Abstract
PML nuclear bodies (NBs) are subnuclear structures whose integrity is compromised in certain human diseases, including leukemia and neurodegenerative disorders. Infection by a number of DNA viruses similarly triggers the reorganization of these structures, suggesting an important role for the NBs in the viral infection process. While expression of the adenovirus E4 ORF3 protein leads to only a moderate redistribution of PML to filamentous structures, the herpes simplex virus (HSV) ICP0 protein and the cytomegalovirus (CMV) IE1 protein both induce a complete disruption of the NB structure. Recently, we and others have shown that the NB proteins PML and Sp100 are posttranslationally modified by covalent linkage with the ubiquitin-related SUMO-1 protein and that this modification may promote the assembly of these structures. Here we show that the HSV ICP0 and CMV IE1 proteins specifically abrogate the SUMO-1 modification of PML and Sp100, whereas the adenovirus E4 ORF3 protein does not affect this process. The potential of ICP0 and IE1 to alter SUMO-1 modification is directly linked to their capacity to disassemble NBs, thus strengthening the role for SUMO-1 conjugation in maintenance of the structural integrity of the NBs. This observation supports a model in which ICP0 and IE1 disrupt the NBs either by preventing the formation or by degrading of the SUMO-1-modified PML and Sp100 protein species. Finally, we show that the IE1 protein itself is a substrate for SUMO-1 modification, thus representing the first viral protein found to undergo this new type of posttranslational modification.
PML nuclear bodies (NBs) are distinct subnuclear structures which appear as dense spherical particles, 0.3 to 0.5 μm in diameter, that are tightly associated with the nuclear matrix (for recent reviews, see references 17 and 26). Although a number of proteins seem to transiently localize to NBs, two nuclear body antigens, PML and Sp100, are considered to build the framework of these structures. PML was first identified as part of a fusion product with the retinoic acid receptor α (RARα), resulting from the t(15,17) chromosomal translocation associated with acute promyelocytic leukemia (APL) (8, 14, 20, 22, 40). PML is a member of the RING finger family of proteins and, within this family, belongs to a subgroup of proteins harboring one or two additional cysteine-rich regions, referred to as the B1 and B2 boxes, as well as an α-helical coiled-coil region (42). The PML protein exhibits some cell growth, and tumor-suppressive properties as well as a proapoptotic activity (41, 49, 50). However its exact mechanism of action in these different cellular processes remains unknown. Recently, it has been shown that a subset of PML is posttranslationally modified by a covalent linkage with the ubiquitin-related SUMO-1 modifier (21, 39, 47). The unmodified form of PML is found in the soluble nucleoplasmic fraction, whereas the SUMO-1-modified forms are compartmentalized exclusively in the PML NBs. The fraction of SUMO-1-modified PML can be drastically augmented by treatment of cells with arsenic, resulting in a complete recruitment of PML to NBs (39, 52). This finding led us to hypothesize that modification of PML by SUMO-1 may be implicated in its targeting to these structures. The Sp100 protein, first described as an autoantigen in certain autoimmune diseases, has been shown to associate with a number of non-histone-type chromosomal proteins, suggesting a possible role of the NBs in chromatin dynamics (28, 43, 48). Strikingly, Sp100 is also modified by covalent linkage to SUMO-1, strengthening the hypothesis that posttranslational modification by SUMO-1 plays an important role in the biological activity of NB-associated proteins and/or may promote the assembly of these structures (47).
Whereas the exact biological function of NBs is still unclear, a striking feature is the delocalization of NB-associated proteins in a number of pathological situations. Among them, the mutant ataxin-1 protein, responsible for spinocerebellar ataxia type 1, was shown to disrupt the PML NBs, and the human T-cell leukemia virus type 1 Tax oncoprotein was shown to delocalize the Int-6 protein from NBs (6, 44). However, the most intensively studied model remains APL. Whereas normal cells contain 10 to 30 NBs per nucleus, in PML-RARα-expressing APL cells, NBs are highly disorganized into numerous and aberrant microstructures containing both PML and PML-RARα (10, 24, 51). Strikingly, the cytodifferentiating and antileukemogenic drug retinoic acid induces the reorganization of the PML NBs back to their normal number and morphology, suggesting that the integrity of NBs is indispensable for critical cellular processes.
