Abstract
Rhesus monkey rhadinovirus (RRV) is a gamma-2 herpesvirus that is most closely related to the human Kaposi’s sarcoma-associated herpesvirus (KSHV). We have identified a distinct open reading frame at the left end of RRV and designated it R1. The position of the R1 gene is equivalent to that of the saimiri transforming protein (STP) of herpesvirus saimiri (HVS) and of K1 of KSHV, other members of the gamma-2 or rhadinovirus subgroup of herpesviruses. The R1 sequence revealed an open reading frame encoding a product of 423 amino acids that was predicted to contain an extracellular domain, a transmembrane domain, and a C-terminal cytoplasmic tail reflective of a type I membrane-bound protein. The predicted structural motifs of R1, including the presence of immunoreceptor tyrosine-based activation motifs, resembled those in K1 of KSHV but were distinct from those of STP. R1 sequences from four independent isolates from three different macaque species revealed 0.95 to 7.3% divergence over the 423 amino acids. Variation was located predominantly within the predicted extracellular domain. The R1 protein migrated at 70 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and was extensively glycosylated. Tagged R1 protein was localized to the cytoplasmic and plasma membranes of transfected cells. Expression of the R1 gene in Rat-1 fibroblasts induced morphologic changes and focus formation, and injection of R1-expressing cells into nude mice induced the formation of multifocal tumors. A recombinant herpesvirus in which the STP oncogene of HVS was replaced by R1 immortalized T lymphocytes to interleukin-2-independent growth. These results indicate that R1 is an oncogene of RRV.
The gammaherpesviruses can be subclassified as either gamma-1 or gamma-2 on the basis of genetic criteria. The Kaposi’s sarcoma-associated herpesvirus (KSHV) of humans (6) and herpesvirus saimiri (HVS) of New World primates (19) are considered gamma-2 herpesviruses, or rhadinoviruses, to distinguish them from Epstein-Barr virus and its close relatives from great apes and Old World primates. A rhadinovirus from an Old World primate, a rhesus monkey, was described only recently (7). The rhesus monkey rhadinovirus (RRV) has a closer relatedness to KSHV than to any other herpesvirus based on 10 kbp of genomic sequence from the initial H26-95 isolate (7). Serologic surveys revealed a high natural prevalence of RRV in rhesus monkeys (7).
KSHV has been consistently associated with Kaposi’s sarcoma and body cavity-based lymphomas in its natural human host and is likely to play a causative role in these diseases (1, 4, 5, 11, 29). Study of the mechanisms of disease induction by KSHV is hampered by several technical considerations. A truly permissive system for replication of KSHV has not been defined (24). This greatly impedes the construction of gene knockouts and viral mutants and the study of the lytic cycle. Also, animal models for the study of KSHV infection and disease have not been identified. Since RRV can be grown lytically in rhesus monkey fibroblast cultures and since it should be possible to infect rhesus monkeys experimentally with RRV, RRV holds promise for use in studies of the contributions of individual genes to the life cycle of the virus both in cell culture and in animals. However, more detailed knowledge of RRV is needed to define the similarities and differences compared to KSHV and HVS.
The first open reading frames (ORFs) of KSHV and HVS encode proteins called K1 and saimiri transforming protein (STP), respectively (15, 16, 21). The KSHV K1 protein is predicted to have an extracellular domain, a transmembrane region, and a short cytoplasmic tail (16). K1 and STP are each able to growth-transform rodent fibroblast cells and to contribute to the immortalization of primary lymphoid cells (8, 10, 16, 17). A recombinant herpesvirus in which the STP oncogene of HVS was replaced by the K1 gene immortalized primary T lymphocytes to interleukin-2-independent growth and induced lymphomas in common marmosets (16). These results demonstrated the oncogenic potential of the K1 gene. In this paper, we report the identification and characterization of the corresponding R1 reading frame of RRV and show that like K1 and STP, R1 is a viral oncoprotein. The organization of structural motifs in R1 is clearly similar to that found in K1 but not in STP.
MATERIALS AND METHODS
Plasmids.
The K29 clone was obtained by cutting virion DNA of the original RRV isolate H26-95 with the restriction enzyme KpnI and randomly cloning KpnI fragments into vector pSP72 (Promega). The KpnI insert of the K29 clone was fully sequenced from both the 5′ and 3′ ends by using T7 and Sp6 primers. For tagging, the R1 gene was amplified from the K29 clone by PCR with primers to the 5′ and 3′ ends of the gene (5′-CTGCAGATGTTTGTGTTGGTTTTA-3′ and 5′-GAATTCTTATATATAGCGATAGGTGTCTTCTAACCAATCATATTGTTC-3′, respectively). The AU1 epitope sequence (shown in boldface type) was added to the 3′ primer, and the product was cloned into the PstI and EcoRI sites of the pFJ mammalian expression vector by using the same restriction enzymes (underlined sequences) contained within the PCR primers (13). The R1 gene was similarly cloned into the BamHI and EcoRI sites of the pBabe-puromycin expression vector.
Analysis of R1 isolates.
Virion DNA was isolated from rhadinovirus isolates from Mm309-95 (a rhesus monkey), Mf23-97 (a cynomolgus monkey), and Mn19545 (a pig-tailed macaque). Virion DNA was used for PCR amplification with primers that flanked the 5′ end (5′-GGATCCCACATTTCTGTGGAAATG-3′) and 3′ end (5′-GAATTCTTTATTGGGTGTTCAATA-3′) of the R1 reading frame of the H26-95 isolate. The amplified R1 genes were cloned into pSP72 vector with the BamHI and EcoRI sites (underlined), and the genes were sequenced with T7 and SP6 primers.
