally approved by the French Ministry of Higher Education and Research (CODECOH: DC-2014-2325) [22]. Cells from seven different donors were used for this study including eight females (11, 13, 14, 14, 15, 15, 16 and 17 years old) and three males (two of 15 years old and one of 16 years old). These cells display a fibroblast-like structure in 2D cell culture and a mesenchymal immunophenotypic profile with the expression of stem/progenitor cell markers CD271, Stro-1, CD146, and/or MSCA-1 22. Their stemness was confirmed by the ability to differentiate into osteoblasts, adipocytes and chondrocytes [23].Transcript levels of selected proteins were validated using DP-MSCs from at least one male and one female. No differences were observed in the induced protein expression by DP-MSCs in fibrin hydrogel between females and males. Dental pulps from teeth between Nolla developmental stages 5 (crown almost completed) and 7 (one third root completed) were gently extirpated from pulp cavities and cut into fragments of about 0.5–2 mm3 as reported previously [22]. Fragments were then seeded as explants on dishes pre-coated with human placental collagens I and III (ABCellBio, Paris, France), and then cultured in chemically defined culture medium SPE- IV/EBM (ABCellBio) supplemented with 100 IU/mL penicillin and 100 μg/mL streptomycin (Life Technologies, Saint Aubin, France). DP-MSCs outgrowing from the explants were passaged four times with xeno-free recombinant protease TrypLe® Select 1X (Life Technologies) for amplification.
2.2. Construction of hydrogels
Fibrin hydrogels were produced as previously reported [34,46]. Briefly, a fibrinogen solution was prepared in a calcium-free phosphate buffered solution pH 7.2 containing 150 mM NaCl, 20 mM CaCl2. The fibrinogen concentration was calculated to get a 10 mg/mL fibrin concentration in the final hydrogel. DP-MSCs were added to the fibrinogen solution to get a final concentration of 2.2e5 cells/mL. A 0.4 U/mL thrombin solution was then added to initiate fibrinogen polymerization into fibrin and meshwork formation. Fibrin hydrogels were poured in home-made inserts conceived from tips. The inserts were used for culture in 96 well plates in SPE- IV/EBM culture medium.
2.3. RNA extraction from hydrogels and RT-qPCR
RNA extraction and RT-qPCR were performed as described previously [34]. After culture, RNA was extracted from DP-MSCs with the Nucleospin® RNA II kit (Macherey Nagel, Düren, Germany) according to the manufacturer protocol. The amount of isolated RNA was determined by measuring the absorbance at 260 nm by a spectrophotometric method (Nanodrop 2000, Thermofischer Scientific). Purity was assessed by determining absorbance ratios of A260/A280 (greater than 1.9) and A260/A230 (between 2.0 and 2.2). When RNAs are extracted and of adequate quality, a retrotranscription step was performed. The cDNA was obtained from 200 ng of total RNA with which the protocol of the Prime Script RT reagent kit (Takara, Ozyme, Montigny-le-Bretonneux, France) was applied. RT-qPCR experiments were performed on 7 donors for ITGA2 (4 females of 13, 14, 15 and 16 years-old and 3 males of 15, 15 and 16 years-old) and 6 donors for ITGA1, ITGAV, ITGB1, ITGB3 (4 females of 11, 14, 16 and 17 years-old and 2 males of 15 and 17 years-old)
2.4. PCR array
Expression of 84 genes involved in cell adhesion, signaling and matrix remodeling was assessed using the RT2 Profiler PCR Array - Human Extracellular Matrix & Adhesion molecules (Qiagen). The PCR array was performed using cells from a single donor (female of 15 years-old). RNA was extracted for the cells just after addition to fibrin hydrogel or after 14 h of culture. Complementary DNA was retrotranscribed from 200 ng RNA and diluted at 1/3. Diluted cDNA (25 μL) was deposited with the ready-to-use RT2 SYBR Green qPCR Master Mix in each well containing a set of primers specific to the gene analyzed. PCR was then performed in the Rotor-GENE Q thermal cycler (Qiagen, Hilden, Germany) as a conventional PCR. The device was programmed for an initial denaturation step at 95 °C for 10 min followed by 50 cycles of rising to 95 °C (denaturation) for 10 s and lowering to 60 °C (hybridization) for 20 s. Each assay was performed in duplicate and a negative control was systematically performed by replacing cDNA with water. The expression levels of each transcript were normalized to the cDNAs of the housekeeping gene RPL13A, encoding the ribosomal protein L13A selected because of its stability in stem cells [47].
The set of fluorescence thresholds noted Ct of each gene contained in the PCR array was calculated using the software of the device. Finally, fold change of gene expressions was calculated by comparing the day 0 (D0) condition with the day 1 (D1) condition using the 2^-ΔΔCt method [48]. Each value obtained was normalized with 5 housekeeping genes (actin, β-microglobulin, GADPH, HPRT1, RPL0) to enhance robustness of the result and compared with 3 controls (human genomic DNA contamination, reverse transcriptase positive control, positive PCR control). Genes of interest for the rest of the study were selected based on three criteria: fold change value, corrected t-test p-value, and relevance of the gene to current literature.
2.5. Primer design and validation
To validate results obtained with the PCR array for the selected genes, their expression was assessed with real-time PCR (qPCR) on a minimum of 3 samples originating from different donors. Primers sequences for ITGA1 were picked from the literature [49], all others were designed. The Ensembl database [50] was queried to obtain the gene and the Primer Blast resource [51] was used to design the primers without off-targets and with the optimal parameters for RT-qPCR: primer length between 70 and 200 base pair, GC ratio between 50 and 60 %, melting temperature between 59 and 65 °C, avoiding exon-exon junction and with the lowest self-complementarity score. Characteristics of all the primers (sequence, %GC, melting temperature, efficiency, NCBI RefSeq code for targeted mRNA sequence) were reported in the supplementary section (S1. A). Primer efficiencies were determined using calibration lines constructed from decreasing concentration ranges of cDNA derived from 200 ng of RNA extracted from DP-MSCs after one day of culture in a fibrin hydrogel.
2.6. Western blot analyses
To extract proteins from DP-MSCs cultured in fibrin hydrogels, protein samples were suspended in Laemmli buffer supplemented with 5 % of βME. Proteins obtained directly from the sample were loaded on a polyacrylamide gel (11 % running/4 % stacking).
A time-course study from D0 to D4 was performed to evaluate the overexpression at the protein level. The protein expression of ITGA2 (antibody MA5-32306, Invitrogen), MMP1 (antibody MAB901, RND Systems) and MMP3 (anti-MMP3 produced in rabbit, HPA007875, Sigma Aldrich) was validated by immunoblot. Proteins were quantified at D0, D2 and D4 using 5 donors for ITGA2 (3 females of 11, 16 and 17 years-old and 2 males of 15 years-old), 4 donors for MMP1 (3 females of 13, 14 and 14 years-old and 1 male of 15 years-old) and 6 donors for MMP3 (4 females of 11, 14, 16 and 17 years-old and 2 males of 15 and 17 years-old). Membranes were reported as supplementary materials (S2-4). To normalize the signal from the tested proteins, actin labeling was done. Antibodies were diluted at 1/1000 (anti-ITGA2), January 2000 (anti-MMP1), 1/500 (anti-MMP3) and 1/1000 (anti-actin) in 5 % milk PBS-Tween.
2.7. Immunofluorescence experiments
Hydrogels were collected from the plastic molds and fixed in 4.5 % paraformaldehyde (PFA) overnight at 4 °C. Samples were then embedded in paraffine and 50 μm thick sections were made with a Leica handheld microtome. Samples were gradually rehydrated and then washed 2 × 5 min with PBS-Tween 20 and then once with PBS for 5min. Citrate buffer pH5 was used to unmask the major antigens and for them to be recognized by the corresponding antibody. Then, a blocking step with 5 % BSA was performed to block aspecific binding of the antibody. The primary anti-MMP3 or anti-ITGA2 antibodies were diluted respectively at 1:50 and 1:1000 in 2.5 % BSA and incubated overnight at 4 °C. Recovered samples were then washed as above. The revelation was then done with the secondary antibody Alexa 546 (A11030, Invitrogen, Carlsbad, USA) diluted at 1/1000 in 2.5 % BSA. Samples were incubated for 1h in a humid chamber protected from light at room temperature. After washing, slides were mounted in Vectashield® Antifade Mounting Medium (H-1200-10, Vector Laboratories) containing the DNA intercalant DAPI. Slides were then observed with a Nikon TiE inverted fluorescence microscope (Nikon instrument inc. Melville, USA) motorized with DAPI filters for nuclei (ex: 358 nm/em: 461 nm) and TRITC (ex: 544 nm/em: 570 nm).
