Abstract
The int3 oncogene was discovered as a frequent target in mouse mammary tumor virus-induced mammary tumors and encodes the intracellular domain of a Notch4/int3 protein. In one spontaneous mammary tumor, no. 9, that developed in a BALB/c mouse, we have found an insertion of a 1.2-kb sequence, consisting of a 5′ long terminal repeat and gag sequences of an intracisternal type A particle (IAP) as well as an extra copy of the Notch4/int3 genomic sequences containing exons 23 and 24, into the intron between exons 24 and 25 of the Notch4/int3 gene. In this tumor, unique splicing events between the IAP and the Notch4/int3 sequences generated two types of IAP-Notch4/int3 fusion transcripts encoding two different portions of the intracellular domain of Notch4/int3 proteins: one with a RAM domain and the other without. Interestingly, these two proteins showed different subcellular localizations in a mouse mammary epithelial cell line, HC-11.
Notch family genes encode transmembrane receptor proteins mediating signals which regulate various cell fate decisions that involve cell-cell interactions (1). To date, four members of this family have been identified in the mouse (6, 8, 12, 15, 21, 22). One member, Notch4/int3, was originally discovered as an oncogene involved in mouse mammary tumor virus (MMTV)-induced mammary tumors (6, 7, 18). In those tumors, MMTV provirus integration at a short region of the Notch4/int3 locus leads to expression of a truncated Notch4/int3 transcript of 2.3 kb, which encodes the intracellular part of the Notch4/int3 protein (7, 18). Loss of the extracellular domain causes constitutive activation of the Notch4/int3 protein, which leads to hyperproliferation of glandular epithelial tissues and development of mammary carcinomas (11). In addition, similar truncations of other Notch gene members represent gain-of-function mutations (10) and thus are known to associate with tumorigenesis (2, 5, 8, 19). Notch4/int3 transcripts are primarily restricted to endothelial cells in embryonic and adult life and encode a protein consisting of 1,964 amino acid residues (6, 21). The extracellular domain contains signal peptide, 29 epidermal growth factor (EGF)-like repeats, and 3 Notch/lin-12 repeats. The intracellular domain is characterized by RAM domain (a strong binding site for RBP-Jκ transcription factor) (20), six tandem copies of a cdc10/ankyrin repeat, and a PEST sequence motif.
Murine intracisternal A particles (IAPs) are defective murine retroviruses encoded by a large family of endogenous proviruses which are present at about 1,000 copies per haploid genome of Mus musculus (14, 17). IAPs undergo transpositions and act as endogenous mutagens. IAPs can affect the expression of an adjacent gene by providing various transcriptional (3, 9) or posttranscriptional (16) regulatory elements for the adjacent gene. Kordon et al. have reported rearrangement of Notch4/int3 gene in a spontaneous Czech II mouse mammary tumor, in which an IAP provirus was integrated in the opposite transcriptional orientation relative to the Notch4/int3 gene and a cryptic promoter in this oppositely oriented 5′ long terminal repeat (LTR) drove expression of a IAP-Notch4/int3 chimeric RNA encoding the intracellular part of the Notch4/int3 protein (13).
Here, we describe a new type of IAP-mediated activation of the Notch4/int3 gene in a mammary tumor that spontaneously developed in a BALB/c mouse. In this tumor, the inserted IAP 5′ LTR drove the transcription of the IAP-Notch4/int3 fusion mRNAs. Furthermore, two types of IAP-Notch4/int3 fusion transcripts were generated by different splicing events between IAP and the Notch4/int3 sequences. These two RNA species encoded two forms of the intracellular domain of Notch4/int3 protein, which showed different subcellular localizations in HC-11 cells.
Rearrangement of the Notch4/int3 gene in a mammary tumor spontaneously developed in a BALB/c mouse.