Cell culture and transfection.
Cos-1, Bosc-23, and Rat-1 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. For puromycin selection of Rat-R1 cells, puromycin at 5 μg/ml was added to Dulbecco’s modified Eagle’s medium. Owl monkey kidney (OMK) cells were cultured in minimal essential medium supplemented with 10% fetal calf serum. Common marmoset peripheral blood mononuclear cells (PBMCs) used for immortalization assays were maintained in RPMI medium supplemented with 10% fetal calf serum. Lipofectamine (Gibco-BRL) was used for transfection of Cos-1 and Bosc-23 cells with the pFJ-R1 vector. To demonstrate glycosylation of the R1 protein, transfected cells were treated with 20 μg of tunicamycin per ml for 20 h.
Immunofluorescence.
Transfected cells were placed on slides by using a cytospin centrifuge. The cells were fixed in acetone-methanol (1:1) at −20°C for 10 min, washed with phosphate-buffered saline (PBS), and incubated with 10% goat serum for 15 min. They were then washed twice and incubated with anti-AU1 antibody (Berkeley Antibody) at a dilution of 1:100 in PBS for 30 min. They were washed three times in PBS, incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody at a dilution of 1:100 in PBS for 30 min, and washed another three times in PBS, and immunofluorescence was detected with an Olympus immunofluorescence microscope.
Transformation assays.
Rat-1 fibroblasts were transfected with the pBabe-R1 vector, and stable cell lines were obtained after selection with puromycin (5 μg/ml). The cells were trypsinized and plated on 100-mm tissue culture dishes. Foci were observed after 2 weeks. Methylene blue staining was performed after the cells were fixed in 10% formaldehyde. A 5% solution of methylene blue in PBS was used to stain the cells for 30 min, and the cells were washed with water for several hours after being stained. For the nude-mouse experiments, 106 Rat-1 or Rat-R1 cells were injected into the hindlimb of nude mice.
Construction of recombinant HVS.
The complete STP coding sequence was deleted from a 3.6-kbp fragment of the left end of L-DNA of HVS C488, and multicloning sites were inserted into the STP locus by PCR as described previously (9, 10). An EcoRI-XbaI fragment containing the R1 gene was cloned into the multicloning site of this 3.1-kbp L-DNA such that the R1 gene was under the control of the STP promoter. Linearized plasmid DNA containing the 3.1 kbp of HVS L-DNA sequence flanking 1.3 kbp of R1 gene sequence was then cotransfected into OMK cells with HVSΔSTP/SEAP virion DNA by the calcium phosphate method. To isolate HVSΔSTP/R1, limiting-dilution and secreted engineered alkaline phosphatase (SEAP) assays were performed as described previously (9, 10). SEAP in the medium was detected with Phospha-Light reagents (Tropix Inc.) as recommended by the manufacturer.
In vitro immortalization of common marmoset lymphocytes.
Immortalization assays used lymphocytes from common marmosets (Callithrix jacchus) as previously described (9, 10). PBMCs were isolated from 2.5 ml of heparinized blood from five different common marmosets by centrifugation through lymphocyte separation medium followed by washing with RPMI medium. The PBMCs were resuspended at 106 cells per ml in RPMI medium and used for immortalization assays in 12-well tissue culture dishes. Aliquots of each preparation of PBMCs were infected separately with the wild-type HVS and three recombinant HVS strains (HVSΔSTP/SEAP, HVSΔSTP/K1, and HVSΔSTP/R1). The medium was changed every 5 days, and immortalization was assayed microscopically.
RESULTS
Genomic organization of the left end of the RRV genome.
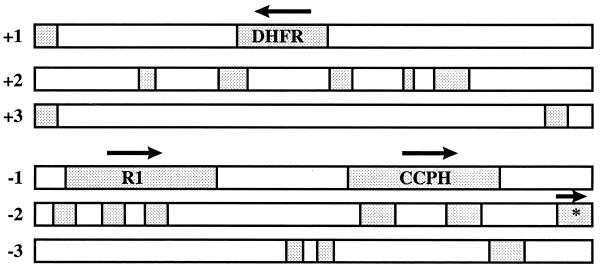
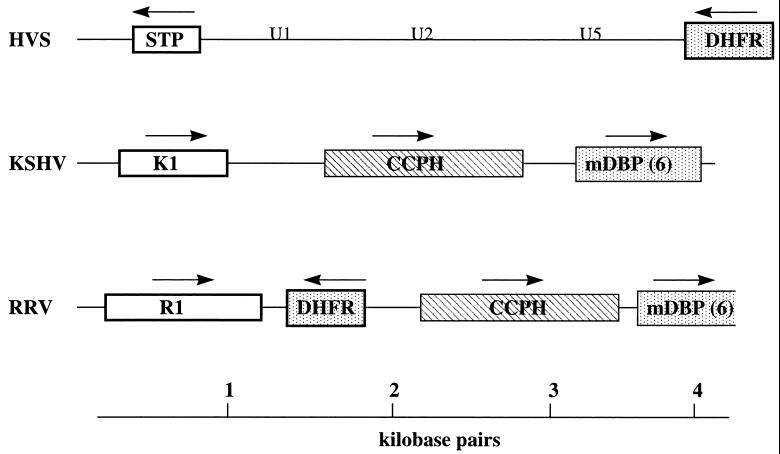
We isolated a 4,228-bp KpnI clone (K29) from the left end of the RRV genome of the original H26-95 isolate. Analysis of the sequences within this 4,228-bp fragment revealed ORFs for a dihydrofolate reductase (DHFR), a complement control protein homolog (CCPH), a protein we have called R1, and a partial reading frame corresponding to the major DNA binding protein (mDBP) (Fig. 1). The organization of ORFs within this region of the RRV genome was compared to that in KSHV and HVS (Fig. 2). The first ORF of HVS encodes STP. The STP oncogene is not required for replication of the virus but is required for transformation of primary lymphoid cells (8, 10). The first ORF of KSHV, called K1, also encodes a transforming protein (16). We have called the first ORF within K29 of RRV R1 because of its positional correspondence with K1 of KSHV and because of other similarities (described below). The direction of the reading frame of R1 is the same as that of K1 but is opposite to that of STP (Fig. 2). R1 is followed by the DHFR ORF, approximately equivalent to where the DHFR ORF is found in HVS and in the same direction of expression as observed in HVS. The DHFR gene is displaced in the KSHV genome nine reading frames further downstream in a cluster of genes homologous to cellular genes (23, 25). The third and fourth ORFs in RRV were a complement control protein homolog (CCPH 4) and the major DNA binding protein (mDBP6), respectively (Fig. 2). The organization of ORFs in this region was equivalent in RRV and KSHV except for the displacement of the DHFR gene.