2.8. Statistical analyses
The Wilcoxon-Mann-Whitney test was used to compare the gene expression levels between two time points (T0 and T24). This test was chosen due to its ability to handle both paired and independent samples. This method allowed us to account for the nature of the samples and conduct an adequate statistical analysis to determine whether there were significant variations across the time periods. Differences were considered significant when the probability value P was lower than 0.05. All analyses were performed using the Graph Pad Prism 9.0.1 software (GraphPad Software, La Jolla, CA).
3. Results
3.1. Screening of extracellular matrix and cell surface proteins
To identify the genes involved during the early steps of the matrix synthesis and remodeling of the tissue, a qPCR array was performed on human DP-MSCs cultured in a fibrin hydrogel. Experiments were focused at D0 and D1 to focus on the genes initiated by DP-MSCs after culture in fibrin hydrogel. Among the 84 genes investigated, 38 (45 %) were significantly over-expressed or under-expressed at least two-fold after 1 day of culture (D1) compared to the day of seeding (D0). A list of genes is presented in Table 1 and shows that almost all of them correspond to a positive regulation. Only the VCAM1 molecule has a log2 fold change of −5.47, i.e., 44 times less expressed at D1 than at D0. Conversely, MMP3, MMP10 and MMP1 genes were strongly upregulated, between 1000 and 8000 times more at D1 (10 < log2 FC < 13) (Table 1).
Table 1.
Main genes with a strong regulation between D0 and D1 identified by RT-qPCR. Sorting of the differentially expressed genes based on the log2FC (fold change), p-value and relevance regarding literature.
| Symbol | Gene description | Log FC | p-value |
|---|---|---|---|
| MMP3 | Matrix metallopeptidase 3 | 13.02 | 0,000000 |
| MMP10 | Matrix metallopeptidase 10 | 10.23 | 0.000002 |
| MMP1 | Matrix metallopeptidase 1 | 10.04 | 0.000015 |
| ITGA2 | Integrin, alpha 2 | 7.89 | 0.000000 |
| SPP1 | Secreted phosphoprotein 1 | 7.81 | 0.000002 |
| MMP8 | Matrix metallopeptidase 8 | 5.69 | 0.000043 |
| TNC | Tenascin C | 5.49 | 0.000000 |
| MMP12 | Matrix metallopeptidase 12 | 5.32 | 0.000054 |
| MMP9 | Matrix metallopeptidase 9 | 4.89 | 0.000005 |
| TIMP1 | TIMP metallopeptidase inhibitor 1 | 4.65 | 0.000000 |
| LAMA3 | Laminin, alpha 3 | 4.41 | 0.007955 |
| PECAM1 | Platelet adhesion molecule | 4.28 | 0.000176 |
| LAMB3 | Laminin, beta 3 | 4.26 | 0.000002 |
| TGFBI | Transforming growth factor | 3.93 | 0.000001 |
| COL7A1 | Collagen, type VII, alpha 1 | 3.92 | 0.000000 |
| CD44 | CD44 molecule | 3.73 | 0.000003 |
| COL8A1 | Collagen, type VIII, alpha 1 | 3.06 | 0.000008 |
| ITGA5 | Integrin, alpha 5 | 3.05 | 0.000014 |
| LAMA1 | Laminin, alpha 1 | 2.97 | 0.003188 |
| VCAM1 | Vascular adhesion molecule 1 | −5.47 | 0.000001 |
A final selection was performed among the 38 genes to confirm the observed upregulation or downregulation. The study was narrowed down to 11 genes based on the log fold change (Log FC) and the p-value (Table 1).
The genes were first sorted into the families to which they belong, and then shown as a heatmap (Fig. 1). This representation gave us a broad overview of all the genes analyzed while also highlighting the ones that were the most regulated.
Fig. 1.
Screening of extracellular and cell surface genes expressed by DP-MSCs in fibrin hydrogel. Extracellular and cell surface genes differentially expressed after 1 day were screened using a PCR array. Results were represented by a heatmap where each box's color is proportional to its fold change value in the logarithm base of two (scale at the right). The genes in both red and bold are the top 10 most significantly up-regulated. Corrected t-test was performed for this analysis. Pvalues < 0.005 for red-colored genes.
Four MMPs molecules were selected because of their very high fold change and their relevance in our study: MMP1, MMP3, MMP10, and MMP12. The majority of compounds with active regulation were found in this category. MMP3 stands out from the other genes with a fold change of 13.02, indicating a strong increase during the first day of culture. It was accordingly selected to be further studied at protein level.