We have analyzed by Southern blotting whether rearrangement of Notch genes occurred in 20 type B mammary adenocarcinomas (4) spontaneously developed in BALB/c mice. DNA fragments encoding the RAM domain of each Notch gene were amplified by PCR as described previously (12) and used as probes for the initial screenings. Although no rearrangement was detected with the Notch-1, -2, and -3 probes, rearrangement of the Notch4/int3 gene was found in a tumor named no. 9 with the Notch4/int3 RAM probe, probe d (Fig. 1A). Thus, the following four Notch4/int3 cDNA fragments were used as probes for detailed analysis by Southern and Northern blottings (Fig. 2A): probe a; a 1.5-kb SpeI-NdeI fragment encoding from the N terminus to the middle region of EGF-like repeats of the Notch4/int3 cDNA: probe b, a 2.7-kb XbaI-XhoI fragment encoding from the 5′ terminus to two-thirds of the EGF-like repeat domain; probe c, a 2.4-kb BamHI fragment encoding from Notch/lin-12 repeats to the C terminus; and probe d, a 0.4-kb fragment encoding a RAM domain amplified by PCR with the sense primer, M2, and the antisense primer, TMCDR (12).
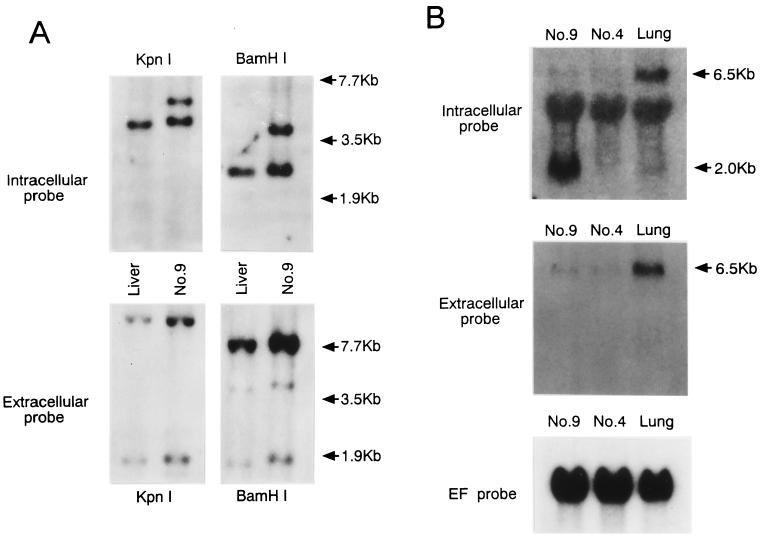
FIG. 1.
Rearrangement of the Notch4/int3 genome leads to marked expression of truncated mRNAs encoding the intracellular domain of Notch4/int3 protein in the no. 9 mammary tumor. (A) Southern blot analysis. Genomic DNAs from the no. 9 mammary tumor and normal BALB/c liver were digested with KpnI or BamHI, Southern blotted, and hybridized as described previously (23) with the intracellular Notch4/int3 probe, probe d (upper panels), or with the extracellular probe, probe b (lower panels). Numbers on the right indicate the migration positions of DNA molecular weight markers. The positions of probes d and b are shown in Fig. 2A. (B) Northern blot analysis. Total RNAs (10 μg) from mammary tumors with (no. 9) or without (no. 4) Notch4/int3 rearrangement, and from normal BALB/c mouse lung were subjected to Northern analyses as described previously (23) with the Notch4/int3 intracellular probe, probe d (upper panel), or with the extracellular probe, probe a (middle panel). The lung RNA was used as a positive control for normal Notch4/int3 mRNA. The 6.5- and 2.0-kb arrows indicate the approximate sizes of the full-length and the truncated Notch4/int3 transcripts, respectively. Probe d weakly hybridized with 28S rRNAs. As a loading control, the blot shown in the upper panel was stripped and rehybridized with the human elongation factor 1α (EF) probe (lower panel).