FIG. 1.
Analysis of ORFs present in the K29 clone. Any ORF larger than 50 amino acids is indicated by stippling. Complete ORFs for CCPH, DHFR, and R1 are depicted, along with the partial ORF of the mDBP gene (encoding the first 109 amino acids of the mDBP) (asterisk). The arrows indicate the direction of expression for each gene.
FIG. 2.
Genomic organization of the left end of the RRV, KSHV, and HVS genomes. The first ORFs, R1, K1, and STP, are depicted. The DHFR gene constitutes the second ORF in RRV and HVS but is displaced further downstream in KSHV. The third and fourth ORFs in the RRV and HSV genomes are the CCPH4 and mDBP6 genes. Because of the displacement of DHFR in KSHV, CCPH and mDBP6 are the second and third ORFs in KSHV. U1, U2, and U5 indicate the locations of coding sequences for small U-RNAs (18, 20). The arrows indicate the direction of gene expression.
Identification of the R1 protein.
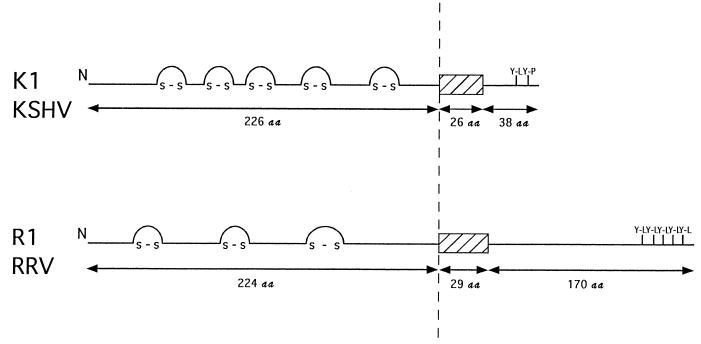
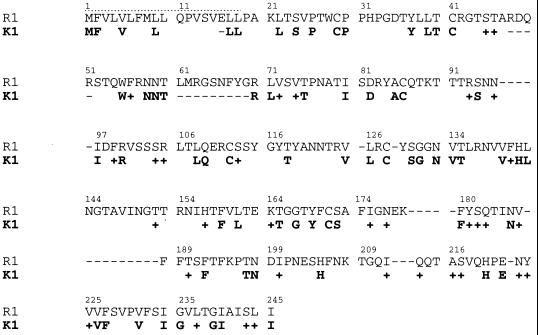
The R1 protein is predicted from the DNA sequence to have a signal peptide sequence at the N terminus, an extracellular domain, a transmembrane domain, and a cytoplasmic tail at the C terminus (Fig. 3). The predicted extracellular domain of R1 shows homology to the variable regions of the lambda chain of immunoglobulin light chains and contains cysteine residues which may be involved in disulfide linkages. While no similarity between R1 and STP was detected, the organization of structural motifs in R1 was similar to that of K1 of KSHV (Fig. 3). The extracellular domains of R1 and K1 are similar in length, and both exhibit homology to the immunoglobulin receptor superfamily. R1 exhibits high homology to a human immunoglobulin G receptor, CD16 (E = 3 × 10−10), which was isolated from human lung and peripheral blood leukocyte cDNA libraries (26). In addition, the cytoplasmic tail of R1 contains five potential SH2-binding motifs (YXXL) which have the potential of serving as immunoreceptor tyrosine-based activation motifs (ITAMs) (3, 27, 28). KSHV K1 also has two potential SH2-binding motifs (YXXP and YXXL) in its cytoplasmic tail, which serve as an ITAM (Fig. 3) (17). The similarity in the organization of structural motifs between R1 and K1 exists despite extremely limited identity in the comparative amino acid sequences (Fig. 4). At most 27% identity and 40% similarity in amino acid sequence could be identified by comparison of the first 245 N-terminal amino acid sequences (Fig. 4). Other than the tyrosine-containing elements described above, little or no similarity was evident between the short (38-amino-acid) cytoplasmic tail of K1 and the 170-amino-acid cytoplasmic tail of R1.
FIG. 3.