This analysis reveals the transcriptional regulation of adhesion molecules such as osteopontin (SPP1) and tenascin (TNC). The main role of these molecules is to interact with cells and proteins of the matrix which will trigger the activation of specific signaling pathways involved in ECM remodeling [52]. VCAM1 gene expression significantly decreased, but no link has been established with regenerative processes and further studies are needed to better understand this result.
3.2. ITGA2 expression by DP-MSCs cultured in a fibrin hydrogel
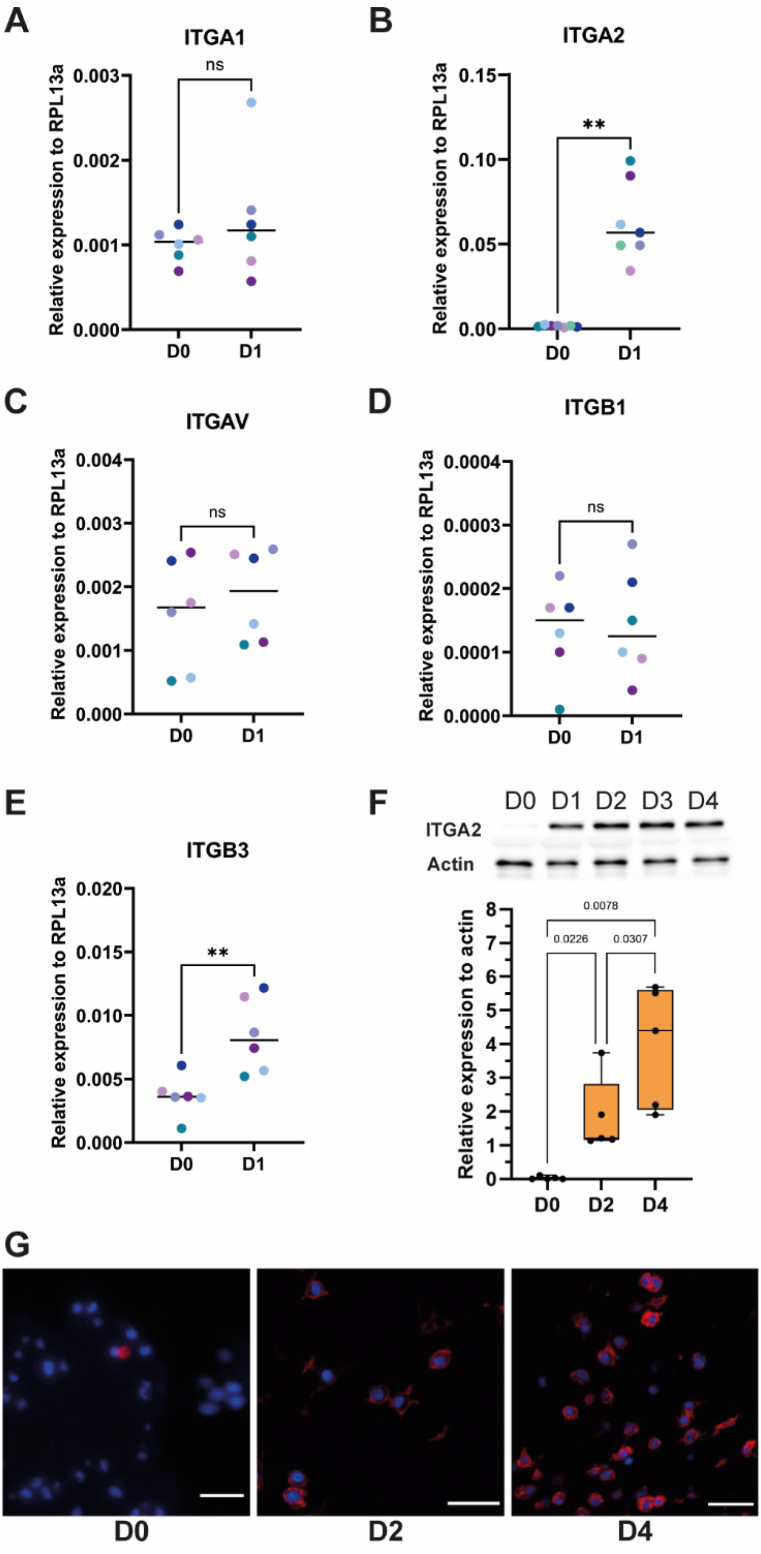
Screening of the cell surface and extracellular genes expressed by DP-MSCs cultured in a fibrin hydrogel was performed. This screening is limited by the use of cells from a single donor. The transcript level of integrin subunits expressed by mesenchymal cells and known to bind collagen (ITGA1, ITGA2, ITGB1) or fibrin (ITGAV, ITGB3) was further assessed by RT-qPCR [53]. While ITGA1, ITGAV, ITGB1 transcript levels are not affected, the transcript levels of ITGA2 and ITGB3 genes were significantly upregulated after 24 h of culture of DP-MSCs in the hydrogel (Fig. 2A–E). Increase of the transcript level did not necessarily reflect the protein level as several mechanisms could regulate the protein expression such as the spatial and temporal availability of mRNA for protein biosynthesis or protein degradation [54]. Protein expression was assessed for ITGA2 for which all the 7 donors show an increase of ITGA2 subunit transcript level (Fig. 2B). Experiments were performed during the four first days to investigate the protein initially expressed by DP-MSCs in fibrin hydrogel. The ITGA2 protein level was quantified using cells from 5 donors at D0, D2 and D4. Upregulation of ITGA2 transcript level resulted in the increase of the ITGA2 protein level (Fig. 2F). The overall ITGA2 protein level dramatically increases at D1 comparing to the starting level observed just after fibrin hydrogel polymerization and further increases between D2 and D4 (Fig. 2F). ITGA2 was observed by immunofluorescence in DP-MSCs with a specific antibody using a single donor. It clearly confirms the overexpression of ITGA2 by DP-MSCs in fibrin hydrogels at D2 and D4 (Fig. 2G).
Fig. 2.
Early expression of integrin-alpha 2 subunit by DP-MSCs in fibrin hydrogel
Transcript level of ITGA1 (A), ITGA2 (B), ITGAV (C), ITGB1 (D); ITGB3 (E) expressed by mesenchymal cells were investigated in DP-MSCs from different donors cultured in fibrin hydrogels for one day (n = 7 for ITGA2 and n = 6 for the other genes). Statistics were performed using a Mann-Whitney test. **pvalue<0.01; ***pvalue<0.001; ns: non-significant. (F) Protein level of ITGA2 assessed by immunoblot using DP-MSCs cultured in a fibrin hydrogel from day 0 (D0) to day 4 (D4). Protein level of actin was used as control. Quantification of ITGA2 protein level at day 0, 2 (D2) and 4 was performed using 5 different donors and reported as relative protein expression to actin. Statistics were performed using a Mann-Whitney test. Pictures of the immunoblot membranes were reported as supplementary material (S2 A-J). The immunoblot membranes used for illustration were reported in the supplemental sections (S2 E) and (S2 J). (G) Immunofluorescence imaging of ITGA2 in DP-MSCs cultured in a fibrin hydrogel at day 0 (D0), day 2 (D2) and day 4 (D4). Scale bar is 25 μm.
3.3. MMP expression by DP-MSCs in fibrin hydrogels
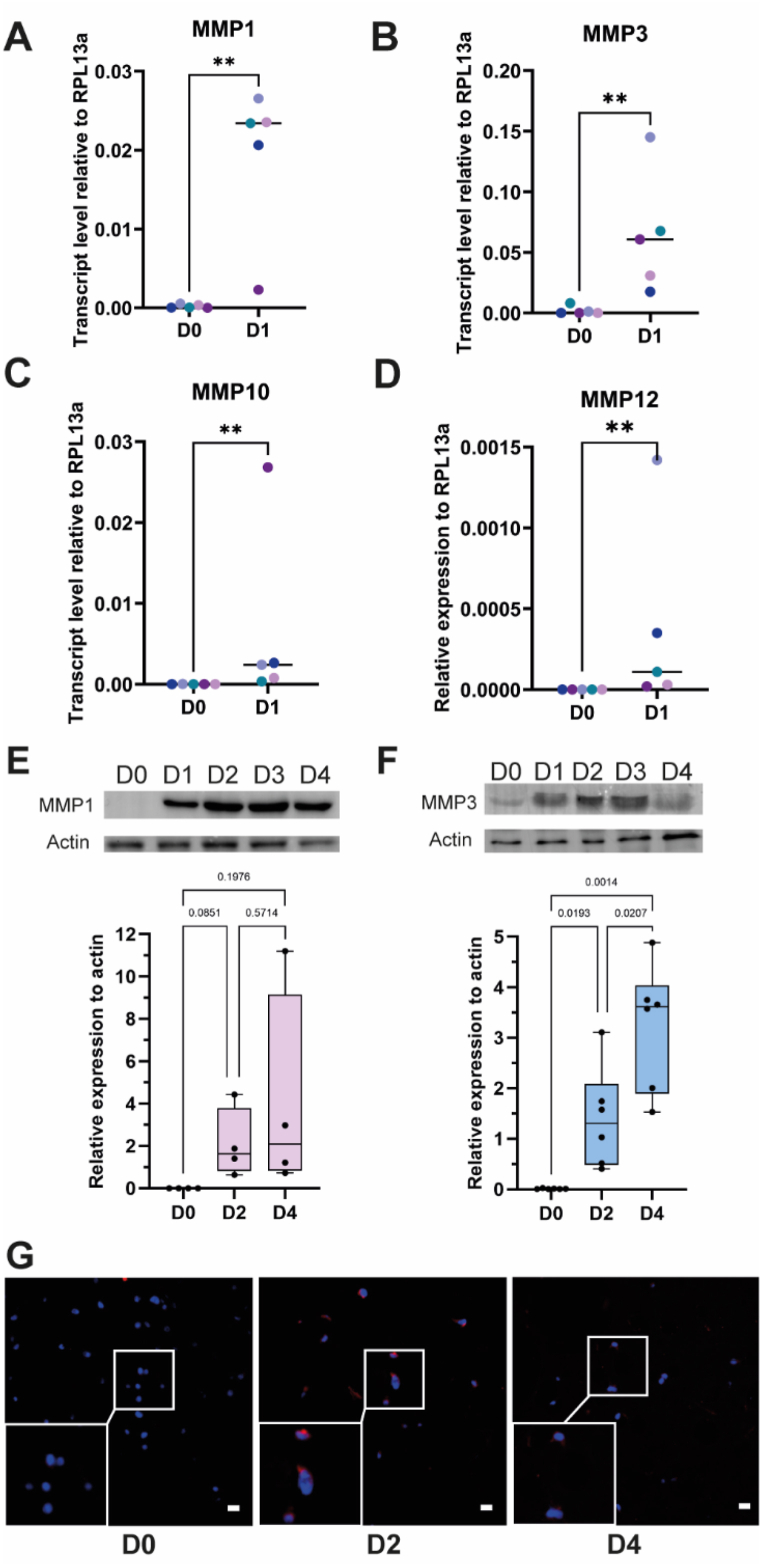
Screening of the extracellular proteases expressed by DP-MSCs in fibrin hydrogel using a single donor shows the upregulation of genes encoding the MMP1 collagenase and the MMP3, MMP10 and MMP12 stromelysins (Table 1). This result was confirmed by RT-qPCR using DP-MSCs isolated from five donors (Fig. 3A–D). MMP3 was the most upregulated MMP followed by MMP1, confirming the results obtained with the PCR array (Fig. 1). The protein level of these two proteases in the fibrin hydrogel was investigated by immunoblot and quantified at D0, D2 and D4 using cells from 4 to 6 different donors respectively for MMP1 and MMP3. Both MMP1 and MMP3 protein levels were increased over time with a burst increase during the first day of culture (Fig. 3E–F). Further investigation was focused on MMP3 which is the highly expressed proteins reported in inflamed dental pulp [55]. Immunofluorescence experiments using a single donor show a punctuate staining of MMP3 within DP-MSCs in the fibrin hydrogel (Fig. 3G). While it was not detected at D0, a clear staining could be observed at D2 which comforts the overexpression of MMP3 at the protein level.