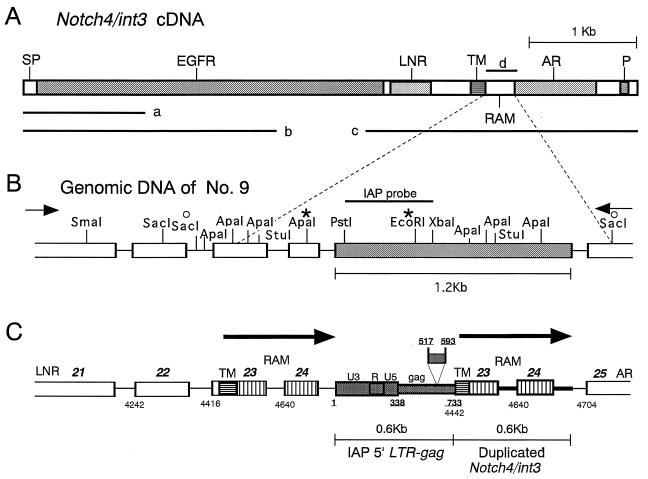
FIG. 2.
Structure of the rearranged Notch4/int3 allele in the no. 9 mammary tumor. (A) Schematic representation of full-length Notch4/int3 cDNA. The major structural elements encoded by Notch4/int3 cDNA are shown. SP, signal peptide; EGFR, EGF-like repeats; LNR, Notch/lin-12 repeats; TM, transmembrane domain; RAM, RAM 23-homologous domain; AR, cdc10/ankyrin repeats; P, a PEST sequence motif. Fragments used for probes are indicated by bars. (B) Restriction map of a rearranged region of Notch4/int3 genome 2.7 kb long. The region of the Notch4/int3 genome containing the rearrangement was amplified by PCR. The right and left arrows indicate the positions of sense (Notch4-sense, which corresponds to nucleotide positions 3881 to 3907 of the Notch4/int3 cDNA sequence) and antisense (TMCDR) primers, respectively. Exons and introns are indicated by open boxes and solid lines, respectively. The shaded box indicates the 1.2-kb DNA sequences inserted. The asterisks indicate the ApaI and EcoRI sites used for construction of the CAT plasmid. The bar indicates the position of the IAP-LTR-gag probe. The two open circles indicate the positions of the fragment whose nucleotide sequence was submitted to the DDBJ database. (C) Schematic representation of a rearranged Notch4/int3 genome in the no. 9 mammary tumor. The relative positions of boxes and lines in panel C correspond to those in panel B. Bold italic numbers above the boxes indicate the exon numbers of the Notch4/int3 gene. Plain and underlined numbers indicate the nucleotide positions of the Notch4/int3 cDNA (6) and a representative IAP provirus (17), respectively. The shaded boxes indicate the IAP 5′ LTR and gag sequences in the insert (a 77-bp deletion in this IAP gag sequence is also shown). The boxes downstream of IAP sequences marked TM, 23, and 24 and the bold lines adjacent to them indicate extra copies of the Notch4/int3 exons and introns, respectively. The boxes hatched horizontally and vertically represent sequences encoding the transmembrane domain and the RAM domain, respectively. The two large arrows indicate the duplicated regions of the Notch4/int3 genome.
Southern blot analyses with the extracellular probe (probe b) failed to reveal Notch4/int3 rearrangement (Fig. 1A, lower panels). However, in addition to the 4.0-kb KpnI and the 2.5-kb BamHI fragments derived from the unaffected Notch4/int3 allele, the intracellular RAM domain probe hybridized with an additional 5.2-kb KpnI and an additional 3.7-kb BamHI fragment in genomic DNA from the no. 9 tumor, indicating that this tumor has a rearrangement in one allele of the Notch4/int3 gene and that the rearrangement occurred close to the RAM domain-encoding sequences (Fig. 1A, upper panels).
Next, we analyzed whether the Notch4/int3 rearrangement affects Notch4/int3 transcripts in the no. 9 mammary tumor. Total RNAs were isolated from the no. 9 tumor, the no. 4 mammary tumor with a normal Notch4/int3 allele, which spontaneously developed in the BALB/c mouse, and normal BALB/c mouse lung. By Northern analyses, the expression of a normal Notch4/int3 RNA species 6.5 kb long was detected in both mammary tumors and lung with the extracellular (Fig. 1B, middle panel) and the intracellular (Fig. 1B, upper panel) Notch4/int3 probes. With the intracellular probe, but not with the extracellular probe, however, a unique Notch4/int3 RNA species of about 2.0 kb was detected only in the no. 9 tumor. Therefore, the rearrangement of the Notch4/int3 allele in the no. 9 tumor activates the expression of RNA species which correspond to the intracellular domain of Notch4/int3.