Apart from being disorganized in a number of human diseases, the PML NBs have been shown to be highly sensitive to environmental stimuli such as response to stress or interferon as well as to viral infection (16, 27, 35, 45; for a recent review, see reference 31). Indeed infection with DNA viruses such as adenovirus, herpes simplex virus (HSV), and cytomegalovirus (CMV) causes a dramatic disturbance of the NBs (2, 5, 9, 12, 13, 23, 25, 32, 33). Thus, in adenovirus-infected cells, a reorganization of the NBs into fibrous PML-containing structures is observed. The product encoded by adenovirus E4 ORF3 was shown to be responsible for this reorganization and to colocalize with PML into these fibers (5, 9). In the case of HSV-infected cells, it has been shown that the viral immediate-early transactivator ICP0 transiently localizes to the PML NBs before disrupting them (12, 13, 32, 33). Similarly, the CMV immediate-early protein IE1 colocalizes with PML in NBs at very early times after infection and subsequently induces the dispersal of both proteins to the nucleoplasm (2, 23, 25). These observations suggest that the two viruses have developed different strategies to interact with and disrupt or reorganize the PML NBs. Together with the fact that PML and Sp100 are highly upregulated by interferons, these data indicate that the NBs may play a critical role in virus-host interactions. Here we report that the HSV ICP0 protein and the CMV IE1 protein can specifically abrogate the SUMO-1 modification of PML and Sp100. With respect to the presumed role of the SUMO-1 modification in NB formation, this may provide a simple explanation for the capacity of these viral proteins to efficiently disrupt NBs.
The HSV ICP0 and CMV IE1 proteins abrogate the covalent linkage of SUMO-1 to PML and Sp100.
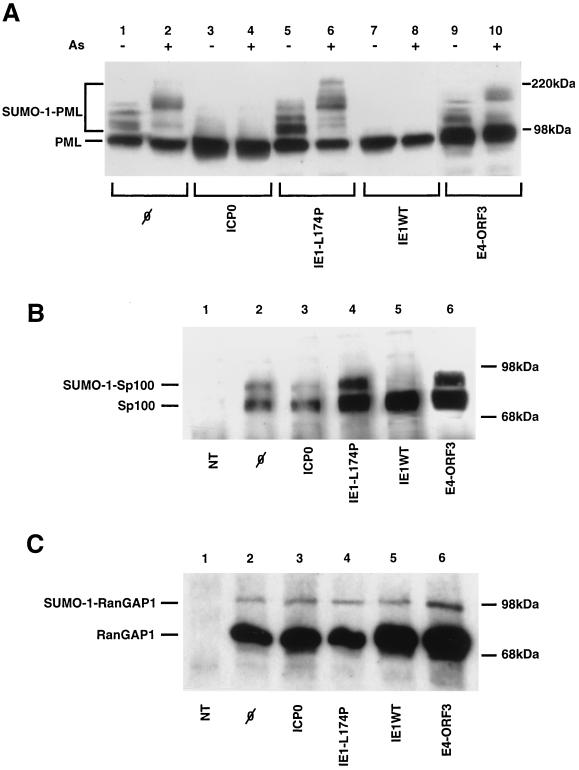
Since the covalent linkage of SUMO-1 to PML is associated with its targeting to the NBs, we wished to determine whether immediate-early viral proteins from DNA viruses, which either completely disrupt nuclear bodies or solely disorganize them, may affect the SUMO-1 modification of PML. To this end, we transfected HeLa cells with a PML expression vector alone or together with plasmids encoding either the HSV ICP0 protein, the CMV IE1 protein, or the adenovirus E4 ORF3 protein. In addition, we used a plasmid encoding a mutated form of IE1 (IE1-L174P) which harbors a leucine-to-proline mutation at amino acid position 174 and is no longer able to disrupt PML NBs (see below). The mutation results from a single base pair exchange created by PCR. To facilitate detection of the exogenous PML, we used a construct expressing an F-epitope-tagged version of PML (22). Cells were directly lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer 36 h after transfection, and both PML and the SUMO-1–PML conjugates were detected by immunoblotting with a monoclonal antibody directed against the F tag (3) (Fig. 1A). In cells transfected with the PML expression vector alone, the antibody detects a major PML(F) form with an apparent molecular mass of 100 kDa (Fig. 1A, lane 1). In addition, three higher-molecular-mass PML species are seen migrating between 20 and 60 kDa above this major form. Using immunoprecipitation followed by Western blotting, we and others had previously shown that these bands correspond to SUMO-1–PML conjugates, where one or more SUMO-1 molecules are covalently attached to PML (21, 39, 47). Treatment of cells with As2O3 was found to induce the conversion of these oligo-SUMO-1-modified PML forms toward poly-SUMO-1-modified species (39), migrating from 160 kDa toward the top of the gel (lane 2). Strikingly, when ICP0 is coexpressed with PML, only the unmodified form of PML can be detected; the higher-molecular-weight SUMO-1–PML conjugates are lost (lane 3). In addition, in the presence of ICP0, arsenic treatment is unable to induce the formation of the poly-SUMO-1-modified PML species (lane 4). Similarly, expression of the wild-type IE1 protein completely abolishes SUMO-1 modification of PML, as