Organization of the structural regions of the R1 and K1 proteins. N represents the amino termini of the two proteins, and the hatched boxes represent their putative transmembrane domains. The N-terminal extracellular domains have approximately the same number of amino acids (aa) and contain cysteines that may form disulfide linkages similar to other members of the immunoglobulin superfamily. The cytoplasmic tail of K1 contains one ITAM in which a YXXL and YXXP are separated by 7 amino acids (16, 17). The cytoplasmic tail of R1 contains five potential YXXL SH2-binding motifs separated by 3 to 9 amino acids.
FIG. 4.
Amino acid identity and similarity between the R1 and K1 proteins. The N-terminal 245 amino acids of R1 were compared to the equivalent region of K1. These two proteins exhibit 27% identity and 40% similarity in their extracellular domains. +, amino acid similarity; −, gaps in amino acid sequence. The dotted lines represent the signal peptide sequence.
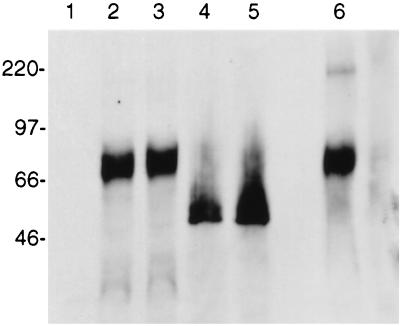
To facilitate the detection of R1 protein in mammalian cells, the R1 gene from the H26-95 RRV isolate was tagged with an AU1 epitope at the C-terminal end of the coding sequence and cloned into expression vector pFJ. At 48 h posttransfection, Cos-1 and Bosc-23 cells were harvested and cellular lysates were used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot detection with an anti-AU1 antibody (Fig. 5). A protein migrating at approximately 70 kDa was specifically detected from whole-cell lysate (Fig. 5, lane 2) or the membrane fraction (lane 3) of Cos-1 cells transfected with a pFJ-R1 expression vector tagged with the AU1 epitope at its terminus. No such protein was detected in control cells lacking the R1 gene (lane 1).
FIG. 5.
Detection of the R1 protein. The R1 gene was tagged with an AU1 epitope and expressed in Cos-1 cells. Lysates were run on SDS-PAGE under reducing conditions (lanes 1 to 5) or nonreducing conditions (lane 6) and transferred to nitrocellulose. Western blotting was performed with an anti-AU1 antibody. Lanes: 1, lysates of cells transfected with the control vector; 2, lysates of Cos-1 cells transfected with the R1 expression vector; 3, membrane fractions from Cos-1 cells transfected with R1; 4, lysates of R1-transfected Cos-1 cells treated with tunicamycin; 5, membrane fractions of R1-transfected Cos-1 cells treated with tunicamycin; 6, lysate of R1-transfected Cos-1 cells (nonreducing condition of SDS-PAGE).
From its sequence, the R1 protein is predicted to have a molecular mass of 48 kDa. The increased size of the R1 protein in SDS-PAGE could be the result of glycosylation. The extracellular domain of R1 contains eight potential sites for N-linked glycosylation (NXT or NXS) (Fig. 4). To examine whether glycosylation was responsible for the decreased mobility, transfected cells were treated with an N-linked glycosylation inhibitor, tunicamycin. The addition of tunicamycin 20 h before the Cos-1 cells were harvested resulted in a faster-migrating R1 protein of approximately 50 kDa (Fig. 5, lanes 4 and 5). This molecular mass corresponds closely to the predicted size of R1, which is 48 kDa. These results indicate that the R1 protein is indeed glycosylated.
Previously, the KSHV K1 protein was shown to exist in oligomeric forms as a result of disulfide bonding (16). Cell lysates containing R1 were resolved under nonreducing conditions, i.e., in the absence of 2-mercaptoethanol or other reducing agents (Fig. 5, lane 6). The vast majority of R1 continued to migrate at 70 kDa under these conditions. In contrast to what has been observed previously with K1, only a very minor proportion of R1 migrated as higher-molecular-weight forms under these nonreducing conditions (lane 6).
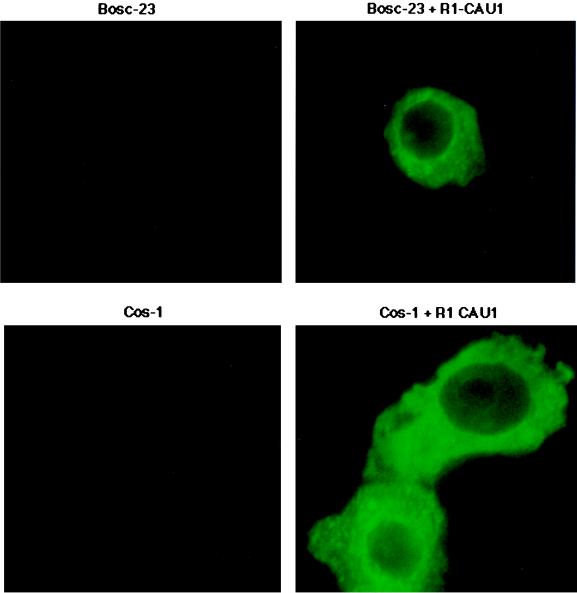
We used immunofluorescence tests to analyze the location of the R1 protein in R1-expressing cells. Cos-1 or Bosc-23 cells were transfected with an AU1-tagged R1 expression vector, and transfected cells were stained with an anti-AU1 antibody (Fig. 6). These experiments demonstrated R1 principally in the cytoplasm, apparently associated with cytoplasmic membranes. No R1 was found in the nuclei of transfected cells (Fig. 6). A B-cell line, BJAB, showed similar R1 localization patterns when transfected with a R1 expressing vector (data not shown).
FIG. 6.
