Abstract
Hypervariable region 5 (HVR5) is a hydrophilic, serotypically nonconserved loop of the hexon monomer which extrudes from the adenovirus (Ad) capsid. We have replaced the HVR5 sequence of Ad5 with that of heterologous peptides and studied their effects on virus viability and peptide accessibility. A poliovirus model epitope was first inserted in a series of nine “isogenic” viruses that differed in their flanking spacers. Whereas virus productivity was not profoundly altered by any of these modifications, immunoprecipitation experiments under nondenaturing conditions demonstrated that epitope recognition by its cognate monoclonal antibody (C3 MAb) was strongly linker dependent and correlated perfectly with the ability of C3 MAb to inhibit transgene delivery and expression. An αv-specific ligand (DCRGDCF) was then inserted in a suitable linker context to investigate whether hexon-modified capsids would enhance the transduction of cells displaying limiting amounts of the virus attachment receptors. Interestingly, although hexon has never been implicated in Ad entry, the modified virus significantly increased the transduction of human vascular smooth muscle cells in vitro. Competition experiments with 293 cells saturated with recombinant knob further indicated that the hexon-modified virus could use an additional, knob-independent pathway for entry. We concluded that genetic engineering of the Ad5 hexon monomer constitutes a novel and feasible approach to equip the virus with additional targeting ligands.
Subgroup C-based adenovirus (Ad) vectors are being used in a growing number of clinical trials for acquired (e.g., cancer) and monogenic hereditary disorders. Ideally, these vectors should deliver and express their transgene only in the targeted cells in vivo. However, barriers exist that limit their efficacy: (i) widespread distribution of the virus receptors makes it difficult to restrict infection to particular cells; (ii) therapeutic targets expressing small amounts of receptors require high vector doses, thereby favoring spreading; and (iii) neutralizing antibodies specific to the capsid proteins can preclude efficient readministration in vivo (7). For example, intratumoral administration of recombinant Ad in murine models has been associated with vector dissemination to distant organs (e.g., lungs, liver, and spleen). Clinical data have also established the presence of the recombinant virus in bodily fluids of cancer patients intratumorally injected with the highest doses (3, 21). Taken together, these data emphasize the need to adapt the vector tropism to particular target cells in order to enhance infection while reducing vector dissemination and shedding.
Virus cell attachment, the initial step that dictates viral tropism, proceeds through high-affinity binding of the distal domain (i.e., the knob) of the capsid fibers to cell surface receptors (2, 10, 19). Accordingly, targeting strategies have focused primarily on the knob-mediated attachment step. For example, incubation of recombinant virus with an anti-fiber neutralizing antibody chemically conjugated to a cell-specific ligand (e.g., folate or fibroblast growth factor) has been shown to enhance the transduction of cells displaying the appropriate receptors (6, 16). A conceptually different strategy relies on the incorporation of chimeric fibers within the capsid to switch the tropism of different serotypes (8, 18), especially that of subgroup B (15). Additional strategies are being investigated, including peptidic insertion and C-terminal extension of the fiber knob (11–13, 23, 24). Although these approaches are not examples of a true retargeting strategy, since the native tropism is maintained, they should reduce viral dissemination by allowing the use of lower doses to reach clinical efficacy. Indeed, a polylysine extension of the fiber C terminus increases transduction both in vitro and in vivo, most probably because virus internalization occurred after virus attachment to heparan sulfate receptors (23, 24). However, genetic engineering of the fiber C terminus is tricky: deletions as small as 2 residues can prevent fiber trimerization in vitro, whereas addition of larger peptides often impairs virus growth (9, 13). Peptide insertion within the knob domain constitutes a more permissive approach, as recently reported for HI loop insertion between residues 546 and 547 (11) and the successful recovery of 15 of 15 HI-modified viruses with a foreign peptide ranging in size from 11 to 19 residues (5a).