demonstrated by the absence of SUMO-1–PML conjugates in extracts from IE1-transfected cells (lane 7). Moreover, IE1 expression prevented the arsenic-induced poly-SUMO-1-modification of PML (lane 8). In contrast, expression of the mutant form of IE1 (IE1-L174P), which leaves the NBs intact (see below), does not affect the SUMO-1 modification pattern of PML (compare lane 5 to lanes 1 and 3). In the presence of the mutated IE1 protein, the ability of arsenic to induce poly-SUMO-1 modification of PML remained unaltered (compare lane 6 to lane 2). Interestingly, expression of the adenovirus E4 ORF3 protein, which induces only a morphological change of the PML nuclear bodies rather than their disruption, does not interfere with the conjugation of SUMO-1 to PML, as the SUMO-1 modification pattern of PML in E4 ORF3-expressing cells is indistinguishable from that in cells expressing PML alone (compare lane 9 to lane 1). In addition, the presence of E4 ORF3 does not hamper the capacity of arsenic to trigger poly-SUMO-1 modification of PML (lane 10). In all experiments, equal expression of the viral proteins was verified by Western blotting (data not shown).
FIG. 1.
The HSV ICP0 and CMV IE1 proteins abrogate the covalent linkage of SUMO-1 to PML and Sp100. (A) Cellular extracts from HeLa cells transiently transfected with a vector expressing PML(F) alone (lanes 1 and 2) or in combination with vectors expressing either ICP0 (lanes 3 and 4), IE1-L174P (lanes 5-6), wild-type IE1 (IE1WT; lanes 7 and 8), or E4 ORF3 (lanes 9 and 10) were separated by SDS-PAGE on a 7.5% gel and transferred to a nitrocellulose membrane, and the blot was immunostained with a monoclonal antibody directed against the F tag (3). Cells were untreated (lanes 1, 3, 5, 7, and 9) or treated with 1 μM As2O3 (lanes 2, 4, 6, 8, and 10) for 3 h. (B) Extracts from cells transfected with a vector expressing Sp100-HA alone (lane 2) or in combination with vectors expressing the different viral proteins as indicated (lanes 3 to 6) were analyzed by immunoblotting with a monoclonal antibody directed against the HA tag (12CA5; Boehringer Mannheim). An extract from nontransfected (NT) cells served as a negative control (lane 1). (C) Extracts from cells transfected with a vector expressing Myc-RanGAP1 alone (lane 2) or in combination with vectors expressing the different viral proteins as indicated (lanes 3 to 6) were analyzed by immunoblotting with a monoclonal antibody directed against the Myc tag (9E10; Pharmingen). An extract from nontransfected cells was used a negative control (lane 1).
Because Sp100 was identified as another SUMO-1 substrate protein (47), we wished to determine whether the viral proteins also act on the SUMO-1 modification of Sp100. To address this question, a hemagglutinin (HA) epitope-tagged Sp100 protein was transiently expressed in HeLa cells either alone or in combination with ICP0, IE1, IE1-L174P, or E4 ORF3. Cellular extracts were analyzed by immunoblotting with a monoclonal anti-HA antibody (Fig. 1B). As shown in lane 2, the antibody specifically detects two major Sp100 bands migrating at 80 and 95 kDa. It has been shown previously that these bands correspond to the free form of Sp100 and the SUMO-1–Sp100 conjugate (47). In contrast to PML, there is a single SUMO-1-modified band, indicating that only one SUMO-1 molecule is attached per Sp100 molecule. When Sp100 is coexpressed with ICP0, the amount of SUMO-1–Sp100 is significantly reduced, making the SUMO-1–Sp100 conjugate barely detectable (lane 3). Furthermore, coexpression of Sp100 with the wild-type IE1 protein completely abrogates SUMO-1 modification of Sp100, as only nonmodified Sp100 can be detected (lane 5). In contrast, expression of the mutated form of IE1 does not change the ratio between the SUMO-1–Sp100 conjugate and free Sp100 compared to cells expressing only Sp100 (compare lane 4 to lane 1). Finally, similar to what has been observed for SUMO-1–PML conjugates, expression of the adenovirus E4 ORF3 protein had no effect on the SUMO-1 modification of Sp100 (lane 6).
