Abstract
The stimulator of interferon genes (STING), an integral adaptor protein in the DNA-sensing pathway, plays a pivotal role in the innate immune response against infections. Additionally, it presents a valuable therapeutic target for infectious diseases and cancer. We observed that fangchinoline (Fan), a bis-benzylisoquinoline alkaloid (BBA), effectively impedes the replication of vesicular stomatitis virus (VSV), encephalomyocarditis virus (EMCV), influenza A virus (H1N1), and herpes simplex virus-1 (HSV-1) in vitro. Fan treatment significantly reduced the viral load, attenuated tissue inflammation, and improved survival in a viral sepsis mouse model. Mechanistically, Fan activates the antiviral response in a STING-dependent manner, leading to increased expression of interferon (IFN) and interferon-stimulated genes (ISGs) for potent antiviral effects in vivo and in vitro. Notably, Fan interacts with STING, preventing its degradation and thereby extending the activation of IFN-based antiviral responses. Collectively, our findings highlight the potential of Fan, which elicits antiviral immunity by suppressing STING degradation, as a promising candidate for antiviral therapy.
Keywords: Fangchinoline, Virus, Type I interferons, Stimulator of interferon genes, Protein degradation
Graphical abstract

Highlights
-
•
Fan strongly inhibits various virus replications.
-
•
Fan protects against VSV infection in vivo.
-
•
Fan initiates innate antiviral immunity in a STING-dependent manner.
-
•
Fan interacts with STING to prevent its degradation and prolong the antiviral immunity.
1. Introduction
In response to cytosolic DNA, stimulator of interferon genes (STING), a crucial adaptor in the DNA-sensing pathway, dissociates from the nuclear periphery endoplasmic reticulum and translocates to the Golgi apparatus. It subsequently recruits the downstream kinase TANK binding kinase 1 (TBK1), initiating the expression of type I interferon (IFN-I) [1,2]. Extracellular IFN-I, bound to IFN-α/β receptor (IFNAR), activates the Janus kinase-signal transducers and activators of transcription (JAK-STAT) pathway, leading to the expression of IFN-stimulated genes (ISGs) [3,4]. To date, over 300 ISG-encoding proteins with direct roles in pathogen control and immune regulation have been identified [5,6]. The development of modulators targeting STING has garnered significant attention for potential applications in treating infectious diseases and cancer, and as vaccine adjuvants [[7], [8], [9], [10]]. Recent studies have elucidated the intracellular regulatory mechanism that modulates the intensity and duration of STING-mediated innate immune responses by inhibiting STING protein degradation [11,12]. To the best of our knowledge, no small-molecule drugs targeting this mechanism have been reported to date.
Bis-benzylisoquinoline alkaloids (BBAs), including tetrandrine, cepharanthine, and berbamine, have demonstrated potent antiviral activities against a broad spectrum of viruses, such as Ebola virus (EBOV), herpes simplex virus 1 (HSV-1), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [13,14]. For example, tetrandrine inhibits the entry of the EBOV into cells by blocking two endosomal calcium channels [15]. Cepharanthine inhibits HSV-1 by promoting cellular autophagy [16], whereas berbamine restricts SARS-CoV-2 replication by disrupting the endolysosomal transport of angiotensin-converting enzyme 2 (ACE2) through transient receptor potential mucolipins (TRPMLs) [17]. However, these pharmacological mechanisms do not fully elucidate the broad antiviral spectrum of BBAs, suggesting that the potential shared antiviral mechanism of BBAs remains unknown.
In this study, we report that fangchinoline (Fan), a BBA isolated from Stephania tetrandra, potently inhibits vesicular stomatitis virus (VSV), encephalomyocarditis virus (EMCV), influenza A virus (H1N1), and HSV-1 replication in vitro. In a viral sepsis model, Fan significantly diminishes viral loads in organs, mitigates the release of inflammatory cytokines, and enhances mouse survival, highlighting Fan as a promising candidate for antiviral therapy. Mechanistically, Fan directly interacts with the STING protein, preventing its degradation and prolonging the activation of the IFN-based antiviral immune pathway. This suggests that the pharmacological maintenance of STING protein expression as a strategy holds the potential for eliciting an antiviral immune response in antiviral therapy.
2. Materials and methods
2.1. Reagents and antibodies
Fan (C37H40N2O6; molecular weight: 608.74; purity: 98.5%) was provided by Chengdu Push Bio-Technology (Chengdu, China). Hoechst 33342 was purchased from Sigma-Aldrich (St. Louis, MO, USA). The 2'3'-cyclic guanosine monophosphate (GMP)-adenosine monophosphate (AMP) (2',3'-cGAMP), TRIzol, poly(dA:dT), and poly(I:C) (low molecular weight) were offered by InvivoGen (Hong Kong, China). 5,6-Dimethylxantheonone-4-acetic acid (DMXAA) was purchased from Macklin Biochemical Technology (Shanghai, China). SeaPlaque™ Agarose was purchased from Lonza Group (Rochester, NY, USA). Fixative solution (4% formaldehyde, methanol-free) was purchased from Biosharp Life Sciences (Hefei, China). Dimethyl sulfoxide (DMSO), 1 % crystal violet stain solution, and Triton X-100 were ordered from Beijing Solarbio Science & Technology (Beijing, China). Cell Counting Kit-8 (CCK-8) was purchased from Dojindo Laboratories (Kumamoto, Japan). Ruxolitinib (Rux) was supplied by TopScience LLC. (Shanghai, China). The antibodies used for immunoblotting and immunofluorescence are displayed in Table S1.
2.2. Mice
The mice we used were all male C57BL/6J (age, 8 weeks) except as otherwise noted. The male C57BL/6J mice were offered by SPF Biotechnology Co., Ltd. (Beijing, China). Before the experimental procedures, all mice were acclimatized in a pathogen-free environment for one week. C57BL/6 Sting−/− mice were provided by Dr. Kunpeng Liu from College of Medicine, Guangxi University, China. The animal experimental procedures and animal care were approved by the Animal Care Committee of the Beijing University of Chinese Medicine, Beijing, China (Approval No.: BUCM–2023112002–4086).
2.3. Cells and viruses
A549 and L929 cell lines were purchased from American Type Culture Collection (ATCC). Immortalized bone marrow-derived macrophages (iBMDMs) were offered by Dr. Feng Shao (National Institute of Biological Sciences, Beijing, China). These cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. The THP-1 cells, provided by the Chinese Academy of Sciences (Kunming, China), were fed with Roswell Park Memorial Institute (RPMI) 1640 medium with 10% FBS and 1% penicillin-streptomycin. The methods to isolate and culture BMDMs and mouse embryonic fibroblast (MEF) were described previously [18,19]. All cells were kept at 37 °C in a humidified 5% CO2 incubator.
All viruses we used were from our laboratory, including VSV (Indiana strain), VSV labeled with enhanced green fluorescent protein (VSV-eGFP), EMCV, H1N1 (PR8), and HSV-1.
2.4. Cell viability assay
A549 and L929 cells were seeded (cultured) in 96-well plates (3.5 × 104/5 × 104 cells per well) and grown for 12 h. Cells were then incubated with Fan for 12 h. Cell viability was measured by calculating the absorbance of the cells at 450 nm, using a CCK-8 (Dojindo Laboratories).
