Abstract
Background
Prognostic markers and biological pathways linked to detrimental clinical outcomes in heart failure with preserved ejection fraction (HFpEF) remain incompletely defined.
Methods and Results
We measured serum levels of 4123 unique proteins in 1117 patients with HFpEF enrolled in the PARAGON‐HF (Efficacy and Safety of LCZ696 Compared to Valsartan, on Morbidity and Mortality in Heart Failure Patients With Preserved Ejection Fraction) trial using a modified aptamer proteomic assay. Baseline circulating protein concentrations significantly associated with the primary end point and the timing and occurrence of total heart failure hospitalization and cardiovascular death were identified by recurrent events regression, accounting for multiple testing, adjusted for age, sex, treatment, and anticoagulant use, and compared with published analyses in 2515 patients with heart failure with reduced ejection fraction from the PARADIGM‐HF (Prospective Comparison of ARNI With ACEI to Determine Impact on Global Mortality and Morbidity in Heart Failure) and ATMOSPHERE (Efficacy and Safety of Aliskiren and Aliskiren/Enalapril Combination on Morbidity–Mortality in Patients With Chronic Heart Failure) clinical trials. We identified 288 proteins that were robustly associated with the risk of heart failure hospitalization and cardiovascular death in patients with HFpEF. The baseline proteins most strongly related to outcomes included B2M (β‐2 microglobulin), TIMP1 (tissue inhibitor of matrix metalloproteinase 1), SERPINA4 (serpin family A member 4), and SVEP1 (sushi, von Willebrand factor type A, EGF, and pentraxin domain containing 1). Overall, the protein–outcome associations in patients with HFpEF did not markedly differ as compared with patients with heart failure with reduced ejection fraction. A proteomic risk score derived in patients with HFpEF was not superior to a previous proteomic score derived in heart failure with reduced ejection fraction nor to clinical risk factors, NT‐proBNP (N‐terminal pro‐B‐type natriuretic peptide), or high‐sensitivity cardiac troponin.
Conclusions
Numerous serum proteins linked to metabolic, coagulation, and extracellular matrix regulatory pathways were associated with worse HFpEF prognosis in the PARAGON‐HF proteomic substudy. Our results demonstrate substantial similarities among serum proteomic risk markers for heart failure hospitalization and cardiovascular death when comparing clinical trial participants with heart failure across the ejection fraction spectrum.
Registration
URL: https://www.clinicaltrials.gov; Unique Identifiers: NCT01920711, NCT01035255, NCT00853658.
Keywords: cardiovascular, ejection fraction, heart failure, HFpEF, HFrEF, proteomics
Subject Categories: Heart Failure
Nonstandard Abbreviations and Acronyms
- ATMOSPHERE
Efficacy and Safety of Aliskiren and Aliskiren/Enalapril Combination on Morbidity–Mortality in Patients With Chronic Heart Failure
- FDR
false discovery rate
- HFH
heart failure hospitalization
- HFpEF
heart failure with preserved ejection fraction
- HFrEF
heart failure with reduced ejection fraction
- PARADIGM‐HF
Prospective Comparison of ARNI With ACEI to Determine Impact on Global Mortality and Morbidity in Heart Failure
- PARAGON‐HF
Efficacy and Safety of LCZ696 Compared to Valsartan, on Morbidity and Mortality in Heart Failure Patients With Preserved Ejection Fraction
- SOMAmer
slow off‐rate modified aptamer
Clinical Perspective.
What Is New?
In this large‐scale proteomic investigation, we systematically characterized associations between serum protein levels and risk for heart failure hospitalization and cardiovascular death in patients with heart failure with preserved ejection fraction, highlighting the importance of proteins involved in coagulation, metabolic disease, and extracellular matrix regulation in heart failure with preserved ejection fraction pathogenesis and disease progression.
What Are the Clinical Implications?
Associations between protein levels and outcomes in patients with heart failure with preserved ejection fraction were largely similar to those in patients with heart failure with reduced ejection fraction, reinforcing that shared pathologic processes are likely important drivers of poor prognosis across the ejection fraction spectrum.
An heart failure with preserved ejection fraction‐specific proteomic risk score did not meaningfully improve risk prediction compared with clinical factors and commonly measured biomarkers.
Heart failure with preserved ejection fraction (HFpEF) is common, rising in prevalence, and associated with high morbidity and death. 1 , 2 , 3 Despite clinical similarities among patients with HFpEF and heart failure with reduced ejection fraction (HFrEF), the greater efficacy of neurohormonal therapies in HFrEF suggests distinct underlying pathophysiology. 4 , 5 Systemic inflammation, nitrosative stress, and myocardial fibrosis, among other mechanisms, are implicated in HFpEF pathogenesis. 6 , 7 A greater understanding of factors that increase the risk of cardiovascular events in HFpEF can facilitate therapeutic development and identification of HFpEF subpopulations that may better respond to specific targeted interventions.
Broad proteomic investigation may identify novel proteins contributing to HF progression, potentially highlighting discrete dysregulated pathways among patients with HFpEF as compared with HFrEF. In patients with HFrEF, we previously identified and replicated 167 serum proteins associated with risk for HF hospitalization (HFH) or cardiovascular death, where an HFrEF‐derived proteomic risk score only modestly improved risk prediction performance. 8 Broad proteomic profiling and discovery efforts in HFpEF populations to date have been limited in size, have not included long‐term clinical outcome data, or have focused on treatment effects on protein levels. 9 , 10
In this post hoc analysis, we performed discovery proteomics using the SomaLogic SomaScan platform to identify associations between serum protein levels and clinical outcomes in a substudy of 1117 patients with HFpEF enrolled in the PARAGON‐HF (Efficacy and Safety of LCZ696 Compared to Valsartan, on Morbidity and Mortality in Heart Failure Patients With Preserved Ejection Fraction) trial. We then compared protein‐outcome associations between patients with HFpEF enrolled in PARAGON‐HF and patients with HFrEF from the ATMOSPHERE (Efficacy and Safety of Aliskiren and Aliskiren/Enalapril Combination on Morbidity–Mortality in Patients With Chronic Heart Failure) and PARADIGM‐HF (Prospective Comparison of ARNI With ACEI to Determine Impact on Global Mortality and Morbidity in Heart Failure) trials to identify proteins with different prognostic relationships between the 2 HF subtypes. Finally, we derived and evaluated the performance of an HFpEF‐specific proteomic risk score.
Methods
The data, analytic methods, and study materials will not be made available to other researchers for purposes of reproducing the results.
Patient Population
The PARAGON‐HF trial was a global, multicenter, randomized clinical trial comparing sacubitril/valsartan and valsartan (1:1) in 4796 patients with chronic HF, left ventricular ejection fraction ≥45%, and elevated NT‐proBNP (N‐terminal pro‐B‐type natriuretic peptide) levels. The design and primary results of PARAGON‐HF have been reported previously. 11 , 12 Key inclusion criteria included New York Heart Association functional class II to IV symptoms, either left ventricular hypertrophy or left atrial enlargement by echocardiography, and diuretic use for at least 30 days. Only patients who tolerated sequential valsartan and sacubitril/valsartan run‐in periods were randomized and included in the protein‐outcome analyses. Sacubitril/valsartan was associated with a 13% lower rate of the primary end point, total (first and recurrent) HFH and cardiovascular death, which did not reach statistical significance compared with valsartan alone (rate ratio [RR], 0.87 [95% CI, 0.75–1.01]; P=0.06). The primary outcome end points were adjudicated by a central clinical events committee.
Measurements of Circulating Proteins
Serum samples for proteomic assays were available for 1117 patients in PARAGON‐HF at the prespecified baseline time point (before the start of the valsartan run‐in period). Availability was determined by patient consent and whether the local site was included in the biomarker substudy. Samples were stored at −80°C and analyzed centrally in a single batch. The SomaScan version 3 proteomic assay was performed at SomaLogic Inc. (Boulder, CO) to quantify levels of 4828 signal SOMAmers (slow off‐rate modified aptamers) targeting 4123 unique circulating proteins in these serum samples. 13 , 14 , 15 Modified aptamers are DNA oligonucleotides, which have been selected to bind specific proteins with high affinity. 16 SOMAmer levels were standardized at SomaLogic Inc. including hybridization normalization (controls for variability in the readout of individual microarrays), plate scaling (accounts for plate‐by‐plate variation), median signal normalization (controls for total signal differences between individual samples), and calibration (removes the variation between assay runs within and across experiments using calibrator samples). Median intra‐ and interassay coefficients of variation are <6%. 17 Relative fluorescence units were transformed to log2 scale, normalized to the median separately by dilution level across all plates, and standardized (mean, 0 and SD, 1). No samples were removed for poor data quality, and there were no missing values in the data.