Fig. 3.
Early expression of MMP1 and MMP3 by DP-MSCs in fibrin hydrogels
Transcript level of MMP1 (A), MMP3 (B), MMP10 (C) and MMP12 (D) expressed by mesenchymal cells were investigated in DP-MSCs from different donors (n = 5) cultured in fibrin hydrogels for one day. The transcript level of MMP3 is not detectable for two donors at day 0. Statistics was performed with a Mann-Whitney test. **pvalue<0.01; ***pvalue<0.001; ns: non-significant. Protein levels of MMP1 (E) and MMP3 (F) assessed by immunoblot from day 0 (D0) to day 4 (D4). Protein quantification at day 0, 2 (D2) and 4 was performed using 4 and 6 different donors respectively for MMP1 and MMP3 and reported as relative protein expression to actin. Statistics were performed using a Mann-Whitney test. Pictures of the immunoblot membranes used for quantification were reported as supplementary material (S3 A-H, S4 A-F and S4 H-M). The immunoblot membranes used for illustration were reported in the supplemental sections (S3 B, S3 F, S4 G and S4 N). (G) Immunofluorescence staining of MMP3 in DP-MSCs cultured in a fibrin hydrogel at day 0 (D0), day 2 (D2) and day 4 (D4). Scale bar is 25 μm.
4. Discussion
This study investigated the molecular mechanisms involved in DP-MSC-based dental pulp regeneration using a fibrin hydrogel-based scaffold. Proper tissue regeneration involves finely tuned degradation of the scaffold or damaged tissue and the neo-synthesis of an extracellular matrix structurally and functionally similar to the original tissue [56,57]. This study was focused on the remodeling of a fibrin-based hydrogel by DP-MSCs for dental pulp regeneration [5,28]. Better knowledge of the fibrin meshwork replacement could allow to adapt the treatment to various pathological conditions. Uncontrolled tissue regeneration mechanisms could lead to fibrotic scaring or mineralization resulting in inappropriate tissue formation [[9], [10], [11], [12], [13], [14],58]. The crucial role of a controlled cross-talk between cells and their extracellular environment for tissue integrity and functionality is notably illustrated by pathophysiological modifications occurring during aging. These indeed result in a disorganized extracellular matrix environment forming disorganized fibrous tissues and calcospherites and subsequent changes in dental pulp cell functions in response to this stressful environment [[59], [60], [61], [62]]. Modification of dental pulp composition affects DP-MSC function which induces a pro-inflammatory and matrix-degrading response [62]. Tissue regeneration requires a battery of cell surface receptors and proteases, such as integrins and metalloproteases (MMPs) to sense the environment and orchestrate ECM remodeling [16,17]. While most of the studies investigating dental pulp regeneration develop materials, evaluate the cyto-bio-compatibility of the materials (inflammation, differentiation) and report histological analyses [[4], [5], [6], [7], [8], [9]]; this study investigates the response of DP-MSCs to the fibrin hydrogel at the biomolecular level, which is crucial for proper dental pulp regeneration. Extracellular and cell surface actors were identified to better understand the early mechanisms of DP-MSCs resulting to cell adhesion and spreading that leads to fibrin hydrogel degradation and progressive replacement.
As a first step, extracellular matrix and cell surface candidates were screened using a PCR array. The PCR array highlighted approximately forty genes with a strong variation of their expression. Four major families were overrepresented: integrins, proteases and their regulators, and adhesion and signaling molecules. We thus identified integrins such as ITGA2, and metalloproteases (MMPs; MMP1, MMP3, MMP10, MMP12) as potential central players in DP-MSC response to the fibrin hydrogel. Some of the selected candidates, with high overexpression level (ITGA2, MMP1 and MMP3), were further investigated validated at protein level by biochemical methods with specific attention paid to ITGA2 and MMP3 that were previously reported to be involved in tissue repair and remodeling [[63], [64], [65], [66]].
Biological regulators and growth factors including TIMP1, TGF-β1 and osteopontin (OPN) were also identified. These may also be involved in DP-MSC-derived tissue construction. Indeed, the main role of TIMP1 is to regulate MMP activities and therefore contribute to tissue homeostasis [67,68]. TIMP-1 inhibits collagenases, gelatinases and the proteoglycanase MMP3 [68]. In this context, it might regulate MMP mechanisms of fibrin hydrogel degradation and, as a result, DP-MSC migration and ECM formation. This regulation is complemented by TGF-β1 which is known to stimulate DP-MSC proliferation [69], and OPN reported to affect MMP secretion profile during wound skin healing [70]. PCR array screening also allowed to identify VCAM1, a receptor involved in the recruitment of regenerative cells once the damaged matrix is removed [71].
This study reports the upregulation of the integrin α2 subunit by DP-MSCs cultured in the fibrin hydrogel. The constant transcript level of ITGAV and ITGB3, observed in the PCR array, could mean that the level of these receptors is already sufficient to interact with fibrin, or that these receptors are not or faintly implicated in DP-MSC adhesion to fibrin in this particular context. Further investigations are needed to precisely assess the protein level of these cell receptors and to identify those interacting and being responsible for DP-MSC adhesion to fibrin. Integrin receptors act as heterodimers composed of α and β subunits as for ITGA2 which forms a receptor in association with ITGB1 [53,63]. The constant transcript level of ITGB1 observed in the PCR array may imply that the level of this subunit is sufficient and that another level of regulation might be involved. This suggests that ITGB1 associates preferentially with ITGA2 during the process of integrin dimer formation or that ITGB1 dissociates from a dimer and swaps to ITGA2. The regulation of integrin receptors by lateral association remains a topic not clearly understood [72]. Clustering of integrins through transmembrane helix association and homomeric association was reported [73]. This mechanism based on integrin clustering was observed in the presence of multivalent substrates such as fibrin or collagen [74,75]. Such a mechanism could be implemented by DP-MSCs in fibrin hydrogels but further precise investigations are needed to precisely decipher the interaction of ITGA2 with the DP-MSC environment. The integrin receptor α2β1 does not seem to be involved in wound healing in vivo but its absence seems to promote angiogenesis and metalloprotease expression [76]. The functional role of α2β1 as assessed in deficient mice or by in vitro assays was associated to dysregulation of the expression of MMPs such as MMP2, MMP3, MMP9, MMP10 and MMP13 65,76,77. The integrin receptors α2β1 is implicated in the transient adhesion of collagen matrix being synthesized by cells remodeling their environment [78]. This study shows a clear overexpression of ITGA2 by DP-MSCs cultured in fibrin hydrogel suggesting that they could develop an early outside-in response from the ECM to the cell [79]. Further investigations are needed to clarify the role of the integrin α2β1 in fibrin hydrogel degradation and dental pulp tissue neoformation.
Matrix metalloproteinases were the most represented proteins among the ECM and cell surface proteins families screened in this study. These include collagenase MMP1, stromelysins MMP3 and MMP10, and MMP12 66. This result was relatively expected as these enzymes degrade a wide variety of substrates, including most of the components of the ECM, and as such are involved in the regulation of many biological processes such as development, branching morphogenesis and tissue remodeling [64,80]. These MMPs may be mainly involved in DP-MSC integration to and remodeling of their environment such as MMP1 which was reported to be secreted when ITGA2 binds to type I collagen in order to facilitate stem cell migration into the ECM [81,82].