Structure of the rearranged Notch4/int3 allele in the no. 9 mouse mammary tumor.
To further characterize the Notch4/int3 rearrangement in the no. 9 tumor, we constructed the restriction map of the rearranged Notch4/int3 allele of the no. 9 tumor by Southern blotting with several probes. The restriction map suggested that a 1.2-kb-long DNA sequence of unknown origin is inserted into the RAM domain of the Notch4/int3 genome (Fig. 2B). To specify the rearrangement of the Notch4/int3 genome in the no. 9 tumor, a 2.7-kb Notch4/int3 genomic sequence containing the 1.2-kb DNA insert was amplified by PCR with the genomic DNA from the no. 9 tumor and the set of primers indicated with arrows in Fig. 2B. Nucleotide sequencing revealed that the 1.2-kb DNA insert consists of a part of the IAP sequence and a part of the Notch4/int3 genomic sequence duplicated and that this 1.2-kb sequence is inserted into an intron between exons 24 and 25 of the Notch4/int3 genome (Fig. 2C). The 5′ 0.6 kb of the inserted DNA shows very close homology to the IAP 5′ LTR (bp 1 to 338) and the gag (bp 339 to 733) region of a typical full-length IAP sequence (17), except for a short deletion (bp 517 to 593). The 3′ 0.6 kb of the inserted DNA adjacent to the IAP sequence consists of an extra set of exon 23 (about 3′ 80% of the exon), exon 24 (the whole exon), and the introns adjoining them, which encode the transmembrane and the RAM domains of the Notch4/int3 protein. The IAP 5′ LTR was inserted in the same direction as for Notch4/int3 gene transcription.
To confirm the IAP 5′ LTR-gag sequence inserted in the Notch4/int3 genome functions as a promoter for these truncated Notch4/int3 transcripts, we performed a chloramphenicol acetyltransferase (CAT) assay. As expected, the 0.6-kb ApaI-EcoRI (Fig. 2B) fragment containing the IAP 5′ LTR drove marked expression of the CAT gene in a mammary epithelial cell line, HC-11 (data not shown).
Generation of truncated Notch4/int3 transcripts by IAP-induced splicing events.
To characterize the truncated Notch4/int3 transcripts and to elucidate the structures of the proteins encoded by them, we cloned the cDNAs of the transcripts and sequenced them. As a first step in this analysis, we amplified cDNAs which correspond to the 5′ part of the IAP-Notch4/int3 fusion transcript by reverse transcription-PCR (RT-PCR) using single-stranded cDNAs synthesized from poly(A)+ RNAs from the no. 9 mammary tumor and a sense (IAP-223) primer, and an antisense (TMCDR) primer, derived from the R region of the IAP LTR and the RAM domain of Notch4/int3, respectively. The 0.6-kb cDNA fragment thus amplified was cloned into pBluescript II KS(+). DNA sequence analysis of a clone designated R2 revealed that it has a cDNA sequence derived from the IAP LTR and the Notch4/int3 cDNA encoding the RAM domain (Fig. 3A). These results suggest that the truncated Notch4/int3 transcripts start from the R region of the LTR (nucleotide position 225 of IAP). Surprisingly, R2 transcript was generated by an unique splicing event using a splicing donor in the IAP provirus (nucleotide position 511 of IAP) and a cryptic splicing acceptor in the exon of the Notch4/int3 gene (which corresponds to nucleotide position 4512 of the RAM domain, Fig. 3A, and B). The splicing donor site used in R2 is commonly used to generate subgenomic IAP RNA and is well conserved in several other IAP sequences (14, 17). However, the splicing acceptor at position 4512 of the Notch4/int3 cDNA is located in the middle of exon 23 and is an unusual acceptor.