Statistical Analysis
The study design and statistical approach are shown in Figure 1. Baseline characteristics of patients with and without available proteomic data were described using number and proportion for categorical variables, mean and SD for normally distributed continuous variables, and median and interquartile range for skewed continuous variables. We evaluated associations between individual serum protein levels and risk of the primary end point, the occurrence, and timing of total HFH and cardiovascular death, with recurrent event regression models using the semiparametric proportional rates method of Lin et al, 18 which was prespecified for the primary end point in PARAGON‐HF. SOMAmers for which the proportional hazards assumption was violated (false discovery rate [FDR] <0.05) were excluded (2% excluded). Two covariate models were used. First, a minimally adjusted regression model included covariates for age, sex, treatment arm, and anticoagulant status (given the observation that anticoagulants affect measurement of serum proteins), and stratified by region. Second, a risk factor–adjusted regression model included 10 additional baseline covariates: diabetes, smoking, history of angiotensin‐converting enzyme intolerance, New York Heart Association functional class, systolic blood pressure, body mass index, left ventricular ejection fraction, total cholesterol, estimated glomerular filtration rate (eGFR), and log‐transformed NT‐proBNP. Protein‐outcome associations with Benjamini–Hochberg FDR <0.05 were considered significant. To avoid excess influence from extreme values that may be unreliable due to technical error, we defined outlier individuals for each SOMAmer as those individuals whose protein levels exceeded 5 SDs from the mean value. Only associations that remained significant after exclusion of the extreme protein levels of outlier individuals were considered robust. Between 0 and 20 outlier individuals were identified and removed per SOMAmer, with a median of 4 outlier individuals per SOMAmer. QIAGEN Ingenuity Pathway Analysis (QIAGEN Inc., https://digitalinsights.qiagen.com/IPA) was underpowered to identify groups of biologically related proteins that were overrepresented in the set of proteins associated with the primary end point from the minimally adjusted model, and the results were inconclusive and therefore omitted. 19 For a conservative sensitivity analysis, patients were randomly partitioned into a discovery cohort (67%) and replication cohort (33%) with the removal of per‐protein outliers, as previously described, such that the event rate was the same in both cohorts. Only proteins associated with the primary endpoint (FDR <0.05) in the discovery cohort were tested in the replication cohort; those also associated (FDR <0.05) with the primary end point in the replication cohort were considered significant. We also conducted regression analyses for the subcomponents of the primary end point: total HFH, cardiovascular death, and time to first HFH.
Figure 1. Study design.
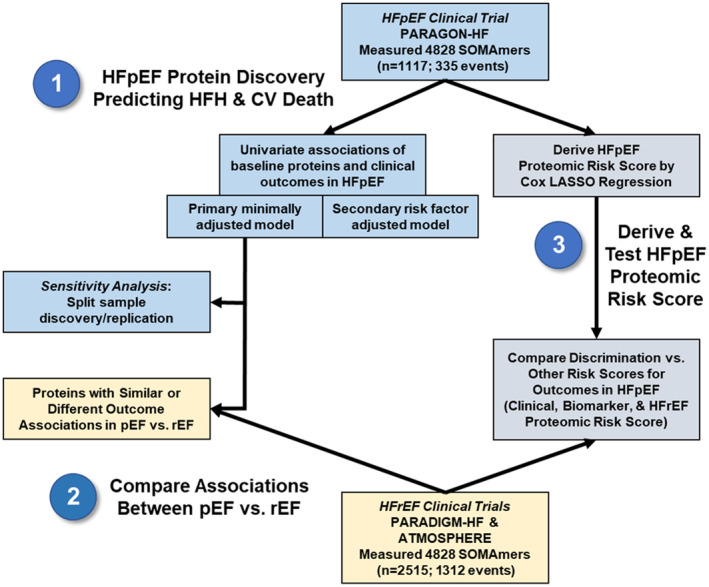
ATMOSPHERE indicates Efficacy and Safety of Aliskiren and Aliskiren/Enalapril Combination on Morbidity–Mortality in Patients With Chronic Heart Failure; CV, cardiovascular; HFH, heart failure hospitalization; HFpEF, heart failure with preserved ejection fraction; HFrEF, heart failure with reduced ejection fraction; LASSO, least absolute shrinkage and selection operator; PARADIGM‐HF, Prospective Comparison of ARNI With ACEI to Determine Impact on Global Mortality and Morbidity in Heart Failure; pEF, preserved ejection fraction; rEF, reduced ejection fraction; and SOMAmer, slow off‐rate modified aptamer.
We then compared protein–outcome associations for the primary end point in patients with HFpEF in PARAGON‐HF to patients with HFrEF in the ATMOSPHERE and PARADIGM‐HF clinical trials whose baseline serum samples were assayed with the same SOMAscan platform. Both ATMOSPHERE and PARADIGM‐HF enrolled a population of patients with chronic, stable HFrEF; ATMOSPHERE compared the direct renin antagonist aliskiren to enalapril, and PARADIGM‐HF compared sacubitril/valsartan with enalapril. Baseline protein–outcome results from these patients have been previously published. 8 The identical end point (total HFH and cardiovascular death), minimally adjusted covariates (age, sex, treatment, anticoagulant status), and regression model (recurrent event regression using the method of Lin et al 18 ) were used in all 3 trials in this study to facilitate comparisons of regression β coefficients. SOMAmers for which the proportional hazards assumption was violated (FDR <0.05) in any of the 3 trials were excluded (6% excluded). To enable a single comparison between HFpEF and HFrEF, summary statistics for the 2 HFrEF trials were meta‐analyzed using the inverse variance‐weighted average method and random‐effects models. To identify proteins whose relationship with the primary end point was statistically different between trials, we calculated a Z‐statistic equal to (βrEF − βpEF)/(SErEF 2 + SEpEF 2). Z scores corresponding to Benjamini–Hochberg FDR <0.05 provided statistical evidence of a difference in the magnitude of the protein–outcome association between the 2 HF populations. As a negative control, we compared β coefficients between the 2 HFrEF populations from the ATMOSPHERE and PARADIGM‐HF clinical trials using the same approach.
We derived a proteomic risk score in PARAGON‐HF using Cox least absolute shrinkage and selection operator regression and compared its discriminating power for the risk of a first HFH or cardiovascular death end point to existing clinical, immunoassay, and proteomic risk prediction models. To avoid overestimating discrimination by evaluating the score on the same patients on which it was derived, we performed a leave‐one‐out cross‐validation: The PARAGON‐HF cohort was split into a testing set with 1 patient and a training set including all other patients. The Cox least absolute shrinkage and selection operator model was derived in the training set and used to calculate a score for the single patient in the test set. This procedure was then repeated for each patient as the test set, such that each patient's proteomic score was derived from the remaining patients. Only proteins, not clinical variables, were included in the proteomic risk score. These proteomic scores were then compared with 4 external risk models also evaluated in PARAGON‐HF, including the Meta‐Analysis Global Group in Chronic Heart Failure clinical risk score (which predicts all‐cause death in HFrEF and HFpEF), 20 NT‐proBNP measured by immunoassay, high‐sensitivity cardiac troponin T measured by immunoassay, and a previously published 64‐protein risk score derived to predict time to first HFH or cardiovascular death from SomaScan data in the ATMOSPHERE trial of chronic HFrEF. 8 , 21 , 22 Discrimination was assessed by Harrell's C‐statistic and the 95% CIs for C‐statistics, C‐statistic differences, and P values for differences in C‐statistics, which were estimated by Somer's D method.