Genetic engineering of the capsid hexon monomer is an attractive approach because this polypeptide is by far the most abundant capsid component (for a review, see reference 17). If the approach is successful, hexon-modified vectors should exhibit profound modifications of their physical and biological properties: target avidity for a particular ligand may reduce vector dissemination following local delivery and/or may alter tropism (see below). The primary sequence of the hexon monomer is well conserved among serotypes; the main differences are clustered within seven stretches (the so-called hypervariable regions [HVRs]) that are localized mostly within the extruding domains of the molecule (1). Some of these HVRs, including HVR5, must be quite flexible, as suggested by the absence of assignable structure in the crystallographic study of the Ad2 hexon monomer (1). Additional arguments prompted us to select HVR5 for peptide insertion. First, in contrast to HVR3, HVR4, and HVR7, HVR5 apparently does not include residues involved in intramolecular interactions within the molecule (1). Second, HVR5 is quite variable in length, ranging from 9 residues (e.g., in Ad30 and Ad37) to 20 residues (e.g., in Ad10, Ad19, and Ad45). Finally, HVR5 displays linear epitopes capable of eliciting anti-Ad2 neutralizing antibodies in vivo (20), and it could be replaced with a vaccinating poliovirus epitope (4). Based on these observations, we performed the present study by substituting HVR5 residues 269 to 281 (TTEAAAGNGDNLT), keeping intact its most highly conserved residues on either side (Ser268 and Pro282).
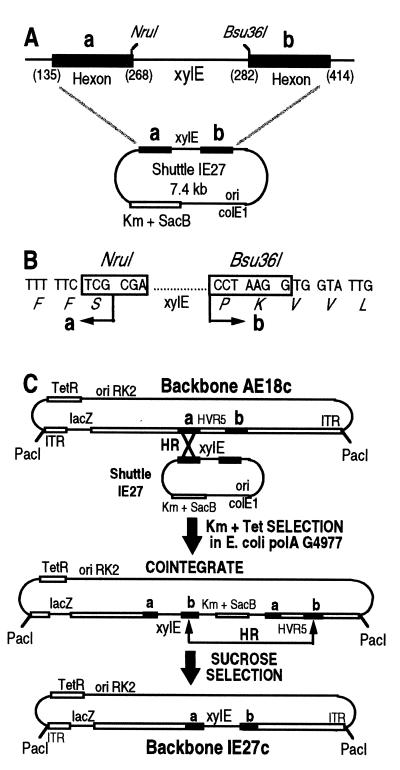
To facilitate construction of the HVR5-modified viruses by recombinational cloning in Escherichia coli (5), we first introduced the xylE marker gene from Pseudomonas putida in place of HVR5, since its phenotypic expression in E. coli is readily detectable in the presence of catechol (26). Shuttle plasmid IE27 was therefore engineered to contain the flanking regions of HVR5 (residues 135 to 268 and 282 to 414) flanking the xylE marker gene (Fig. 1A). This ColE1-based episome was introduced into E. coli G4977, a polymerase A (polA)-deficient strain that cannot replicate ColE1-derived plasmids, and recombined with plasmid AE18c (an RK2-based plasmid which does not require the polA gene product for replication). Following a two-step recombinational gene replacement in E. coli (5), plasmid IE27c was recovered, which contains a PacI-excisable ΔE1ΔE3 viral backbone with a cytomegalovirus-lacZ expression cassette in place of the E1 genes (the deletion extends from Ad5 nucleotides 382 to 3512), together with the xylE phenotypic marker inserted within HVR5 (Fig. 1C).
FIG. 1.