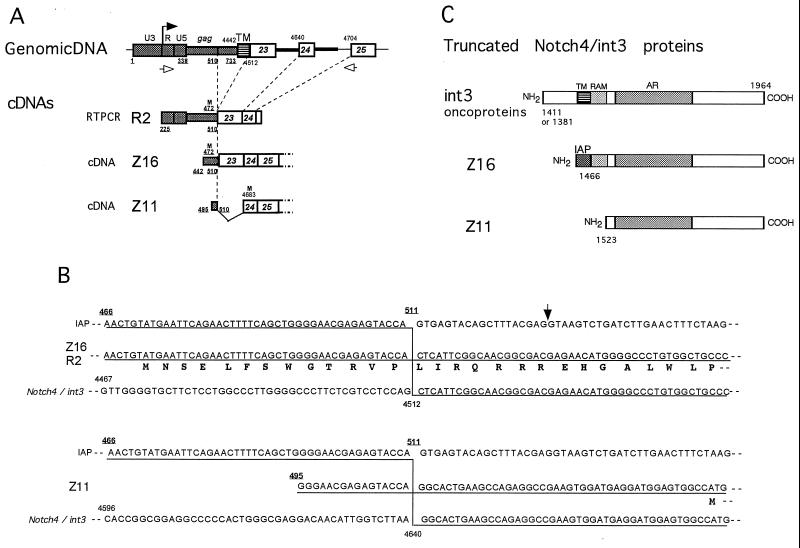
FIG. 3.
Structural analyses of the truncated Notch4/int3 transcripts and their proteins. (A) Schematic representation of the structures of cDNA clones generated from the truncated Notch4/int3 transcripts. The structures of the truncated Notch4/int3 cDNA clones are compared with that of rearranged Notch4/int3 genome (symbols as in Fig. 2C). Only regions of interest are shown. The black arrow on the top indicates the start of transcription. The Z16 and the Z11 cDNA clones were screened from the no. 9 tumor cDNA library, and the R2 clone was obtained by RT-PCR using the sense and antisense primers indicated by the open arrows. The R2 and the Z16 cDNAs lack the transmembrane domain but have the complete RAM domain. Plain and underlined numbers indicate nucleotide positions of the Notch4/int3 cDNA and the IAP nucleotide positions, respectively. An M in the R2, Z16, and Z11 clones indicates potential initiating methionines. Numbers under M indicate nucleotide positions of the ATG codons in the IAP or Notch4/int3 sequences. (B) IAP-induced splicing events generating two types of truncated Notch4/int3 transcripts. (Upper panel) The nucleotide sequence of a selected region of the Z16 or the R2 cDNA clones (middle sequence) is shown, together with that of an unspliced IAP proviral (upper) and normal Notch4/int3 (lower) transcript. Lines under the sequences of IAP and the Notch4/int3 cDNA indicate homology with the Z16 or the R2 cDNA sequence. A vertical line indicates a splicing event generating the Z16 and R2 clones. The deduced amino acid sequence appears below the nucleotide sequence of the Z16 or the R2 cDNA. The ATG codon for a potential initiating methionine is located at nucleotide position 472 in the IAP gag gene. The arrow indicates the position of the previously reported splicing donor site used in the generation of IAP-interleukin 3 or IAP-granulocyte-macrophage colony-stimulating factor fusion transcripts (16). (Lower panel) Splicing event generating the Z11 transcript. Symbols are as described above. M indicates the potential initiating methionine corresponding to nucleotide position 4683 of the Notch4/int3 cDNA. (C) Schematic structures of the int3 oncoprotein and the truncated Notch4/int3 proteins encoded by the Z16 and the Z11 cDNA clones. The 13 amino acids encoded by the IAP sequence are shown as IAP in the Z16-encoded protein. Numbers shown to the lower left of each protein indicate the amino acid residue of the wild-type Notch4/int3 protein, which corresponds to the amino terminus of each truncated Notch4/int3 protein.