Informed consent was obtained from all patients in PARAGON‐HF, PARADIGM‐HF, and ATMOSPHERE. The study protocol was approved by local institutional review boards and ethics committees. Strengthening the Reporting of Observational Studies in Epidemiology cohort reporting guidelines were used. 23 Analyses were conducted in R version 4.0 (R Foundation for Statistical Computing, Vienna, Austria) and STATA version 16 (StataCorp, College Station, TX). 24 , 25 Recurrent events regression and meta‐analysis of the HFrEF trials were performed using the “survival” and “meta” packages in R, respectively. 26 , 27
Results
Baseline Characteristics and Outcomes
Baseline characteristics of patients in PARAGON‐HF with or without baseline proteomic data are shown in Table 1. Patients with proteomic data were slightly older, more likely White race, less likely to have been enrolled in Asia or Latin America, and had slightly lower eGFR. Patients with proteomic data experienced 335 instances of the primary end point, total HFH or cardiovascular death, over a mean follow‐up of 2.7 years. The primary end point event rate was lower in patients with proteomic data compared with those without (11.0 versus 14.5 events per 100 patient‐years; P=0.02). Patients with proteomic data experienced 262 total HFH (150 patients) and 73 deaths.
Table 1.
Baseline Characteristics of PARAGON‐HF Participants With and Without Proteomic Data
| Available proteomic data | No proteomic data | P value | |
|---|---|---|---|
| n | n=1117 | n=3679 | |
| Age, y | 73.5±8.0 | 72.5±8.5 | <0.001 |
| Female sex | 578 (51.7) | 1901 (51.7) | 0.97 |
| Race | <0.001 | ||
| White | 1035 (92.7) | 2872 (78.1) | |
| Asian | 37 (3.3) | 570 (15.5) | |
| Black | 15 (1.3) | 87 (2.4) | |
| Other | 30 (2.7) | 150 (4.1) | |
| Region | <0.001 | ||
| Asia/Pacific and other | 106 (9.5) | 656 (17.8) | |
| Central Europe | 532 (47.6) | 1183 (32.2) | |
| Latin America | 13 (1.2) | 357 (9.7) | |
| North America | 136 (12.2) | 423 (11.5) | |
| Western Europe | 330 (29.5) | 1060 (28.8) | |
| Diabetes | 466 (41.7) | 1596 (43.4) | 0.33 |
| Stroke | 119 (10.7) | 389 (10.6) | 0.92 |
| Hypertension | 1072 (96.0) | 3512 (95.5) | 0.47 |
| Prior MI | 237 (21.2) | 846 (23.0) | 0.21 |
| Ischemic pathogenesis | 412 (36.9) | 1311 (35.6) | 0.45 |
| Atrial fibrillation/flutter | 359 (32.2) | 1193 (32.6) | 0.83 |
| Prior HF hospitalization | 402 (36.0) | 1904 (51.8) | <0.001 |
| NYHA functional class | 0.17 | ||
| I or II | 884 (79.2) | 2959 (80.4) | |
| III or IV | 233 (20.9) | 718 (19.5) | |
| Medications | |||
| ACEi/ARB | 993 (88.9) | 3146 (85.5) | 0.004 |
| MRA | 272 (24.4) | 967 (26.3) | 0.2 |
| Diuretic | 1082 (96.9) | 3503 (95.2) | 0.018 |
| β Blocker | 919 (82.3) | 2902 (78.9) | 0.014 |
| Anticoagulant | 366 (32.8) | 1177 (32.0) | 0.63 |
| Systolic BP, mm Hg | 130.8±15.3 | 130.5±15.5 | 0.5 |
| Diastolic BP, mm Hg | 74.5±10.3 | 74.3±10.6 | 0.59 |
| BMI, kg/m2 | 30.7±4.8 | 30.1±5.0 | <0.001 |
| LVEF, % | 57.3±7.5 | 57.6±8.0 | 0.18 |
| Potassium, mmol/L | 4.5±0.4 | 4.5±0.5 | 0.76 |
| eGFR, mL/min per 1.73 m2 | 60.2±17.9 | 63.3±19.4 | <0.001 |
| NT‐proBNP, pg/mL | 945 (506– 1551) | 892 (452–1643) | 0.19 |
| hs‐TNT, ng/L | 17 (11–26) | 24 (11–37) | 0.045 |
Values are n (%) for categorical variables or median (interquartile range) for continuous variables. ACEi indicates angiotensin‐converting enzyme inhibitor; ARB, angiotensin receptor blocker; BMI, body mass index; BP, blood pressure; eGFR, estimated glomerular filtration rate; HF, heart failure; hs‐TNT, high‐sensitivity cardiac troponin T; LVEF, left ventricular ejection fraction; MI, myocardial infarction; MRA, mineralocorticoid receptor antagonist; NT‐proBNP, N‐terminal pro‐B‐type natriuretic peptide; NYHA, New York Heart Association; and PARAGON‐HF, Efficacy and Safety of LCZ696 Compared to Valsartan, on Morbidity and Mortality in Heart Failure Patients With Preserved Ejection Fraction.
Individual Proteins Predicting Risk of Heart Failure Events
Associations between baseline serum levels of individual proteins and the risk of the primary end point are shown in Figure 2. We identified 381 SOMAmers representing 366 unique proteins to be significantly associated with total HFH and cardiovascular death in the minimally adjusted model (FDR <0.05), of which 301 SOMAmers representing 288 proteins remained significant after removal of outlier protein values >5 SD from the mean (Figure 2A). Higher levels of 207 proteins were associated with a greater risk of the primary end point (Table 2 and Table S1). The strongest positive associations included B2M (β‐2 microglobulin; RR, 1.74 [95% CI, 1.45–2.10] per SD; P=3.9 × 10−9), TIMP1 (tissue inhibitor of matrix metalloproteinase 1; RR, 1.49 [95% CI, 1.30–1.70] per SD; P=6.5 × 10−9), and SVEP1 (sushi, von Willebrand factor type A, EGF, and pentraxin domain containing 1; RR, 1.46 [95% CI, 1.28–1.67] per SD; P=1.3 × 10−8). Higher levels of 81 proteins were associated with a lower risk of the primary end point (Table 3 and Table S1). The strongest negative associations included SERPINA4 (serpin family A member 4; RR, 0.73 [95% CI, 0.66–0.81] per SD; P=1.1 × 10−9), AFM (afamin; RR 0.65 [95% CI 0.56–0.77] per SD, P=2.3 × 10−7), and growth hormone receptor (RR, 0.68 [95% CI, 0.59–0.80] per SD; P=9.4 × 10−6). A sensitivity analysis in which PARAGON‐HF patients were randomly split into discovery (67%) and replication (33%) cohorts identified a conservative subset of 23 of the 288 proteins from the full discovery population, which included SVEP1, TIMP1, SERPINA4, and GDF15 (growth differentiation factor 15) (Table S2).
Figure 2. Serum proteins associated with risk of total heart failure hospitalization and cardiovascular death.
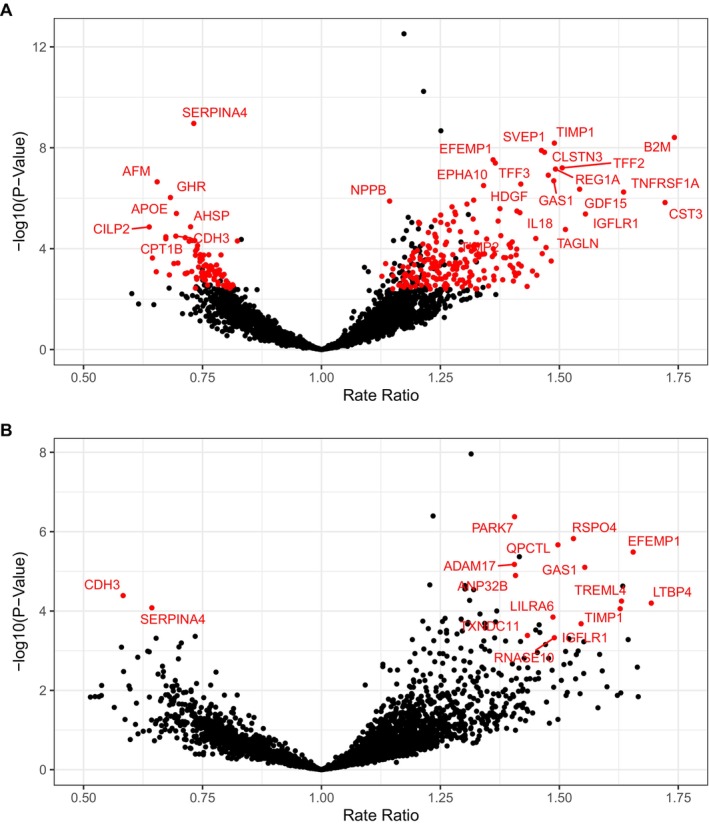
Circulating proteins significantly associated with the primary end point, total heart failure hospitalization and cardiovascular death. Associations were considered significant (red label) if the P value was lower than the false discovery rate threshold of 0.05 both with and without inclusion of outlier values >5 SDs from the mean. (A) Minimally adjusted model includes age, sex, treatment, and anticoagulant use. (B) Risk factor adjusted model is further adjusted for ten clinical risk factors: diabetes, smoking, history of angiotensin‐converting enzyme intolerance, New York Heart Association functional class, systolic blood pressure, body mass index, left ventricular ejection fraction, total cholesterol, estimated glomerular filtration rate, and log‐transformed N‐terminal pro‐B‐type natriuretic peptide.