2.5. Flow cytometry and fluorescence analysis
Before assay, A549 or L929 cells were seeded in a 24-well plate at a density of 2 × 105/well, and cultured overnight before the assay. After VSV-eGFP infection and treatment with Fan, the cells were washed after removing the medium. The samples were trypsinized, centrifuged, and resuspended in cold staining buffer (phosphate-buffered saline (PBS) with 1% FBS), and analyzed by flow cytometry (Beckman Coulter, Brea, CA, USA).
For the assay of fluorescence microanalysis, after cotreatment of VSV-eGFP (MOI 0.1) and Fan for 12 h, A549 cells in a 24-well plate were observed using a fluorescence microscope (Olympus IX2-UCB, Tokyo, Japan).
2.6. Plaque assay
The Fan was incubated with VSV in A549 cells for 12 h, and the supernatant was collected. The appropriate dilution ratio for the plaque assay was determined. After incubating A549 cells in a 24-well plate with the diluted virus supernatant at 37 °C for 2 h, the supernatant was removed, and melted agarose diluted in DMEM was added. After solidification of the agarose, the plate was inverted and cultured at 37 °C for three days. The 4% formaldehyde was used to fix cells and stained cells with a crystal violet solution for counting visible plaques.
2.7. Virus infection assay
Cells were infected with VSV-eGFP (MOI 0.1), and viruses were absorbed on the cell surface at 4 °C for 2 h. The supernatant was removed and replaced with fresh medium. Cells were kept for 10 h at 37 °C with Fan at different stages: pretreatment, attachment, entry, and post-entry. The intensity of GFP was quantified with flow cytometry.
2.8. Immunoblotting
As mentioned previously [19], Western blot analysis was performed according to standard procedures. Briefly, proteins from cells or animal tissues were fully lysed and boiled. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to concentrate and segregate proteins. Then proteins were wet transferred to a nitrocellulose membrane, and sealed in 1× tris buffered saline with Tween 20 (TBST) with 5% bovine serum albumin (BSA) for 1 h. With overnight incubation with relevant primary antibodies at 4 °C, the membranes were sealed in the appropriate horseradish peroxidase (HRP)-conjugated antibodies for 1 h. The membranes were exposed using the Immobilon enhanced chemiluminescence (ECL) kit (Beyotime, Beijing, China) and detected on the Tanon-5200 Imaging System (YPH-Bio, Beijing, China). The primary and secondary antibodies are listed in Table S1.
2.9. RNA isolation and real-time quantitative polymerase chain reaction (qRT-PCR) analysis
Total RNA from cells and tissues was extracted using total RNA extractor (TRIzol) solution (Invitrogen, Carlsbad, CA, USA). RNA was quantified and reverse-transcribed using an Evo M-MLV RT kit (Accurate Biology, Changsha, China). The complementary DNA (cDNA) was diluted to perform qRT-PCR using the SYBR® Green Premix Pro Taq HS qRT-PCR Kit (Accurate Biology, Changsha, China). The CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) was used to measure the relative messenger RNA (mRNA) level. The relative level of mRNA was normalized by the β-actin expression in each sample. Relative expression changes were calculated by the method of . The primer sequences are listed in Table S2.
2.10. Enzyme-linked immunosorbent assay (ELISA)
Blood was collected from the euthanized mice. After centrifugation at 4 °C, the blood was separated and mouse serum samples were collected. The level of interleukin-6 (IL-6) and tumour necrosis factor-alpha (TNF-α) in serum samples was determined following the instructions of the manufacturer (Proteintech, Wuhan, China).
2.11. Transfection of stimulation
For poly(I:C) stimulation, A549 cells were transfected with poly(I:C)/poly(dA:dT) (1 μg/mL) delivered by polyethyleneimine (2 μg/mL) for 5 h. After stimulation, the media was changed completely and the samples were treated or collected for further analysis.
2.12. Fluorescent imaging
Cells were infected with H1N1 or EMCV and co-treated with Fan. The cells were fixed, permeabilized, and blocked. Then appropriate primary antibodies were chosen to overnight incubate them at 4 °C, followed by secondary fluorescent antibodies. Hoechst 33342 was used to show the shape and position of the nucleus. Images were viewed on an Olympus FV1000 confocal microscope.
2.13. Molecular docking
Molecular docking assays were performed using PyMol 2.5 software and MOE 2019 software. The PubChem database was used to download the small-molecule Fan three-dimensional (3D) structure. After removing the water molecule, and adding hydrogen atoms, the crystal structure of STING (Protein Data Bank (PDB) ID: 6MX3) for molecular docking was generated.
2.14. Cellular thermal shift assay (CETSA)
iBMDM cells were lysed by using a liquid-nitrogen freezing system. At room temperature, the total proteins were incubated with DMSO or Fan (80 μM) for 1 h. The soluble proteins were separated into indicated PCR tubes and heated at the temperatures respectively (37, 41, 44, 47, 50, 53, 56, 59, 63, and 67 °C) for 3 min in a PCR instrument. After heating, the proteins were cooled and centrifuged at 4 °C for 40 min by centrifugation at 15,000 g. The supernatant was prepared to perform an immunoblotting analysis.
2.15. STING target engagement by drug affinity responsive target stability (DARTS) assay
DARTS assay was an effective method to assess target engagement. iBMDM cell protein was harvested, washed with chilled PBS, and lysed on ice for 1 h. The supernatant was transferred into 72.2 μL of reaction buffer (50 mM tris-HCl (pH 8.0), 50 mM NaCl, 10 mM CaCl2. The concentration of protein was quantitated and diluted to 5 μg/μL. Then the protein was incubated with Fan (80 μM) or DMSO for 1 h. The protein samples were then proteolyzed with pronase as indicated for 30 min. Finally, the cell lysates were subjected to Western blot to determine the abundance of STING.
2.16. STING degradation assay
iBMDM cells were pre-treated with DMSO or Fan (10 μM) for 3 h and then co-incubated with DMXAA (15 μM) for different time. Western blot analysis was then performed according to standard procedures to detect the expression levels of STING and phosphorylated TBK1.
2.17. Animal model of VSV infection
Before the experimental procedures, C57BL/6J mice were acclimatized at the condition of specific pathogen free. After intraperitoneal injection with Fan (10 or 30 mg/kg/day) or DMXAA (10 mg/kg/day) for 12 h, the mice were treated with an intraperitoneal injection of VSV (2 × 108 plaque forming unit (PFU) per mouse). Before analyzing the data by the Kaplan-Meier method, the infected mice were monitored to record survival and weight. The viral loads or cytokine expressions in livers, lungs, and spleens were detected after 24 h infection.
2.18. Hematoxylin and eosin (H&E) staining
Lungs were collected from all groups, rinsed with pre-chilled PBS, and fixed overnight, followed by dewaxed, rehydrated, and then stained with H&E. Tissue sections were prepared and performed microscopic pathology analysis.
2.19. Transcriptome data analysis
Both wild-type (WT) and IFNAR1−/− A549 cells were treated with DMSO or Fan for a duration of 24 h. Trizol was used for cellular RNA extraction. Then sequencing libraries were established. Illumina NovaSeq platform was used to obtain paired-end reads of 150 bp by sequencing the libraries. Clean data (clean reads) were acquired by eliminating reads that contained adapters, poly-N, or were of low quality from the raw data. The reads were aligned to the reference genome using the hisat2 tool. Differential expression analysis was conducted on the raw data counts of all genes in each sample, considering genes with a P value < 0.05 and log2(fold change) > 1 as significantly differential expression.
