Abstract
Islands are crucial evolutionary hotspots, providing unique opportunities for differentiation of novel biodiversity and long-term segregation of endemic species. Islands are also fragile ecosystems, where biodiversity is more exposed to environmental and anthropogenic pressures than on continents. The Ponza grayling, Hipparchia sbordonii, is an endemic butterfly species that is currently found only in two tiny islands of the Pontine archipelago, off the coast of Italy, occupying an area smaller than 10 km2. It has been classified as Endangered (IUCN) because of the extremely limited area of occurrence, population fragmentation, and the recent demographic decline. Thanks to a combination of different assemblers of long and short genomic reads, bulk transcriptome RNAseq, and synteny analysis with phylogenetically close butterflies, we produced a highly contiguous, chromosome-scale annotated reference genome for the Ponza grayling, including 28 autosomes and the Z sexual chromosomes. The final assembly spanned 388.61 Mb with a contig N50 of 14.5 Mb and a BUSCO completeness score of 98.5%. Synteny analysis using four other butterfly species revealed high collinearity with Hipparchia semele and highlighted 10 intrachromosomal inversions longer than 10 kb, of which two appeared on the lineage leading to H. sbordonii. Our results show that a chromosome-scale reference genome is attainable also when chromatin conformation data may be impractical or present specific technical challenges. The high-quality genomic resource for H. sbordonii opens up new opportunities for the accurate assessment of genetic diversity and genetic load and for the investigations of the genomic novelties characterizing the evolutionary path of this endemic island species.
Keywords: conservation genomics, island biogeography, endemic species, Endemixit, Nymphalidae
Significance.
Islands are biodiversity hotspots often harboring unique endemic species that are inherently more vulnerable to environmental and anthropogenic factors. The Ponza grayling, Hipparchia sbordonii, is an endemic butterfly found in only two islands of the Pontine archipelago, along the west coast of Italy. In this study, we provide a highly contiguous chromosome-scale reference genome of the Ponza grayling that will be a cornerstone of genomics-informed conservation actions in this species and a useful resource supporting investigations into butterfly evolution in general.
Introduction
Although islands contribute only 6.7% of land surface area, they harbor ∼20% of the earth's biodiversity (Kier et al. 2009; Sayre et al. 2019). Unfortunately, they also account for ∼50% of the threatened species and 75% of the known extinctions since European expansion around the globe (Russell and Kueffer 2019). Due to their geological and geographical history and characteristics, islands act simultaneously as cradles of evolutionary diversity and museums of formerly widespread lineages, achieving outstanding endemicity (Cronk 1997). Nevertheless, the majority of these endemic species are inherently vulnerable due to genetic and demographic factors linked with the way islands are colonized (Fernández-Palacios et al. 2021). Additionally, island populations can be small in size, cannot easily move to track their habitat, and, therefore, are more often at risk of extinction (Frankham 1997). Small populations are characterized by reduced genetic diversity, higher effects of genetic drift, and, hence, higher realized genetic load (Bertorelle et al. 2022).
In this study, we present the high-quality chromosome-level genome of the endangered island endemic Ponza grayling, Hipparchia sbordonii (Nymphalidae: Satyrinae). This species is found only in the Pontine archipelago, located west of Naples, with an extremely restricted range of occurrence (Fig. 1a). The historical total area of occupancy is limited to the three islands of Ponza, Palmarola, and Zannone and has been estimated to be as small as 16 km2 (https://www.iucnredlist.org/species/173231/64640021). However, the butterfly has not been found in Palmarola and Zannone in recent years (Bonelli et al. 2018), thus further reducing its distribution. As a result, the status of the Ponza grayling as Endangered in the Red List of Endangered Species of the IUCN (2009) could be reviewed for the worse. The main threat to its survival appears to be improper land and biodiversity management (Bonelli et al. 2018; Sbordoni 2018). The high-quality reference genome of the Ponza grayling is a valuable resource to investigate the genomic consequences of thriving at small (and further reducing) population size, and it also constitutes the basis to explore the genomic features characterizing the unique evolutionary pathways of this butterfly, a natural experiment of island biodiversity.