Table 2.
Top 10 Proteins Associated With Greater Risk of Total Heart Failure Hospitalization and Cardiovascular Death in HFpEF
| Protein symbol | Protein name | Minimally adjusted | Risk factor adjusted | ||
|---|---|---|---|---|---|
| Rate ratio (95% CI) | P value | Rate ratio (95% CI) | P value | ||
| B2M | β‐2 microglobulin | 1.74 (1.45–2.10) | 3.9 × 10−9 | 2.12 (1.64–2.74) | 1.2 × 10−8 |
| TIMP1 | Metalloproteinase inhibitor 1 | 1.49 (1.30–1.70) | 6.5 × 10−9 | 1.66 (1.28–2.15) | 1.4 × 10−4 |
| SVEP1 | Sushi, von Willebrand factor type A, EGF and pentraxin domain‐containing protein 1 | 1.46 (1.28–1.67) | 1.3 × 10−8 | 1.55 (1.21–1.99) | 6.0 × 10−4 |
| CLSTN3 | Calsyntenin‐3 | 1.47 (1.29–1.68) | 1.5 × 10−8 | 1.17 (0.89–1.53) | 2.5 × 10−1 |
| EFEMP1 | EGF‐containing fibulin‐like extracellular matrix protein 1 | 1.36 (1.22–1.52) | 3.0 × 10−8 | 1.66 (1.34–2.05) | 3.3 × 10−6 |
| TFF3 | Trefoil factor 3 | 1.37 (1.22–1.53) | 4.0 × 10−8 | 1.19 (0.83–1.72) | 3.4 × 10−1 |
| TFF2 | Trefoil factor 2 | 1.51 (1.30–1.75) | 6.3 × 10−8 | 1.41 (1.05–1.91) | 2.4 × 10−2 |
| REG1A | Lithostathine‐1‐α | 1.49 (1.29–1.73) | 7.0 × 10−8 | 1.43 (0.99–2.07) | 5.8 × 10−2 |
| SPON1 | Spondin‐1 | 1.48 (1.28–1.71) | 1.2 × 10−7 | 1.44 (1.02–2.04) | 3.7 × 10−2 |
| GAS1 | Growth arrest‐specific protein 1 | 1.49 (1.28–1.73) | 2.0 × 10−7 | 1.55 (1.28–1.88) | 7.9 × 10−6 |
The 10 proteins most strongly associated with greater risk of the primary end point, total heart failure hospitalization and cardiovascular death, in PARAGON‐HF in a recurrent events regression model adjusted for age, sex, treatment, and anticoagulant use. Only associations that remained statistically significant at false discovery rate <0.05 after removal of outlier values >5 SDs from the mean were considered significant. Results from the risk factors adjusted model are included for reference. The full lists of protein associations are provided in Tables S1 and S3. HFpEF indicates heart failure with preserved ejection fraction; and PARAGON‐HF, Efficacy and Safety of LCZ696 Compared to Valsartan, on Morbidity and Mortality in Heart Failure Patients With Preserved Ejection Fraction.
Table 3.
Top 10 Proteins Associated With Lower Risk of Heart Failure Hospitalization and Cardiovascular Death in HFpEF
| Protein symbol | Protein name | Minimally adjusted | Risk factor adjusted | ||
|---|---|---|---|---|---|
| Rate ratio (95% CI) | P value | Rate ratio (95% CI) | P value | ||
| SERPINA4 | Kallistatin | 0.73 (0.66–0.81) | 1.1 × 10−9 | 0.7 (0.59–0.84) | 1.2 × 10−4 |
| AFM | Afamin | 0.65 (0.56–0.77) | 2.2 × 10−7 | 0.64 (0.49–0.84) | 1.1 × 10−3 |
| GHR | Growth hormone receptor | 0.68 (0.59–0.8) | 9.4 × 10−7 | 0.7 (0.54–0.92) | 9.7 × 10−3 |
| APOE | Apolipoprotein E (isoform E4) | 0.70 (0.60–0.81) | 4.0 × 10−6 | 0.72 (0.53–0.97) | 3.2 × 10−2 |
| CILP2 | Cartilage intermediate layer protein 2 | 0.64 (0.52–0.78) | 1.4 × 10−5 | 0.84 (0.64–1.11) | 2.1 × 10−1 |
| AHSP | α‐Hemoglobin‐stabilizing protein | 0.72 (0.63–0.84) | 1.4 × 10−5 | 0.87 (0.66–1.16) | 3.4 × 10−1 |
| CDH3 | Cadherin‐3 | 0.69 (0.58–0.82) | 3.2 × 10−5 | 0.58 (0.45–0.75) | 4.1 × 10−5 |
| CPT1B | Carnitine O‐palmitoyltransferase 1, muscle isoform | 0.67 (0.56–0.81) | 3.4 × 10−5 | 0.85 (0.61–1.2) | 3.7 × 10−1 |
| CLEC4M | C‐type lectin domain family 4 member M | 0.71 (0.61–0.84) | 3.7 × 10−5 | 0.67 (0.51–0.87) | 3.4 × 10−3 |
| CLEC3B | Tetranectin | 0.67 (0.56–0.81) | 4.0 × 10−5 | 0.69 (0.51–0.94) | 1.9 × 10−2 |
The 10 proteins most strongly associated with lower risk of the primary end point, total heart failure hospitalization and cardiovascular death, in PARAGON‐HF in a recurrent events regression model adjusted for age, sex, treatment, and anticoagulant use. Only associations that remained statistically significant at false discovery rate <0.05 after removal of outlier values >5 SDs from the mean were considered significant. Results from the risk factors adjusted model are included for reference. The full lists of protein associations are provided in Tables S1 and S3. HFpEF indicates heart failure with preserved ejection fraction; and PARAGON‐HF, Efficacy and Safety of LCZ696 Compared to Valsartan, on Morbidity and Mortality in Heart Failure Patients With Preserved Ejection Fraction.
Further adjustment for clinical risk factors, including eGFR and NT‐proBNP, identified a smaller set of 60 SOMAmers corresponding to 59 proteins associated with the primary end point at FDR <0.05, of which 19 SOMAmers representing 18 proteins remained significant after removal of outlier protein values >5 SDs from the mean (Figure 2B, Table S3). Eleven of the 18 proteins from the risk factor–adjusted model were also significantly associated with the primary endpoint in the minimally adjusted model, including TIMP1 and SERPINA4. Notably, B2M, a known marker of renal function, remained strongly associated with the primary end point despite the adjustment for clinical risk factors and eGFR (RR, 2.12 [95% CI, 1.64–2.74] per SD; P=1.2 × 10−8).
Associations between baseline serum levels of individual proteins and the risk of the subcomponents of the primary end point, total HFH, cardiovascular death, and time to first HFH suggested similar results (Tables S4–S7). The number of events for each of the subcomponent outcomes was limiting, and there were no proteins significantly associated with cardiovascular death in the risk factor–adjusted model or time to first HFH in the risk factor–adjusted model.
Comparison of Protein–Outcome Associations Between HFpEF and HFrEF
Overall, associations between protein levels and total HFH and cardiovascular death were similar between patients with HFpEF in PARAGON‐HF and patients with HFrEF in ATMOSPHERE and PARADIGM‐HF (Figure 3). For example, SVEP1 levels were strongly associated with total HFH and cardiovascular death in both HFpEF (RR, 1.46 [95% CI, 1.28–1.67] per SD) and HFrEF (RR, 1.64 [95% CI, 1.52–1.76] per SD) populations.
