Abstract
The live-attenuated respiratory syncytial virus vaccine candidate cpts530/1009 was previously shown to contain two separate amino acid changes in the L protein, mutations 530 and 1009 (Phe-521→Leu and Met-1169→Val, respectively, according to the amino acid sequence of the L protein). Each mutation independently specifies temperature-sensitive (ts) and attenuation phenotypes. In this study, we examined the effects of these mutations on transcription and RNA replication, using complete infectious recombinant virus as well as a plasmid-based minireplicon system, the latter under conditions in which effects on replication and transcription are uncoupled. In comparison with recombinant wild-type virus, the 530 and 1009 viruses were partially restricted at 37°C for RNA replication, mRNA synthesis, and virus growth. The 1009 virus was partially restricted for RNA synthesis and virus growth even at 32°C, which suggested that the 1009 mutation has a non-ts component in addition to the ts component. Interestingly, the synthesis of polycistronic readthrough mRNAs was elevated 1.6- to 3.8-fold for the 1009 virus, and this defect was non-ts. Studies with the minigenome system showed that the 530 and 1009 mutations each directly affect both replication and transcription, that the effect on replication was marginally greater than on transcription for the 530 mutation, and that the increase in readthrough mRNA associated with the 1009 mutation also was observed with the minigenome system.
Human respiratory syncytial virus (RSV), a pneumovirus of the family Paramyxoviridae of the nonsegmented negative-strand RNA viruses, is one of the leading causes of serious viral bronchiolitis and pneumonia in young children (3). Unfortunately, an RSV vaccine is not yet available. To meet this need, we have been developing live-attenuated mutants of subgroup A RSV strain A2 as vaccine candidates (5–7, 9). Two such vaccine candidates are the cold-passaged (cp) temperature-sensitive (ts) viruses cpts530 and cpts530/1009. The parent for these viruses was cpRSV, a cp virus made in the 1960s which contains five amino acid substitutions that confer attenuation in chimpanzees but do not confer significant growth restriction or temperature sensitivity in cell culture (11, 12, 16).
The cpts530 virus was derived from cpRSV by chemical mutagenesis (7). Compared to cpRSV, cpts530 had acquired the ts phenotype and exhibited an increased level of attenuation in vivo (7). Sequence analysis showed that it had sustained one additional mutation in the L protein at amino acid 521, namely, a phenylalanine-to-leucine substitution (15). The cpts530/1009 virus was derived from cpts530 by a second round of chemical mutagenesis and was found to be more ts and attenuated than its cpts530 parent (8). cpts530/1009 sustained one additional mutation in the L protein, namely, a methionine-to-valine substitution at amino acid position 1169 (14). The two mutations were introduced separately into infectious recombinant RSV (rRSV), yielding r-530 and r-1009. Genetic and phenotypic analysis confirmed that each mutation independently conferred the ts and attenuation phenotypes, although the 1009 mutation was somewhat less ts than was the 530 mutation. The introduction of both mutations together with the five cp mutations resulted in r-cp530/1009, which was phenotypically indistinguishable from its biologically derived version, cpts530/1009. It was also shown that these two mutations were additive with regard to attenuation in the upper respiratory tract of mice, while in the lower respiratory tract the 1009 mutation was the main contributor to the attenuation phenotype of cpts530/1009 (14).
Multicycle growth of rRSVs.
The ts phenotype of rRSV bearing the 530 or 1009 mutation was previously characterized by determining the efficiency of plaque formation at various temperatures from 32 to 40°C (14, 15). This analysis showed that the shutoff temperature (the lowest restrictive temperature at which there is a 100-fold or greater reduction in the efficiency of plaque formation compared to 32°C) of recombinant virus bearing the 1009 or 530 mutation was 39°C. At this temperature, recombinant virus bearing the 530 mutation was 800-fold more restricted in plaque formation than one bearing the 1009 mutation, indicating that the 530 mutation specifies a slightly greater degree of temperature sensitivity than does the 1009 mutation (14). The shutoff temperature for r-cp530/1009 was 37°C, at which there was a 6,000-fold reduction in plaque formation, compared to only 8- to 10-fold for recombinants bearing the 530 or 1009 mutation (14).