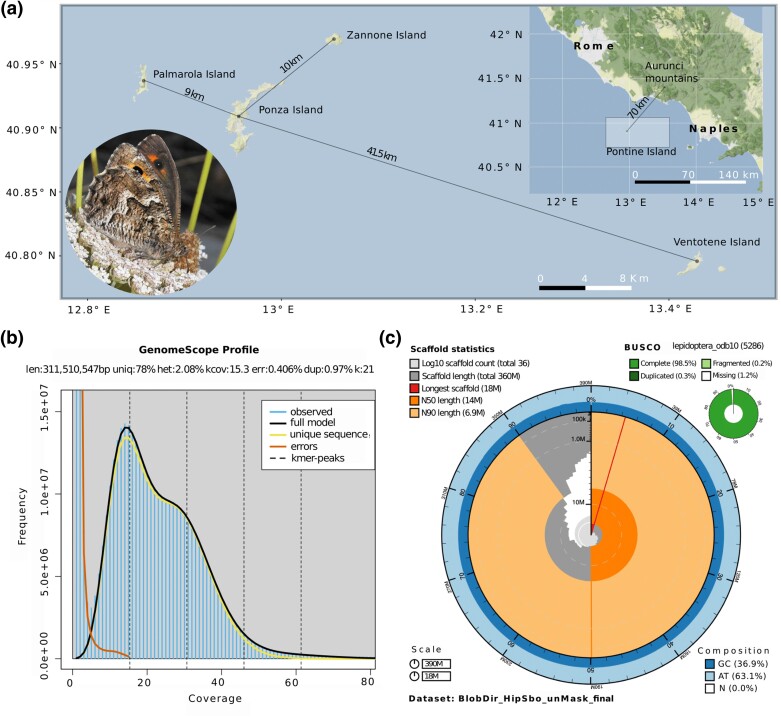
Fig. 1.
(a) Hipparchia sbordonii is present in the Pontine islands with the exception of Ventotene (large map), about 70 km from the west coast of Italy (inset map). A picture of one individual from Ponza Island (round inset; photo credit: Valerio Sbordoni). (b) K-mer spectra, genome size, and heterozygosity estimated with GenomeScope 2.0. (c) A BlobToolKit Snail plot showing a graphical representation of the quality metrics for the H. sbordonii primary assembly.
Results
The final genome size of H. sbordonii (388.61 Mb) is consistent with the size predicted by the k-mer spectra with Genomescope2.0 (Fig. 1b) and closely resembles the genome size of Hipparchia semele for which a reference genome is available (403 Mb; NCBI Accession: GCA_933228805.2). The k-mer spectrum shows a bimodal distribution with two major peaks, at ∼15- and ∼30-fold coverage, corresponding to heterozygous and homozygous states, respectively. Based on Illumina reads, we estimated a 2.08% nucleotide heterozygosity rate (Fig. 1b). The mitochondrial genome size is 15,321 bp, which is in agreement with the mitochondrial genome size of its sister species H. semele (15,223 bp; OW121739.2; see supplementary table S1 and fig. S1, Supplementary Material online). The primary assembly of H. sbordonii contains 36 scaffolds with an N50 of 14.5 Mb and the longest scaffold of 17.7 Mb (Fig. 1c; supplementary table S2, Supplementary Material online). The alternative assembly contains 1,606 scaffolds spanning 352.9 Mb, having an N50 of 409.7 kb. The completeness of the primary assembly is very high, with a BUSCO completeness score of 98.5% ([single copy: 98.2%, duplicated: 0.3%], fragmented: 0.2%, missing: 1.3%) using the Lepidoptera gene set, high-read back-mapping rates (91.91% and 93.76% from genomic and transcriptomic libraries, respectively), a k-mer completeness score of 83.01%, and a per-base quality value (QV) of 40.87. In total, 16,346 protein-coding genes were predicted. The BUSCO completeness score of the gene annotation using the Lepidoptera gene set was 97% ([single copy: 96.2%, duplicated: 0.8%], fragmented: 1.2%, missing: 2.3%), and gene annotation rates were 73.66% and 58.88% for InterPro and gene ontology (GO) terms, respectively. Following the annotation of noncoding RNA, we identified 6,068 putative transfer RNAs (tRNAs), of which 24 are potential suppressor tRNAs, 614 unknown isotypes, and 5,430 standard tRNAs. These results are consistent with those obtained on H. semele (see supplementary table S3, Supplementary Material online). The identification of repetitive elements resulted in a 40.46% repeat content. The major class of repetitive elements was constituted by DNA transposons (see supplementary table S4, Supplementary Material online). The alignment of the genomes of H. sbordonii and H. semele revealed 10 chromosomal inversions exceeding a size of 10 kb (supplementary table S5 and fig. S2, Supplementary Material online). The conservation of the same karyotype between this species and Hipparchia (Wiemers et al. 2020) allowed us to confirm that 23 out of the 28 autosomes expected to be present in H. sbordonii were correctly assembled to their full length (Fig. 2), with the exception of the presence of some unassembled telomeric ends, which we estimated to account for ∼2.1 Mb (supplementary table S6, Supplementary Material online). Chromosomes 15, 18, 23, and 26 were split between two scaffolds in the H. sbordonii assembly. Chromosome 27 displayed the highest level of fragmentation, corresponding to three contigs in the Ponza grayling. Concerning the two chromosomes involved in sex determination, W was not present in the H. sbordonii reference genome due to the fact that the sequenced individual was a male. On the other hand, the Z chromosome was present and matched two contigs in H. sbordonii (supplementary table S2, Supplementary Material online).