MMP3 was found to be the most upregulated gene on the PCR array, making it a candidate of choice for elucidating DP-MSC-dependent mechanisms of fibrin hydrogel degradation. MMP3 is a stromelysin that was originally called proteoglycanase as it degrades several types of proteoglycans such as aggrecan [66,68,80]. MMP3 is involved in extracellular matrix turnover occurring in normal and pathological conditions such as development or wound healing [64,80]. Fibrinogen is cleaved by MMP3 and MMP1, which suggests that their overexpression by DP-MSCs may participate to fibrin hydrogel degradation [43,83,84].
In the literature, ITGA2, MMP1 and MMP3 were related to inflammation [77,85,86]. In particular, MMP3 is overexpressed in the dental pulp of damaged rat incisors, which suggests that it has an anti-inflammatory role and stimulates the regeneration of dental pulp in damaged teeth [55]. We also report the overexpression of MMP3 suggesting a potential role in DP-MSC apoptosis [[87], [88], [89]]. How MMP3 overexpression affects the stability of the hydrogels, the composition of the produced ECM and the inflammatory state are totally unknown and might be further investigated to better drive dental pulp regeneration. Another important parameter to consider is the stability of the hydrogel, i.e. the time needed to be replaced in a controlled manner by a DP-MSC-derived ECM. Precise mechanisms of fibrin hydrogel degradation or contraction, which is classically attributed to the presence of fibroblasts, and affects tissue formation [90] still remains unknown and needs to be further characterized.
Cell-derived matrix is crucial for tissue neoformation. A recent study using wide spectrum approaches (quantitative proteomics) highlighted the crucial role of the cell-dependent organizing ECM in hepatic commitment. A 3D ECM derived from confined human pluripotent stem cells or exogenous ECM stimuli was shown to promote the ability of hepatic progenitor cells to differentiate into functional hepatocytes [91]. The identification of ITGA2, MMP1 and MMP3 as early response suggests that DP-MSCs rapidly degrade the fibrin cell-surrounding environment. These proteins might be therapeutic targets to act on in order to regulate regenerative endodontic procedures based on DP-MSCs and fibrin hydrogels. The negative regulation of some of these proteases could be a way to control the kinetic of degradation of fibrin and its replacement by a dental pulp-like matrix. Their overexpression or proteolytic products could be early biomarkers of the quality of the neo-tissue tissue formed that could be useful to control the quality of the biomaterials developed for endodontic regeneration.
5. Conclusion
This study aimed at investigating the early response of DP-MSCs to a 3D fibrin hydrogel, in order to better understand the biological mechanisms of dental pulp regeneration. Taken together, these results demonstrate the early expression of ITGA2, MMP1 and MMP3 by DP-MSCs in the fibrin hydrogel. These extracellular and cell surface proteins provide potential therapeutic targets opening the way to new research towards the rational control of stem-cell-based tissue regeneration. Understanding biological mechanisms responsible for hydrogel degradation and replacement with a new ECM able to promote tissue regeneration could lead to improve endodontic treatment protocols of the irreversibly inflamed dental pulp.
Funding Sources
This work was supported by the French National Centre for Scientific Research (CNRS), the French Institute for Odontological Research (IFRO), the “Fondation de l’Avenir” and the French National Research Agency (Tridentomic projet, N°ANR-21-CE44-0015-01 and Endonanobiotic project N°ANR-21-CE19-0001). The research developed by MBe is supported by a grant (AAP Accueil EC) from the University Lyon 1 (UCBL).
Data availability statement
Data included in article/supp. material/referenced in article.
CRediT authorship contribution statement
David Tong: Writing – original draft, Visualization, Investigation. Stéphanie Gobert: Writing – original draft, Supervision, Methodology, Investigation. Alicia Reuzeau: Writing – original draft, Investigation. Jean-Christophe Farges: Writing – review & editing, Supervision. Marianne Leveque: Writing – review & editing, Investigation. Marie Bolon: Writing – original draft, Investigation. Arthur Costantini: Visualization, Investigation. Marielle Pasdeloup: Writing – review & editing, Methodology, Investigation. Jérôme Lafont: Writing – review & editing, Methodology. Maxime Ducret: Writing – review & editing, Supervision, Funding acquisition, Conceptualization. Mourad Bekhouche: Writing – review & editing, Writing – original draft, Supervision, Project administration, Methodology, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e32891.
Contributor Information
David Tong, Email: david.tong@outlook.fr.
Stéphanie Gobert, Email: stephanie.gobert@univ-lyon1.fr.
Alicia Reuzeau, Email: alicia.reuzeau99@gmail.com.
Jean-Christophe Farges, Email: jean-christophe.farges@univ-lyon1.fr.
Marianne Leveque, Email: marianne.leveque@ibcp.fr.
Marie Bolon, Email: marie.bolon@ibcp.fr.
Arthur Costantini, Email: arthur.costantini@gmail.com.
Marielle Pasdeloup, Email: marielle.pasdeloup@ibcp.fr.
Jérôme Lafont, Email: jerome.lafont@ibcp.fr.
Maxime Ducret, Email: maxime.ducret@univ-lyon1.fr.
Mourad Bekhouche, Email: mourad.bekhouche@univ-lyon1.fr.
ABBREVIATIONS
- DP
dental pulp;
- DP-MSC
dental pulp mesenchymal stem cells
- REP
regenerative endodontic procedures
- ECM
extracellular matrix
- MMP
matrix metalloproteinase
- TGF-β1
transforming growth factor beta1
- ITGA
integrin alpha
- RT-qPCR
Real time quantitative polymerase chain reaction
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Diogenes A., Ruparel N.B., Shiloah Y., Hargreaves K.M. Regenerative endodontics: a way forward. J. Am. Dent. Assoc. 2016;147(5):372–380. doi: 10.1016/j.adaj.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Godoy F., Murray P.E. Recommendations for using regenerative endodontic procedures in permanent immature traumatized teeth. Dent. Traumatol. 2012;28(1):33–41. doi: 10.1111/j.1600-9657.2011.01044.x. [DOI] [PubMed] [Google Scholar]
- 3.Richert R., Ducret M., Alliot-Licht B., Bekhouche M., Gobert S., Farges J.-C. A critical analysis of research methods and experimental models to study pulpitis. Int. Endod. J. 2022;55(Suppl 1):14–36. doi: 10.1111/iej.13683. [DOI] [PubMed] [Google Scholar]
- 4.Siddiqui Z., Acevedo-Jake A.M., Griffith A., Kadincesme N., Dabek K., Hindi D., Kim K.K., Kobayashi Y., Shimizu E., Kumar V. Cells and material-based strategies for regenerative endodontics. Bioact. Mater. 2022;14:234–249. doi: 10.1016/j.bioactmat.2021.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ducret M., Fabre H., Celle A., Mallein-Gerin F., Perrier-Groult E., Alliot-Licht B., Farges J.-C. Current challenges in human tooth revitalization. Bio Med. Mater. Eng. 2017;28(s1):S159–S168. doi: 10.3233/BME-171637. [DOI] [PubMed] [Google Scholar]
- 6.Galler K.M., D'Souza R.N., Hartgerink J.D., Schmalz G. Scaffolds for dental pulp tissue engineering. Adv. Dent. Res. 2011;23(3):333–339. doi: 10.1177/0022034511405326. [DOI] [PubMed] [Google Scholar]
- 7.Galler K.M., Brandl F.P., Kirchhof S., Widbiller M., Eidt A., Buchalla W., Göpferich A., Schmalz G. Suitability of different natural and synthetic biomaterials for dental pulp tissue engineering. Tissue Eng. 2018;24(3–4):234–244. doi: 10.1089/ten.TEA.2016.0555. [DOI] [PubMed] [Google Scholar]