To define the complete structure of the truncated Notch4/int3 transcripts and to characterize the mechanism of splicing, the no. 9 mammary tumor cDNA library prepared with the ZAP Express cDNA synthesis kit (Stratagene) was first screened with a 32P-labeled Notch4/int3 intracellular probe (probe d) for the cDNA clones derived from the IAP-Notch4/int3 fusion transcripts. The clones obtained were then screened again with an IAP LTR-gag probe (Fig. 2B), but none of the clones hybridized with this probe strongly. Therefore, clones that hybridized with Notch4/int3 probe were rescued as pBK-CMV plasmids by in vivo excisions, and the cDNA inserts in these plasmids were sequenced. Eventually, two-types of cDNA clones with IAP-derived sequences and a poly(A) tail at their 5′ and 3′ ends, respectively, were obtained. According to ZAP Express, the plasmid clones containing 2.1- and 1.9-kb cDNA insert of IAP-Notch4/int3 fusion sequence were named Z16 and Z11, respectively (Fig. 3A). Sequence analysis revealed that these cDNAs correspond to different transcripts generated by different alternative splicings (Fig. 3A and B). The Z16 clone was generated by the same splicing event that produced the R2 clone. However, the Z11 clone was generated by another type of splicing using nucleotide position 511 of the IAP provirus as a splicing donor and the normal splicing acceptor at the intron-exon 24 boundary of the Notch4/int3 genome as a splicing acceptor (corresponding to nucleotide position 4640). Compared with Z16, Z11 lacks the 3′ part of exon 23 encoding the N-terminal portion of the RAM domain.
Truncated Notch4/int3 transcripts encode the intracellular domain of Notch4/int3 protein.
Sequence analysis revealed that both the Z16 and the Z11 clones can encode the truncated Notch4/int3 proteins (Fig. 3B and C). Using ATG codon at nucleotide position 472 of IAP as an initiation methionine, we found that the Z16 clone encodes a protein in which the N-terminal 13 amino acid residues are derived from the IAP sequence and the following carboxyl-terminal 499 amino acid residues correspond to the intracellular domain of the Notch4/int3 protein. On the other hand, the Z11-encoded protein was shown to consist of 442 carboxyl-terminal amino acids of the Notch4/int3 protein (from ankyrin repeats to the C terminus) by using the ATG codon at nucleotide position 4683 in exon 24 as an initiation methionine. Therefore, the Z16-encoded protein is 70 amino acids larger than the Z11-encoded one. The schematic protein structures of these two truncated Notch4/int3 proteins and the MMTV-induced int3 oncoproteins are compared in Fig. 3C. Two types of int3 oncoprotein so far reported (6) have a short stretch of the extracellular and transmembrane domains of the Notch4/int3 protein. In contrast, both the Z16- and Z11-encoded proteins have no extracellular or transmembrane domains. While both of these proteins contain the intracellular domains from ankyrin repeats to the carboxy terminus, only the Z16 protein has the strong binding site for RBP-Jκ, located from amino acid 1470 to amino acid 1513 in the RAM domain (12, 20).
Subcellular localization of the Z16- and Z11-encoded proteins.
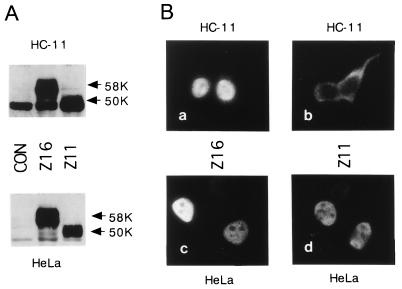
To express the truncated Notch4/int3 proteins in Flag-tagged forms, cDNA inserts of the Z16 and Z11 plasmids were cloned into the EcoRI- and XbaI-cleaved pFlag-CMV (cytomegalovirus)-2 expression vector (Eastman Kodak Co.). For immunoblot and subcellular localization analyses of the truncated Notch4/int3 proteins, HC-11 or HeLa cells were transfected with the Notch4/int3 expression constructs by using the Lipofectamine Plus reagent (Gibco/BRL). Forty-eight hours after transfection, the lysates of transfected cells were prepared and subjected to Western blot analysis (23) with mouse monoclonal anti-Flag-M2 antibody (Eastman Kodak Co.). In both HC-11 and