Modification of the Ad5 hexon monomer by recombinational cloning in E. coli. (A) Shuttle plasmid IE27 (the detailed construction strategy is available upon request) is a kanamycin (Km)-selectable plasmid in which the xylE phenotypic marker has been inserted in place of HVR5 of Ad5 (residues 268 to 282). Its replication requires the polA gene product of E. coli. SacB is a suicide gene for E. coli in the presence of sucrose as the carbon source. a and b refer to HVR5-flanking sequences from Ad5 (numbers in parentheses refer to hexon amino acid numbering). (B) The xylE phenotypic marker of shuttle plasmid IE27 is bordered by unique NruI and Bsu36I sites to facilitate subsequent HVR5 modifications. (C) Principle of HVR5 modification by homologous recombination in E. coli (adapted from reference 5). Homologous recombination (HR) between identical sequences (black bars a and b) from a suicide ColE1 shuttle plasmid (e.g., IE27) and an RK2-derived (replication is polA independent) plasmid (e.g., AE18c) in a polA mutant of E. coli generates a kanamycin (Km)- and tetracycline (Tet)-selectable cointegrate. Resolution of the cointegrate by HR leads to the loss of the sacB suicide gene, which is selected by using sucrose as a carbon source. Depending on the recombination pathway, resolution of the cointegrate generates either the starting backbone AE18c or the xylE-containing IE27c backbone, which can be differentiated upon catechol addition (26). IE27c has been used as a recipient backbone to generate all HVR5-modified backbones and viruses used in this study (see the text).
The DNPASTTNKDK model peptide, a well-studied neutralizing epitope from poliovirus type 1 (22), was then inserted in HVR5: shuttle plasmids IE35, IE37, IE39, IE40, IE43, IE44, IE45, IE46, and IE47, which are identical to IE27 except that the xylE marker had been replaced with the poliovirus epitope bordered by connecting linkers of different sizes and/or sequences, were thus constructed (Table 1). These plasmids were recombined with backbone IE27c in E. coli G4977 by using the xylE screening to generate a serie of PacI-excisable ΔE1ΔE3 lacZ-expressing viral backbones (Table 1). Plasmid IE31c contains a control viral backbone in which peptide NLDSLEQPTTRAQKPRLD was inserted in place of HVR5 residues 269 to 285 as reported previously (4). IE30c, IE32c, and IE33c are additional controls in which the HVR5 Ad5 sequence was replaced with that from Ad2, Ad30, and Ad19, respectively (Table 1). In all cases, the substitutions within HVR5 were checked by nucleotide sequencing.
TABLE 1.
Characteristics of the HVR5 substitutions
| Virus | Upstream HVR5 se-quence | Upstream linker | Inserted peptide | Down-stream linker | Downstream HVR5 se-quence |
|---|---|---|---|---|---|
| AdIE30 | FFS | NTTSLNDRQGNATK | PKVVLYS | ||
| AdIE32 | FFS | STTINI | PKVVLYS | ||
| AdIE33 | FFS | TPGANPPAGGSGNEEYK | PKVVLYS | ||
| AdIE31 | FFS | NLDSLEQPTTRAQKPRLD | LYS | ||
| AdIE35 | FFS | DNAPASTTNKDK | L | PKVVLYS | |
| AdIE37 | FFS | G | DNAPASTTNKDK | PKVVLYS | |
| AdIE39 | FFS | GS | DNAPASTTNKDK | PKVVLYS | |
| AdIE40 | FFS | S | DNAPASTTNKDK | PKVVLYS | |
| AdIE43 | FFS | G | DNAPASTTNKDK | LGG | PKVVLYS |
| AdIE44 | FFS | G | DNAPASTTNKDK | LGS | PKVVLYS |
| AdIE45 | FFS | G | DNAPASTTNKDK | LS | PKVVLYS |
| AdIE46 | FFS | G | DNAPASTTNKDK | LSG | PKVVLYS |
| AdIE47 | FFS | G | DNAPASTTNKDK | LSS | PKVVLYS |
| AE57 | FFS | GS | DCRGDCF | GS | PKVVLYS |
All viral backbones were excised by PacI restriction and transfected in 293 cells as described previously (5). With the exception of IE31c, they all induced a cytopathic effect that could be progressively amplified. Laboratory-scale batches (3 × 108 infected cells; one assay per virus) were prepared 48 h after infection at a multiplicity of infection (MOI) ranging from 5 to 10 transducing units, and viral productivity was assessed by counting the number of virus particles produced (VP/cell). The Ad5/Ad2, Ad5/Ad19, or Ad5/Ad30 chimeric viruses exhibited a correct productivity (i.e., 2,000 to 6,600 VP/cell), as did all nine HVR5-modified viruses containing the poliovirus epitope (i.e., 8,500 ± 4,700 VP/cell). HVR5 is thus quite permissive for insertion, since peptides ranging in size from 6 to 17 residues can be included with no obvious deleterious effect. The finding that virus AdIE31 could not be recovered despite repeated attempts was not expected. Possibly, the original virus of Crompton et al., which was constructed by homologous recombination between an Ad5-based chromosome and a recombinant Ad2-based HVR5 mutant plasmid, did exhibit extragenic compensatory mutations (4). In fact, the sole deletion of residues 269 to 285 in our construct most probably affected viability, since residues 281 to 285 have been assigned to a β-strand located immediately downstream of HVR5 (1).