Figure 3. Associations between protein levels and total heart failure hospitalization and cardiovascular death in HFpEF vs HFrEF.
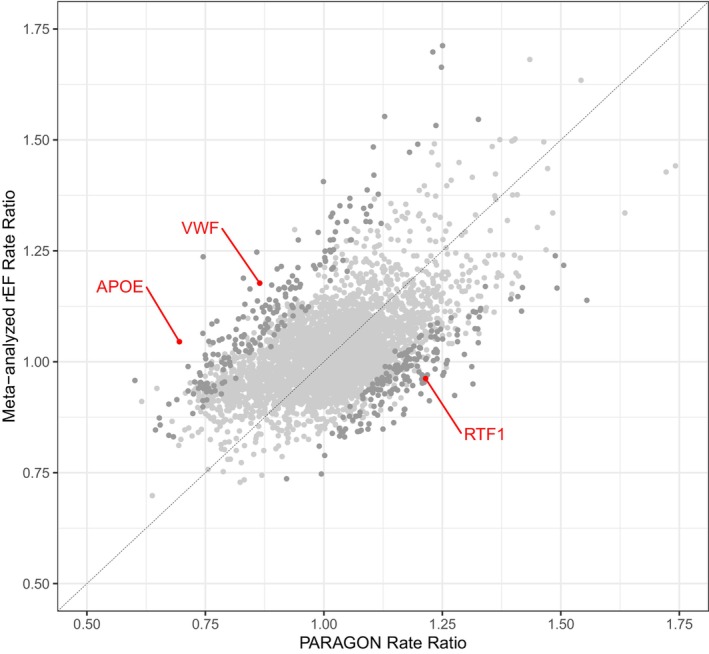
Only 3 proteins (red dots) demonstrated different associations with outcomes with HFrEF vs HFpEF at a significance level of Benjamini–Hochberg FDR <0.05. Light gray dots indicate proteins with no evidence of differential association between ejection fraction subtypes. Dark gray dots indicate proteins for which the difference in association was statistically significant by nominal P value <0.05 but not FDR <0.05. APOE indicates apolipoprotein E; FDR, false discovery rate; HFpEF, heart failure with preserved ejection fraction; HFrEF, heart failure with reduced ejection fraction; rEF, reduced ejection fraction; and VWF, von Willebrand factor.
RRs were compared between the patients with HFpEF and patients with HFrEF using recurrent event regression models that included identical covariates and clinical outcomes. Three proteins had statistically significant differences in RRs between the 2 HF subtypes after adjustment for multiple testing (FDR <0.05) (Figure 3 and Table S8). Apolipoprotein E was inversely associated with the primary outcome in HFpEF (RR, 0.70 [95% CI, 0.54–0.85]) but neutral in HFrEF (RR, 1.05 [95% CI, 0.96–1.14]; P=6.7 × 10−6 for difference). RNA polymerase‐associated protein RTF1 homolog (RTF1) was positively associated with the primary outcome in HFpEF (RR, 1.21 [95% CI 1.16–1.27)] but neutral in HFrEF (RR, 0.96 [95% CI, 0.89–1.05]; P=6.2 × 10−6 for difference). von Willebrand factor was inversely associated with the primary outcome in HFpEF (RR, 0.86 [95% CI 0.76–0.96]) and positively associated in HFrEF (RR, 1.18 [95% CI, 1.07–1.29]; P=1.1 × 10−5 for difference). However, in the negative control comparison between the 2 HFrEF populations (ATMOSPHERE and PARADIGM‐HF), there was a similar number of proteins with nominal and multiple‐testing adjusted significant differences in the magnitude of the protein‐outcome association (357 proteins in the HFpEF versus HFrEF analysis as compared with 446 proteins in the HFrEF versus HFrEF analysis). These negative control findings suggest that much of the difference between the protein–outcome associations in the HFpEF and HFrEF populations may be explained by chance or nonbiological factors, rather than physiological differences.
Comparison of the Performance of Clinical and Proteomic Risk Scores
A 46‐protein proteomic risk score for the time to first HFH or cardiovascular death was developed in PARAGON‐HF and assessed using leave‐one‐out cross‐validation to prevent overfitting. In the HFpEF test set, discrimination of the HFpEF‐derived PARAGON‐HF proteomic score (C‐statistic, 0.66 [95% CI, 0.62–0.70]) was similar to that of the HFrEF‐derived ATMOSPHERE proteomic score (C‐statistic, 0.67 [95% CI, 0.63–0.71]; P=0.8 for difference between C‐statistics). The HFpEF‐derived PARAGON‐HF proteomic score provided only modestly higher discrimination than the Meta‐Analysis Global Group in Chronic Heart Failure clinical risk score (C‐statistic, 0.61 [95% CI, 0.57–0.65]; P=0.03 versus PARAGON‐HF proteomic score) and no significant improvement as compared with log‐transformed NT‐proBNP alone (C‐statistic, 0.62 [95% CI, 0.58–0.66]; P=0.06 versus PARAGON‐HF score) or high‐sensitivity cardiac troponin T alone (C‐statistic, 0.65 [95% CI, 0.61–0.71]; P=0.8 versus PARAGON‐HF score; Figure 4). In a sensitivity analysis, a PARAGON‐HF proteomic risk developed with Cox least absolute shrinkage and selection operator with a more stringent variable selection penalty (λ 1 SE) included 6 proteins and provided similar discrimination in cross‐validation (C‐statistic, 0.64 [95% CI, 0.60–0.68]); this result suggests any overfitting did not impair the discrimination of the HFpEF‐specific proteomic risk score.
Figure 4. Risk discrimination of clinical, biomarker, and proteomic risk scores for time to first heart failure hospitalization or cardiovascular death in PARAGON‐HF.
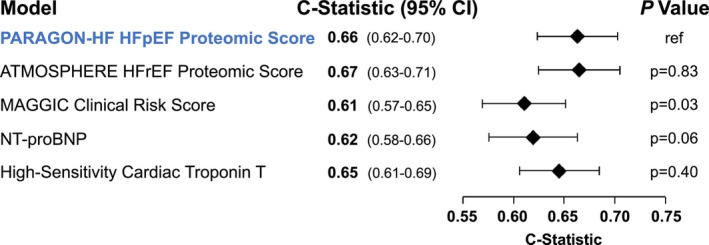
Risk discrimination was evaluated in PARAGON‐HF. The PARAGON‐HF HFpEF proteomic risk score was derived and evaluated in PARAGON‐HF using a leave‐one‐out cross‐validation method to avoid overfitting. P‐values evaluate the null hypothesis that each C‐statistic is significantly different from the C‐statistic for the PARAGON‐HF HFpEF proteomic score. ATMOSPHERE indicates Efficacy and Safety of Aliskiren and Aliskiren/Enalapril Combination on Morbidity–Mortality in Patients With Chronic Heart Failure; HFpEF, heart failure with preserved ejection fraction; HFrEF, heart failure with reduced ejection fraction; MAGGIC, Meta‐Analysis Global Group in Chronic Heart Failure; NT‐proBNP, N‐terminal pro‐B‐type natriuretic peptide; and PARAGON‐HF, Efficacy and Safety of LCZ696 Compared to Valsartan, on Morbidity and Mortality in Heart Failure Patients With Preserved Ejection Fraction.
Discussion
In this broad proteomic study of 1117 patients with HFpEF enrolled in the PARAGON‐HF trial, we identified 288 serum proteins that were significantly associated with total HFH and cardiovascular death after correction for multiple testing and outlier samples. Proteins and protein pathways associated with these clinical events implicate coagulation, metabolic processes, and extracellular matrix regulation as important in the progression of HFpEF. For example, SVEP1, a novel HF biomarker identified in a similar study of patients with HFrEF, was also among the proteins most strongly associated with clinical outcomes in patients with preserved ejection fraction (EF). Notably, associations between baseline protein levels and outcomes in HFpEF patients in PARAGON‐HF were similar to those in patients with HFrEF. Three proteins were differentially associated with outcomes in HFpEF as compared with HFrEF (apolipoprotein E, RTF1, and von Willebrand factor). However, we note that a similar number of proteins were differentially associated with outcomes in a comparison of 2 HFrEF trials (ATMOSPHERE and PARADIGM‐HF), suggesting the possibility for false discovery. Despite enthusiasm for improved risk prediction in HFpEF with combinations of multiple proteins, a new proteomic risk score derived in patients with HFpEF was not meaningfully superior to clinical risk factors, circulating levels of NT‐proBNP, or high‐sensitivity cardiac troponin. Surprisingly, the internally derived HFpEF proteomic risk score did not outperform an externally derived HFrEF proteomic risk score in the PARAGON‐HF HFpEF population. Taken together, these results suggest that physiological processes that are reflected in circulating protein levels similarly influence risk of adverse cardiovascular outcomes in both patients with HFpEF and patients with HFrEF.