Localization of R1 protein. Bosc-23 (top panels) or Cos-1 (bottom panels) cells were transfected with the expression vector alone or a vector expressing a C-terminal AU1-tagged R1 protein (R1 CAU1). At 48 h posttransfection, immunofluorescence was performed with an anti-AU1 antibody. The R1 protein was localized to the cytoplasmic and plasma membranes.
Natural antibodies to R1.
We previously described enzyme-linked immunosorbent assay procedures with whole lysed virions to identify monkeys with antibodies to RRV (7). A total of 10 RRV-positive monkey serum samples and 10 RRV-negative monkey serum samples were tested for antibody reactivity to the R1 protein by Western blotting with lysates from transfected Cos-1 cells. A total of 7 of the 10 RRV-positive serum samples and 0 of the 10 RRV-negative serum samples had antibodies reactive with R1 in these tests (data not shown).
R1 sequences from independent virus isolates.
The R1 sequences described above were derived from the original H26-95 isolate from a rhesus monkey (Macaca mulatta) (7). We used PCR primers from outside the R1 reading frame to amplify R1 sequences from virion DNA of three additional, independent, macaque rhadinovirus isolates. The three additional isolates for sequence comparisons were obtained from a cynomolgus macaque (Macaca fascicularis), a pig-tailed macaque (Macaca nemestrina), and another rhesus monkey. These three isolates are referred to as H-Mf23-97, H-Mn19545, and H-Mm309-95, respectively, and the original isolate is referred to as H-Mm26-95. Comparisons of the R1 genes from these four isolates revealed that the R1 protein sequences were well conserved over the amino-terminal 250 amino acids and the carboxy-terminal 173 amino acids of the R1 protein (Fig. 7). Moreover, the sequences did not cluster according to species of origin in this limited survey. As shown in Table 1, H-Mm309-95 and H-Mm26-95, both derived from M. mulatta, were 5.9% divergent, while H-Mm26-95 versus H-Mf23-97 and H-Mm26-95 versus H-Mn19545 exhibited only 0.95 and 2.8% divergence, respectively. The greatest degree of amino acid divergence among the R1 isolates was observed in the putative extracellular domain of the R1 protein (Table 1).
FIG. 7.
Amino acid sequence of the R1 gene from four different macaque rhadinovirus isolates. H-Mm26-95 is the original M. mulatta isolate. H-Mm309-95 was also isolated from M. mulatta (a different animal). H-Mf23-97 was isolated from M. fascicularis, and H-Mn19545 was isolated from M. nemestrina. The dotted lines represent the signal peptide sequence, the box represents the transmembrane domain, and the five putative SH2-binding motifs are underlined in bold. The R1 protein shows a high degree of homology to the immunoglobulin receptor superfamily in the N-terminal 220 amino acids.
TABLE 1.
Amino acid divergence in R1 from four independent macaque rhadinovirus isolates
| Rhadinovirus isolate and protein | Amino acid divergence with respect to:
|
||
|---|---|---|---|
| H-Mm309-95 | H-Mn19545 | H-Mf23-97 | |
| Entire R1 proteina | |||
| H-Mm26-95 | 25 (5.9%) | 12 (2.8%) | 4 (0.95%) |
| H-Mf23-97 | 27 (6.4%) | 16 (3.8%) | |
| H-Mn19545 | 31 (7.3%) | ||
| Extracellular domain of R1b | |||
| H-Mm26-95 | 17 (7.6%) | 9 (4%) | 3 (1.3%) |
| H-Mf23-97 | 20 (8.9%) | 12 (5%) | |
| H-Mn19545 | 22 (9.8%) | ||
Results are the number (percentage) of amino acids different in full-length R1.
Results are the number (percentage) of amino acids different in 224 amino acids of the R1 extracellular domain.
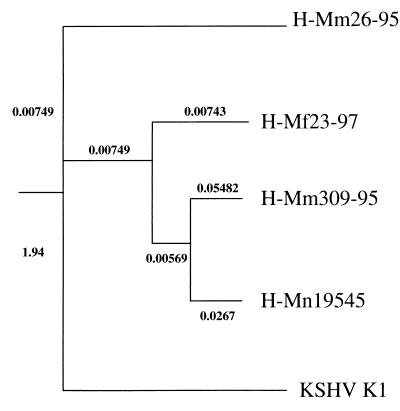
A phylogenetic analysis of the four R1 genes was performed by the distance method, using the neighbor-joining program. The phylogenetic phenogram tree clusters the four R1 isolates together and demonstrates that R1 is most closely related to K1 of KSHV (Fig. 8). The phenogram also shows that the two R1 genes from M. mulatta do not cluster together (Fig. 8).
FIG. 8.
Phylogenetic analysis of R1 sequences. An evolutionary tree based on the amino acid sequences encoded by the four R1 genes from four macaque rhadinovirus isolates (H-Mm26-95, H-Mm309-95, H-Mf23-97, and H-Mn19545) is shown. The phenogram was obtained by the distance method, using the neighbor-joining program. The R1 sequences cluster together and are more closely related to each other than to K1.
Growth transformation of Rat-1 cells by the R1 gene.