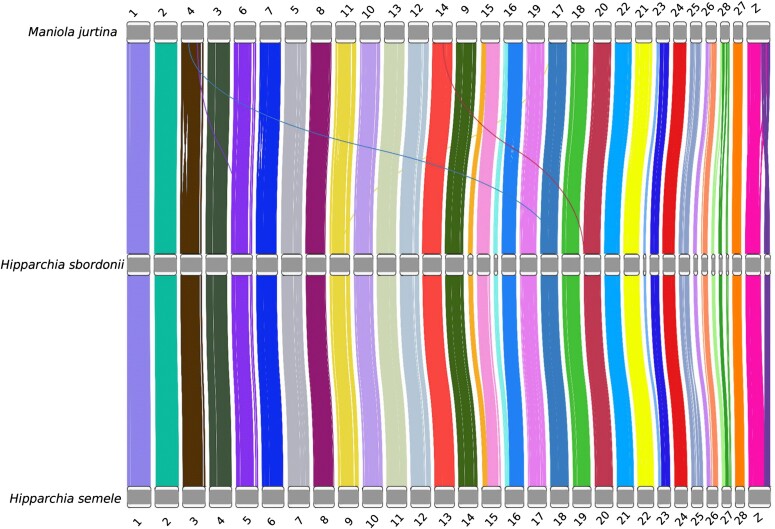
Fig. 2.
Conserved synteny between the chromosome-scale assemblies of M. jurtina (version ilManJurt1.1, top), H. semele (ilHipSeme1.2, bottom), and the genome assembly of H. sbordonii reported in this study (middle). Synteny blocks are highlighted by lines connecting the orthologous genes identified in the three species, colored based on their placement on each of the 36 scaffolds obtained in H. sbordonii. Note that the W chromosome is not reported in this plot due to its absence in the H. sbordonii reference genome, obtained from a male individual.
Discussion
Island endemics have a greater susceptibility to anthropogenic changes due to their small range size, geographic isolation, and a peculiar evolutionary history characterized by a potentially low initial founding size and long-term maintenance of small populations. Island endemics are, therefore, predisposed to lower genetic diversity and higher rates of inbreeding (Frankham 1997). We presented the high-quality chromosome-scale genome assembly for the Ponza grayling (36 scaffolds, N50: 14.5 Mb), a beneficial asset to investigate the genomic peculiarities of this endangered endemic island butterfly. The alignment between the genomes of H. sbordonii and H. semele showed a very high synteny, suggesting that both assemblies are structurally accurate and that the two species share a very similar chromosomal organization (Fig. 2). Despite missing Hi-C data, our genomic reconstruction exhibited notable integrity. While Hi-C data are a powerful tool for refining genome structures, our strategy showcased the efficacy of alternative methodologies in achieving chromosomal-scale assemblies, especially in cases where Hi-C might be impractical to obtain or might present technical challenges.