Reassuringly, we reproduced many known directly measured biomarkers of HF prognosis, including NT‐proBNP and GDF15. Proteins within the coagulation system and extracellular matrix proteins such as TIMP1, ADAM17, and LTBP4 were associated with HFpEF outcomes, particularly after adjustment for clinical risk factors, consistent with prior work. 28 , 29 The inverse association between kallistatin/SERPINA4 levels and clinical events in our study has been observed with other forms of cardiovascular disease, and some animal studies support a causal role for this protein in reducing oxidative stress and adverse cardiac remodeling. 30 , 31 , 32 B2M, a known renal function marker, emerged as the most strongly associated biomarker after adjustment for clinical covariates, including eGFR and NT‐proBNP, highlighting that known pathologic processes, such as poor renal function, are key markers of adverse HFpEF outcomes.
Compared with traditional biomarker immunoassays, large‐scale proteomic assays offer the ability to characterize associations between many proteins and clinical outcomes, and they may identify new associations using an unbiased hypothesis‐free approach. SVEP1, a novel HF biomarker identified in our previous study of patients with HFrEF in PARADIGM‐HF and ATMOSPHERE, 8 was found to be among the strongest predictors of clinical cardiovascular outcomes in HFpEF. SVEP1 has not been previously identified as an HFpEF prognosis marker. This finding provides additional validation of SVEP1 and indicates a potential clinical and biological role across the EF spectrum. The specificity of the SomaScan SVEP1 aptamer for the correct target protein is supported by data from the deCODE study, in which aptamer‐measured SVEP1 levels were strongly associated with a genetic variant in the SVEP1 gene on chromosome 9. 33 SVEP1 is an extracellular matrix protein expressed in vascular smooth muscle cells that promotes inflammation and atherosclerosis through integrin, notch, and fibroblast growth factor receptor signaling in animal studies. 34 Plasma levels of SVEP1 have been causally linked to human platelet activation and cardiovascular disease. 35 The mechanism linking circulating SVEP1 and HFpEF outcomes merits further investigation.
Contrary to our initial hypothesis, the vast majority of proteins showed similar associations with clinical outcomes in patients with HFpEF versus HFrEF. Consistent outcome associations between HFpEF and HFrEF are also observed for most traditional prognostic biomarkers (creatinine, NT‐proBNP, troponin, collagen precursors). 29 , 36 These results underscore that substantial pathophysiology of the HF syndrome is shared regardless of ejection fraction. Given the number of proteins tested, numerical differences between the magnitude of protein–outcome associations in the HFpEF and HFrEF populations were expected. We used a rigorous statistical approach to distinguish robust evidence of true differential associations in the 2 HF subtypes. Despite a total sample size of >3600 patients HF with across the ejection fraction spectrum, only 3 proteins (apolipoprotein E, RTF1, and von Willebrand factor) showed statistically significant heterogeneity. While these 3 proteins may validly identify distinct biology between ejection fraction subtypes, they are currently speculative, given the absence of external validation, possible effects of outliers, potential for residual confounding by anticoagulants for von Willebrand factor, and the fact that our approach similarly identified proteins with differential outcome associations in the negative control analysis between the 2 HFrEF trial populations.
Finally, we evaluated an HFpEF‐specific proteomic risk score, which did not meaningfully improve risk discrimination as compared with commonly ascertained clinical parameters and biomarkers like NT‐proBNP or high‐sensitivity cardiac troponin. There has been substantial excitement that combining information from thousands of protein levels rather than 1 or 2 would improve clinical risk prediction. Despite this potential, we observe only small improvements in discrimination with multiprotein risk scores that are marginally statistically significant and would be unlikely to meaningfully impact clinical care. Our data do not support a role for proteomic risk scores in the clinical care of patients with HFpEF at this time. The strong prognostic value of NT‐proBNP and troponin may explain marginal improvement with the inclusion of more proteins, many of which are correlated with clinical biomarkers. We acknowledge that the relatively small number of HFH or cardiovascular death events in PARAGON‐HF and the limitations of the SomaScan platform may have prevented development of an accurate HFpEF‐specific proteomic risk score.
The results of our study should be considered in the context of several limitations. The SomaScan version 3 platform contains a wide spectrum of >4000 circulating proteins. However, this captures only a modest proportion of all human proteins, protein isoforms, and protein modifications. Many important pathways of cellular physiology and tissue‐specific biology are not reflected in the circulating proteome. Even when present, protein levels may not reflect the activation of relevant cellular processes or HFpEF physiology. Our methodology does not define the tissue from which serum proteins originated. The SomaScan platform has not been validated for all proteins, and further antibody‐based or mass spectrometry methods may be needed to fully determine aptamer accuracy. The protein quantitative trait loci identified for many of the proteins strongly support aptamer specificity. Cis‐protein quantitative trait loci were identified in the deCODE study for 8 of the top 10 proteins significantly associated with total HFH and cardiovascular death in the minimally adjusted model, including SERPINA4, TIMP1, SVEP1, EFEMP1 (EGF‐containing fibulin‐like extracellular matrix protein 1), TFF3 (trefoil factor 3), TFF2 (trefoil factor 2), REG1A (lithostathine‐1‐α), and SPON1 (spondin‐1). Trans‐protein quantitative trait loci were reported for the remaining 2 of the top 10 significant proteins, B2M and CLSTN3 (calsyntenin‐3). 33 The PARAGON‐HF clinical trial had strict inclusion criteria, which may impact the generalizability of the findings. For example, the inclusion criteria may have homogenized protein levels, biasing results toward the null. Proteomic data were collected on the basis of patient consent, and the subset of individuals with proteomic data may not be fully representative of patients with clinical HFpEF. Our analyses do not account for concomitant medications, other than anticoagulants, taken by patients before or during the clinical trial. These concomitant medications may impact individual protein levels and outcomes, and they may modify the effect of protein levels on outcome. Our comparison between HFpEF and HFrEF trials lacked statistical power to identify subtle differences in protein–outcome associations between these EF subtypes. However, our analysis represents the largest sample of patients with HF with large‐scale proteomic and clinical outcome data currently available. Because only 1 cohort of patients with HFpEF with proteomic and cardiovascular outcome data was available, external replication was not possible. Other methodological strengths of our study include rigorous inclusion criteria defining HFpEF, centrally adjudicated clinical outcomes, and performance of the SomaScan assay in a single batch for all PARAGON‐HF participants. Consortium efforts and meta‐analysis with additional cohorts may identify additional proteins more strongly associated with outcomes in HFpEF or HFrEF.
In this large‐scale proteomic investigation, we systemically characterized associations between serum protein levels and risk for total HFH and cardiovascular death in patients with HFpEF. Proteins involved in coagulation, metabolic disease, and extracellular matrix regulation were associated with adverse outcomes, highlighting the importance of these pathways in HFpEF pathogenesis and disease progression. SVEP1, a novel HF biomarker identified in a similar study of patients with HFrEF, was also among the proteins most strongly associated with clinical cardiovascular outcomes in patients with preserved ejection fraction. Associations between protein levels and outcomes in HFpEF patients in PARAGON‐HF were largely similar to those in patients with HFrEF. An HFpEF‐specific proteomic risk score did not meaningfully improve risk prediction compared with clinical factors and commonly measured biomarkers. These results identify and prioritize proteins associated with adverse outcomes in HFpEF, including substantial overlap with HFrEF. While the continued search for effective therapies unique to HFpEF is warranted, our results reinforce that shared pathologic processes are likely important drivers of poor HF prognosis across the EF spectrum.
Sources of Funding
The PARAGON‐F, PARADIGM‐HF, and ATMOSPHERE trials, and this proteomic study, were sponsored by Novartis. J.W.C. is supported by the KL2/Harvard Catalyst Medical Research Investigator Training program and the American Heart Association (23CDA1052151).