HeLa cells, Flag-tagged proteins with apparent molecular masses of 58 and 50 kDa were expressed, which correspond to the open reading frames in the Z16 and the Z11 constructs, respectively (Fig. 4A). Next, HC-11 or HeLa cells transiently transfected with the Notch4/int3 expression constructs were immunostained with anti-Flag antibody (Fig. 4B). The samples, prepared as described previously (23), were observed with a confocal microscope. Both the Z16- and the Z11-encoded proteins localize exclusively in the nucleus in HeLa cells (Fig. 4B, panels c and d). In HC-11 cells, however, the Z11-encoded protein localized mainly in the cytoplasm, while the Z16-encoded protein localized in the nucleus (Fig. 4B, panels a and b). These differences in subcellular localization may be due to the presence and absence of the RAM domain in the Z16- and the Z11-encoded proteins, respectively, because the IAP-derived 13 amino acids (Fig. 3B) are unlikely to cause these differences. All of the MMTV and IAP proviruses were inserted so that the truncated Notch4/int3 proteins containing the entire intracellular domain, but lacking a portion of the extracellular domain amino terminus to the Notch/lin-12 repeats, can be expressed (6). Therefore, to date, there has been no report of MMTV integration which has led to expression of the truncated Notch4/int3 protein consisting of a region from cdc10/ankyrin repeats to the carboxy terminus. These results suggest that the entire intracellular domain is essential to the function of a Notch4/int3 oncoprotein. Thus, we speculate that the Z16-encoded, but not the Z11-encoded, protein functioned as an oncoprotein. The observation that the entire intracellular domain of the Notch4/int3 protein (Z16) translocates into the nucleus more efficiently than the protein consisting of a region from cdc10/ankyrin repeats to the carboxy terminus (Z11) in mammary epithelial cells is consistent with the idea that the entire intracellular domain is necessary for mammary tumor induction.
FIG. 4.
Subcellular localization of truncated Notch4/int3 proteins in HeLa and HC-11 cells. (A) Western blots showing expression of the Flag-tagged truncated Notch4/int3 proteins encoded by the Z16 or the Z11 clones. Transient expression of the truncated Notch4/int3 proteins in HC-11 cells (upper panel) and HeLa cells (lower panel) is shown. Arrows indicate the molecular weights of (58,000 [58K] and 50,000 [50K]) of these truncated Notch4/int3 proteins. CON indicates that the pFlag-CMV (cytomegalovirus)-2 vector, a negative control plasmid, was transfected. Intense 48-kDa nonspecific bands are present in the blot with HC-11 cell lysate. (B) Immunofluorescence analyses showing the distribution of truncated Notch4/int3 proteins. The Flag-tagged truncated Notch4/int3 proteins encoded by Z16 (a and c) or Z11 (b and d) were overexpressed in HC-11 (a and b) or HeLa (c and d) cells and were visualized with anti-Flag monoclonal antibody M2 and fluorescein isothiocyanate-labeled antimouse immunoglobulin G.
Nucleotide sequence accession number.
The nucleotide sequence of the 2.0-kb SacI genomic fragment in the rearranged Notch4/int3 allele (Fig. 2) can be accessed with accession no. AB016771 of the DDBJ database (DNA Data Bank of Japan). The nucleotide sequences of the regions of interest of the R2, Z16, and the Z11 cDNA clones (Fig. 3A) can be accessed with no. AB016772, AB016773, and AB016774, respectively.
Acknowledgments
We thank Y. Sirayoshi (National Institute of Genetics, Japan), S. Nagata (Osaka University), B. Groner (San Tumorforschungs, Gmbh), and H. Hiai (Kyoto University) for the plasmids containing the Notch4/int3 cDNA, the plasmid containing the human EF-1α cDNA, the HC-11 cell line, and the histological analysis of the mammary tumors, respectively.
This work was supported by a grant-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan to S.Y.