The ClusterProfiler R package was used to implement Gene Ontology (GO) enrichment analysis based on the differentially expressed genes. The gene set enrichment analysis (GSEA) software was used to carry out GSEA using curated gene sets (C2) obtained from the Molecular Signatures Database (MSigDB) database.
2.20. Statistics
All of the data were reported as mean ± standard deviation (SD) as indicated. For two groups, comparisons were performed by Student's t-test. Multiple comparisons were tested by one-way analysis of variance (ANOVA) with a Bonferroni post-test. Experiment data were analyzed using GraphPad Prism 9.0 software. P < 0.05 is considered to be statistical significant.
3. Results
3.1. Fan inhibits VSV in vitro
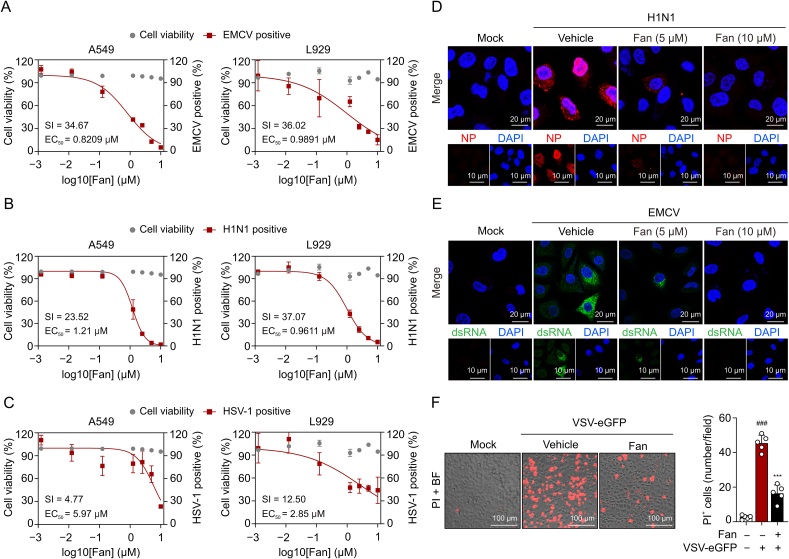
A549 lung epithelial cells, known for their susceptibility to viruses including VSV, EMCV, H1N1, and HSV-1, were subjected to immunofluorescence staining screening to assess the efficacy of 317 compounds from a traditional Chinese medicine monomer library. Among these compounds, Fan, a BBA extracted from Stephaniae tetrandra (Fig. 1A), exhibited the most pronounced reduction of VSV glycoprotein (VSV-G) levels, as indicated by reduced green immunofluorescence staining (Fig. 1B). Furthermore, Fan demonstrated a dose-dependent reduction in plaque formation, indicating decreased viral yield in cell culture supernatants (Fig. 1C). Additionally, qRT-PCR and immunoblotting were employed to assess the antiviral activity of Fan by monitoring the expression of VSV RNA and G protein, respectively. The results demonstrated that Fan exerted potent inhibition on VSV replication in dose- and time-dependent manners (Figs. 1D and E).
Fig. 1.
Fangchinoline (Fan) inhibits vesicular stomatitis virus (VSV) in vitro. (A) Molecular structure of Fan. (B) Fluorescence microscopy imaging of VSV labeled with enhanced green fluorescent protein (VSV-eGFP) (multiplicity of infection (MOI) 0.02) infected A549 cells under different doses of Fan treatment for 12 h. (C) Plaque assay analysis of infectious VSV viral particle count. Fan-L: low-dose Fan (5 μM); Fan-H: high-dose Fan (10 μM). (D) Real-time quantitative polymerase chain reaction (qRT-PCR) analysis of VSV glycoprotein (VSV-G) and the large protein of VSV polymerase (VSV-L) messenger RNA (mRNA) levels. (E) Western blot analysis of VSV-G expression levels. VSV-eGFP (MOI 0.05) infected A549 and L929 cells were co-incubated with increasing doses of Fan (2.5, 5, and 10 μM) for 12 h, or incubated with Fan for different durations (0, 8, 16, and 24 h). (F–H) Flow cytometry analysis of viral replication at different stages in Fan treated A549 cells: illustration of the stages of drug treatment analysis (F), flow cytometry histograms of VSV-eGFP (MOI 0.02) infected A549 cells (G), and quantification of the cells that are GFP-positive (H). The “+” in Fig. 1G means VSV-eGFP infection. The data above are presented as mean ± standard deviation (SD) (n = 3). ###P < 0.001, compared to control group; ∗P < 0.05 and ∗∗∗P < 0.001, compared to VSV-eGFP group. ns: not significant. BF: bright field image; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; SSC-A: side scattering-area.
The antiviral effect of Fan was evaluated at different stages using a time-of-drug addition assay in a single infectious cycle (Fig. 1F). Flow cytometry revealed that Fan primarily inhibited VSV infection at the post-entry step, as indicated by decreased viral replication when Fan was added 3.5 h after infection (Figs. 1G and H). Notably, pre-treatment with Fan before viral attachment significantly suppressed viral replication, albeit with a less pronounced effect during viral attachment, suggesting that Fan may modulate host factors post-viral entry to exert inhibitory effects on viral replication.
3.2. Fan is a pan-viral inhibitor in vitro
To investigate the potential cross-protection of Fan against other viruses, we conducted viral-load reduction assays for H1N1, EMCV, and HSV-1 in various virus-permissive cell lines. Fan exhibited minimal cytotoxicity in both A549 and L929 cell types, with a half maximal inhibitory concentration (IC50) value of approximately 40 μM. Subsequent experiments were performed using concentrations that maintained cell viability above 90%. qRT-PCR analysis revealed that Fan treatment led to a notable decline in the viral mRNA levels of EMCV, H1N1, and HSV-1 (Figs. 2A−C). Immunofluorescence staining indicated reduced H1N1 nucleoprotein (NP)-positive cells and attenuated fluorescence intensity within these cells following Fan treatment (Fig. 2D). Furthermore, Fan treatment substantially decreased double-stranded RNA (dsRNA) formation during EMCV infection [20] (Fig. 2E) and reduced the percentage of dead cells induced by viral infection (Fig. 2F). Collectively, Fan demonstrated broad-spectrum antiviral efficacy, inhibiting the replication of EMCV, H1N1, and HSV-1 in both human and mouse cells.
Fig. 2.
Fangchinoline (Fan) inhibits encephalomyocarditis virus (EMCV), influenza A virus (H1N1), and herpes simplex virus-1 (HSV-1) in vitro. (A–C) Dose-response curves showing the inhibitory effect of Fan in the range of 0.00125–10 μM on viral replication in A549 and L929 cells. Real-time quantitative polymerase chain reaction (qRT-PCR) analysis of viral genome encoding the EMCV 1C proteins (A), the H1N1 hemagglutinin (HA) proteins (B), and the HSV-1 I27 proteins (C). (D) Confocal microscopy of the nucleoprotein (NP) (red) in A549 cells infected with H1N1 and treated with Fan (5 and 10 μM). Hoechst 33342 dye was used to stain the nuclei of cells (blue). (E) Immunofluorescence images of the double-stranded RNA (dsRNA) (green) in A549 cells infected with EMCV and treated with Fan (5 and 10 μM). Nuclei (blue) are shown by Hoechst 33342. (F) Fluorescence images of vesicular stomatitis virus labeled with enhanced green fluorescent protein (VSV-eGFP) (multiplicity of infection (MOI) 0.1) infected A549 cells with the treatment of dimethyl sulfoxide (DMSO) or Fan for 24 h. Propidium iodide (PI) and Hoechst 3334 were used to stain the cells (n = 5). The data above are presented as mean ± standard deviation (SD). ###P < 0.001, compared to control group; ∗∗∗P < 0.001, compared to VSV-eGFP group. SI: selectivity index; EC50: concentration for 50% of maximal effect; DAPI: 4',6-diamidino-2-phenylindole; BF: bright field image.