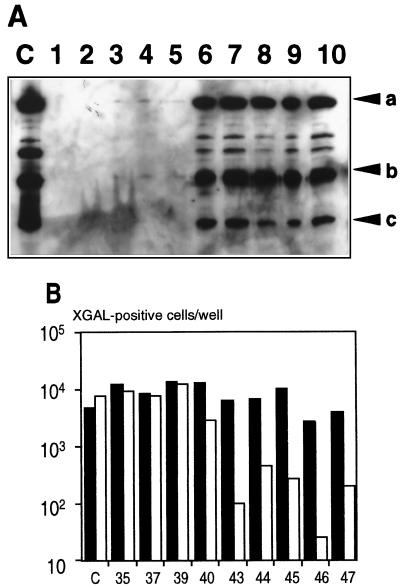
That the physical properties of the virus capsid were modified after HVR5 insertion was first evidenced by its different elution profile during anion-exchange chromatography (2a). The accessibility of the inserted epitope was first assessed by immunoprecipitating nondenatured capsids with C3 MAb, a monoclonal antibody (MAb) specific to the poliovirus epitope (22). Briefly, 1010 particles of CsCl-purified virus were resuspended in 400 μl of 50 mM Tris-HCl (pH 7.5)–150 mM NaCl–0.05% Nonidet P-40 prior to C3 MAb incubation for 1 h at 4°C. A 400-μl volume of protein A-Sepharose complex equilibrated in the same buffer was then added, and the mixture was incubated for another 1 h at 4°C. The complex was then collected by centrifugation, washed twice, and further incubated at 4°C for 20 min in 1 ml of 10 mM Tris (pH 7.5)–0.1% NP40. It was then resuspended in 50 μl of Laemmli buffer, boiled for 2 min, and subjected to Western analysis (enhanced chemiluminescence [Amersham]) with an anti-Ad5 rabbit serum. The presence of a spacer larger than 2 residues downstream of the poliovirus epitope drastically affected the outcome of this assay: whereas AdIE43, AdIE44, AdIE45, AdIE46, and AdIE47 were efficiently immunoprecipitated by C3 MAb, apparently with a similar efficacy, this was not the case for the other viruses (Fig. 2A).
FIG. 2.
(A) C3 MAb-mediated virus immunoprecipitation. HVR5-modified Ads were immunoprecipitated under nondenaturing conditions before being subjected to Western analysis with a polyclonal serum against Ad5 (see the text). In lane C, 109 VP of control AE18 virus was boiled for 2 min before being loaded. Lanes 1 to 10 correspond to AdIE30, AdIE35, AdIE37, AdIE39, AdIE40, AdIE43, AdIE44, AdIE45, AdIE46, and AdIE47, respectively. Arrows a, b, and c indicate the positions of the hexon, pV, and pVI/pVII polypeptides, respectively. (B) Virus opsonization inhibits lacZ transduction of W162 reporter cells. Control virus AE18 (lane C) or the indicated HVR5-modified viruses were incubated for 1 h at 37°C in the absence (solid bars) or presence (open bars) of C3 MAb before being used for infection. The number of X-Gal-positive cells was determined after 48 h.