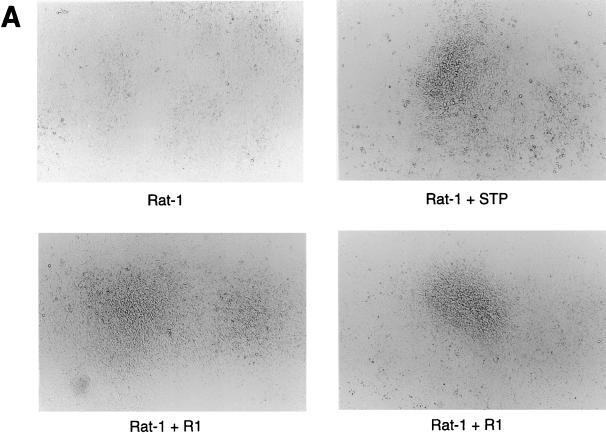
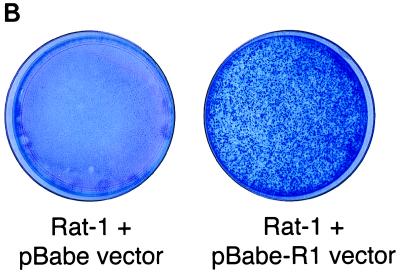
Since the R1 gene is located at a position equivalent to the STP gene of HVS and the K1 gene of KSHV, we investigated the ability of the R1 gene to transform rodent fibroblasts. The R1 gene was cloned into a retroviral vector, pBabe-puromycin, and expressed in Rat-1 cells. After selection with puromycin, rat fibroblasts stably expressing R1 were isolated. To investigate the effects of R1 expression on cell growth, the growth properties of Rat-R1 cells were compared with those of control cells and cells transfected with STP C488, a known viral oncoprotein of HVS (Fig. 9). The Rat-R1 cells, like the Rat-STP C488-expressing cells, were markedly different in morphology from what was observed with control Rat-1 fibroblasts (Fig. 9A). In 100-mm tissue culture dishes, control Rat-1 fibroblasts grew in flat monolayers, in sharp contrast to the Rat-R1-expressing fibroblasts, which formed hundreds of foci (Fig. 9B). Thus, similar to the STP oncogene of HVS and K1 of KSHV, the R1 gene is capable of transforming Rat-1 cells, resulting in morphological changes and focus formation.
FIG. 9.
Transforming activity of the R1 protein. (A) Puromycin-resistant cells obtained after transfection with a pBabe control retroviral vector (top left) or a pBabe-STP (top right) or pBabe-R1 (bottom)-expressing vector were plated at 106 cells per 100-mm tissue culture dish. After 14 days of incubation, the cells were photographed to show morphologic changes. (B) The cells were stained with methylene blue to show foci of transformed cells.
To further test the transforming potential of R1, we injected nude mice with R1-expressing Rat-1 cells. The cells were injected into the right hindlimb of these mice. By day 15, four of the four nude mice injected with the R1-expressing cells developed large (5- to 10-mm), multifocal, disseminated tumors whereas nude mice injected with the control Rat-1 fibroblasts showed only small nodules at the site of injection. These results demonstrated that expression of R1 in rodent fibroblasts resulted in tumorigenicity in nude mice.
Construction of HVSΔSTP/R1 recombinant virus.
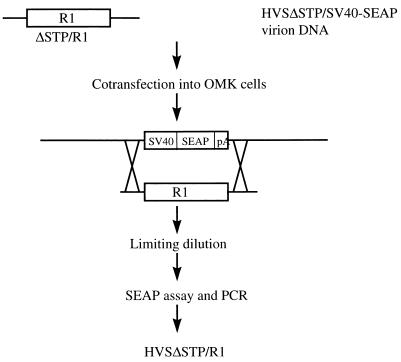
To examine the transforming ability of R1 in the context of a virus, we used a recombinant HVS system which has also been used for the study of the KSHV K1 gene (16). We used virion DNA from the nononcogenic HVSΔSTP/SV40-SEAP to isolate a recombinant HVSΔSTP containing the R1 gene. The R1 ORF was used instead of the STP ORF of HVS strain C488, and the R1 gene was thus under the control of the STP promoter. STP is not required for viral replication of HVS but is required for in vitro immortalization and in vivo oncogenicity (10). HVSΔSTP/SV40-SEAP virion DNA was cotransfected into permissive OMK cells together with a cloned 3.9-kbp HVS C488 within which the STP ORF was replaced by R1 (Fig. 10). Homologous recombination within the cotransfected OMK cells resulted in the production of an HVSΔSTP/R1. To isolate the recombinant HVSΔSTP/R1, SEAP-negative viruses were recovered from limiting dilutions as shown in Fig. 10. The presence of the R1 gene and the absence of STP in the HVSΔSTP/R1 recombinant were confirmed by PCR and DNA sequencing analysis.
FIG. 10.
Construction of recombinant HVSΔSTP/R1. The diagram shows the strategy used to make the recombinant HVSΔSTP/R1. The detailed procedure has been described previously (9).
Immortalization of T lymphocytes with recombinant HVSΔSTP/R1.
Primary T lymphocytes from common marmosets were isolated and incubated with HVSΔSTP/R1. HVSΔSTP and HVSΔSTP/SV40-SEAP were used as negative controls in this assay, and HVSΔSTP/K1 and wild-type HVS C488 were used as positive controls. Each virus was tested individually with unstimulated PBMCs from each of five different common marmosets. HVSΔSTP/R1 and HVSΔSTP/K1 immortalized PBMCs from all five marmosets to growth in an interleukin-2-independent fashion similar to wild-type HVS C488. HVSΔSTP/SEAP-SV40 and HVSΔSTP did not immortalize the PBMCs. The HVSΔSTP/R1-immortalized lymphocytes were analyzed by flow cytometry and were shown to be CD3+ CD4− CD8+ CD56+ CD20− T cells (data not shown).