With respect to the results described above, we wished to examine whether the expression of ICP0 and IE1 could exert a general effect on SUMO-1 modification of any substrate protein or whether the effect is restricted to NB-associated proteins. To this end, we examined the effects of the different viral proteins on the SUMO-1 modification of RanGAP1, the first identified SUMO-1 target (29, 30). RanGAP1 is a component of the nuclear import machinery which is targeted from the cytosol to the nuclear pore complex by modification with SUMO-1. HeLa cells were transfected with a vector expressing an N-terminally Myc-tagged RanGAP1 protein alone or in combination with ICP0, IE1, IE1-L174P, or E4 ORF3. Extracts prepared from transfected cells were analyzed by immunoblotting with a monoclonal antibody directed against the Myc tag (Fig. 1C). In cells expressing Myc-RanGAP1 alone, we detected two specific anti-Myc-reactive bands migrating with relative molecular masses of 75 and 90 kDa, representing the unmodified and SUMO-1-modified forms of RanGAP1 (lane 2). In extracts from cells expressing Myc-RanGAP1 together with ICP0 (lane 3), IE1-L174P (lane 4), IE1 (lane 5), or E4 ORF3 (lane 6), none of the viral proteins had a significant effect on the conjugation of SUMO-1 to RanGAP1. These results suggest that the alteration in SUMO-1 modification mediated by ICP0 and IE1 is specific for the protein components of the NBs.
The CMV IE1 protein delocalizes PML and SUMO-1 from NBs.
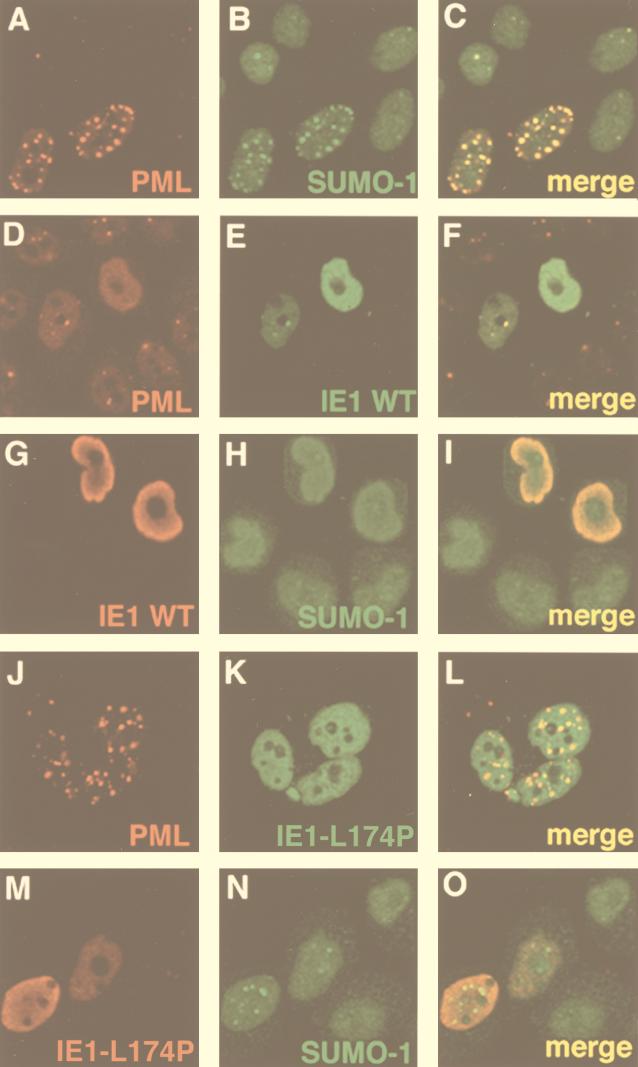
Having established that the CMV IE1 protein completely abrogates the SUMO-1 modification of PML, we wished to compare the subnuclear localization of SUMO-1 in cells expressing PML alone to its localization in cells coexpressing PML and IE1. To do so, HeLa cells were transfected with a PML expression vector either alone or in combination with a vector expressing an HA-tagged IE1 protein; 36 h after transfection, indirect immunofluorescence microscopy was performed by a standard procedure (39) using a polyclonal anti-PML antibody (51), an anti-SUMO-1 monoclonal antibody (30), and either a polyclonal or a monoclonal anti-HA antibody. Localization of the proteins was analyzed by confocal laser microscopy (Fig. 2). As has been reported before (39), cells overexpressing PML have a higher number of NBs than normal cells and thus show a more intense nuclear punctate staining of PML than surrounding nontransfected cells (Fig. 2A). In these nontransfected cells, the endogenous SUMO-1 protein shows predominantly a nuclear diffuse staining (Fig. 2B). However, in PML-overexpressing cells, SUMO-1 labeling gives rise to an intense nuclear punctate pattern due to the recruitment of SUMO-1–PML