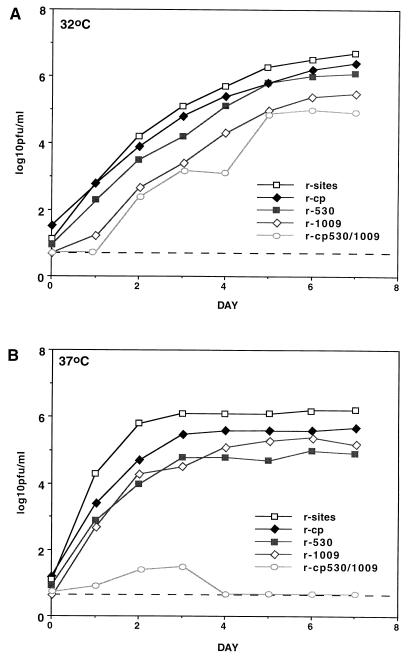
In the present study, the following recombinant viruses were compared: r-sites, which is a wild-type (wt) recombinant virus identical to that described previously (2) except that six translationally silent restriction enzyme sites have been introduced into the large polymerase (L) gene, which also are present in all of the other recombinant viruses described here (15, 21); r-530 or r-1009, containing the 530 or 1009 amino acid substitution; r-cp, which contains the five amino acid substitutions which specify the attenuation phenotype of cpRSV; and r-cp530/1009, which contains the five cp changes as well as the 530 and 1009 amino acid substitutions (14, 15). The effect of the 530 and 1009 mutations on multicycle virus replication at 32 or 37°C was examined first since these are the temperatures at which the effects of the mutations on genome replication and transcription were carried out. Monolayers of HEp-2 cells were infected at a multiplicity of infection (MOI) of 0.01 and incubated at 32 or 37°C; medium samples were harvested at 24-h intervals, and virus was quantitated by plaque assay.
r-cp was only marginally restricted at both 32 and 37°C compared to r-sites (0.2 and 0.5 log10 reductions, respectively, on day 7 [Fig. 1]), consistent with previous results that showed that it is not a ts virus (21). In contrast, r-cp530/1009 was highly restricted in growth at 37°C compared to r-sites (>6.0 log10 reduction on day 7), as expected. r-cp530/1009 also exhibited reduced growth at 32°C (1.7 log10 reduction), which was not previously appreciated. Compared to r-sites, r-530 was clearly restricted at 37°C but only marginally restricted at 32°C (1.3 and 0.5 log10 reductions, respectively). In these comparisons, differences on the order of 0.5 log10 or less, although reproducible, were considered to be marginal, and differences on the order of 1.0 log10 or more were considered to be clear. In contrast to r-530, r-1009 was clearly restricted at both 32 and 37°C (1.1 and 1.0 log10 reductions, respectively). The finding that r-1009 was clearly restricted at 32°C, which was not previously appreciated, suggested that its reduced replication involves a non-ts component in addition to the previously described ts component. The restriction of replication of r-530 at 37°C was equivalent to, or slightly greater than, that of r-1009 (1.3 versus 1.0 log10 reduction). The finding that the 1009 mutation has a non-ts component would provide an explanation for previous biological findings, namely, that a recombinant bearing the 1009 mutation, although less ts than one bearing the 530 mutation, is more attenuated in the lower respiratory tract of mice (14).
FIG. 1.
Kinetics of growth of infectious, recombinant viruses bearing the cp, 530, and 1009 mutations alone and in combination. Triplicate monolayers of HEp-2 cells were infected with the indicated virus at an MOI of 0.01 at 32°C (A) or 37°C (B). Culture fluids were harvested at 24-h intervals for 7 days. Samples were analyzed by plaque assay in duplicate, using an immunoperoxidase staining procedure as previously described (17). Standard errors (n = 6) from days 0 to 7 at both temperatures for all viruses ranged from 0.0 to 0.18. Dashed line indicates the minimum limit of detection (0.7 log10 PFU/ml).
Levels of transcription and replication of rRSVs.