REFERENCES
- 1.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 2.Capobianco A J, Zagouras P, Blaumueller C M, Artavanis-Tsakonas S, Bishop J M. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol Cell Biol. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christy R J, Huang R C C. Functional analysis of the long terminal repeats of intracisternal A-particle genes: sequences within the U3 region determine both the efficiency and direction of promoter activity. Mol Cell Biol. 1988;8:1093–1102. doi: 10.1128/mcb.8.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn T B. Morphology of mammary tumors in mice. In: Homburger F, Fishman W, editors. The physiopathology of cancer. New York, N.Y: Hoeber Co.; 1955. pp. 38–84. [Google Scholar]
- 5.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 6.Gallahan D, Callahan R. The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4) Oncogene. 1997;14:1883–1890. doi: 10.1038/sj.onc.1201035. [DOI] [PubMed] [Google Scholar]
- 7.Gallahan D, Kozak C, Callahan R. A new common integration region (int-3) for mouse mammary tumor virus on mouse chromosome 17. J Virol. 1987;61:218–220. doi: 10.1128/jvi.61.1.218-220.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girard L, Hanna Z, Beaulieu N, Hoemann C D, Simard C, Kozak C A, Jolicoeur P. Frequent provirus insertional mutagenesis of Notch1 in the thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 9.Horowitz M, Luria S, Rechavi G, Givol D. Mechanism of activation of the mouse c-mos oncogene by the LTR of an intracisternal A-particle gene. EMBO J. 1984;3:2937–2941. doi: 10.1002/j.1460-2075.1984.tb02235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 11.Jhappan C, Gallahan D, Stahle C, Chu E, Smith G H, Merlino G, Callahan R. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 1992;6:345–355. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- 12.Kato H, Sakai T, Tamura K, Minoguchi S, Shirayoshi Y, Hamada Y, Tsujimoto Y, Honjo T. Functional conservation of mouse Notch receptor family members. FEBS Lett. 1996;395:221–224. doi: 10.1016/0014-5793(96)01046-0. [DOI] [PubMed] [Google Scholar]
- 13.Kordon E C, Smith G H, Callahan R, Gallahan D. A novel non-mouse mammary tumor virus activation of the int-3 gene in a spontaneous mouse mammary tumor. J Virol. 1995;69:8066–8069. doi: 10.1128/jvi.69.12.8066-8069.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuff E D, Lueders K K. The intracisternal A-particle gene family: structure and functional aspects. Adv Cancer Res. 1988;51:183–276. doi: 10.1016/s0065-230x(08)60223-7. [DOI] [PubMed] [Google Scholar]
- 15.Lardelli M, Dahlstrand J, Lendahl U. The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech Dev. 1994;46:123–136. doi: 10.1016/0925-4773(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 16.Leslie K B, Lee F, Schrader J W. Intracisternal A-type particle-mediated activations of cytokine genes in a murine myelomonocytic leukemia: generation of functional cytokine mRNAs by retroviral splicing events. Mol Cell Biol. 1991;11:5562–5570. doi: 10.1128/mcb.11.11.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mietz J A, Grossman Z, Lueders K K, Kuff E L. Nucleotide sequence of a complete mouse intracisternal A-particle genome: relationship to known aspects of particle assembly and function. J Virol. 1987;61:3020–3029. doi: 10.1128/jvi.61.10.3020-3029.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robbins J, Blondel B J, Gallahan D, Callahan R. Mouse mammary tumor gene int-3: a member of the notch gene family transforms mammary epithelial cells. J Virol. 1992;66:2594–2599. doi: 10.1128/jvi.66.4.2594-2599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohn J L, Lauring A S, Linenberger M L, Overbaugh J. Transduction of Notch2 in feline leukemia virus-induced thymic lymphoma. J Virol. 1996;70:8071–8080. doi: 10.1128/jvi.70.11.8071-8080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-Jk/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 21.Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Notch4/int3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122:199–205. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 22.Weinmaster G, Roberts V J, Lemke G A. Notch 2: a second mammalian Notch gene. Development. 1992;116:2251–2259. doi: 10.1242/dev.116.4.931. [DOI] [PubMed] [Google Scholar]
- 23.Yanagawa S, Lee J-S, Haruna T, Oda H, Uemura T, Takeichi M, Ishimoto A. Accumulation of Armadillo induced by Wingless, Dishevelled and dominant negative Zeste-white 3 leads to elevated DE-cadherin in Drosophila wing disc cells. J Biol Chem. 1997;272:25243–25251. doi: 10.1074/jbc.272.40.25243. [DOI] [PubMed] [Google Scholar]