conjugates into NBs (Fig. 2B). Superimposition of the PML and the SUMO-1 signals demonstrates the colocalization of PML and SUMO-1 in almost all of the NBs (Fig. 2C). When the IE1 protein is coexpressed together with PML, the vast majority of transfected cells show exclusively a nuclear diffuse distribution of both PML (Fig. 2D) and IE1 (Fig. 2E). However, in a very small number (<5%) of cells where the expression of IE1 is low, residual NBs (Fig. 2D), and colocalization of IE1 and PML in these residual structures (Fig. 2F) can be detected. Similar to what we observed with PML, high expression of IE1 leads to the redistribution of Sp100 into a nuclear diffuse form (data not shown). Interestingly, when the localization of endogenous SUMO-1 is investigated in cells coexpressing PML and IE1, the punctate SUMO-1 staining seen in Fig. 2B is no longer detectable, and both IE1 (Fig. 2G) and SUMO-1 (Fig. 2H) show a nuclear diffuse distribution.
FIG. 2.
The CMV IE1 protein delocalizes PML and SUMO-1 from NBs. HeLa cells were transfected with a PML expression vector either alone (A to C) or in combination with a vector expressing the HA-tagged wild-type IE1 protein (D to I) or the HA-tagged IE1-L174P mutant (J to O). Indirect immunofluorescence microscopy was performed with an anti-PML polyclonal antibody (51), an anti-SUMO-1 monoclonal antibody (30), and either a polyclonal or a monoclonal anti-HA antibody (Y-11 [Santa Cruz Biotechnology] or 12CA5 [Boehringer Mannheim]). Localization of the proteins was analyzed by confocal laser microscopy. The red and green signals were obtained with anti-rabbit immunoglobulin Ig Texas red-conjugated and anti-mouse Ig fluorescein-conjugated secondary antibodies, respectively. Superimposing the two colors (merge) results in a yellow signal, where the two proteins colocalize.
An analogous set of cotransfection experiments was performed with a vector expressing the IE1-L174P mutant protein (Fig. 2J to O). IE1-L174P exhibited exclusively a nuclear diffuse staining pattern (Fig. 2K and M), and in contrast to the wild-type IE1 protein, it was never detected in NBs even at low expression levels (Fig. 2M). Strikingly, IE1-L174P was not able to disorganize the NB, as evidenced by the intense nuclear punctate labeling of overexpressed PML (Fig. 2J). In cells coexpressing IE1-L174P and PML, SUMO-1 showed the typical mixture of nuclear diffuse and speckled pattern which was indistinguishable from that seen in cells expressing PML only (compare Fig. 2N to Fig. 2B). Superimposition of IE1-L174P distribution (Fig. 2M) and SUMO-1 staining (N) confirms the absence of the mutant IE1 form from NBs (Fig. 2O).
The CMV IE1 protein is itself covalently modified by SUMO-1.