Since both the 530 and 1009 mutations are in the L gene, the growth restriction that they confer probably would be due to effects on replication and/or transcription. To evaluate this, monolayers of HEp-2 cells were infected with r-sites, r-530, or r-1009 at an MOI of 4 and incubated at 32 or 37°C. Total RNA was harvested 24 h postinfection and analyzed by Northern blot hybridization with double-stranded cDNA probes specific for the NS1, nucleocapsid (N), or fusion (F) gene of RSV. Hybridization was quantified by phosphorimager analysis (Table 1). At 32°C, RNA replication and transcription by r-530 was equivalent to, or somewhat greater than, that of the r-sites wt control. In contrast, both processes were reduced for r-1009. This result is consistent with the contribution of a non-ts component to the attenuation phenotype of r-1009 but not r-530. Normalized against r-sites, levels of RNA replication and transcription by r-1009 at 37°C were on average 48 and 63%, respectively, of those at 32°C, showing that a contribution by a ts component was evident at the higher temperature. In comparison, levels of RNA replication and transcription by r-530 (normalized against r-sites) at 37°C were 29 and 37%, respectively, of those at 32°C, which is consistent with the level of temperature sensitivity of r-530 being somewhat greater than that of r-1009. These data illustrate a significant restriction of RNA synthesis at this intermediate temperature, which is fully 2°C below the shutoff temperature of r-530 and r-1009.
TABLE 1.
Percent RNA replication (synthesis of genome and antigenome combined) or transcription (synthesis of the indicated mRNA) at 32 or 37°C by r-530 or r-1009 compared to wt r-sitesa
| Probe | Process measured | Temp (°C) of incu-bation | No. of independent expts | Mean % RNA synthesis compared to wt for the indicated rRSV
|
|
|---|---|---|---|---|---|
| r-530 | r-1009 | ||||
| NS1 | Replication | 32 | 2 | 114.3 ± 23 | 24.8 ± 13.2 |
| 37 | 2 | 34.5 ± 5.5 | 5.6 ± 1.2 | ||
| Transcription | 32 | 2 | 138.5 ± 8.5 | 47.6 ± 7.6 | |
| 37 | 2 | 47.1 ± .05 | 30 ± 5.4 | ||
| N | Replication | 32 | 3 | 114 ± 22 | 10.2 ± 1.2 |
| 37 | 3 | 42 ± 7.2 | 9.2 ± 1.5 | ||
| Transcription | 32 | 3 | 130 ± 0.5 | 25.7 ± 8.2 | |
| 37 | 3 | 57 ± 9.9 | 27.7 ± 2.6 | ||
| F | Replication | 32 | 3 | 162.2 ± 21 | 49.3 ± 17.2 |
| 37 | 3 | 31.1 ± 4.6 | 14.7 ± 4.4 | ||
| Transcription | 32 | 3 | 156 ± 6.2 | 46.6 ± 2.2 | |
| 37 | 3 | 52 ± 7.7 | 30.7 ± 1.3 | ||
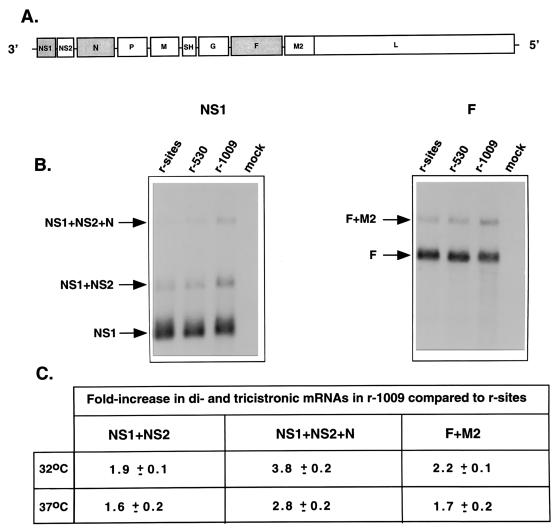
Total intracellular RNA was isolated from HEp-2 cells infected with wt r-sites or r-530 or r-1009 and analyzed by Northern blot hybridization with the indicated double-stranded cDNA probe (Fig. 2A). RNA replication or transcription was measured by phosphorimagery of the amount of probe hybridized to the genome and antigenome band (RNA replication) or the appropriate mRNA band (transcription).