DISCUSSION
The R1 gene of RRV is located at a position equivalent to that of the K1 gene of KSHV and the STP gene of HVS. Although R1 shows no homology to STP, it contains sequence motifs and an organization of structural features that are very similar to K1. The R1 and K1 proteins exhibit approximately 27% amino acid identity in their extracellular domains, and both extracellular domains resemble those of the immunoglobulin receptor superfamily. R1 and K1 differ significantly in the length of their cytoplasmic domains, but both contain YXXL motifs that resemble SH2-binding sequences. R1 has five YXXL elements near the C terminus of its cytoplasmic tail. The third and fourth YXXL motifs of R1 show the conserved pattern for a putative ITAM: (D/E)X0–3YX2LX7–12YX2L (3). The fourth and fifth YXXL motifs could also possibly function as an ITAM, since the YXXL motifs are separated by 8 amino acids and are preceded by negatively charged amino acids. ITAMs are present in B-cell, T-cell and Fc immunoreceptors and interact with both Src and Syk family protein tyrosine kinases (3, 27, 28). In addition to the YXXL motifs, there are several other tyrosine residues that could serve as potential phosphorylation sites. These include two YXXP motifs, one YXXA motif, and one YXXV motif. Such motifs have also been shown to interact with SH2 domains present in a variety of tyrosine kinases and other signaling molecules (3, 27, 28). Hence, R1 has rich potential for affecting cellular signaling through sequence motifs present within its cytoplasmic domain.
The finding of R1 localization predominantly in the cytoplasm was surprising. Much of the staining was in punctate form near the perimeter of the cytoplasm, suggestive of possible endosomal localization. YXXH, where H is a hydrophobic amino acid, can also serve as a signal for internalization of plasma membrane proteins (30). The cytoplasmic domain of R1 also contains a dileucine repeat at residues 318 and 319 (Fig. 7), which can also serve as a signal for endocytosis (30). Further work is needed to delineate the effects of specific sequence motifs in the cytoplasmic domain of R1 on endocytosis and signaling.
We have sequenced four independent R1 genes. The four rhadinovirus isolates from which the sequences were derived were obtained from different sources. Three of the macaques (Mm26-95, Mm309-95, and Mf23-97) were born and raised at the New England Regional Primate Research Center, and Mn19545 was obtained from the Oregon Regional Primate Research Center. The R1 genes from the two M. mulatta animals were the most divergent. This suggests that R1 may exhibit polymorphic divergence independent of the species of origin or that one or more of the viruses was derived from cross-species transmission. Although there is some divergence among the R1 sequences, the percent divergence is lower than was observed with the K1 gene of KSHV (16, 22). Comparison of the primary amino acid sequences of K1 genes isolated from two body cavity-based lymphoma cell lines, BCBL-1 and BC-1, and a Kaposi’s sarcoma biopsy specimen revealed many amino acid substitutions around the transmembrane region of K1, but the carboxy-terminal sequences were the most conserved among the K1 isolates (16, 17, 22). Despite the divergence seen among individual K1 and R1 sequences, the carboxy-terminal ITAMs in both the R1 and K1 genes are well conserved, suggesting that these motifs may play an important role in the function of these proteins.
STP of HVS and K1 of KSHV both transform rodent fibroblasts and contribute to oncogenic transformation of T lymphocytes from common marmosets (10, 16). The STP gene of HVS C488 interacts with cellular Ras (12). K1 has recently been shown to interact with the Syk family of protein kinases (17). We have now demonstrated that R1 can also transform rodent fibroblasts and induce tumors in nude mice. In addition, an HVSΔSTP/R1 recombinant was capable of immortalizing primary lymphocytes. Demonstration of a role for R1 in the natural history of primary RRV infection, persistence, or disease is dependent on further animal model development.
ACKNOWLEDGMENTS
We thank Susan Czajak and Danny Silva for providing the macaque rhadinovirus isolates and virion DNA. We thank Amanda Knapp and Kim Deary for helping to sequence the K29 clone, and we thank Marcy Auerbach and Welkin Johnson for assisting with the phylogenetic analysis.
This work was supported by U.S. Public Health Service grants RR00168 and AI38131. B. Damania is a fellow of the Cancer Research Institute.