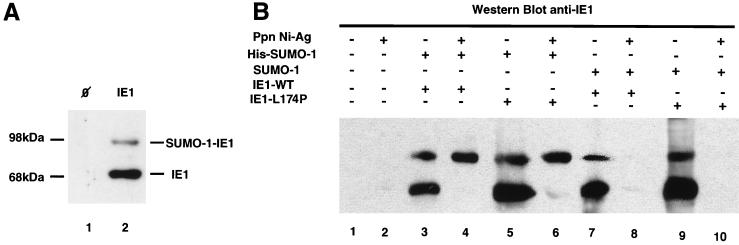
When cellular extracts from HeLa cells transiently transfected with an IE1 expression vector were examined by immunoblotting with the mouse monoclonal antibody E-13, directed against IE1 (36), we consistently detected two IE1-immunoreactive bands (Fig. 3A, lane 2). The major band migrates with an apparent molecular mass of 68 kDa, fairly consistent with the calculated molecular mass of the protein (46). Above this band, a second IE1-reactive band migrating at ∼90 kDa was visible. Neither of these two bands could be detected in extracts from untransfected HeLa cells (Fig. 3A, lane 1). This observation led us to wonder whether the 90-kDa band might represent a SUMO-1-modified IE1 species. To test this hypothesis, HeLa cells were cotransfected with a vector expressing a His-tagged SUMO-1 protein with a vector expressing either wild-type IE1 or the mutated IE1-L174P form described above; 36 h after transfection, cells were lysed in 6 M guanidine-HCl and the extracts were subjected to precipitation with nickel-charged agarose beads to recover the putative His–SUMO-1–IE1 conjugates. The Ni-agarose precipitates were separated by SDS-PAGE and analyzed by Western blotting with the anti-IE1 monoclonal antibody (Fig. 3B). Nontransfected cells served as a negative control (lanes 1 and 2). In unprecipitated extracts from cells transfected with either the wild-type IE1 protein or the mutant IE1-L174P, the 68- and 90-kDa IE1-reactive bands were detected (lanes 3 and 5). Of these two bands, only the 90-kDa band was retained on Ni-agarose beads, demonstrating that this band corresponds to an IE1 form covalently attached to His–SUMO-1 (lanes 4 and 6). To exclude any artifact which might be due to nonspecific binding of the upper band to the agarose beads, we performed an analogous experiment replacing the vector expressing the His–SUMO-1 protein by a vector expressing an untagged SUMO-1 protein. In unprecipitated extracts, the untagged SUMO-1 still could form conjugates with both wild-type IE1 and the IE1-L174P mutant (lanes 7 and 9), but these conjugates which lack the His tag were not retained on the nickel-charged agarose beads (lanes 8 and 10). It is noteworthy that in our experiments the SUMO-1–IE1 conjugates could be detected only when cells were lysed under denaturing conditions, indicating that such modified species are rapidly cleaved in vitro by a demodifying activity. A similar in vitro reversal of the SUMO-1 modification has been observed for other SUMO-1 substrates (7, 29, 30, 39).
FIG. 3.
The CMV IE1 protein itself is covalently modified by SUMO-1. (A) Extracts from nontransfected HeLa cells (lane 1) and from cells transfected with a vector expressing the IE1 protein (lane 2) were analyzed by Western blotting with monoclonal antibody E-13, directed against IE1 (36). (B) Extracts from HeLa cells which had been cotransfected with a vector expressing a His-tagged SUMO-1 protein or the untagged SUMO-1 protein together with either a vector expressing wild-type IE1 (IE1-WT; lane 4 and 8) or the mutated IE1-L174P (lane 6 and 10) were subjected to precipitation with Ni-agarose beads (Ppn Ni-Ag), and the precipitates were analyzed by Western blotting with the anti-IE1 monoclonal antibody. Aliquots of the corresponding unprecipitated extracts are loaded in lane 3, 5, 7, and 9. Nontransfected cells (lanes 1 and 2) served as a negative control.
Discussion.
Taken together, our results demonstrate that the HSV ICP0 and the CMV IE1 proteins abrogate SUMO-1 modification of the NB components PML and Sp100. This effect is specific to NB-associated proteins, since the modification of a non-NB protein such as RanGAP1 is not affected by expression of the two viral proteins. The potential to abrogate SUMO-1 modification of PML and Sp100 is directly linked to their capacity to disassemble NBs, as a mutant IE1 protein, which does not abolish SUMO-1 modification of PML and Sp100, is no more able to disrupt NBs. In addition, the adenovirus E4 ORF3 gene product, which reorganizes moderately the PML NBs but cannot completely disrupt them, did not affect the SUMO-1 modification of PML and Sp100. Taken together, these results are consistent with the idea that the posttranslational modification of PML and Sp100 with SUMO-1 may either direct the assembly of NBs or stabilize these structures. In agreement with our data, Everett et al. have recently shown that the disruption of NBs upon HSV infection is accompanied by the loss of high-molecular-weight PML isoforms and suggested that these forms may correspond to SUMO-1–PML conjugates (11). The biological significance of the destruction of NBs upon viral infection is still unclear. The use of HSV and CMV mutants has shown that ICP0 and IE1 are not essential for virus replication at high multiplicity of infection. However, an ICP0 mutant HSV or an IE1 mutant CMV exhibits a marked deficiency in replication at a low multiplicity of infection (13, 15, 37). Both ICP0 and IE1 (together with IE2) seem to play a particular role in triggering efficient initiation of the lytic cycle, and ICP0 is essential for reactivation from the latent state. Intriguingly, mutations in ICP0 which alter its interaction with NBs also affect its role in the initiation of viral infection (13). Thus, the disruption of NBs may serve to recruit host proteins participating in viral transcription or replication (18, 19, 34). In support of this hypothesis, it has been shown recently that in the course of HSV infection, PML is recruited to viral replication compartments in the presence of viral DNA polymerase (4).