In addition to the effects on the quantity of RNA replication and transcription by the rRSVs described above, we noticed a small but reproducible increase in the level of polycistronic readthrough mRNAs produced by r-1009 (Fig. 2). The ratios between mono- and polycistronic mRNAs were quantitated by phosphorimagery in three independent experiments performed at 32 and 37°C. Results for the NS1 and F probes are illustrated in Fig. 2B and summarized in Fig. 2C. A 1.6- to 3.8-fold increase in readthrough mRNAs was found with r-1009 compared to wt r-sites, and this increase was not ts under these circumstances (Fig. 2C). The magnitude of the increase in readthrough mRNA was dependent on the specific gene junction involved and was consistent between experiments.
FIG. 2.
(A) RSV gene map, showing the relative locations of the NS1, N, and F probes. (B) Northern blot analysis of intracellular RSV mRNAs with the NS1 and F probes. HEp-2 cells were infected with r-sites, r-530, or r-1009 at an MOI of 4 or were mock infected, and the cultures were incubated at 32 or 37°C. At 24 h postinfection, total RNA was harvested by using TRIzol (Life Technologies) and was purified with additional phenol-chloroform extraction and ethanol precipitation. Total RNA of each preparation was electrophoresed in 1.5% agarose gel containing 0.44 M formaldehyde, transferred to nitrocellulose, fixed by UV cross-linking, and hybridized to a cDNA probe of the NS1 or F gene. For the sake of brevity, blots are shown for the 37°C samples but not those of 32°C nor for the N probe. (C) Quantitation of the amount of readthrough mRNA detected at 32 and 37°C with probes against the NS1 and F genes. The N probe yielded similar results.
Effects of the 530 and 1009 mutations on reconstituted RNA replication and transcription by minigenome C75.
Analysis of infectious mutant rRSV as described above does not distinguish clearly between effects of the mutations on RNA replication versus transcription. This is because for infectious virus, the two processes are interdependent: any diminution in transcription reduces the supply of helper proteins, which in turn reduces RNA replication, and any reduction in RNA replication reduces the amount of template, which in turn reduces transcription. However, the two processes can be dissociated in a minigenome system using a mutant plasmid-supplied negative-sense minigenome, C75 (Fig. 3A; see the Figure legend for a detailed description). Minigenome C75 encodes two small chloramphenicol acetyltransferase (CAT) mRNAs and contains a C→G (negative-sense) substitution at the penultimate nucleotide in the trailer region. This mutation has no apparent effect on encapsidation or on the synthesis of positive-sense RNA, but synthesis of progeny minigenome is inhibited very strongly, presumably because the antigenome promoter has been inactivated (10, 17a). Therefore, the only intracellular minigenome would be that synthesized directly from the plasmid. Because the minigenome template and support proteins are supplied in trans from transfected plasmids and are independent of minigenome transcription and replication, the two processes are no longer interdependent and each process can be manipulated without indirect effects on the other.
FIG. 3.
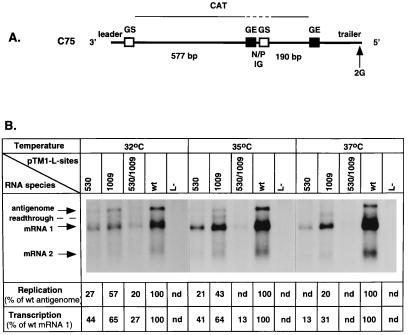
Effects of the 530 and 1009 mutations on reconstituted RNA replication and transcription by minigenome C75. (A) Diagram of the cDNA-encoded, negative-sense minigenome C75. This minigenome, like the previously described C2 minigenome (2) from which it was derived, contains a negative-sense copy of the 660-nucleotide CAT gene, which is flanked on the upstream end by the 3′-extragenic leader region and part of the upstream nontranslated region of the nonstructural protein (NS)1 gene including the gene start (GS) transcription signal; the CAT sequence is flanked on the downstream end by the downstream nontranslated region of the L gene including the transcription GE signal and the 5′ extragenic trailer region. C75 differs from C2 in two features. First, the CAT gene was broken into two transcriptional units by the introduction, into a unique NcoI site, of a sequence cassette containing the junction between the N and P genes of strain A2. This gene junction consists of the N gene GE signal, the single-nucleotide N-P intergenic region (N/P IG), and the P gene GS signal. The resulting minigenome encodes two polyadenylated mRNAs: an upstream one, mRNA 1, of 577 nucleotides exclusive of polyadenylate and a downstream one, mRNA 2, of 190 nucleotides. Second, the penultimate nucleotide position from the 5′ end contains a C→G transition (negative sense) which restricts the minigenome to the synthesis of positive-sense RNA (see the text). (B) Northern blot analysis of intracellular RNAs encoded by minigenome C75. Transfections were performed as described previously (4, 10). HEp-2 cells in six-well dishes were infected with a vaccinia virus-T7 recombinant at an MOI of 5 and transfected at the same time with 0.4 μg of C75 plasmid along with support plasmids pTM1-N (0.4 μg), pTM1-P (0.3 μg), pTM1-M2(ORF1) (0.1 μg), and empty pTM1 vector (0.1 μg) instead of pTM1-L as a polymerase-minus control (lanes L-) or the same N, P, and M2-1 plasmids together with 0.1 μg of the indicated pTM1-L wt or mutant plasmid. Transfections and incubations were performed at the indicated temperature; total intracellular RNA was harvested at 72 h (32°C), 60 h (35°C), or 48 h (37°C) posttransfection and analyzed by Northern blot hybridization with negative-sense CAT riboprobe. nd = not detectable.