Materials and Methods
Sampling, Genomic DNA Extraction and Sequencing
One specimen of H. sbordonii was sampled in Ponza Island in June 2019. The individual was immediately frozen in liquid nitrogen to preserve the integrity of nucleic acids. High-molecular-weight DNA was isolated from the head and the thorax using the Nanobind Tissue big DNA kit (Circulomics Inc., Baltimore, MD, USA). DNA quality and fragment length were checked by employing pulse-field gel electrophoresis, and DNA concentration was measured with fluorometric and spectrophotometric assays using a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA) and a TECAN Nanoquant Infinite 200 Pro (Tecan Mannedorf, Switzerland), respectively. Fragments of 30 kb length were selected using a Blue Pippin device (Sage Science, Beverly, MA, USA). Isolated fragments were used to prepare the DNA library with a SMRTbell express template prep kit 2.0 (Pacific Biosciences, Menlo Park, CA, USA), according to the manufacturer's protocols. The library was run on four PacBio SMRT Cells 1 M in continuous long-read (CLR) sequencing mode on a PacBio Sequel platform. Extracted DNA was also used to construct a short-read genomic library using a Illumina DNA PCR-Free Prep Kit (Illumina), according to the manufacturer's protocol. The target coverage was 10 to 15×. The library was sequenced paired-end on an Illumina NovaSeq 6000 System using a 300-cycle Reagent Kit v1.5.
RNA Extraction and Sequencing
Upon sagittal dissection, approximately half of the body of an adult male individual was placed in a plastic tube with a 1 ml RNA-Solv reagent (Omega Bio-tek, Norcross, GA, USA) and five paramagnetic beads. RNA extraction was performed following the manufacturer's instructions after grinding the tissues for 1 min with a bead-beater homogenizer. RNA was further purified using a Direct-zol RNA Miniprep kit (Zymo Research, Irvine, CA, USA), with an additional DNaseI treatment to remove residual genomic DNA contamination. Total extracted RNA was used as an input for the preparation of a poly(A)-selected library with a TruSeq library preparation kit (Illumina, San Diego, CA, USA), which was subjected to RNA sequencing on an Illumina NovaSeq 6000 platform at the Genomic Core Facility of AREA Science Park (Trieste, Italy), using a 2 × 150 bp paired-end sequencing strategy. Raw reads were trimmed with fastp (Chen et al. 2018), removing sequencing adapters and nucleotides characterized by poor-quality scores. After trimming, reads shorter than 75 nucleotides were discarded.
Genome Assembly
In this study, we applied a multiassembler approach to reconstruct a chromosomal-scale genome without using Hi-C data (supplementary table S7 and fig. S3, Supplementary Material online). Firstly, PacBio CLRs and Illumina reads were filtered to remove remnant adapter sequences. After trimming with Trimmomatic (Bolger et al. 2014), Illumina short reads were used to estimate the genome size using a k-mer based approach with Jellyfish (Marçais and Kingsford 2011). The distribution of k-mers (k = 31) was calculated, and GenomeScope 2.0 (Ranallo-Benavidez et al. 2020) was then used to infer the genome size, repeat content, and genome-wide heterozygosity. The genome of H. sbordonii was assembled with Canu (Koren et al. 2017) using PacBio CLRs, as a set of primary contigs, representing the initial haploid assembly, and separating alternative haplotypic variants. Following the assembly, PacBio subreads were mapped to assembly and sorted using pbmm2 package (https://github.com/PacificBiosciences/pbmm2/) from SMRT analysis software (https://github.com/PacificBiosciences/pbbioconda). The mapped PacBio subreads were then used for polishing the contigs with GCpp v 2.0.2 (https://github.com/PacificBiosciences/gcpp) that use Arrow algorithm (https://github.com/PacificBiosciences/gcpp). Following polishing with GCpp, we carried out two rounds of polishing using the Illumina reads to further fix the indel errors in the contigs with POLCA (POLishing by Calling Alternatives; Zimin and Salzberg 2020). Primary contigs were then processed with purge_dups (Guan et al. 2020) to identify residual haplotype duplication in the assembly. Such duplicated sequences were moved to the alternate assembly to remain with a final set of nonredundant haplotypic variants. The contigs included in the primary assembly were anchored to scaffolds, exploiting long-read information, using LRscaf (Qin et al. 2019). TGS-GapCloser was then used to fill the gaps that originated during the scaffolding step due to the presence of repeats or regions characterized by low coverages (Xu et al. 2020). After gap filling, a final polishing step was performed with POLCA, using Illumina reads, to correct any errors made in the scaffolding and gap filling steps. The assembly was subjected to a second round of analysis with Purge_dups to remove any duplicates that may have been added during the last scaffolding and gap filling steps, obtaining the haploid primary genome assembly. The two haplotype fasta files generated by the purging process, representing duplicated genomic sequences most likely ascribable to the alternative haplotype, were merged. The GetOrganelle and MITOS toolkits were then used to assemble and annotate the mitochondrial genome sequence, respectively (Bankevich et al. 2012; Bernt et al. 2013; Jin et al. 2020). The genome was also assembled using PacBio long reads with three other distinct assemblers, Haslr (Haghshenas et al. 2020), miniasm (Li 2016), and Raven (Vaser and Šikić 2021; supplementary fig. S3, Supplementary Material online).