Virus opsonization by C3 MAb also inhibited lacZ transduction of W162 reporter cells (Fig. 2B). Briefly, C3 MAb was incubated in phosphate-buffered saline for 1 h at 37°C with 105 CsCl-purified infectious particles (i.e., 105 lacZ-transducing units [TDU]) (25), at which time subconfluent W162 cells were infected at 37°C for 1 h at an MOI of 0.1 TDU/cell. After the cells were washed, fresh culture medium was added, and the number of lacZ-expressing cells was quantified 2 days later by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining. Again, the flanking spacers had a major influence in this assay: whereas lacZ transduction was inhibited between 97 and 99% upon incubation of AdIE43, AdIE44, AdIE45, AdIE46, and AdIE47 with a 10−3 dilution of C3 MAb, that of the other HVR5-modified viruses remained unchanged and paralleled that of virus AE18, a nonmodified “isogenic” ΔE1ΔE3 virus expressing lacZ from the cytomegalovirus promoter (Fig. 1C). C3 MAb binding to the HVR5-modified capsids therefore correlated perfectly with the inclusion of linker sequences on both sides of the inserted epitope (Table 1).
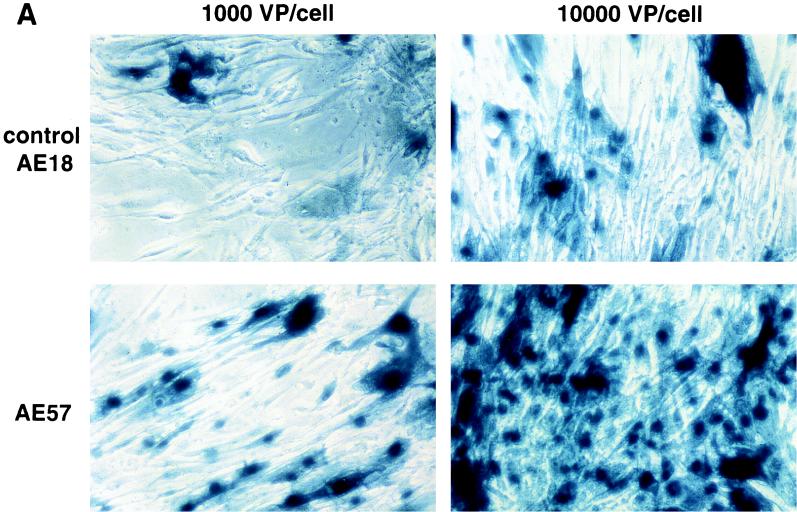
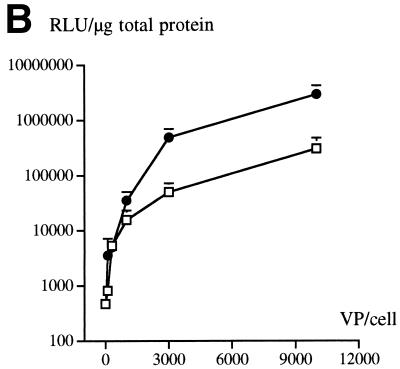
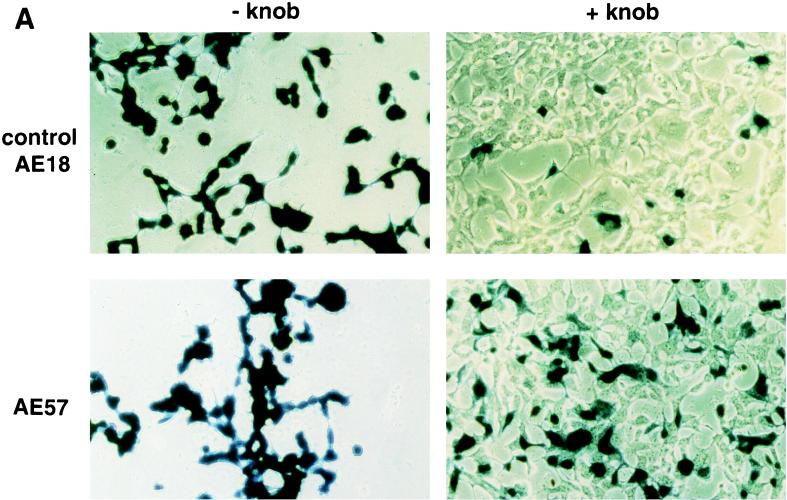
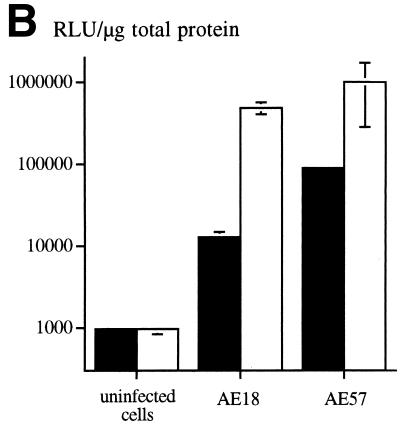
An RGD-containing peptide specific to αv integrins (14) was then flanked by GS linkers (GSDCRGDCFGS) and inserted into HVR5 by recombinational cloning in E. coli (virus AE57 [Table 1]). Again, CsCl-purified batches were prepared with a correct productivity (6,000 VP/cell). We then asked whether capsid inclusion of this targeting ligand would facilitate the infection of human cells naturally refractory to Ad5 infection. Human vascular smooth muscle cells were selected for this assay because they display αv integrins at the cellular surface but have only small numbers of knob primary attachment receptors (24). Interestingly, 30% of the smooth muscle cells stained positive for X-Gal following infection with 10-fold less of the RGD-containing virus (AE57) (1,000 VP/cell) compared to its unmodified counterpart (AE18) (Fig. 3A). In this assay, X-Gal-positive cells were scored in five fields for each condition, i.e., AE18 (MOI 1,000; 14 ± 2 per field), AE18 (MOI 10,000; 113 ± 17), AE57 (MOI 1,000; 90 ± 14), and AE57 (MOI 10,000; 337 ± 107), and correlated with detectable levels of