Disclosures
N.L. Patel‐Murray, L. Zhang, P. Serrano‐Fernandez, S. Wandel, C.‐W. Chen, D. Xu, G.M. Turner, Dr Chutkow, D.P. Yates, Dr O'Donnell, M.F. Prescott, Dr Lefkowitz, Dr Gimpelewicz, M.T. Beste, Dr Mendelson, and M. Healey are employees of Novartis and may own stock of Novartis. J. Jacob, L. Gou, and F. Zhao are former employees of Novartis and may own stock of Novartis. B.L. Claggett has received consulting fees from Bristol Myers Squibb, Cardurion, Corvia, Cytokinetics, Intellia, Novartis, and Rocket. Dr Desai reports institutional grant support from Abbott, Alnylam, AstraZeneca, Bayer, Novartis, and Pfizer; and consulting fees from Abbott, Alnylam, AstraZeneca, Avidity, Axon Therapeutics, Bayer, Biofourmis, Boston Scientific, Cytokinetics, GlaxoSmithKline, Medpace, Merck, New Amsterdam, Novartis, Parexel, Regeneron, River2Renal Roche, Veristat, Verily, and Zydus. P.S. Jhund reports speakers' fees from AstraZeneca, Novartis, Alkem Metabolics, ProAdWise Communications, and Sun Pharmaceuticals; advisory board fees from AstraZeneca, Boehringer Ingelheim, and Novartis; research funding from AstraZeneca, Boehringer Ingelheim, and Analog Devices Inc. P.S. Jhund's employer, the University of Glasgow, has been remunerated for clinical trial work from AstraZeneca, Bayer AG, Novartis, and NovoNordisk; Director, Global Clinical Trial Partners. Dr Packer reports personal fees from AbbVie, Actavis, Altimmune, Amgen, Amarin, Ardelyx, AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Caladrius, Casana, CSL Behring, Cytokinetics, Eli Lilly, Johnson & Johnson, Imara, Moderna, Novartis, ParatusRx, Pfizer, Reata, Regeneron, Relypsa, Salamandra, Synthetic Biologics, and Theravance. Dr Pfeffer has received research grant support through his institution from Novartis, has been a consultant to Alnylam, AstraZeneca, Boehringer Ingelheim, and Eli Lilly Alliance, Corvidia, DalCor, GlaxoSmithKline, Lexicon, National Heart, Lung, and Blood Institute CONNECTs (Collaborating Network of Networks for Evaluating COVID‐19 and Therapeutic Strategies; Master Protocol Committee), Novartis, Novo Nordisk, Peerbridge, and Sanofi, and has equity in DalCor. Dr Redfield is a nonpaid consultant for Novartis. Dr Rouleau has been a consultant to Bayer, Novartis, AstraZeneca, and Bristol Myers Squibb. Dr Zannad reports personal fees from Applied Therapeutics, Bayer, Boehringer, BMS, CVRx, Cardior, Cereno Pharmaceutical, Cellprothera, CEVA, Merck, Novartis, NovoNordisk, Owkin, Pfizer, and Servier, has stock options at G3Pharmaceutical and equities at Cereno Pharmaceutical, Cardiorenal, and Eshmoun Clinical Research, and is the founder of Cardiovascular Clinical Trialists. Dr Zile has received research funding from Novartis and has been a consultant for Novartis, Abbott, Boston Scientific, CVRx, EBR, Endotronics, Ironwood, Merck, Medtronic, and Myokardia V Wave. Dr McMurray has received payments through Glasgow University for work on clinical trials, consulting, and other activities from Alnylam, Amgen, AstraZeneca, Bayer, Boehringer Ingelheim, BMS, Cardurion, Cytokinetics, Dal‐Cor, GSK, Ionis, KBP Biosciences, Novartis, Pfizer, and Theracos; and personal lecture fees from the Corpus, Abbott, Hikma, Sun Pharmaceuticals, Medscape/Heart.Org, Radcliffe Cardiology, Servier Director, and Global Clinical Trial Partners. Dr Solomon has received research grants from Actelion, Alnylam, Amgen, AstraZeneca, Bellerophon, Bayer, BMS, Celladon, Cytokinetics, Eidos, Gilead, GSK, Ionis, Lilly, Mesoblast, MyoKardia, NIH/NHLBI, Neurotronik, Novartis, NovoNordisk, Respicardia, Sanofi Pasteur, Theracos, and US2.AI; and has consulted for Abbott, Action, Akros, Alnylam, Amgen, Arena, AstraZeneca, Bayer, Boeringer‐Ingelheim, BMS, Cardior, Cardurion, Corvia, Cytokinetics, Daiichi‐Sankyo, GSK, Lilly, Merck, Myokardia, Novartis, Roche, Theracos, Quantum Genomics, Cardurion, Janssen, Cardiac Dimensions, Tenaya, Sanofi‐Pasteur, Dinaqor, Tremeau, CellProThera, Moderna, American Regent, Sarepta, Lexicon, Anacardio, and Akros. Dr Cunningham has consulted for Roche Diagnostic, Occlutech, and KCK. The remaining authors have no disclosures to report.
Supporting information
Tables S1–S8
This manuscript was sent to Sula Mazimba, MD, MPH, Associate Editor, for review by expert referees, editorial decision, and final disposition.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.123.033544
For Sources of Funding and Disclosures, see page 11 and 12.
References
- 1. Heidenreich PA, Albert NM, Allen LA, Bluemke DA, Butler J, Fonarow GC, Ikonomidis JS, Khavjou O, Konstam MA, Maddox TM, et al. Forecasting the impact of heart failure in the United States: a policy statement from the American Heart Association. Circ Heart Fail. 2013;6:606–619. doi: 10.1161/HHF.0b013e318291329a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tsao CW, Lyass A, Enserro D, Larson MG, Ho JE, Kizer JR, Gottdiener JS, Psaty BM, Vasan RS. Temporal trends in the incidence of and mortality associated with heart failure with preserved and reduced ejection fraction. JACC Heart Fail. 2018;6:678–685. doi: 10.1016/j.jchf.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lam CS, Donal E, Kraigher‐Krainer E, Vasan RS. Epidemiology and clinical course of heart failure with preserved ejection fraction. Eur J Heart Fail. 2011;13:18–28. doi: 10.1093/eurjhf/hfq121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Solomon SD, Vaduganathan M, Claggett BL, Packer M, Zile M, Swedberg K, Rouleau J, Pfeffer MA, Desai A, Lund LH, et al. Sacubitril/valsartan across the spectrum of ejection fraction in heart failure. Circulation. 2020;141:352–361. doi: 10.1161/CIRCULATIONAHA.119.044586 [DOI] [PubMed] [Google Scholar]
- 5. Dewan P, Jackson A, Lam CSP, Pfeffer MA, Zannad F, Pitt B, Solomon SD, McMurray JJV. Interactions between left ventricular ejection fraction, sex and effect of neurohumoral modulators in heart failure. Eur J Heart Fail. 2020;22:898–901. doi: 10.1002/ejhf.1776 [DOI] [PubMed] [Google Scholar]
- 6. Schiattarella GG, Altamirano F, Tong D, French KM, Villalobos E, Kim SY, Luo X, Jiang N, May HI, Wang ZV, et al. Nitrosative stress drives heart failure with preserved ejection fraction. Nature. 2019;568:351–356. doi: 10.1038/s41586-019-1100-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paulus WJ, Zile MR. From systemic inflammation to myocardial fibrosis. Circ Res. 2021;128:1451–1467. doi: 10.1161/CIRCRESAHA.121.318159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang L, Cunningham JW, Claggett BL, Jacob J, Mendelson MM, Serrano‐Fernandez P, Kaiser S, Yates DP, Healey M, Chen C‐W, et al. Aptamer proteomics for biomarker discovery in heart failure with reduced ejection fraction. Circulation. 2022;146:1411–1414. doi: 10.1161/CIRCULATIONAHA.122.061481 [DOI] [PubMed] [Google Scholar]