- 8.Rosa V., Della Bona A., Cavalcanti B.N., Nör J.E. Tissue engineering: from research to dental clinics. Dent. Mater. 2012;28(4):341–348. doi: 10.1016/j.dental.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mangione F., EzEldeen M., Bardet C., Lesieur J., Bonneau M., Decup F., Salmon B., Jacobs R., Chaussain C., Opsahl-Vital S. Implanted dental pulp cells fail to induce regeneration in partial pulpotomies. J. Dent. Res. 2017;96(12):1406–1413. doi: 10.1177/0022034517725523. [DOI] [PubMed] [Google Scholar]
- 10.Moussa D.G., Aparicio C. Present and future of tissue engineering scaffolds for dentin-pulp complex regeneration. Journal of Tissue Engineering and Regenerative Medicine. 2019;13(1):58–75. doi: 10.1002/term.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu X., Wang Y., Liu Y., Huang G.T.-J., Zhang C. Immunohistochemical and histochemical analysis of newly formed tissues in root canal space transplanted with dental pulp stem cells plus platelet-rich plasma. J. Endod. 2014;40(10):1573–1578. doi: 10.1016/j.joen.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Antunes L.S., Salles A.G., Gomes C.C., Andrade T.B., Delmindo M.P., Antunes L.A.A. The effectiveness of pulp revascularization in root formation of necrotic immature permanent teeth: a systematic review. Acta Odontol. Scand. 2016;74(3):161–169. doi: 10.3109/00016357.2015.1069394. [DOI] [PubMed] [Google Scholar]
- 13.Wang X., Thibodeau B., Trope M., Lin L.M., Huang G.T.-J. Histologic characterization of regenerated tissues in canal space after the revitalization/revascularization procedure of immature dog teeth with apical periodontitis. J. Endod. 2010;36(1):56–63. doi: 10.1016/j.joen.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 14.Arango-Gómez E., Nino-Barrera J.L., Nino G., Jordan F., Sossa-Rojas H. Pulp revascularization with and without platelet-rich plasma in two anterior teeth with horizontal radicular fractures: a case report. Restor Dent Endod. 2019;44(4):e35. doi: 10.5395/rde.2019.44.e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willyard C. Unlocking the secrets of scar-free skin healing. Nature. 2018;563(7732):S86–S88. doi: 10.1038/d41586-018-07430-w. [DOI] [PubMed] [Google Scholar]
- 16.Rohani M.G., Parks W.C. Matrix remodeling by MMPs during wound repair. Matrix Biol. 2015;44–46:113–121. doi: 10.1016/j.matbio.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Heissig B., Dhahri D., Eiamboonsert S., Salama Y., Shimazu H., Munakata S., Hattori K. Role of mesenchymal stem cell-derived fibrinolytic factor in tissue regeneration and cancer progression. Cell. Mol. Life Sci. 2015;72(24):4759–4770. doi: 10.1007/s00018-015-2035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakashima M., Iohara K., Murakami M., Nakamura H., Sato Y., Ariji Y., Matsushita K. Pulp regeneration by transplantation of dental pulp stem cells in pulpitis: a pilot clinical study. Stem Cell Res. Ther. 2017;8(1):61. doi: 10.1186/s13287-017-0506-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xuan K., Li B., Guo H., Sun W., Kou X., He X., Zhang Y., Sun J., Liu A., Liao L., Liu S., Liu W., Hu C., Shi S., Jin Y. Deciduous autologous tooth stem cells regenerate dental pulp after implantation into injured teeth. Sci. Transl. Med. 2018;10(455) doi: 10.1126/scitranslmed.aaf3227. [DOI] [PubMed] [Google Scholar]
- 20.Ducret M., Fabre H., Degoul O., Atzeni G., McGuckin C., Forraz N., Alliot-Licht B., Mallein-Gerin F., Perrier-Groult E., Farges J.-C. Manufacturing of dental pulp cell-based products from human third molars: current strategies and future investigations. Front. Physiol. 2015;6:213. doi: 10.3389/fphys.2015.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marei M.K., El Backly R.M. Dental mesenchymal stem cell-based translational regenerative dentistry: from artificial to biological replacement. Front. Bioeng. Biotechnol. 2018;6:49. doi: 10.3389/fbioe.2018.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ducret M., Fabre H., Farges J.-C., Degoul O., Atzeni G., McGuckin C., Forraz N., Mallein-Gerin F., Perrier-Groult E. Production of human dental pulp cells with a medicinal manufacturing approach. J. Endod. 2015;41(9):1492–1499. doi: 10.1016/j.joen.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 23.Fabre H., Ducret M., Degoul O., Rodriguez J., Perrier-Groult E., Aubert-Foucher E., Pasdeloup M., Auxenfans C., McGuckin C., Forraz N., Mallein-Gerin F. Characterization of different sources of human MSCs expanded in serum-free conditions with quantification of chondrogenic induction in 3D. Stem Cell. Int. 2019;2019 doi: 10.1155/2019/2186728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gharaei M.A., Xue Y., Mustafa K., Lie S.A., Fristad I. Human dental pulp stromal cell conditioned medium alters endothelial cell behavior. Stem Cell Res. Ther. 2018;9(1):69. doi: 10.1186/s13287-018-0815-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fracaro L., Senegaglia A.C., Herai R.H., Leitolis A., Boldrini-Leite L.M., Rebelatto C.L.K., Travers P.J., Brofman P.R.S., Correa A. The expression profile of dental pulp-derived stromal cells supports their limited capacity to differentiate into adipogenic cells. Int. J. Mol. Sci. 2020;21(8):E2753. doi: 10.3390/ijms21082753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eckhard U., Marino G., Abbey S.R., Matthew I., Overall C.M. TAILS N-terminomic and proteomic datasets of healthy human dental pulp. Data Brief. 2015;5:542–548. doi: 10.1016/j.dib.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janmey P.A., Winer J.P., Weisel J.W. Fibrin gels and their clinical and bioengineering applications. J. R. Soc. Interface. 2009;6(30):1–10. doi: 10.1098/rsif.2008.0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ducret M., Costantini A., Gobert S., Farges J.-C., Bekhouche M. Fibrin-based scaffolds for dental pulp regeneration: from biology to nanotherapeutics. Eur. Cell. Mater. 2021;41:1–14. doi: 10.22203/eCM.v041a01. [DOI] [PubMed] [Google Scholar]
- 29.Galler K.M. Clinical procedures for revitalization: current knowledge and considerations. Int. Endod. J. 2016;49(10):926–936. doi: 10.1111/iej.12606. [DOI] [PubMed] [Google Scholar]
- 30.Roura S., Gálvez-Montón C., Bayes-Genis, Fibrin A. The preferred scaffold for cell transplantation after myocardial infarction? An old molecule with a new life. J Tissue Eng Regen Med. 2017;11(8):2304–2313. doi: 10.1002/term.2129. [DOI] [PubMed] [Google Scholar]
- 31.Thompson W.D., Campbell R., Evans T. Fibrin degradation and angiogenesis: quantitative analysis of the angiogenic response in the chick chorioallantoic membrane. J. Pathol. 1985;145(1):27–37. doi: 10.1002/path.1711450103. [DOI] [PubMed] [Google Scholar]
- 32.McKenzie R., Pepper D.S., Kay A.B. The generation of chemotactic activity for human leukocytes by the action of plasmin on human fibrinogen. Thromb. Res. 1975;6(1):1–8. doi: 10.1016/0049-3848(75)90145-0. [DOI] [PubMed] [Google Scholar]