Manual Curation and Synteny Analysis
The primary genome assembly obtained with Canu was selected as the reference genome for manual curation due to its superior quality compared with the three other assemblies. Orthologous genes among the four assemblies were identified using BUSCO (Manni et al. 2021) with Lepidoptera ODB v.10 database and OrthoFinder (Emms and Kelly 2019). OrthoFinder was further used to identify 1:1 orthologous genes between H. sbordonii and other closely related butterflies’ chromosome-scale genomes (H. semele—GCA_933228805.1; Pararge aegeria—GCF_905163445.1; Maniola jurtina—GCF_905333055.1; and Maniola hyperantus—GCF_902806685.1) in order to leverage synteny and orthologous gene information from closely related species and improve the genome assembly of H. sbordonii. The Whole Genome Alignment plugin in CLC Genomics Workbench21 (Qiagen, Hilden, Germany) was used to align the genome assemblies by setting the seed value to 115 and by not allowing mismatches. We merged distinct scaffolds of the primary assembly of H. sbordonii into super-scaffolds only if the following criteria were met: (i) the joining of the two neighboring scaffolds was supported by at least one of the three available auxiliary assemblies; (ii) the relative placement of the two joined scaffolds was corroborated by synteny data from at least one chromosome-scale genome assembly of a species belonging to the Satyrinae subfamily.
Genome Assemblies’ Quality Assessment
Prior to gene annotation, we assessed the quality of the genome assembly using (i) BUSCO v5.2.2 in the genome mode with default parameters against the lepidopteran dataset included in ODB v.10 (lepidoptera_odb10) and (ii) Merqury v1.3 (Rhie et al. 2020) to estimate the base-level accuracy (QV) and the assembly completeness, by comparing the k-mers represented in the assembly and those observed in the Illumina reads. All assembly metrics were computed using FASTA tools (https://github.com/b-brankovics/fasta_tools; see supplementary table S8, Supplementary Material online). To have a summarized graphical representation of the quality of the genome assembly, we employed BlobToolKit2 (Challis et al. 2020).
Repetitive Elements, Gene Models, and Noncoding RNA Annotation
To identify and annotate repetitive elements, we first generated a de novo repeat library using the Extensive de novo TE Annotator v1.9.9 (Ou et al. 2019). Subsequently, we refined the library using DeepTE (Yan et al. 2020), which employs convolutional neural networks to classify unknown elements at the order and superfamily levels. Then, we used RepeatMasker v4.1.2 (Smit et al. 2015) with the final library to mask the genome and parsed the RepeatMasker output file with RM_TRIPS script (https://github.com/clbutler/RM_TRIPS). Transposable element landscapes were generated using the RepeatMasker script calcDivergenceFromAlign.pl (supplementary fig. S4, Supplementary Material online). The final version of the H. sbordonii genome underwent gene model annotation using BRAKER2 (Hoff et al. 2019), with the proteomes of 28 lepidopteran species (from NCBI) as a custom reference protein database for homology detection. Moreover, Illumina paired-end RNAseq data generated from the whole body of a single H. sbordonii individual were supplied to provide transcriptomic evidence. AGAT tools (Dainat et al. 2022) were employed to adjust the output of BRAKER. The final set of annotated proteins was evaluated using BUSCO. The protein sequences generated from the in silico translation of annotated gene models were subjected to InterProScan analysis (Jones et al. 2014), to assign PFAM functional domain (Mistry et al. 2021), and GO terms (Ashburner et al. 2000). Additionally, INFERNAL (Kalvari et al. 2018) with cmscan was used to annotate the most conserved classes of noncoding RNAs, and tRNAscan-SE was used to predict tRNA genes.
Supplementary Material
Acknowledgments
This work is dedicated to Valerio Sbordoni, who passed away on February 6, 2024. Valerio trained and motivated generations of scholars with his passion for exploring the biodiversity and evolution of butterflies and many other species in Italy and around the world. The genome we are presenting here, of the species that bears his name, will also be a way to remember him.
Contributor Information
Sebastiano Fava, Department of Life and Environmental Sciences, Marche Polytechnic University, Ancona, Italy.
