Despite advances, cardiovascular disease remains the leading killer in women. Compared with men, the risk is underrecognized and diagnosis is delayed, contributing to overall worse coronary artery disease (CAD) outcomes. 1 The mechanisms for elevated risk in women remain incompletely explained by conventional risk factors. The role of varying genetic effects in this elevation remains uncertain. Genome‐wide association studies have enabled the discovery of genetic variants associated with CAD and other complex diseases. A CAD polygenic risk score (PRS) sums individually modest effects from CAD risk alleles and can be used with traditional risk calculators to independently predict and jointly improve CAD risk discrimination. 2
Characterizing and understanding generalizability of a novel biomarker is critical to ensure disparities are not exacerbated. Analyses in the UK Biobank indicate older CAD PRS performs worse in women relative to men. 1 Whether similar performance differences exist in modern US populations with novel CAD PRS is unknown, and strategies to mitigate this bias are unclear.
We calculated a new multiancestry CAD PRS, GPSmult, which was shown to outperform other CAD PRSs, in the Mass General Brigham Biobank, a health care–associated biobank containing genetic, environmental, and electronic health record information from patients in the Mass General Brigham health care system. 2 Participants were excluded if they had (1) samples with >1% genotype missingness, (2) mismatch between self‐reported and genetically inferred sex, or (3) sample overlap with the training cohort. 2 The remaining 37 641 participants were 82.2% White race, 5.1% non‐Hispanic Black race and ethnicity, 3.8% non‐Hispanic Asian or Pacific Islander race and ethnicity, 2.1% Hispanic or Latino ethnicity, 6.9% other or unidentified race, and 55.6% women, with mean (SD) age of 52.4 (17.2) years. CAD was defined as presence of International Classification of Diseases, Ninth Revision (ICD‐9), or International Classification of Diseases, Tenth Revision (ICD‐10), codes, as previously described, and occurred in 7.9% of individuals (1108 women). 3 The Mass General Brigham Institutional Review Board approved this study, and informed consent was waived; data are available on request. Adjusting for age, sex, batch, and 5 principal components of ancestry, GPSmult demonstrated a strong association (odds ratio [OR], 1.45 [SD, 1.39–1.51]) with CAD. However, the association was lower among women than men (OR, 1.35 [SD, 1.27–1.45] versus OR, 1.51 [SD, 1.43–1.59]; P interaction=0.04). GPSmult performance diverged particularly at higher percentiles of risk (Figure). Although absolute risks were similar for women and men at the bottom 10th percentile (0.43% and 1.14%, respectively), they were notably deflated for women relative to men in the top decile of risk (17.9% and 29.0%, respectively).
Figure . Sex‐specific curves showing predicted prevalence of coronary artery disease (CAD) with 95% CI, according to percentile of the multiancestry CAD polygenic risk score (GPSMult) in the Mass General Brigham (MGB) Biobank data set consisting of n=37 641.
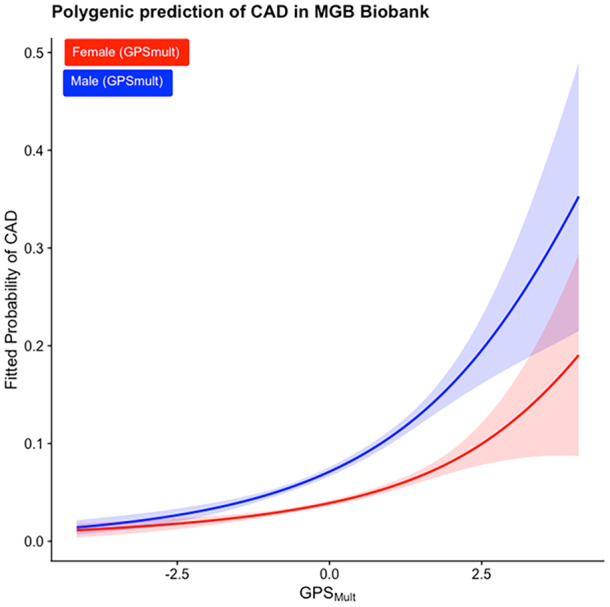
GPSMult was expanded to a natural spline with 3 knots, and a logistic regression was fit to this expanded basis. Initial GPSMult was mapped back to this CAD prediction.
We hypothesized that alleles exhibiting significant sex‐differential effects for CAD might reduce this bias. To test this, we leveraged previously calculated summary statistics from sex‐stratified genome‐wide association studies performed by the CARDIoGRAMplusC4D consortium involving 77 080 CAD cases and 550 952 controls, including participants from the UK Biobank and other similar data sets. 3 Using PLINK2, sex‐differential weights from 10 loci demonstrating between‐sex heterogeneity in the CARDIoGRAMplusC4D sex‐specific analysis were used to create a sex‐differential polygenic risk score, PRSsexdiff. The weights represent the difference in CAD effects in women relative to men. A linear combination of GPSmult, the novel PRSsexdiff, and the base model of age, batch, and first 5 principal components of ancestry yielded improved CAD risk association effects (OR, 1.39 [SD, 1.29–1.49]) in female participants but did not fully resolve the disparity. However, CAD association was heightened for women aged <55 years (OR, 1.44 [SD, 1.25–1.65]; P interaction=3.32×10−7), emphasizing greater utility identifying actionable early risk.