- 33.Ruangsawasdi N., Zehnder M., Weber F.E. Fibrin gel improves tissue ingrowth and cell differentiation in human immature premolars implanted in rats. J. Endod. 2014;40(2):246–250. doi: 10.1016/j.joen.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 34.Ducret M., Montembault A., Josse J., Pasdeloup M., Celle A., Benchrih R., Mallein-Gerin F., Alliot-Licht B., David L., Farges J.-C. Design and characterization of a chitosan-enriched fibrin hydrogel for human dental pulp regeneration. Dent. Mater. 2019;35(4):523–533. doi: 10.1016/j.dental.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 35.Höök P., Litvinov R.I., Kim O.V., Xu S., Xu Z., Bennett J.S., Alber M.S., Weisel J.W. Strong binding of platelet integrin αIIbβ3 to fibrin clots: potential target to destabilize thrombi. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-12615-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gailit J., Clarke C., Newman D., Tonnesen M.G., Mosesson M.W., Clark R.A. Human fibroblasts bind directly to fibrinogen at RGD sites through integrin alpha(v)Beta3. Exp. Cell Res. 1997;232(1):118–126. doi: 10.1006/excr.1997.3512. [DOI] [PubMed] [Google Scholar]
- 37.Flick M.J., Du X., Witte D.P., Jiroušková M., Soloviev D.A., Busuttil S.J., Plow E.F., Degen J.L. Leukocyte engagement of fibrin(ogen) via the integrin receptor αmβ2/mac-1 is critical for host inflammatory response in vivo. J. Clin. Invest. 2004;113(11):1596–1606. doi: 10.1172/JCI20741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swieringa F., Spronk H.M.H., Heemskerk J.W.M., van der Meijden P.E.J. Integrating platelet and coagulation activation in fibrin clot formation. Res Pract Thromb Haemost. 2018;2(3):450–460. doi: 10.1002/rth2.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosesson M.W. Fibrinogen and fibrin structure and functions. J. Thromb. Haemostasis. 2005;3(8):1894–1904. doi: 10.1111/j.1538-7836.2005.01365.x. [DOI] [PubMed] [Google Scholar]
- 40.Roberts I.V., Bukhary D., Valdivieso C.Y.L., Tirelli N. Fibrin matrices as (injectable) biomaterials: formation, clinical use, and molecular engineering. Macromol. Biosci. 2020;20(1) doi: 10.1002/mabi.201900283. [DOI] [PubMed] [Google Scholar]
- 41.Collen D., Lijnen H.R. Basic and clinical aspects of fibrinolysis and thrombolysis. Blood. 1991;78(12):3114–3124. [PubMed] [Google Scholar]
- 42.Deryugina E.I., Quigley J.P. Cell surface remodeling by plasmin: a new function for an old enzyme. J. Biomed. Biotechnol. 2012;2012 doi: 10.1155/2012/564259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bini A., Itoh Y., Kudryk B.J., Nagase H. Degradation of cross-linked fibrin by matrix metalloproteinase 3 (stromelysin 1): hydrolysis of the γ gly 404−Ala 405 peptide bond. Biochemistry. 1996;35(40):13056–13063. doi: 10.1021/bi960730c. [DOI] [PubMed] [Google Scholar]
- 44.Hiller O., Lichte A., Oberpichler A., Kocourek A., Tschesche H. Matrix metalloproteinases collagenase-2, macrophage elastase, collagenase-3, and membrane type 1-matrix metalloproteinase impair clotting by degradation of fibrinogen and factor XII. J. Biol. Chem. 2000;275(42):33008–33013. doi: 10.1074/jbc.M001836200. [DOI] [PubMed] [Google Scholar]
- 45.Lelongt B., Bengatta S., Delauche M., Lund L.R., Werb Z., Ronco P.M. Matrix metalloproteinase 9 protects mice from anti-glomerular basement membrane nephritis through its fibrinolytic activity. J. Exp. Med. 2001;193(7):793–802. doi: 10.1084/jem.193.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bekhouche M., Bolon M., Charriaud F., Lamrayah M., Da Costa D., Primard C., Costantini A., Pasdeloup M., Gobert S., Mallein-Gerin F., Verrier B., Ducret M., Farges J.-C. Development of an antibacterial nanocomposite hydrogel for human dental pulp engineering. J. Mater. Chem. B. 2020;8(36):8422–8432. doi: 10.1039/d0tb00989j. [DOI] [PubMed] [Google Scholar]
- 47.Dang W., Zhang X., Ma Q., Chen L., Cao M., Miao J., Cui Y., Zhang X. Selection of reference genes suitable for normalization of RT-qPCR data in glioma stem cells. Biotechniques. 2020;68(3):130–137. doi: 10.2144/btn-2019-0098. [DOI] [PubMed] [Google Scholar]
- 48.Rao X., Huang X., Zhou Z., Lin X. An improvement of the 2^(–delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 2013;3(3):71–85. [PMC free article] [PubMed] [Google Scholar]
- 49.Wei L., Yin F., Zhang W., Li L. ITGA 1 and cell adhesion-mediated drug resistance in ovarian cancer. Int. Clin. Exp. Pathol. 2017;10:5522–55229. [Google Scholar]
- 50.Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J., Billis K., Boddu S., Charkhchi M., Cummins C., Da Rin Fioretto L., Davidson C., Dodiya K., El Houdaigui B., Fatima R., Gall A., Garcia Giron C., Grego T., Guijarro-Clarke C., Haggerty L., Hemrom A., Hourlier T., Izuogu O.G., Juettemann T., Kaikala V., Kay M., Lavidas I., Le T., Lemos D., Gonzalez Martinez J., Marugán J.C., Maurel T., McMahon A.C., Mohanan S., Moore B., Muffato M., Oheh D.N., Paraschas D., Parker A., Parton A., Prosovetskaia I., Sakthivel M.P., Salam A.I.A., Schmitt B.M., Schuilenburg H., Sheppard D., Steed E., Szpak M., Szuba M., Taylor K., Thormann A., Threadgold G., Walts B., Winterbottom A., Chakiachvili M., Chaubal A., De Silva N., Flint B., Frankish A., Hunt S.E., Iisley G.R., Langridge N., Loveland J.E., Martin F.J., Mudge J.M., Morales J., Perry E., Ruffier M., Tate J., Thybert D., Trevanion S.J., Cunningham F., Yates A.D., Zerbino D.R., Flicek P. Ensembl 2021. Nucleic Acids Res. 2021;49(D1):D884–D891. doi: 10.1093/nar/gkaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 2012;13(1):1–11. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh M., Foster C.R., Dalal S., Singh K. Osteopontin: role in extracellular matrix deposition and myocardial remodeling post-MI. J. Mol. Cell. Cardiol. 2010;48(3):538–543. doi: 10.1016/j.yjmcc.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bachmann M., Kukkurainen S., Hytönen V.P., Wehrle-Haller B. Cell adhesion by integrins. Physiol. Rev. 2019;99(4):1655–1699. doi: 10.1152/physrev.00036.2018. [DOI] [PubMed] [Google Scholar]
- 54.Liu Y., Beyer A., Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165(3):535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 55.Eba H., Murasawa Y., Iohara K., Isogai Z., Nakamura H., Nakamura H., Nakashima M. The anti-inflammatory effects of matrix metalloproteinase-3 on irreversible pulpitis of mature erupted teeth. PLoS One. 2012;7(12) doi: 10.1371/journal.pone.0052523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nugud A., Alghfeli L., Elmasry M., El-Serafi I., El-Serafi A.T. Biomaterials as a vital frontier for stem cell-based tissue regeneration. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.713934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao J., Yu X., Wang X., He Y., Ding J. Biomaterial–related cell microenvironment in tissue engineering and regenerative medicine. Engineering. 2022;13:31–45. doi: 10.1016/j.eng.2021.11.025. [DOI] [Google Scholar]
- 58.Minic S., Vital S., Chaussain C., Boukpessi T., Mangione F. Tissue characteristics in endodontic regeneration: a systematic review. Int. J. Mol. Sci. 2022;23(18) doi: 10.3390/ijms231810534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernick S., Nedelman C. Effect of aging on the human pulp. J. Endod. 1975;1(3):88–94. doi: 10.1016/S0099-2399(75)80024-0. [DOI] [PubMed] [Google Scholar]
- 60.Carvalho T.S., Lussi A. Age-related morphological, histological and functional changes in teeth. J. Oral Rehabil. 2017;44(4):291–298. doi: 10.1111/joor.12474. [DOI] [PubMed] [Google Scholar]
- 61.Maeda H. Aging and senescence of dental pulp and hard tissues of the tooth. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.605996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iezzi I., Pagella P., Mattioli-Belmonte M., Mitsiadis T.A. The effects of ageing on dental pulp stem cells, the tooth longevity elixir. Eur. Cell. Mater. 2019;37:175–185. doi: 10.22203/eCM.v037a11. [DOI] [PubMed] [Google Scholar]