RNA replication (synthesis of antigenome) and transcription (synthesis of mRNA) by the C75 minigenome supported by the N, phosphoprotein (P), M2-1, and L proteins was examined at 32, 35, and 37°C (Fig. 3). Total intracellular RNA was analyzed by Northern blot hybridization, in this case using strand-specific negative-sense CAT riboprobe. The C75-encoded RNAs included the miniantigenome, mRNAs 1 and 2, and a small amount of mRNA 1-mRNA 2 readthrough mRNA (Fig. 3B). Unexpectedly, the synthesis of these RNAs by the reconstituted RSV system using wt support plasmids was found to be ts, with almost no products detectable at 39°C (data not shown). Therefore, 37°C was selected as the highest restrictive temperature for analysis, and even at this temperature the level of RNA synthesized was at least 10-fold lower than that synthesized at 32°C (data not shown). A human parainfluenza virus 3 minigenome system which used the same cell line and vaccinia virus-T7 recombinant virus was not ts, indicating that an RSV-specific component was responsible for the unexpected ts effect (19). But the ts effect did not appear to be due to a ts support protein, since these particular cDNAs also had been used to reconstitute complete recombinant virus which was not ts. Because of the inherent ts nature of the reconstituted RSV system, comparisons were made between mutants versus wt at a given temperature but not between temperatures.
The 530 and 1009 mutations were analyzed by their incorporation, singly and in combination, into the L support plasmid, which was then examined in transfections in place of the wt L support plasmid. Total cellular RNA was analyzed by Northern blot hybridization with the negative-sense riboprobe (Fig. 3B). The ts phenotypes of the 530 and 1009 mutations appeared to be exaggerated in the reconstituted system, as had been described above for the wt reconstituted system. For example, minigenome-templated RNA synthesis supported by the 530-L plasmid appeared to be more ts at each of the three temperatures compared to that supported by 1009-L and was inhibited even at 32°C, a temperature which was not restrictive for growth or RNA synthesis of r-530 (Fig. 1; Table 1). In addition, minigenome-templated RNA synthesis supported by the double mutant 530/1009-L was strongly inhibited at 32°C, whereas growth and RNA synthesis by the r-cp530/1009 virus were only moderately inhibited at this temperature. The exaggerated temperature sensitivity of the reconstituted system would account for the finding that the 530 mutation, which is the more ts of the two mutations, was more inhibitory than the 1009 mutation in the reconstituted system at 32°C, whereas in virus the 1009 mutation, which has the non-ts component, was more inhibitory at 32°C. At 35 and 37°C, both RNA replication and transcription were affected by the 530 and 1009 mutations alone and in combination. The effect seemed to be somewhat greater for replication than for transcription, especially for the 530 mutant. For example, antigenome synthesis by the 530 and 1009 polymerases was barely detectable at 35°C and was not detectable at 37°C. However, phosphorimager analysis indicated that the magnitude of the difference between the effect on transcription and replication was not great: approximately twofold for the 530 mutant and less than twofold for the 1009 mutant. Finally, the increase in levels of readthrough mRNA observed with the r-1009 virus also was observed for the reconstituted minigenome system (Fig. 3B) and was independent of temperature.