3.3. Fan protects against VSV infection in mice
Given that both tetrandrine and Fan are BBAs derived from Stephaniae tetrandra, we administered 30 mg/kg Fan, an equivalent concentration of tetrandrine used clinically to treat pneumoconiosis and rheumatoid arthritis (30–50 mg/kg) [21,22]. The antiviral activity of Fan was assessed in a murine VSV-induced viral sepsis model, with DMXAA, a known STING agonist, serving as a positive control [23] (Fig. 3A). The results demonstrated that pre-treatment with Fan or DMXAA significantly prolonged the survival time of mice (Fig. 3B) and effectively alleviated virus-induced weight loss, with weight recovery observed on day 6 of treatment (Fig. 3C).
Fig. 3.
Fangchinoline (Fan) inhibits the replication of vesicular stomatitis virus (VSV) and alleviates VSV-induced inflammation in vivo. (A) Model diagram of Fan treatment in the C57BL/6 mouse disease model of VSV intraperitoneal infection. Each group of mice except control mice was pre-treated with intraperitoneal injections of Fan and 5,6-dimethylxanthenone-4-acetic acid (DMXAA), followed by intraperitoneal infection with a virus at a dose of 2 × 108 plaque forming unit (PFU) per mouse. (B) Survival curves of mice intraperitoneally infected with VSV under Fan (30 mg/kg/day) and DMXAA (10 mg/kg/day) treatment (n = 5 in each group). (C) Curves of the changes in body weight of mice intraperitoneally infected with VSV under Fan (30 mg/kg/day) and DMXAA (10 mg/kg/day) treatment (n = 5). (D) Real-time quantitative polymerase chain reaction (qRT-PCR) analysis was performed to detect the transcript levels of RNA encoding VSV glycoprotein (VSV-G) in the liver, lung, and spleen tissues of VSV infected mice (n = 5). (E) Immunoblotting of VSV-G levels in lung and spleen tissues. (F) Super-resolution microscopy imaging of lung sections of VSV infected mice. (G) Enzyme linked immunosorbent assay (ELISA) was employed to measure the concentrations of tumour necrosis factor-α (TNF-α) and interleukin-6 (IL-6) in serum (n = 5). Fan-L: low-dose group (10 mg/kg/day); Fan-H: high-dose group (30 mg/kg/day). The data above are shown as mean ± standard deviation (SD). ###P < 0.001, compared to control group; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001, compared to VSV group. GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
Moreover, viral loads were significantly reduced in liver, lung, and spleen tissues in drug-treated mice compared with that in VSV-infected mice (Fig. 3D); furthermore, expression levels of VSV-G were decreased in lung and spleen tissues (Fig. 3E). H&E staining demonstrated that Fan treatment effectively mitigated virus-induced inflammatory cell infiltration in lung tissues (Fig. 3F). Additionally, Fan significantly inhibited mRNA expression of inflammation-related genes, including Tnf-a, Il-6, and C−X−C motif chemokine ligand 10 (Cxcl10) (Fig. S1), and led to decreased serum secretion of TNF-α and IL-6 (Fig. 3G). Collectively, our findings suggest that Fan protects against VSV challenge in a mouse model by suppressing viral replication and mitigating associated inflammatory dysregulation in the host.
3.4. Treatment with Fan initiates innate antiviral immunity and ISG expression
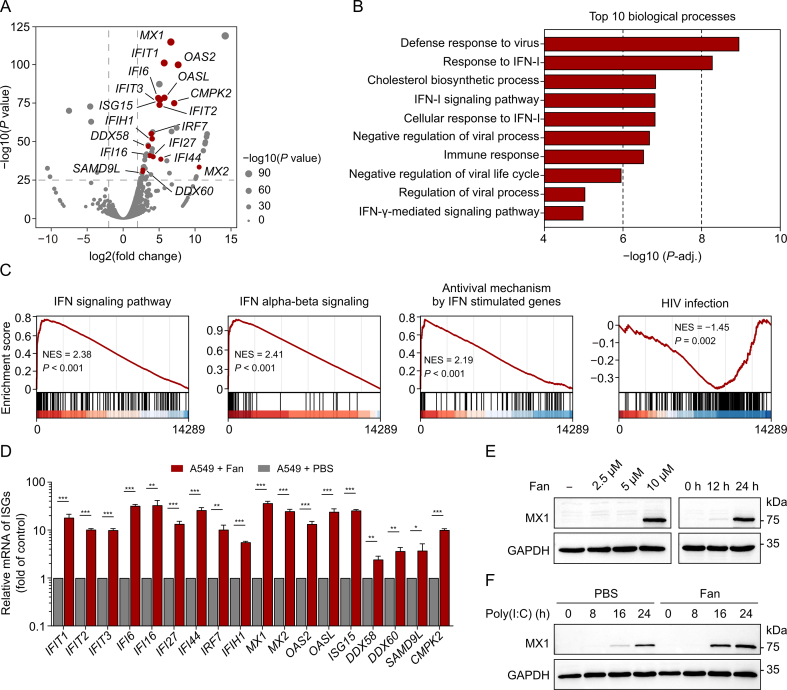
We used RNA sequencing (RNA-seq) to examine global transcriptome changes in human A549 cells after Fan treatment. Bioinformatics analysis revealed 896 differentially expressed genes (DEGs), including 663 upregulated and 233 downregulated genes, in Fan-treated cells compared with that in the control group. The upregulated DEGs included several ISGs, including myxovirus resistance 1 (MX1), IFN induced protein with tetratricopeptide repeats 1 (IFIT1), and 2'-5'-oligoadenylate synthetase 2 (OAS2) (highlighted in red in Fig. 4A). For these upregulated genes, GO enrichment analysis revealed significant enrichment of pathways associated with antiviral defence response and IFN-I signalling (Fig. 4B). GSEA using the Reactome Pathway Database as a reference gene set indicated substantial upregulation of IFN-related pathways in Fan-treated cells. Conversely, the process of human immunodeficiency virus (HIV) infection was markedly downregulated, consistent with previous findings on the inhibitory effects of Fan against HIV [24] (Fig. 4C). Additionally, qRT-PCR analysis validated the RNA seq results, showing a significant induction of all annotated ISGs upon treatment with Fan, exhibiting a 5–10-fold increase compared with that in the control group (Fig. 4D). Furthermore, immunoblotting confirmed the upregulation of the most prominent ISG, MX1, at the protein level. Treatment with Fan alone enhanced MX1 expression in a dose- and time-dependent manner (Fig. 4E). We observed that this treatment further augmented the induction of MX1 expression by the dsRNA mimic, poly(I:C) (Fig. 4F), a potent activator of the innate antiviral immune response. These findings suggest that Fan induces and enhances the expression of ISGs involved in antiviral immunity.