- 63.Barczyk M., Carracedo S., Gullberg D. Integrins. Cell Tissue Res. 2010;339(1):269–280. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Page-McCaw A., Ewald A.J., Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 2007;8(3):221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ravanti L., Heino J., López-Otín C., Kähäri V.M. Induction of collagenase-3 (MMP-13) expression in human skin fibroblasts by three-dimensional collagen is mediated by P38 mitogen-activated protein kinase. J. Biol. Chem. 1999;274(4):2446–2455. doi: 10.1074/jbc.274.4.2446. [DOI] [PubMed] [Google Scholar]
- 66.Steffensen B., Häkkinen L., Larjava H. Proteolytic events of wound-healing — coordinated interactions among matrix metalloproteinases (MMPs), integrins, and extracellular matrix molecules. Crit. Rev. Oral Biol. Med. 2001;12(5):373–398. doi: 10.1177/10454411010120050201. [DOI] [PubMed] [Google Scholar]
- 67.Stetler-Stevenson W.G. Tissue inhibitors of metalloproteinases in cell signaling: metalloproteinase-independent biological activities. Sci. Signal. 2008;1(27):re6. doi: 10.1126/scisignal.127re6. re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brew K., Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim. Biophys. Acta. 2010;1803(1):55–71. doi: 10.1016/j.bbamcr.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joo C.-K., Seomun Y. Matrix metalloproteinase (MMP) and TGF-β1-stimulated cell migration in skin and cornea wound healing. Cell Adhes. Migrat. 2008;2(4):252–253. doi: 10.4161/cam.2.4.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Standal T., Borset M., Sundan A. Role of osteopontin in adhesion, migration, cell survival and bone remodeling. Exp. Oncol. 2004;26(3):179–184. [PubMed] [Google Scholar]
- 71.Choo H.-J., Canner J.P., Vest K.E., Thompson Z., Pavlath G.K. A tale of two niches: differential functions for VCAM-1 in satellite cells under basal and injured conditions. Am. J. Physiol. Cell Physiol. 2017;313(4):C392–C404. doi: 10.1152/ajpcell.00119.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo B.-H., Carman C.V., Springer T.A. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li R., Mitra N., Gratkowski H., Vilaire G., Litvinov R., Nagasami C., Weisel J.W., Lear J.D., DeGrado W.F., Bennett J.S. Activation of integrin αIIbß3 by modulation of transmembrane helix associations. Science. 2003;300(5620):795–798. doi: 10.1126/science.1079441. [DOI] [PubMed] [Google Scholar]
- 74.Buensuceso C., de Virgilio M., Shattil S.J. Detection of integrin αIIbβ3Clustering in living cells. J. Biol. Chem. 2003;278(17):15217–15224. doi: 10.1074/jbc.M213234200. [DOI] [PubMed] [Google Scholar]
- 75.Kim M., Carman C.V., Yang W., Salas A., Springer T.A. The primacy of affinity over clustering in regulation of adhesiveness of the integrin αLβ2. JCB (J. Cell Biol.) 2004;167(6):1241–1253. doi: 10.1083/jcb.200404160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grenache D.G., Zhang Z., Wells L.E., Santoro S.A., Davidson J.M., Zutter M.M. Wound healing in the Α2β1 integrin-deficient mouse: altered keratinocyte biology and dysregulated matrix metalloproteinase expression. J. Invest. Dermatol. 2007;127(2):455–466. doi: 10.1038/sj.jid.5700611. [DOI] [PubMed] [Google Scholar]
- 77.Peters M.A., Wendholt D., Strietholt S., Frank S., Pundt N., Korb-Pap A., Joosten L.A.B., van den Berg W.B., Kollias G., Eckes B., Pap T. The loss of Α2β1 integrin suppresses joint inflammation and cartilage destruction in mouse models of rheumatoid arthritis. Arthritis Rheum. 2012;64(5):1359–1368. doi: 10.1002/art.33487. [DOI] [PubMed] [Google Scholar]
- 78.Jokinen J., Dadu E., Nykvist P., Käpylä J., White D.J., Ivaska J., Vehviläinen P., Reunanen H., Larjava H., Häkkinen L., Heino J. Integrin-mediated cell adhesion to type I collagen fibrils. J. Biol. Chem. 2004;279(30):31956–31963. doi: 10.1074/jbc.M401409200. [DOI] [PubMed] [Google Scholar]
- 79.Moreno-Layseca P., Icha J., Hamidi H., Ivaska J. Integrin trafficking in cells and tissues. Nat. Cell Biol. 2019;21(2):122–132. doi: 10.1038/s41556-018-0223-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bonnans C., Chou J., Werb Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014;15(12):786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pilcher B.K., Dumin J.A., Sudbeck B.D., Krane S.M., Welgus H.G., Parks W.C. The activity of collagenase-1 is required for keratinocyte migration on a type I collagen matrix. J. Cell Biol. 1997;137(6):1445–1457. doi: 10.1083/jcb.137.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guan S.P., Lam A.T.L., Newman J.P., Chua K.L.M., Kok C.Y.L., Chong S.T., Chua M.L.K., Lam P.Y.P. Matrix metalloproteinase-1 facilitates MSC migration via cleavage of IGF-2/IGFBP2 complex. FEBS Open Bio. 2018;8(1):15–26. doi: 10.1002/2211-5463.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bini A., Wu D., Schnuer J., Kudryk B.J. Characterization of stromelysin 1 (MMP-3), matrilysin (MMP-7), and membrane type 1 matrix metalloproteinase (MT1-MMP) derived fibrin(ogen) fragments D-dimer and D-like monomer: NH2-terminal sequences of late-stage digest fragments. Biochemistry. 1999;38(42):13928–13936. doi: 10.1021/bi991096g. [DOI] [PubMed] [Google Scholar]
- 84.Kumar L., Planas-Iglesias J., Harms C., Kamboj S., Wright D., Klein-Seetharaman J., Sarkar S.K. Activity-dependent interdomain dynamics of matrix metalloprotease-1 on fibrin. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-77699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wan J., Zhang G., Li X., Qiu X., Ouyang J., Dai J., Min S. Matrix metalloproteinase 3: a promoting and destabilizing factor in the pathogenesis of disease and cell differentiation. Front. Physiol. 2021;12 doi: 10.3389/fphys.2021.663978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen Q., Jin M., Yang F., Zhu J., Xiao Q., Zhang L. Matrix metalloproteinases: inflammatory regulators of cell behaviors in vascular formation and remodeling. Mediat. Inflamm. 2013;2013 doi: 10.1155/2013/928315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jobin P.G., Butler G.S., Overall C.M. New intracellular activities of matrix metalloproteinases shine in the moonlight. Biochim. Biophys. Acta, Mol. Cell Res. 2017;1864(11 Pt A):2043–2055. doi: 10.1016/j.bbamcr.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 88.Bassiouni W., Ali M.A.M., Schulz R. Multifunctional intracellular matrix metalloproteinases: implications in disease. FEBS J. 2021;288(24):7162–7182. doi: 10.1111/febs.15701. [DOI] [PubMed] [Google Scholar]
- 89.Si-Tayeb K., Monvoisin A., Mazzocco C., Lepreux S., Decossas M., Cubel G., Taras D., Blanc J.-F., Robinson D.R., Rosenbaum J. Matrix metalloproteinase 3 is present in the cell nucleus and is involved in apoptosis. Am. J. Pathol. 2006;169(4):1390–1401. doi: 10.2353/ajpath.2006.060005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Montero A., Acosta S., Hernández R., Elvira C., Jorcano J.L., Velasco D. Contraction of fibrin-derived matrices and its implications for in vitro human skin bioengineering. J. Biomed. Mater. Res. 2021;109(4):500–514. doi: 10.1002/jbm.a.37033. [DOI] [PubMed] [Google Scholar]
- 91.Michielin F., Giobbe G.G., Luni C., Hu Q., Maroni I., Orford M.R., Manfredi A., Di Filippo L., David A.L., Cacchiarelli D., De Coppi P., Eaton S., Elvassore N. The microfluidic environment reveals a hidden role of self-organizing extracellular matrix in hepatic commitment and organoid formation of hiPSCs. Cell Rep. 2020;33(9) doi: 10.1016/j.celrep.2020.108453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.