A striking observation was the detection of a SUMO-1-modified subfraction of IE1, thus identifying IE1 as the first viral protein undergoing this new type of covalent modification. This process seems to be restricted to IE1, as we have been unable to detect any SUMO-1–ICP0 conjugates in our Ni-agarose precipitation assay (data not shown). The modification of the IE1-L174P mutant, which is not targeted to NBs, strongly suggests that the attachment of SUMO-1 to IE1 takes place in the nucleoplasm and not in the PML NBs. In addition, these data make it unlikely that the attachment of SUMO-1 to IE1 is implicated in the initial targeting of IE1 to NBs. In agreement with Ahn et al. (1), we could detect a specific interaction between PML and the wild-type IE1 protein in a yeast two-hybrid assay. Interestingly, in this assay the IE1-L174P mutant could not interact with PML, suggesting that the interaction with PML is responsible for targeting of IE1 to NBs (38). Besides its role in protein targeting, SUMO-1 modification has recently shown to be implicated in the stabilization of proteins by generating proteins resistant to degradation (7). Although we have no experimental evidence for a similar effect of SUMO-1 on IE1, these findings open up new perspectives in the study of this major CMV regulatory protein.
Acknowledgments
We thank Marie-Pierre Gaub, Daniel Metzger, Pierre Chambon, Roger Everett, Michael J. Matunis, Günter Blobel, Susan Michelson, and Marie-Christine Mazeron for the generous gifts of antibodies and plasmids used in these experiments. We are grateful to Emmanuelle Perret for excellent help with confocal microscopy. We thank Pierre Tiollais for support and all members of our group for stimulating discussions and for providing reagents.
This work was supported by grants from the European Economic Community (EEC), the Association pour la Recherche contre le Cancer, and la Fondation pour la Recherche Médicale et le Ministère de la Recherche et de la Technologie. S.M. was supported by a TMR fellowship from the EEC.
REFERENCES
- 1.Ahn J H, Brignole III E J, Hayward G S. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol. 1998;18:4899–4913. doi: 10.1128/mcb.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali S, Lutz Y, Bellocq J P, Chenard-Neu M P, Rouyer N, Metzger D. Production and characterization of monoclonal antibodies recognising defined regions of the human oestrogen receptor. Hybridoma. 1993;12:391–405. doi: 10.1089/hyb.1993.12.391. [DOI] [PubMed] [Google Scholar]
- 4.Burkham J, Coen D M, Weller S K. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J Virol. 1998;72:10100–10107. doi: 10.1128/jvi.72.12.10100-10107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carvalho T, Seeler J S, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desbois C, Rousset R, Bantignies F, Jalinot P. Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I Tax oncoprotein. Science. 1996;273:951–953. doi: 10.1126/science.273.5277.951. [DOI] [PubMed] [Google Scholar]
- 7.Desterro J M, Rodriguez M S, Hay R T. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 8.De Thé H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RARα fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 9.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 10.Dyck J A, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 11.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett R, O’Hare P, O’Rourke D, Barlow P, Orr A. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J Virol. 1995;69:7339–7344. doi: 10.1128/jvi.69.11.7339-7344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goddard A D, Borrow J, Freemont P S, Solomon E. Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science. 1991;254:1371–1374. doi: 10.1126/science.1720570. [DOI] [PubMed] [Google Scholar]
- 15.Greaves R F, Mocarski E S. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guldner H H, Szostecki C, Grötzinger T, Will H. IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J Immunol. 1992;149:4067–4073. [PubMed] [Google Scholar]
- 17.Hodges M, Tissot C, Howe K, Grimwade D, Freemont P S. Structure, organization, and dynamics of promyelocytic leukemia protein nuclear bodies. Am J Hum Genet. 1998;63:297–304. doi: 10.1086/301991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishov A M, Stenberg R M, Maul G G. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kakizuka A, Miller W H, Jr, Umesono K, Warrell R P, Jr, Frankel S R, Murty V V, Dmitrovsky E, Evans R M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 21.Kamitani T, Nguyen H P, Kito K, Fukuda-Kamitani T, Yeh E T. Covalent modification of PML by the sentrin family of ubiquitin-like proteins. J Biol Chem. 1998;273:3117–3720. doi: 10.1074/jbc.273.6.3117. [DOI] [PubMed] [Google Scholar]