We demonstrate that the latest CAD PRS in a contemporary US cohort still exhibits disparities by sex, underscoring the importance of sex in reporting. Our work builds on prior descriptions in the UK Biobank of differences in risk discrimination by sex and demonstrates that some of this difference is related to gene‐sex interactions. 1 , 4 CAD PRS distribution across sex was similar, making sex‐participation bias an unlikely driver. 5 Efforts to understand and address barriers toward improved and less biased representation of women in genomic studies are imperative to resolve PRS performance discrepancies.
Our study has notable limitations to consider in interpretation. First, as Mass General Brigham Biobank is a contemporary cohort with limited follow‐up, we focused on prevalent CAD to maximize power. As germline genetics are fixed from conception, cross‐sectional analysis may be reasonable to evaluate sex disparities in CAD PRS performance. Second, our sex‐differential PRS approach may be underpowered, and generalizability remains limited. Novel PRS methods use numerous variants to optimize performance, yet few substantive data sets are available to enable sex‐stratified training and validation as many established repositories, such as UK Biobank, overlap with data used in training here. However, using a 10‐loci score may facilitate portability and implementation. Finally, whether similar sex differences and CAD PRS performance improvements are observed in non‐European populations requires study.
Further investigations discovering and incorporating larger numbers of sex‐differential variants may help bridge the polygenic risk score performance gap in CAD and may be applied broadly across conditions to address sex disparities widely.
Sources of Funding
This study was supported by the Massachusetts General Hospital Executive Committee on Research Fund for Medical Discovery Population Healthcare Sciences Research Fellowship Award (awarded to Dr Paruchuri) and the National Human Genome Research Institute U01HG011719 (awarded to Dr Natarajan).
Disclosures
Dr Paruchuri reports grant support from Genentech, AstraZeneca, Novartis, and Allelica, unrelated to this work. Dr. Natarajan reports research grants from Allelica, Amgen, Apple, Boston Scientific, Genentech / Roche, and Novartis, personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Creative Education Concepts, CRISPR Therapeutics, Eli Lilly & Co, Foresite Labs, Genentech / Roche, GV, HeartFlow, Magnet Biomedicine, Merck, and Novartis, scientific advisory board membership of Esperion Therapeutics, Preciseli, TenSixteen Bio, and Tourmaline Bio, scientific co‐founder of TenSixteen Bio, equity in MyOme, Preciseli, and TenSixteen Bio, and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. The remaining authors have no disclosures to report.
This manuscript was sent to Jacquelyn Y. Taylor, PhD, PNP‐BC, RN, Associate Editor, for review by expert referees, editorial decision, and final disposition.
For Sources of Funding and Disclosures, see page 2.
References
- 1. Surakka I, Wolford BN, Ritchie SC, Hornsby WE, Sutton NR, Elvenstad Gabrielsen M, Skogholt AH, Thomas L, Inouye M, Hveem K, et al. Sex‐specific survival bias and interaction modeling in coronary artery disease risk prediction. Circ Genomic Precis Med. 2023;16:e003542. doi: 10.1161/CIRCGEN.121.003542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Patel AP, Wang M, Ruan Y, Koyama S, Clarke SL, Yang X, Tcheandjieu C, Agrawal S, Fahed AC, Ellinor PT, et al. A multi‐ancestry polygenic risk score improves risk prediction for coronary artery disease. Nat Med. 2023;29:1793–1803. doi: 10.1038/s41591-023-02429-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aragam KG, Jiang T, Goel A, Kanoni S, Wolford BN, Atri DS, Weeks EM, Wang M, Hindy G, Zhou W, et al. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nat Genet. 2022;54:1803–1815. doi: 10.1038/s41588-022-01233-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oliva M, Muñoz‐Aguirre M, Kim‐Hellmuth S, Wucher V, Gewirtz ADH, Cotter DJ, Parsana P, Kasela S, Balliu B, Viñuela A, et al. The impact of sex on gene expression across human tissues. Science. 2020;369:eaba3066. doi: 10.1126/science.aba3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pirastu N, Cordioli M, Nandakumar P, Mignogna G, Abdellaoui A, Hollis B, Kanai M, Rajagopal VM, Parolo PDB, Baya N, et al. Genetic analyses identify widespread sex‐differential participation bias. Nat Genet. 2021;53:663–671. doi: 10.1038/s41588-021-00846-7 [DOI] [PMC free article] [PubMed] [Google Scholar]