Fig. 4.
The treatment with fangchinoline (Fan) induces upregulation of interferon (IFN)-stimulated genes (ISGs) associated with antiviral protection. (A) Volcano plots of differentially expressed genes (DEGs) between A549 cells treated with Fan for 24 h and the control cells. ISGs were indicated in red (only points with −log10(P value) > 25 were labeled). To identify the DEGs, a threshold of log2(fold change) > 1 and P value < 0.05 was used. (B) Gene Ontology (GO) (biological process) enrichment analysis of upregulated genes of DEGs. (C) Gene set enrichment analysis (GSEA) plot of gene expression spectrum from Fan treated A549 cell using the Reactome Pathway Database. (D) Real-time quantitative polymerase chain reaction (qRT-PCR) analysis of the ISGs expression levels. A549 cells were treated with 10 μM Fan for 24 h. (E) Immunoblotting of myxovirus resistance 1 (MX1) protein levels. A549 cells were incubated with escalating doses of Fan (2.5, 5, and 10 μM) for 24 h or with 10 μM Fan for different durations (12 and 24 h). (F) Immunoblotting analysis of MX1 protein expression levels. A549 cells were transfected with 1 μg/mL poly(I:C) for 5 h, and then treated with phosphate-buffered saline (PBS) or 10 μM Fan for 0, 8, 16, and 24 h. The data above are presented as mean ± standard deviation (SD) (n = 3). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001, compared to control group. NES: normalized enrichment score; HIV: human immunodeficiency virus; mRNA: messenger RNA; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
3.5. Fan elicits antiviral responses in dose- and time-dependent manners
To assess the effect of Fan on IFN and ISG expression in different cell types, qRT-PCR was performed to examine the effect of Fan treatment on THP-1, MEF, BMDM, and L929 cells. The results revealed a noticeable time-dependent increase in the mRNA levels of IFN beta 1 (IFNB1), MX1, IFIT1, and other ISGs (Fig. 5A). Furthermore, Fan treatment enhanced the mRNA levels of IFNB1 and ISGs induced by the treatment with the dsRNA mimic poly(I:C), dsDNA mimic poly(dA:dT), and the STING agonist cGAMP in a time-dependent manner in A549 and L929 cells (Figs. 5B−D). Transcription factor enrichment analysis of the DEGs revealed significant enrichment of transcription factors involved in IFN pathways, including STAT1, IFN regulatory factor 3 (IRF3), IRF9, and nuclear factor kappa-B p65 (RELA) (Fig. S2). Therefore, immunoblotting was performed to assess the impact of drug treatment on transcription factor activation. These results indicated that Fan markedly induced phosphorylation of TBK1, STAT1, and STAT2 in THP-1 and MEF cells in dose- and time-dependent manners (Figs. 5E and F). Additionally, Fan treatment directly facilitated the nuclear translocation of IRF3 and STAT1 (Figs. 5G, 5H, and S3). Collectively, these results indicate that Fan induces antiviral responses in dose- and time-dependent manners.
Fig. 5.
Fangchinoline (Fan) elicits type I interferon (IFN-I) signalling in multiple cell types. (A) Real-time quantitative polymerase chain reaction (qRT-PCR) assay of IFN beta 1 (IFNB1) and IFN-stimulated genes (ISGs) expression levels. THP-1, mouse embryonic fibroblast (MEF), bone marrow-derived macrophages (BMDM), and L929 cells were stimulated with Fan for specific times. (B–D) qRT-PCR assay of IFNB1 and ISGs in A549 and L929 cells. A549 cells were stimulated with 1 μg/mL poly(I:C) (B) or 1 μg/mL poly(dA:dT) (C) for 5 h followed by Fan incubation at 0, 4, 8, 16, and 24 h, and L929 cells were either stimulated with cyclic guanosine monophosphate (GMP)-adenosine monophosphate (AMP) (cGAMP) (5 μM) alone or stimulated and then treated with Fan for 0, 4, 8, and 12 h (D). (E, F) Immunoblotting analysis of phosphorylated TANK binding kinase 1 (p-TBK1) and total TBK1 (TBK1) (E) as well as phosphorylated signal transducer and activator of transcription 1 (p-STAT1), total STAT1 (STAT1), p-STAT2, and total STAT2 (STAT2) (F). THP-1 and MEF cells were treated with different doses of Fan (2.5, 5, and 10 μM) for 12 h or 10 μM Fan for 0, 3, 6, 9, and 12 h. (G, H) Fluorescence microscopy imaging of A549 cells incubated with dimethyl sulfoxide (DMSO) or Fan for 4 h to assess the nuclear localization of IRF3 (G) and STAT1 (H). The data above are presented as mean ± standard deviation (SD). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. ns: not significant. MX1: myxovirus resistance 1; IFIT1: IFN induced protein with tetratricopeptide repeats 1; ISG15: interferon-stimulating gene 15; MX1: myxovirus resistance 1; OAS2: 2'-5'-oligoadenylate synthetase 2; CMPK2: cytidine/uridine monophosphate kinase 2; mRNA: messenger RNA; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; IRF3: IFN regulatory factor 3.
3.6. Antiviral effects of Fan depend on IFN-I-based antiviral immunity
To explore the relationship between the antiviral effect of Fan and the IFN pathway, cells were co-treated with the JAK-specific inhibitor Rux [19]. Significant inhibition of ISG expression was observed following Fan treatment (Fig. 6A). Subsequently, the expression of antiviral-related genes in WT and IFNAR1−/− A549 cells following drug treatment was assessed. Fan upregulated the expression of multiple ISGs, including MX1, IFIT1, and OAS2 in WT cells. However, this upregulation was nearly eliminated in the heatmap in IFNAR1−/− A549 (Fig. 6B), and qRT-PCR experiments yielded similar results (Fig. 6C). Flow cytometry and immunoblotting results further demonstrated that the inhibitory capacity of Fan against viruses was compromised in IFNAR1−/− cells (Figs. 6D and E). We also assessed the knockout efficiency of IFNAR1, as shown in Fig. 6F. The band on the right almost disappeared, demonstrating a successful knockout (Fig. 6F). These data suggest that Fan activates antiviral immunity that relies on the IFN-I pathway.
Fig. 6.