- 22.Kastner P, Perez A, Lutz Y, Rochette-Egly C, Gaub M P, Durand B, Lanotte M, Berger R, Chambon P. Structure, localization and transcriptional properties of two classes of retinoic acid receptor α fusion proteins in acute promyelocytic leukemia (APL): structural similarities with a new family of oncoproteins. EMBO J. 1992;11:629–642. doi: 10.1002/j.1460-2075.1992.tb05095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly C, van Driel R, Wilkinson G W. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J Gen Virol. 1995;76:2887–2893. doi: 10.1099/0022-1317-76-11-2887. [DOI] [PubMed] [Google Scholar]
- 24.Koken M H, Puvion-Dutilleul F, Guillemin M C, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, Degos L, Puvion E, de Thé H. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korioth F, Maul G G, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 26.Lamond A I, Earnshaw W C. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 27.Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi P P, Pelicci P G, Dejean A. The acute-promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene. 1995;11:871–876. [PubMed] [Google Scholar]
- 28.Lehming N, Le Saux A, Schuller J, Ptashne M. Chromatin components as part of a putative transcriptional repressing complex. Proc Natl Acad Sci USA. 1998;95:7322–7326. doi: 10.1073/pnas.95.13.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 30.Matunis M J, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maul G. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 32.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 33.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 34.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 35.Maul G G, Yu E, Ishov A M, Epstein A L. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem. 1995;59:498–513. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- 36.Mazeron M C, Jahn G, Plachter B. Monoclonal antibody E-13 (M-810) to human cytomegalovirus recognizes an epitope encoded by exon 2 of the major immediate early gene. J Gen Virol. 1992;73:2699–2703. doi: 10.1099/0022-1317-73-10-2699. [DOI] [PubMed] [Google Scholar]
- 37.Mocarski E S, Kemble G W, Lyle J M, Greaves R F. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller, S., and A. Dejean. Unpublished data.
- 39.Müller S, Matunis M J, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandolfi P P, Grignani F, Alcalay M, Mencarelli A, Biondi A, Lo Coco F, Grignani F, Pelicci P G. Structure and origin of the acute promyelocytic leukemia myl/RARα cDNA and characterization of its retinoid-binding and transactivation properties. Oncogene. 1991;6:1285–1292. [PubMed] [Google Scholar]
- 41.Quignon F, De Bels F, Koken M, Feunteun J, Ameisen J C, de Thé H. PML induces a novel caspase-independent death process. Nat Genet. 1998;20:259–265. doi: 10.1038/3068. [DOI] [PubMed] [Google Scholar]
- 42.Reddy B A, Etkin L D, Freemont P S. A novel zinc finger coiled-coil domain in a family of nuclear proteins. Trends Biochem Sci. 1992;17:344–345. doi: 10.1016/0968-0004(92)90308-v. [DOI] [PubMed] [Google Scholar]
- 43.Seeler J S, Marchio A, Sitterlin D, Transy C, Dejean A. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skinner P J, Koshy B T, Cummings C J, Klement I A, Helin K, Servadio A, Zoghbi H Y, Orr H T. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature. 1997;389:971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- 45.Stadler M, Chelbi-Alix M K, Koken M H M, Venturini L, Lee C, Saïb A, Quignon F, Pelicano L, Guillemin M C, Schindler C, de Thé H. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene. 1995;11:2565–2573. [PubMed] [Google Scholar]
- 46.Stenberg R M, Thomson D R, Stinski M F. Structural analysis of the major immediate early gene of human cytomegalovirus. J Virol. 1984;49:190–199. doi: 10.1128/jvi.49.1.190-199.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szostecki C, Guldner H H, Netter H J, Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- 49.Wang Z G, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi P P. Pml is essential for multiple apoptotic pathways. Nat Genet. 1998;20:266–272. doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z G, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi P P. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- 51.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 52.Zhu J, Koken M H, Quignon F, Chelbi-Alix M K, Degos L, Wang Z Y, Chen Z, de Thé H. Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:3978–3983. doi: 10.1073/pnas.94.8.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]