intracellular vector DNA following infection with virus AE57 but not AE18 (results not shown). Transgene expression was also significantly increased when the RGD virus was used in place of virus AE18 (Fig. 3B). We next assessed the behavior of virus AE57 toward cells readily infectable by Ad5. AE57 infection of 293 cells incubated with saturating amounts of competitor knob (i.e., amounts almost completely abolishing lacZ transduction when virus AE18 was used) led to the transduction of a significant proportion of cells (Fig. 4A). In this assay, quantitation of X-Gal-positive cells in the presence of knob was associated with 128 ± 9 positive cells per field with AE57 versus 32 ± 9 for the AE18 control virus (n = 5) and correlated with a significant increase in β-galactosidase specific activity (Fig. 4B). That 293 cells remained amenable to transduction following inhibition of the normal attachment pathway supports a direct recruitment of the inserted RGD peptide during the initial step of infection. Possibly, the inserted RGD peptide also contributed to the endocytic internalization process.
FIG. 3.
RGD inclusion in HVR5 enhances the transduction of cells naturally refractory to Ad5 infection. (A) Human primary aortic smooth muscle cells (Clonetics, San Diego, Calif.) were infected with control AE18 (top) or the RGD-containing virus AE57 (bottom) at an MOI of 1,000 or 10,000 VP/cell as indicated. X-Gal staining was carried out 48 h post-infection. (B) lacZ specific activity following infection with AE18 (□) or AE57 (●) at the indicated MOI. Extracts were prepared 48 h postinfection, at which time total protein and β-galactosidase activity were quantified. Data represent the means and standard deviations of duplicate experiments.
FIG. 4.
The RGD virus can use a fiber knob-independent pathway for infection. (A) 293 cells incubated without (left) or with (right) 100 μg of recombinant Ad5 knob per ml were subsequently exposed for 1 h to virus AE18 (top) or AE57 (bottom) at an MOI of 200 VP/cell. Unbound virus was then removed, fresh medium was added, and the cells were stained with X-Gal after 24 h. (B) Same as for panel A, except that the lacZ-specific activity in the cellular extracts was quantified in the absence (open bars) or presence (solid bars) of recombinant knob. Data represent the means and standard deviations of duplicate experiments.