Fangchinoline (Fan) activates the expression of interferon (IFN)-stimulated genes (ISGs) depending on the type I IFN (IFN-I) pathway. (A) Real-time quantitative polymerase chain reaction (qRT-PCR) analysis determining the messenger RNA (mRNA) levels of IFN beta 1 (IFNB1) and ISGs. A549 cells were treated with dimethyl sulfoxide (DMSO) or ruxolitinib (Rux) for 2 h and then co-incubated with Fan (10 μM) for 24 h. (B) Heatmaps showing the expression profile of ISGs in wild type (WT) and IFN α/β receptor 1 (IFNAR1)−/− A549 cells treated with either DMSO or Fan. (C) qRT-PCR analysis of mRNA levels of IFNB1 and ISGs in WT and IFNAR1−/− A549 cells treated with Fan for 24 h. (D) Flow cytometry analysis quantifying the percentage of green fluorescent protein (GFP)-positive cells in vesicular stomatitis virus labeled with enhanced green fluorescent protein (VSV-eGFP) (multiplicity of infection (MOI) 0.02) infected WT and IFNAR1−/− A549 cells and subsequently co-incubated with Fan. The line represents the inhibitory rate at different concentrations. (E) Immunoblotting of VSV glycoprotein (VSV-G) levels in VSV-eGFP (MOI 0.1) infected WT and IFNAR1−/− A549 cells under Fan treatment. (F) Immunoblotting analysis showing the expression level of IFNAR1 in A549 cells. The data above are presented as mean ± standard deviation (n = 3). ∗∗P < 0.01 and ∗∗∗P < 0.001. ns: not significant. MX1: myxovirus resistance 1; IFIT1: IFN induced protein with tetratricopeptide repeats 1; OAS2: 2'-5'-oligoadenylate synthetase 2; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
3.7. Fan binds STING to prevent its protein degradation and initiate antiviral responses
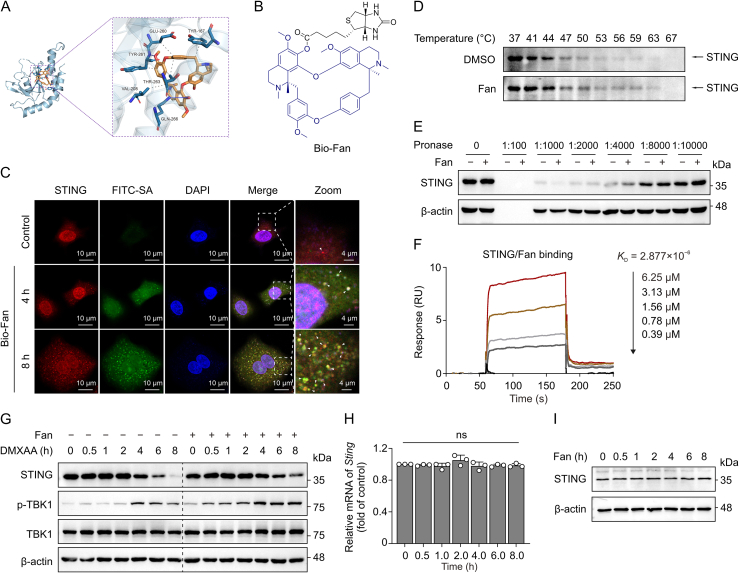
STING, a crucial adaptor molecule in the innate antiviral immune response, plays a pivotal role in activating downstream IFN-I expression via both cyclic GMP-AMP synthase (cGAS)-dependent and -independent mechanisms [25,26]. Significant advancements have been made in elucidating the regulatory mechanisms governing STING degradation to activate and prolong antiviral immune responses [27,28]. Our molecular docking analysis revealed that Fan binds to STING with a free binding energy of −8.0 kcal/mol, forming hydrogen bonds with the GLN-266 and TYR-261 residues of the STING protein and participating in hydrophobic interactions with the VAL-208, THR-263, TYR-261, GLU-260, and TYR-167 residues (Fig. 7A). These results indicate a potential interaction between Fan and STING.
Fig. 7.
Fangchinoline (Fan) induces innate immunity by inhibiting the degradation of stimulator of interferon genes (STING). (A) Docking model showing the interaction between Fan and STING protein. (B) Molecular structure of biotin-Fan (Bio-Fan). (C) Immunofluorescence showing the interaction of Bio-Fan to STING. (D) Cellular thermal shift assay (CETSA) of STING in immortalized bone marrow-derived macrophages (iBMDM) cells incubated with either dimethyl sulfoxide (DMSO) or Fan (50 μM). (E) Drug affinity responsive target stability (DARTS) experiment showing the binding of Fan (50 μM) to STING in iBMDM cells. (F) Binding affinity analysis of STING with Fan determined by surface plasmon resonance (SPR). (G) Immunoblotting of STING and phosphorylated TANK binding kinase 1 (p-TBK1) expression levels in iBMDM cells pre-treated with DMSO or Fan (10 μM) for 3 h and then co-incubated with 5,6-dimethylxanthenone-4-acetic acid (DMXAA) (15 μM) for the specified time. (H) Real-time quantitative polymerase chain reaction (qRT-PCR) assay of Sting expression levels in iBMDM cells stimulated with Fan (10 μM). (I) Immunoblotting of STING expression levels in A549 cells treated with Fan for indicated times. The data above are shown as mean ± standard deviation (SD) (n = 3). ns: not significant. FITC-SA: fluorescein isothiocyanate-streptavidin; DAPI: 4',6-diamidino-2-phenylindole mRNA: messenger RNA.
Biotin-Fan (Bio-Fan) was synthesised to investigate its binding to STING (Fig. 7B). Immunofluorescence confocal microscopy revealed that treatment with Bio-Fan led to the time-dependent formation of STING puncta (Fig. 7C), which is a crucial characteristic associated with the recruitment and activation of downstream proteins [29,30]. Additionally, Bio-Fan was evidently co-localised with STING puncta inside the cells, which gradually increased during drug treatment, suggesting a direct interaction between Fan and STING in the cells (Figs. 7C and S4). Additionally, CETSA and DARTS assay demonstrated that Fan enhanced the stability of STING under high-temperature and protease digestion in vitro (Figs. 7D and E). Consistent with these results, surface plasmon resonance (SPR) indicated that Fan bound to STING in a concentration-dependent manner with a dissociation constant of 2.88 μM, suggesting a strong binding ability of Fan to STING (Fig. 7F). Importantly, we observed that treatment of iBMDM cells with DMXAA led to progressive degradation of STING protein, whereas treatment with Fan significantly attenuated this degradation process, thereby sustaining TBK1 activation (Fig. 7G). qRT-PCR and immunoblotting revealed that treatment with Fan did not alter the mRNA and protein levels of STING (Figs. 7H and I). These findings suggest that Fan directly interacts with STING to inhibit its degradation and sustain the activation of TBK1.
3.8. Fan inhibits virus replication in a STING-dependent manner in vitro and in vivo
To examine the relationship between the antiviral capacity of Fan and STING both in vitro and in vivo, we generated Sting−/− mice and Sting−/− MEF cells (Fig. 8A). Fan treatment in WT cells significantly increased the expression of Ifnb1, Mx1, Ifit1, and Oas2 (Fig. 8B). In contrast, this upregulation was nearly entirely suppressed in Sting−/− MEF cells (Fig. 8B). Similar results were observed in the liver, lung, and spleen tissues of mice intraperitoneally injected with Fan, as determined using qRT-PCR analysis (Figs. 8C and S5). Moreover, the antiviral effect of Fan was weakened in Sting−/− MEF and the protective effect of Fan against VSV infection was eliminated in Sting−/− mice (Figs. 8D and E). These findings indicate that Fan induces an antiviral response and inhibits viral replication both in vitro and in vivo in a STING-dependent manner.
Fig. 8.