REFERENCES
- 1.Boshoff C, Whitby D, Hatziannou T, Fisher C, Van der Walt J, Hatzakis A, Weiss R A, Schultz T F. Kaposi’s sarcoma associated herpesvirus in HIV-negative Kaposi sarcoma. Lancet. 1995;345:1043–1044. doi: 10.1016/s0140-6736(95)90780-7. [DOI] [PubMed] [Google Scholar]
- 2.Burkhardt A L, Bolen J B, Kieff E, Longnecker R. An Epstein-Barr virus transformation-associated membrane protein interacts with src family tyrosine kinases. J Virol. 1992;66:5161–5167. doi: 10.1128/jvi.66.8.5161-5167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cambier J C. Antigen and Fc receptor signaling. The awesome power of the immunoreceptor tyrosine-based activation motif (ITAM) J Immunol. 1995;155:3281–3285. [PubMed] [Google Scholar]
- 4.Cesarman E, Nador R, Knowles D M. Body cavity based lymphoma in an HIV-seronegative patient without Kaposi’s sarcoma associated herpesvirus-like DNA sequences. N Engl J Med. 1996;334:273. doi: 10.1056/NEJM199601253340417. [DOI] [PubMed] [Google Scholar]
- 5.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 6.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 7.Desrosiers R C, Sasseville V G, Czajak S, Zhang X, Mansfield K G, Kaur A, Johnson R P, Lackner A A, Jung J U. A herpesvirus of rhesus monkeys related to the human Kaposi’s sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desrosiers R C, Silva D, Waldron L M, Letvin N L. Nononcogenic deletion mutants of herpesvirus saimiri are defective for in vitro immortalization. J Virol. 1986;57:701–705. doi: 10.1128/jvi.57.2.701-705.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duboise S M, Guo J, Desrosiers R C, Jung J U. Use of virion DNA as a cloning vector for the construction of mutant and recombinant herpesviruses. Proc Natl Acad Sci USA. 1996;93:11389–11394. doi: 10.1073/pnas.93.21.11389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duboise S M, Guo J, Czajak S, Desrosiers R C, Jung J U. STP and Tip are essential for Herpesvirus saimiri oncogenicity. J Virol. 1998;72:1308–1313. doi: 10.1128/jvi.72.2.1308-1313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y Q, Li J J, Kaplan M H, Poiesz B, Katabira E, Zhang W C, Feiner D, Friedman-Kien A E. Human herpesvirus-like nucleic acid in various forms of Kaposi’s sarcoma. Lancet. 1995;345:759–761. doi: 10.1016/s0140-6736(95)90641-x. [DOI] [PubMed] [Google Scholar]
- 12.Jung J U, Desrosiers R C. Association of the viral oncoprotein STP-C488 with cellular ras. Mol Cell Biol. 1995;15:6506–6512. doi: 10.1128/mcb.15.12.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung J U, Lang S, Friedrich U, Jun T, Roberts T, Desrosiers R C, Biesinger B. Identification of lck-binding elements in Tip of herpesvirus saimiri. J Biol Chem. 1995;270:20660–20667. doi: 10.1074/jbc.270.35.20660. [DOI] [PubMed] [Google Scholar]
- 14.Jung J U, Trimble J J, King N W, Biesinger B, Fleckenstein B W, Desrosiers R C. Identification of transforming genes of subgroup A and C strains of herpesvirus saimiri. Proc Natl Acad Sci USA. 1991;88:7051–7055. doi: 10.1073/pnas.88.16.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagunoff M, Ganem D. The structure and coding organization of the genomic termini of Kaposi’s sarcoma-associated herpesvirus. Virology. 1997;236:147–154. doi: 10.1006/viro.1997.8713. [DOI] [PubMed] [Google Scholar]
- 16.Lee H, Veazey R, Williams K, Li M, Guo J, Neipel F, Fleckenstein B, Lackner A, Desrosiers R C, Jung J U. Deregulation of cell growth by the K1 gene of Kaposi’s sarcoma-associated herpesvirus. Nat Med. 1998;4:435–440. doi: 10.1038/nm0498-435. [DOI] [PubMed] [Google Scholar]
- 17.Lee H, Guo J, Li M, Choi J K, DeMaria M, Rosenzweig M, Jung J U. Identification of an immunoreceptor tyrosine-based activation motif of K1 transforming protein of Kaposi’s sarcoma-associated herpesvirus. Mol Cell Biol. 1998;9:5219–5228. doi: 10.1128/mcb.18.9.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S I, Murthy S, Trimble J, Desrosiers R C, Steitz J A. Four novel U RNAs are encoded by a herpesvirus. Cell. 1988;54:599–607. doi: 10.1016/s0092-8674(88)80004-7. [DOI] [PubMed] [Google Scholar]
- 19.Melendez L V, Hunt R D, Daniel M D, Garcia F G, Fraser C E. Herpesvirus saimiri. II. Experimentally induced malignant lymphoma in primates. Lab Anim Care. 1969;19:378–386. [PubMed] [Google Scholar]
- 20.Murthy S, Kamine J, Desrosiers R C. Viral-encoded small RNAs in herpes virus saimiri induced tumors. EMBO J. 1986;5:1625–1632. doi: 10.1002/j.1460-2075.1986.tb04405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murthy S C, Trimble J J, Desrosiers R C. Deletion mutants of herpesvirus saimiri define an open reading frame necessary for transformation. J Virol. 1989;63:3307–3314. doi: 10.1128/jvi.63.8.3307-3314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholas J, Zong J-C, Alcendor D, Ciufo D, Poole L, Sarisky R, Chiou C-J, Zhang X, Wan X, Guo H-G, Reitz M S, Hayward G S. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J Natl Cancer Inst Monogr. 1998;23:79–88. doi: 10.1093/oxfordjournals.jncimonographs.a024179. [DOI] [PubMed] [Google Scholar]
- 23.Nicholas J, Ruvolo V, Zong J, Ciufo D, Guo H G, Reitz M S, Hayward G S. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J Virol. 1997;71:1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scallon B J, Scigliano E, Freedman V H, Miedel M C, Pan Y C, Unkeless J C, Kochan J P. A human immunoglobulin G receptor exists in both polypeptide-anchored and phosphatidylinositol-glycan-anchored forms. Proc Natl Acad Sci USA. 1989;86:5079–5083. doi: 10.1073/pnas.86.13.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Songyang Z, Shoelson S E, Glade J M, Olivier P, Pawson T, Bustelo X R, Barbacid M, Sabe H, Hanafusa H, Yi T, Ren R, Baltimore D, Ratnofsky S, Feldman R A, Cantley L C. Specific motifs recognized by the SH2 domains of Csk, 3BP2, Fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol. 1994;14:2777–2785. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleiden R J, Neel B G, Birge R B, Fajardo J E, Chou M M, Hanafusa H, Schaffhausen B, Cantley L C. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 29.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay M F, Clauvel J P, Raphael M, Degos L. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 30.Trowbridge I S, Collawn J F. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu Rev Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]