In conclusion, the purpose of this work was to provide a further characterization of two mutations, called 530 and 1009, which specify most of the attenuation phenotype of the live-attenuated cpts530/1009 vaccine candidate and are currently being combined with other attenuating mutations to produce new recombinant live-attenuated vaccines for RSV subgroup A (1, 5, 9, 14, 15, 20, 21). The 530 mutation, a Phe-to-Leu substitution at amino acid position 521 in the L protein, is a replacement of a nonpolar aromatic with a nonpolar aliphatic amino acid at a position which is well conserved (with >50% identity) among rhabdoviruses and paramyxoviruses and falls between the first and second conserved regions of the L protein. The 530 mutation is a ts attenuating mutation which affects both transcription and RNA replication, with the effect on replication being somewhat greater. The use of the minigenome system confirmed that both processes were directly affected. The 1009 mutation, a Met-to-Val substitution at amino acid position 1169 in the L protein, is a conservative substitution and occurs downstream of the major conserved domains in the L protein. The 1009 mutation was found to be somewhat less ts than 530, as has been observed previously (14), and contains a non-ts component which was observed in virus infections and the reconstituted minigenome system. The ts and non-ts components of the 1009 mutation resulted in decreased RNA replication and transcription. The non-ts restriction was associated with a small (1.6- to 3.8-fold) but reproducible increase in the accumulation of readthrough transcripts. An increase in the frequency of readthrough of the gene end (GE) signals would be expected to result in a reduction in the amount of monocistronic mRNAs and encoded proteins for upstream genes and a relative increase for downstream genes. This latter effect would be expected because increased readthrough would be expected to deliver more polymerase to downstream genes, making the gradient of transcriptional polarity less steep. This effect would not be large, given the relative low abundance of the readthrough mRNAs. It is possible that this readthrough effect is sufficient to account for the reduction in RNA synthesis observed at 32°C for r-1009. Alternatively, the non-ts component of the 1009 mutation might also involve some other aspect of polymerase function, and the increase in transcriptional readthrough might be an incidental effect. This possibility is suggested by the finding that both RNA replication and transcription by the 1009 polymerase, rather than transcription alone, were affected at 32°C in the minigenome system. However, since ts effects appeared to be exaggerated in the minigenome system and were prominent even at 32°C, it is difficult to reliably assess the non-ts component of the 1009 polymerase. Finally, it was recently shown that the M2-1 protein mediates antitermination at GE signals (9a, 13), and it is interesting to find in the present study that antitermination also can be enhanced by a single amino acid change in the L protein.
Acknowledgments
We acknowledge Rachel Fearns and Michael N. Teng for their intellectual input and Jennifer M. Biggs, Myron Hill, and Ena Camargo for technical help.
REFERENCES
- 1.Bukreyev A, Whitehead S S, Murphy B R, Collins P L. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J Virol. 1997;71:8973–8982. doi: 10.1128/jvi.71.12.8973-8982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins P L, McIntosh K, Chanock R M. Respiratory syncytial virus. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1313–1352. [Google Scholar]
- 4.Collins P L, Mink M A, Hill M G, Camargo E, Grosfeld H, Stec D S. Rescue of a 7502-nucleotide (49.3% of full-length) synthetic analog of respiratory syncytial virus genomic RNA. Virology. 1993;195:252–256. doi: 10.1006/viro.1993.1368. [DOI] [PubMed] [Google Scholar]
- 5.Crowe J E., Jr Current approaches to the development of vaccines against disease caused by respiratory syncytial virus (RSV) and parainfluenza virus (PIV). A meeting report of the WHO Programme for Vaccine Development. Vaccine. 1995;13:415–421. doi: 10.1016/0264-410x(95)98266-d. [DOI] [PubMed] [Google Scholar]