Taken together, our data demonstrate that the Ad5 hexon monomer can accommodate a foreign peptide within HVR5 with no dramatic effect on virus viability and growth. Conditions can also be found that allow the inserted peptide to recognize its cognate receptor either in a soluble phase or at the cellular surface. In particular, the observations that hexon-modified capsids can be efficiently internalized by cells lacking the virus primary receptors or when the initial attachment step has been artificially abolished should open up new avenues to adapt the vector tropism to particular cellular subsets. Genetic engineering of the hexon monomer may also constitute a useful prerequisite to design truly retargeted vectors following ablation of the native entry pathway.
Acknowledgments
We thank F. Blanche, J. Crouzet, J.-D. Guitton, M. Latta, and J. F. Mayaux (CRVA, Vitry-Sur-Seine) for their support and expertise. J. Douglas, V. Krasnykh, and D. Curiel (Gene Therapy Program, University of Alabama at Birmingham) are acknowledged for sharing unpublished data and material, and R. Crainic (Institut Pasteur, Paris, France) is thanked for providing C3 MAb.
This work initiated under the BioAvenir program was supported by Rhône-Poulenc, the French Ministry of Research, and the French Ministry of Industry.
REFERENCES
- 1.Athapilly F K, Murali R, Rux J J, Cai Z, Burnett R M. The refined crystal structure of hexon, the major coat protein of adenovirus type 2, at 2.9 A resolution. J Mol Biol. 1994;242:430–455. doi: 10.1006/jmbi.1994.1593. [DOI] [PubMed] [Google Scholar]
- 2.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 2a.Blanche, F. Personal communication.
- 3.Clayman G L, el-Naggar A K, Lippman S M, Henderson Y C, Frederick M, Merritt J A, Zumstein L A, Timmons T M, Liu T J, Ginsberg L, Roth J A, Hong W K, Bruso P, Goepfert H. Adenovirus-mediated p53 gene transfer in patients with advanced recurrent head and neck squamous cell carcinoma. J Clin Oncol. 1998;16:2221–2232. doi: 10.1200/JCO.1998.16.6.2221. [DOI] [PubMed] [Google Scholar]
- 4.Crompton J, Toogood C I A, Wallis N, Hay R T. Expression of a foreign epitope on the surface of the adenovirus hexon. J Gen Virol. 1994;75:133–139. doi: 10.1099/0022-1317-75-1-133. [DOI] [PubMed] [Google Scholar]
- 5.Crouzet J, Naudin L, Orsini C, Vigne E, Ferrero L, Le Roux A, Benoit P, Latta M, Torrent C, Denèfle P, Mayaux J-F, Perricaudet M, Yeh P. Recombinational construction in Escherichia coli of infectious adenoviral genomes. Proc Natl Acad Sci USA. 1997;94:1414–1419. doi: 10.1073/pnas.94.4.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Dedieu, J.-F., and E. Vigne. Unpublished data.
- 6.Douglas J T, Rogers B E, Rosenfeld M E, Michael S I, Feng M, Curiel D T. Targeted gene delivery by tropism-modified adenoviral vectors. Nat Biotechnol. 1996;14:1574–1578. doi: 10.1038/nbt1196-1574. [DOI] [PubMed] [Google Scholar]
- 7.Gahéry-Ségard H, Farace F, Godfrin D, Gaston J, Lengagne R, Tursz T, Boulanger P, Guillet J-G. Immune response to recombinant capsid proteins of adenovirus in human: antifiber and anti-penton base antibodies have a synergistic effect on neutralizing activity. J Virol. 1998;72:2388–2397. doi: 10.1128/jvi.72.3.2388-2397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gall J, Kass-Eisler A, Leinwand L, Falck-Pedersen E. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J Virol. 1996;70:2116–2123. doi: 10.1128/jvi.70.4.2116-2123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong J S, Engler J A. Domains required for assembly of adenovirus type 2 fiber trimers. J Virol. 1996;70:7071–7078. doi: 10.1128/jvi.70.10.7071-7078.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong S S, Karayan L, Tournier J, Curiel D T, Boulanger P A. Adenovirus type 5 fiber knob binds to MHC I α2 domain at the surface of human epithelial and B lymphoblastoid cells. EMBO J. 1997;16:2294–2306. doi: 10.1093/emboj/16.9.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krasnykh V, Dmitriev I, Mikheeva G, Ryan Miller C, Belousova N, Curiel D T. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J Virol. 1998;72:1844–1852. doi: 10.1128/jvi.72.3.1844-1852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krasnykh V N, Mikheeva G V, Douglas J T, Curiel D T. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J Virol. 1996;70:6839–6846. doi: 10.1128/jvi.70.10.6839-6846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latta, M. Unpublished results.