Fangchinoline (Fan) inhibits virus relies on stimulator of interferon genes (STING). (A) Immunoblotting of the STING protein level in mouse embryonic fibroblast (MEF) cells. (B) Real-time quantitative polymerase chain reaction (qRT-PCR) analysis showing the messenger RNA (mRNA) levels of interferon (IFN) beta 1 (Ifnb1) and IFN-stimulated genes (ISGs) in Fan (10 μM for 12 h) treated wild type (WT) and Sting−/− MEF cells. (C) qRT-PCR assay of Ifnb1 and ISGs expression levels in lung tissues of WT and Sting−/− mice treated with Fan (30 mg/kg/day) for 24 and 48 h (n = 3). (D) Statistical analysis of the inhibitory rates of Fan on vesicular stomatitis virus (VSV) in WT and Sting−/− MEF cells. Viral RNA was measured by qRT-PCR. (E) qRT-PCR analysis of the gene levels of VSV glycoprotein (VSV-G) and the large protein of VSV polymerase (VSV-L) in the lung and spleen tissues of WT and Sting−/−mice infected with VSV and treated with Fan (n = 5 in each group). The data above are shown as mean ± standard deviation (SD). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. ns: not significant. Mx1: myxovirus resistance 1; Ifit1: IFN induced protein with tetratricopeptide repeats 1; Oas2: 2'-5'-oligoadenylate synthetase 2; eGFP: enhanced green fluorescent protein.
4. Discussion
Previous studies have shown that cepharanthine, tetrandrine, and berbamine, which are BBAs components of plants of the genus Stephania, have potent antiviral activities [[31], [32], [33]]. However, the reported antiviral mechanisms to date do not comprehensively elucidate the broad antiviral spectrum of BBAs, indicating a need to uncover a potential shared mechanism underlying their antiviral effects. Fan, also a BBA and a major active therapeutic constituent of Stephania tetrandra, was investigated in this study. We observed that Fan effectively inhibited the replication of VSV, EMCV, H1N1, and HSV-1 in vitro. Additionally, in a viral infection model, Fan demonstrated the capacity to reduce viral loads in organs, mitigate virus-induced inflammation, and extend the survival of infected mice.
Transcriptome sequencing analysis of Fan-treated cells revealed a significant upregulation of genes associated with antiviral innate immunity, the IFN-I pathway, and negative regulation of viral replication. Furthermore, Fan activated ISGs such as Mx1, Ifit1, and Oas2 in both primary cells and mice. Fan treatment induced the phosphorylation of the downstream protein kinase TBK1 and transcription factors IRF3, STAT1, and STAT2. However, the antiviral and immune-activating activities of Fan were markedly suppressed by JAK inhibitor treatment and in IFNAR1−/− cells, indicating that Fan exerts its antiviral effects through an IFN-I-based immune response. Based on their similar chemical structures, we speculated that BBAs might inhibit viral replication via a shared mechanism similar to that of Fan.
STING, a key adaptor protein in the IFN-I pathway, recruits TBK1 to activate the antiviral innate immune response by inducing the expression of IFN-I and proinflammatory cytokines. It plays an indispensable role in host defence against viral infections caused by both DNA and RNA viruses [2,[34], [35], [36]]. Our experiments revealed that Fan was unable to promote the expression of ISGs and significantly attenuated the antiviral function, both in vivo and in vitro, when STING was knocked out. This suggests that Fan activates the STING-dependent immune response.
STING agonists, including DMXAA, cGAMP, and 5-hydroxytryptamine-DNA (HT-DNA), have demonstrated promising antiviral and anti-tumour activities in drug development [[37], [38], [39], [40]]. STING gradually degrades upon activation by its agonists, such as DMXAA, cGAMP, and HT-DNA [41,42]. Notably, inhibiting or slowing down the degradation of STING can extend and amplify STING-mediated immune responses [43]. The sustained expression of STING promotes the expression of ISGs and serves as an effective defence mechanism for the host against viral invasion [44]. Despite their therapeutic potential, certain small-molecule agonists of STING, such as DMXAA, exhibit dose-limiting toxicity and species limitations in early-phase clinical trials [45]. Our findings reveal that Fan substantially inhibits STING degradation and prolongs the maintenance of innate immune signalling through direct interaction with STING. The administration of Fan may help leverage this innovative pharmacological mechanism to augment immune activation and antiviral effects with minimal toxicity, thereby offering promising clinical applications.
5. Conclusion
Collectively, our findings highlight the potential of Fan as a natural small molecule that enhances the maintenance of antiviral innate immune activation by inhibiting STING degradation. Fan exhibits antiviral activity both in vivo and in vitro, positioning it as a potential drug candidate against viral infections. The findings suggest that the pharmacological maintenance of STING protein expression and activation of the antiviral immune response could be a viable strategy to combat viral infections and develop new therapeutics.
CRediT author statement
Jinyong Wang and Fang Xie: Conceptualization, Investigation, Formal analysis, Validation, Writing - Original draft preparation, Reviewing and Editing; Xin Jia: Funding acquisition, Supervision; Xuejiao Wang, Lingdong Kong, and Yiying Li: Methodology, Visualization, Investigation; Xue Liang and Meiqi Zhang: Software; Yuting He, Wandi Feng, and Tong Luo: Validation, Data curation; Yao Wang: Writing - Reviewing and Editing, Resources, Project administration; Anlong Xu: Supervision, Funding acquisition, Project administration.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the Beijing Nova Program, China (Grant No.: 20230484342), the Young Elite Scientists Sponsorship Program by China Association of Chinese Medicine (CACM), China (Grant No.: 2023-QNRC2-A02), and the Joint Fund of Beijing University of Traditional Chinese Medicine and USANA Health Sciences corporation, China (Grant No.: BUCM2023-JS-KF-032).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2024.100972.
Contributor Information
Yao Wang, Email: yaowang@bucm.edu.cn.