Marco Sollitto, Department of Life Sciences, University of Trieste, Trieste, Italy.
Mbarsid Racaku, Department of Life Sciences, University of Trieste, Trieste, Italy.
Alessio Iannucci, Department of Biology, University of Florence, Florence, Italy.
Andrea Benazzo, Department of Life Sciences and Biotechnology, University of Ferrara, Ferrara, Italy.
Lorena Ancona, Department of Life and Environmental Sciences, Marche Polytechnic University, Ancona, Italy.
Paolo Gratton, Department of Biology, University of Rome “Tor Vergata”, Rome, Italy.
Fiorella Florian, Department of Life Sciences, University of Trieste, Trieste, Italy.
Alberto Pallavicini, Department of Life Sciences, University of Trieste, Trieste, Italy.
Claudio Ciofi, Department of Biology, University of Florence, Florence, Italy.
Donatella Cesaroni, Department of Biology, University of Rome “Tor Vergata”, Rome, Italy.
Marco Gerdol, Department of Life Sciences, University of Trieste, Trieste, Italy.
Valerio Sbordoni, Department of Biology, University of Rome “Tor Vergata”, Rome, Italy.
Giorgio Bertorelle, Department of Life Sciences and Biotechnology, University of Ferrara, Ferrara, Italy.
Emiliano Trucchi, Department of Life and Environmental Sciences, Marche Polytechnic University, Ancona, Italy.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online.
Funding
This work was supported by the University of Ferrara (Italy) and funded by the MIUR PRIN 2017 grant 201794ZXTL to G.B.
Data Availability
Raw sequencing data, primary genome assembly, and mitochondrial DNA sequence are available under NCBI BioProject PRJNA1089943. Gene annotations and RepeatMasker output are available at https://zenodo.org/records/11204878.
Literature Cited
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000:25(1):25–29. 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012:19(5):455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 2013:69(2):313–319. 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- Bertorelle G, Raffini F, Bosse M, Bortoluzzi C, Iannucci A, Trucchi E, Morales HE, van Oosterhout C. Genetic load: genomic estimates and applications in non-model animals. Nat Rev Genet. 2022:23(8):492–503. 10.1038/s41576-022-00448-x. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014:30(15):2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonelli S, Casacci LP, Barbero F, Cerrato C, Dapporto L, Sbordoni V, Scalercio S, Zilli A, Battistoni A, Teofili C, et al. The first red list of Italian butterflies. Insect Conserv Divers. 2018:11(5):506–521. 10.1111/icad.12293. [DOI] [Google Scholar]
- Challis R, Richards E, Rajan J, Cochrane G, Blaxter M. BlobToolKit–interactive quality assessment of genome assemblies. G3 (Bethesda). 2020:10(4):1361–1374. 10.1534/g3.119.400908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018:34(17):i884–i890. 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronk QC. Islands: stability, diversity, conservation. Biodivers Conserv. 1997:6(3):477–493. 10.1023/A:1018372910025. [DOI] [Google Scholar]
- Dainat J, Hereñú D; LucileSol; pascal-git . 2022. NBISweden/AGAT: AGAT-v0.8.1. Zenodo. https://zenodo.org/record/5834795. software site: https://agat.readthedocs.io/en/latest/how_to_cite.html
- Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019:20(1):238. 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Palacios JM, Kreft H, Irl SDH, Norder S, Ah-Peng C, Borges PAV, Burns KC, de Nascimento L, Meyer J-Y, Montes E, et al. Scientists’ warning—the outstanding biodiversity of islands is in peril. Global Ecol Conserv. 2021:31:e01847. 10.1016/j.gecco.2021.e01847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. Do island populations have less genetic variation than mainland populations? Heredity (Edinb). 1997:78(Pt 3):311–327. 10.1038/hdy.1997.46. [DOI] [PubMed] [Google Scholar]