- 9. Zannad F, Ferreira JP, Butler J, Filippatos G, Januzzi JL, Sumin M, Zwick M, Saadati M, Pocock SJ, Sattar N, et al. Effect of empagliflozin on circulating proteomics in heart failure: mechanistic insights into the EMPEROR programme. Eur Heart J. 2022;43:4991–5002. doi: 10.1093/eurheartj/ehac495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sanders‐van Wijk S, Tromp J, Beussink‐Nelson L, Hage C, Svedlund S, Saraste A, Swat SA, Sanchez C, Njoroge J, Tan R‐S, et al. Proteomic evaluation of the comorbidity‐inflammation paradigm in heart failure with preserved ejection fraction. Circulation. 2020;142:2029–2044. doi: 10.1161/CIRCULATIONAHA.120.045810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Solomon SD, Rizkala AR, Gong J, Wang W, Anand IS, Ge J, Lam CSP, Maggioni AP, Martinez F, Packer M, et al. Angiotensin receptor neprilysin inhibition in heart failure with preserved ejection fraction. Rationale and design of the PARAGON‐HF trial. JACC Heart Fail. 2017;5:471–482. doi: 10.1016/j.jchf.2017.04.013 [DOI] [PubMed] [Google Scholar]
- 12. Solomon SD, McMurray JJV, Anand IS, Ge J, Lam CSP, Maggioni AP, Martinez F, Packer M, Pfeffer MA, Pieske B, et al. Angiotensin–neprilysin inhibition in heart failure with preserved ejection fraction. N Engl J Med. 2019;381:1609–1620. doi: 10.1056/NEJMoa1908655 [DOI] [PubMed] [Google Scholar]
- 13. Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, Fitzwater T, et al. Aptamer‐based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ganz P, Heidecker B, Hveem K, Jonasson C, Kato S, Segal MR, Sterling DG, Williams SA. Development and validation of a protein‐based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. 2016;315:2532–2541. doi: 10.1001/jama.2016.5951 [DOI] [PubMed] [Google Scholar]
- 15. Jacob J, Ngo D, Finkel N, Pitts R, Gleim S, Benson MD, Keyes MJ, Farrell LA, Morgan T, Jennings LL, et al. Application of large‐scale aptamer‐based proteomic profiling to planned myocardial infarctions. Circulation. 2018;137:1270–1277. doi: 10.1161/CIRCULATIONAHA.117.029443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L, Janjic N. Nucleic acid ligands with protein‐like side chains: modified aptamers and their use as diagnostic and therapeutic agents. Mol Ther Nucleic Acids. 2014;3:e201. doi: 10.1038/mtna.2014.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Williams SA, Kivimaki M, Langenberg C, Hingorani AD, Casas JP, Bouchard C, Jonasson C, Sarzynski MA, Shipley MJ, Alexander L, et al. Plasma protein patterns as comprehensive indicators of health. Nat Med. 2019;25:1851–1857. doi: 10.1038/s41591-019-0665-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin DY, Wei LJ, Yang I, Ying Z. Semiparametric regression for the mean and rate functions of recurrent events. J R Stat Soc Ser B (Stat Methodol). 2000;62:711–730. doi: 10.1111/1467-9868.00259 [DOI] [Google Scholar]
- 19. Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pocock SJ, Ariti CA, McMurray JJV, Maggioni A, Køber L, Squire IB, Swedberg K, Dobson J, Poppe KK, Whalley GA, et al. Predicting survival in heart failure: a risk score based on 39 372 patients from 30 studies. Eur Heart J. 2013;34:1404–1413. doi: 10.1093/eurheartj/ehs337 [DOI] [PubMed] [Google Scholar]
- 21. McMurray JJV, Krum H, Abraham WT, Dickstein K, Køber LV, Desai AS, Solomon SD, Greenlaw N, Ali MA, Chiang Y, et al. Aliskiren, enalapril, or aliskiren and enalapril in heart failure. N Engl J Med. 2016;374:1521–1532. doi: 10.1056/NEJMoa1514859 [DOI] [PubMed] [Google Scholar]
- 22. Krum H, Massie B, Abraham WT, Dickstein K, Kober L, McMurray JJV, Desai A, Gimpelewicz C, Kandra A, Reimund B, et al. Direct renin inhibition in addition to or as an alternative to angiotensin converting enzyme inhibition in patients with chronic systolic heart failure: rationale and design of the Aliskiren Trial to Minimize OutcomeS in Patients with HEart failuRE (ATMOSPHERE) study. Eur J Heart Fail. 2011;13:107–114. doi: 10.1093/eurjhf/hfq212 [DOI] [PubMed] [Google Scholar]
- 23. von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. J Clin Epidemiol. 2008;61:344–349. doi: 10.1016/j.jclinepi.2007.11.008 [DOI] [PubMed] [Google Scholar]
- 24. R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2022. First Accessed Jan 2022; https://www.R‐project.org/ [Google Scholar]
- 25. StataCorp . Stata Statistical Software: Release 16. College Station, TX: StataCorp LLC; 2019. First Accessed Jan 2022. https://www.stata.com/ [Google Scholar]
- 26. Therneau TM. A Package for Survival Analysis in R . R package version 3.4‐0. 2022. First Accessed Aug 2022. https://CRAN.R‐project.org/package=survival.
- 27. Balduzzi S, Rücker G, Schwarzer G. How to perform a meta‐analysis with R: a practical tutorial. Evid Based Ment Health. 2019;22:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Egerstedt A, Berntsson J, Smith ML, Gidlöf O, Nilsson R, Benson M, Wells QS, Celik S, Lejonberg C, Farrell L, et al. Profiling of the plasma proteome across different stages of human heart failure. Nat Commun. 2019;10:5830. doi: 10.1038/s41467-019-13306-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cunningham JW, Claggett BL, O'Meara E, Prescott MF, Pfeffer MA, Shah SJ, Redfield MM, Faiez Z, Chiang L‐M, Rizkala AR, et al. Effect of sacubitril/valsartan on biomarkers of extracellular matrix regulation in patients with HFpEF. J Am Coll Cardiol. 2020;76:503–514. doi: 10.1016/j.jacc.2020.05.072 [DOI] [PubMed] [Google Scholar]
- 30. Yao Y, Li B, Liu C, Fu C, Li P, Guo Y, Ma G, Liu N, Chao L, Chao J. Reduced plasma kallistatin is associated with the severity of coronary artery disease, and kallistatin treatment attenuates atherosclerotic plaque formation in mice. J Am Heart Assoc. 2018;7:e009562. doi: 10.1161/JAHA.118.009562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xie J, Yu Q‐G, Yang L‐L, Sun Y‐Y. Kallistatin alleviates heart failure in rats by inhibiting myocardial inflammation and apoptosis via regulating sirt1. Eur Rev Med Pharmacol Sci. 2020;24:6390–6399. [DOI] [PubMed] [Google Scholar]
- 32. Gao L, Yin H, Smith S, Chao LR, Chao J. Role of kallistatin in prevention of cardiac remodeling after chronic myocardial infarction. Lab Investig. 2008;88:1157–1166. doi: 10.1038/labinvest.2008.85 [DOI] [PubMed] [Google Scholar]
- 33. Ferkingstad E, Sulem P, Atlason BA, Sveinbjornsson G, Magnusson MI, Styrmisdottir EL, Gunnarsdottir K, Helgason A, Oddsson A, Halldorsson BV, et al. Large‐scale integration of the plasma proteome with genetics and disease. Nat Genet. 2021;53:1712–1721. doi: 10.1038/s41588-021-00978-w [DOI] [PubMed] [Google Scholar]
- 34. Jung I‐H, Elenbaas JS, Alisio A, Santana K, Young EP, Kang CJ, Kachroo P, Lavine KJ, Razani B, Mecham RP, et al. SVEP1 is a human coronary artery disease locus that promotes atherosclerosis. Sci Transl Med. 2021;13:eabe0357. doi: 10.1126/scitranslmed.abe0357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elenbaas JS, Pudupakkam U, Ashworth KJ, Kang CJ, Patel V, Santana K, Jung I‐H, Lee PC, Burks KH, Amrute JM, et al. SVEP1 is an endogenous ligand for the orphan receptor PEAR1. Nat Commun. 2023;14:850. doi: 10.1038/s41467-023-36486-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zile MR, O'Meara E, Claggett B, Prescott MF, Solomon SD, Swedberg K, Packer M, McMurray JJV, Shi V, Lefkowitz M, et al. Effects of sacubitril/valsartan on biomarkers of extracellular matrix regulation in patients with HFrEF. J Am Coll Cardiol. 2019;73:795–806. doi: 10.1016/j.jacc.2018.11.042 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S8