Anlong Xu, Email: xuanlong@bucm.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Kato K., Omura H., Ishitani R., et al. Cyclic GMP-AMP as an endogenous second messenger in innate immune signaling by cytosolic DNA. Annu. Rev. Biochem. 2017;86:541–566. doi: 10.1146/annurev-biochem-061516-044813. [DOI] [PubMed] [Google Scholar]
- 2.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazear H.M., Schoggins J.W., Diamond M.S. Shared and distinct functions of type I and type III interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu X., Li J., Fu M., et al. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021;6 doi: 10.1038/s41392-021-00791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Yuan S., Jia X., et al. Mitochondria-localised ZNFX1 functions as a dsRNA sensor to initiate antiviral responses through MAVS. Nat. Cell Biol. 2019;21:1346–1356. doi: 10.1038/s41556-019-0416-0. [DOI] [PubMed] [Google Scholar]
- 7.Sali T.M., Pryke K.M., Abraham J., et al. Characterization of a novel human-specific STING agonist that elicits antiviral activity against emerging alphaviruses. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y., Crowe W.N., Wang L., et al. An inhalable nanoparticulate STING agonist synergizes with radiotherapy to confer long-term control of lung metastases. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-13094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan B., Perera S.A., Piesvaux J.A., et al. An orally available non-nucleotide STING agonist with antitumor activity. Science. 2020;369 doi: 10.1126/science.aba6098. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z., Zhou J., Xu W., et al. A novel STING agonist-adjuvanted pan-sarbecovirus vaccine elicits potent and durable neutralizing antibody and T cell responses in mice, rabbits and NHPs. Cell Res. 2022;32:269–287. doi: 10.1038/s41422-022-00612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuchitsu Y., Mukai K., Uematsu R., et al. STING signalling is terminated through ESCRT-dependent microautophagy of vesicles originating from recycling endosomes. Nat. Cell Biol. 2023;25:453–466. doi: 10.1038/s41556-023-01098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji Y., Luo Y., Wu Y., et al. SEL1L-HRD1 endoplasmic reticulum-associated degradation controls STING-mediated innate immunity by limiting the size of the activable STING pool. Nat. Cell Biol. 2023;25:726–739. doi: 10.1038/s41556-023-01138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pizzorno A., Padey B., Dubois J., et al. In vitro evaluation of antiviral activity of single and combined repurposable drugs against SARS-CoV-2. Antiviral Res. 2020;181 doi: 10.1016/j.antiviral.2020.104878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khalifa S.A.M., Yosri N., El-Mallah M.F., et al. Screening for natural and derived bio-active compounds in preclinical and clinical studies: One of the frontlines of fighting the coronaviruses pandemic. Phytomedicine. 2021;85 doi: 10.1016/j.phymed.2020.153311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakurai Y., Kolokoltsov A.A., Chen C., et al. Ebola virus. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science. 2015;347:995–998. doi: 10.1126/science.1258758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y., Tang Q., Rao Z., et al. Inhibition of herpes simplex virus 1 by cepharanthine via promoting cellular autophagy through up-regulation of STING/TBK1/P62 pathway. Antiviral Res. 2021;193 doi: 10.1016/j.antiviral.2021.105143. [DOI] [PubMed] [Google Scholar]
- 17.Huang L., Yuen T.T., Ye Z., et al. Berbamine inhibits SARS-CoV-2 infection by compromising TRPMLs-mediated endolysosomal trafficking of ACE2. Signal Transduct. Target. Ther. 2021;6 doi: 10.1038/s41392-021-00584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng W., Wang Y., Luo T., et al. Scoparone suppresses mitophagy-mediated NLRP3 inflammasome activation in inflammatory diseases. Acta Pharmacol. Sin. 2023;44:1238–1251. doi: 10.1038/s41401-022-01028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia X., Zhang M., Wang H., et al. ZNFX1 antisense RNA1 promotes antiviral innate immune responses via modulating ZNFX1 function. J. Med. Virol. 2023;95 doi: 10.1002/jmv.28637. [DOI] [PubMed] [Google Scholar]
- 20.Weber F., Wagner V., Rasmussen S.B., et al. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song M., Wang J., Sun Y., et al. Tetrandrine alleviates silicosis by inhibiting canonical and non-canonical NLRP3 inflammasome activation in lung macrophages. Acta Pharmacol. Sin. 2022;43:1274–1284. doi: 10.1038/s41401-021-00693-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong Z., Qian Z., Zhang X., et al. Tetrandrine prevents bone loss in ovariectomized mice by inhibiting RANKL-induced osteoclastogenesis. Front. Pharmacol. 2020;10 doi: 10.3389/fphar.2019.01530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng G., Wang L.-C., Fridlender Z.G., et al. Pharmacologic activation of the innate immune system to prevent respiratory viral infections. Am. J. Respir. Cell Mol. Biol. 2011;45:480–488. doi: 10.1165/rcmb.2010-0288OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan Z., Lu Y., Liao Q., et al. Fangchinoline inhibits human immunodeficiency virus type 1 replication by interfering with gp160 proteolytic processing. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domizio J.D., Gulen M.F., Saidoune F., et al. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature. 2022;603:145–151. doi: 10.1038/s41586-022-04421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu T., Tu X., Yang K., et al. Tonic prime-boost of STING signalling mediates Niemann-Pick disease type C. Nature. 2021;596:570–575. doi: 10.1038/s41586-021-03762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L., Wei X., Wang Z., et al. NF-κB activation enhances STING signaling by altering microtubule-mediated STING trafficking. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.112185. [DOI] [PubMed] [Google Scholar]
- 28.Gentili M., Liu B., Papanastasiou M., et al. ESCRT-dependent STING degradation inhibits steady-state and cGAMP-induced signalling. Nat. Commun. 2023;14 doi: 10.1038/s41467-023-36132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S., Luo M., Wang Z., et al. Prolonged activation of innate immune pathways by a polyvalent STING agonist. Nat. Biomed. Eng. 2021;5:455–466. doi: 10.1038/s41551-020-00675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J., Canham S.M., Wu H., et al. Activation of human STING by a molecular glue-like compound. Nat. Chem. Biol. 2024;20:365–372. doi: 10.1038/s41589-023-01434-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi D., Li Q., Wang H., et al. Repurposing of berbamine hydrochloride to inhibit Ebola virus by targeting viral glycoprotein. Acta Pharm. Sin. B. 2022;12:4378–4389. doi: 10.1016/j.apsb.2022.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D.E., Min J.S., Jang M.S., et al. Natural bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus OC43 infection of MRC-5 human lung cells. Biomolecules. 2019;9 doi: 10.3390/biom9110696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leng L., Xu Z., Hong B., et al. Cepharanthine analogs mining and genomes of Stephania accelerate anti-coronavirus drug discovery. Nat. Commun. 2024;15 doi: 10.1038/s41467-024-45690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M., Ferretti M., Ying B., et al. Pharmacological activation of STING blocks SARS-CoV-2 infection. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abi9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia M., Qin D., Zhao C., et al. Redox homeostasis maintained by GPX4 facilitates STING activation. Nat. Immunol. 2020;21:727–735. doi: 10.1038/s41590-020-0699-0. [DOI] [PubMed] [Google Scholar]
- 36.King C.R., Liu Y., Amato K.A., et al. Pathogen-driven CRISPR screens identify TREX1 as a regulator of DNA self-sensing during influenza virus infection. Cell Host Microbe. 2023;31:1552–1567.e8. doi: 10.1016/j.chom.2023.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erttmann S.F., Swacha P., Aung K.M., et al. The gut microbiota prime systemic antiviral immunity via the cGAS-STING-IFN-I axis. Immunity. 2022;55:847–861.e10. doi: 10.1016/j.immuni.2022.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Xu N., Palmer D.C., Robeson A.C., et al. STING agonist promotes CAR T cell trafficking and persistence in breast cancer. J. Exp. Med. 2021;218 doi: 10.1084/jem.20200844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chin E.N., Yu C., Vartabedian V.F., et al. Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic. Science. 2020;369:993–999. doi: 10.1126/science.abb4255. [DOI] [PubMed] [Google Scholar]
- 40.Gui X., Yang H., Li T., et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567:262–266. doi: 10.1038/s41586-019-1006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haag S.M., Gulen M.F., Reymond L., et al. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559:269–273. doi: 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- 42.Balka K.R., Venkatraman R., Saunders T.L., et al. Termination of STING responses is mediated via ESCRT-dependent degradation. EMBO J. 2023;42 doi: 10.15252/embj.2022112712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han J., Hu S., Hu Y., et al. Discovery of podofilox as a potent cGAMP-STING signaling enhancer with antitumor activity. Cancer Immunol. Res. 2023;11:583–599. doi: 10.1158/2326-6066.CIR-22-0483. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Lian Q., Yang B., et al. TRIM30α is a negative-feedback regulator of the intracellular DNA and DNA virus-triggered response by targeting STING. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li N., Wang C., Zhao Y., et al. STING controls opioid-induced itch and chronic itch via spinal tank-binding kinase 1-dependent type I interferon response in mice. J. Neuroinflammation. 2023;20 doi: 10.1186/s12974-023-02783-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.