- Guan D, McCarthy SA, Wood J, Howe K, Wang Y, Durbin R. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 2020:36(9):2896–2898. 10.1093/bioinformatics/btaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghshenas E, Asghari H, Stoye J, Chauve C, Hach F. HASLR: fast hybrid assembly of long reads. Iscience. 2020:23(8):101389. 10.1016/j.isci.2020.101389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lomsadze A, Borodovsky M, Stanke M. Whole-genome annotation with BRAKER. Methods Mol Biol. 2019:1962:65–95. 10.1007/978-1-4939-9173-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020:21(1):241. 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014:30(9):1236–1240. 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, Nawrocki EP, Argasinska J, Quinones-Olvera N, Finn RD, Bateman A, Petrov AI. Non-coding RNA analysis using the Rfam database. Curr Protoc Bioinformatics. 2018:62(1):e51. 10.1002/cpbi.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kier G, Kreft H, Lee TM, Jetz W, Ibisch PL, Nowicki C, Mutke J, Barthlott W. A global assessment of endemism and species richness across island and mainland regions. Proc Natl Acad Sci U S A. 2009:106(23):9322–9327. 10.1073/pnas.0810306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017:27(5):722–736. 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics. 2016:32(14):2103–2110. 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Zdobnov EM. BUSCO: assessing genomic data quality and beyond. Curr Protoc. 2021:1(12):e323. 10.1002/cpz1.323. [DOI] [PubMed] [Google Scholar]
- Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011:27(6):764–770. 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021:49(D1):D412–D419. 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Su W, Liao Y, Chougule K, Agda JRA, Hellinga AJ, Lugo CSB, Elliott TA, Ware D, Peterson T, et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 2019:20(1):275. 10.1186/s13059-019-1905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M, Wu S, Li A, Zhao F, Feng H, Ding L, Ruan J. LRScaf: improving draft genomes using long noisy reads. BMC Genomics. 2019:20(1):955. 10.1186/s12864-019-6337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo-Benavidez TR, Jaron KS, Schatz MC. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 2020:11(1):1432. 10.1038/s41467-020-14998-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, Walenz BP, Koren S, Phillippy AM. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020:21(1):245. 10.1186/s13059-020-02134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JC, Kueffer C. Island biodiversity in the anthropocene. Annu Rev Environ Resour. 2019:44(1):31–60. 10.1146/annurev-environ-101718-033245. [DOI] [Google Scholar]
- Sayre R, Noble S, Hamann S, Smith R, Wright D, Breyer S, Butler K, Van Graafeiland K, Frye C, Karagulle D, et al. A new 30 meter resolution global shoreline vector and associated global islands database for the development of standardized ecological coastal units. J Oper Oceanogr. 2019:12(sup2):S47–S56. 10.1080/1755876X.2018.1529714. [DOI] [Google Scholar]
- Sbordoni Valerio. Aspetti genetici ed ecologici del declino di popolazioni di farfalle e altri insetti. Atti Accademia Nazionale Italiana di Entomologia, LXVI; 2018. p. 159–168. [Google Scholar]
- Smit A, Hubley R, Green P. RepeatMasker Open-4.0; 2015. 2013–2015.
- Vaser R, Šikić M. Time-and memory-efficient genome assembly with Raven. Nat Comput Sci. 2021:1(5):332–336. 10.1038/s43588-021-00073-4. [DOI] [PubMed] [Google Scholar]
- Wiemers M, Chazot N, Wheat CW, Schweiger O, Wahlberg N. A complete time-calibrated multi-gene phylogeny of the European butterflies. ZooKeys. 2020:938:97–124. 10.3897/zookeys.938.50878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Guo L, Gu S, Wang O, Zhang R, Peters BA, Fan G, Liu X, Xu X, Deng L, et al. TGS-GapCloser: a fast and accurate gap closer for large genomes with low coverage of error-prone long reads. GigaScience. 2020:9(9):giaa094. 10.1093/gigascience/giaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Bombarely A, Li S. DeepTE: a computational method for de novo classification of transposons with convolutional neural network. Bioinformatics. 2020:36(15):4269–4275. 10.1093/bioinformatics/btaa519. [DOI] [PubMed] [Google Scholar]
- Zimin AV, Salzberg SL. The genome polishing tool POLCA makes fast and accurate corrections in genome assemblies. PLoS Comput Biol. 2020:16(6):e1007981. 10.1371/journal.pcbi.1007981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Dainat J, Hereñú D; LucileSol; pascal-git . 2022. NBISweden/AGAT: AGAT-v0.8.1. Zenodo. https://zenodo.org/record/5834795. software site: https://agat.readthedocs.io/en/latest/how_to_cite.html
Supplementary Materials
Data Availability Statement
Raw sequencing data, primary genome assembly, and mitochondrial DNA sequence are available under NCBI BioProject PRJNA1089943. Gene annotations and RepeatMasker output are available at https://zenodo.org/records/11204878.