- 14.Pasqualini R, Koivunen E, Ruoslahti E. alpha v integrins as receptors for tumor targeting by circulating ligands. Nat Biotechnol. 1998;15:542–546. doi: 10.1038/nbt0697-542. [DOI] [PubMed] [Google Scholar]
- 15.Roelvink P W, Lizonova A, Lee J G M, Li Y, Bergelson J M, Finberg R W, Brough D E, Kovesdi I, Wickham T J. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol. 1998;72:7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogers B E, Douglas J T, Sosnowski B A, Ying W, Pierce G, Buchsbaum D J, Della Manna D, Baird A, Curiel D T. Enhanced in vivo gene delivery to human ovarian cancer xenografts utilizing a tropism-modified adenovirus vector. Tumor Targeting. 1998;3:25–31. [Google Scholar]
- 17.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2111–2148. [Google Scholar]
- 18.Stevenson S C, Rollence M, Marshall-Neff J, McLelland A. Selective targeting of human cells by a chimeric adenovirus vector containing a modified fiber protein. J Virol. 1997;71:4782–4790. doi: 10.1128/jvi.71.6.4782-4790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomko R P, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toogood C I A, Crompton J, Hay R T. Antipeptide antisera define neutralizing epitopes on the adenovirus hexon. J Gen Virol. 1992;73:1429–1435. doi: 10.1099/0022-1317-73-6-1429. [DOI] [PubMed] [Google Scholar]
- 21.Tursz T, Le Cesne A, Baldeyrou P, Gautier E, Opolon P, Schatz C, Pavirani A, Courtney M, Lamy D, Ragot T, Saulnier P, Andremont A, Monier R, Perricaudet M, Le Chevalier T. Phase I study of a recombinant adenovirus-mediated gene transfer in lung cancer patients. J Natl Cancer Inst. 1996;88:1857–1863. doi: 10.1093/jnci/88.24.1857. [DOI] [PubMed] [Google Scholar]
- 22.van der Werf S, Wychowski C, Bruneau P, Blondel B, Crainic R, Horodniceanu F, Girard M. Localization of a poliovirus type 1 neutralization epitope in viral capsid polypeptide VP1. Proc Natl Acad Sci USA. 1983;80:5080–5084. doi: 10.1073/pnas.80.16.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wickham T J, Roelvink P W, Brough D E, Kovesdi I. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat Biotechnol. 1996;14:1570–1573. doi: 10.1038/nbt1196-1570. [DOI] [PubMed] [Google Scholar]
- 24.Wickham T J, Tzeng E, Shears II L L, Roelvink P W, Li Y, Lee G M, Brough D E, Lizonova A, Kovesdi I. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J Virol. 1997;71:8221–8229. doi: 10.1128/jvi.71.11.8221-8229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeh P, Dedieu J F, Orsini C, Vigne E, Denèfle P, Perricaudet M. Efficient dual transcomplementation of adenovirus E1 and E4 regions from a 293-derived cell line expressing a minimal E4 functional unit. J Virol. 1996;70:559–565. doi: 10.1128/jvi.70.1.559-565.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zukowski M M, Gaffney D F, Speck D, Kauffmann M, Findell A, Wisecup A, Lecocq J P. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned pseudomonas gene. Proc Natl Acad Sci USA. 1983;80:1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]