- 6.Crowe J E, Jr, Bui P T, Davis A R, Chanock R M, Murphy B R. A further attenuated derivative of a cold-passaged temperature-sensitive mutant of human respiratory syncytial virus retains immunogenicity and protective efficacy against wild-type challenge in seronegative chimpanzees. Vaccine. 1994;12:783–790. doi: 10.1016/0264-410x(94)90286-0. [DOI] [PubMed] [Google Scholar]
- 7.Crowe J E, Jr, Bui P T, London W T, Davis A R, Hung P P, Chanock R M, Murphy B R. Satisfactorily attenuated and protective mutants derived from a partially attenuated cold-passaged respiratory syncytial virus mutant by introduction of additional attenuating mutations during chemical mutagenesis. Vaccine. 1994;12:691–699. doi: 10.1016/0264-410x(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 8.Crowe J E, Jr, Bui P T, Siber G R, Elkins W R, Chanock R M, Murphy B R. Cold-passaged, temperature-sensitive mutants of human respiratory syncytial virus (RSV) are highly attenuated, immunogenic, and protective in seronegative chimpanzees, even when RSV antibodies are infused shortly before immunization. Vaccine. 1995;13:847–855. doi: 10.1016/0264-410x(94)00074-w. [DOI] [PubMed] [Google Scholar]
- 9.Crowe J E, Jr, Collins P L, Chanock R M, Murphy B R. Vaccines against respiratory syncytial virus and parainfluenza virus type 3. In: Levine M M, Woodrow G C, Kaper J B, Cobon G S, editors. New generation vaccines. 2nd ed. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 711–725. [Google Scholar]
- 9a.Fearns, R., and P. L. Collins. Unpublished data.
- 10.Fearns R, Peeples M E, Collins P L. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology. 1997;236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 11.Firestone C Y, Whitehead S S, Collins P L, Murphy B R, Crowe J E., Jr Nucleotide sequence analysis of the respiratory syncytial virus subgroup A cold-passaged (cp) temperature sensitive (ts) cpts-248/404 live attenuated virus vaccine candidate. Virology. 1996;225:419–422. doi: 10.1006/viro.1996.0618. [DOI] [PubMed] [Google Scholar]
- 12.Friedewald W T, Forsyth B R, Smith C B, Gharpure M A, Chanock R M. Low-temperature-grown RS virus in adult volunteers. JAMA. 1968;203:690–694. [PubMed] [Google Scholar]
- 13.Hardy R W, Wertz G W. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J Virol. 1998;72:520–526. doi: 10.1128/jvi.72.1.520-526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juhasz K, Whitehead S S, Boulanger C A, Firestone C-Y, Collins P L, Murphy B R. The two amino acid substitutions in the L protein of cpts530/1009, a live-attenuated respiratory syncytial virus candidate vaccine, are independent temperature-sensitive and attenuation mutations. Vaccine. 1999;17:1416–1424. doi: 10.1016/s0264-410x(98)00381-8. [DOI] [PubMed] [Google Scholar]
- 15.Juhasz K, Whitehead S S, Bui P T, Biggs J M, Boulanger C A, Collins P L, Murphy B R. The temperature-sensitive (ts) phenotype of a cold-passaged (cp) live attenuated respiratory syncytial virus vaccine candidate, designated cpts530, results from a single amino acid substitution in the L protein. J Virol. 1997;71:5814–5819. doi: 10.1128/jvi.71.8.5814-5819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H W, Arrobio J O, Pyles G, Brandt C D, Camargo E, Chanock R M, Parrott R H. Clinical and immunological response of infants and children to administration of low-temperature adapted respiratory syncytial virus. Pediatrics. 1971;48:745–755. [PubMed] [Google Scholar]
- 17.Murphy B R, Sotnikov A V, Lawrence L A, Banks S M, Prince G A. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3-6 months after immunization. Vaccine. 1990;8:497–502. doi: 10.1016/0264-410x(90)90253-i. [DOI] [PubMed] [Google Scholar]
- 18.Peeples, M. E., and P. L. Collins. Unpublished data.
- 19.Skiadopoulos, M. Unpublished data.
- 20.Whitehead S S, Firestone C Y, Collins P L, Murphy B R. A single nucleotide substitution in the transcription start signal of the M2 gene of respiratory syncytial virus vaccine candidate cpts248/404 is the major determinant of the temperature-sensitive and attenuation phenotypes. Virology. 1998;247:232–239. doi: 10.1006/viro.1998.9248. [DOI] [PubMed] [Google Scholar]
- 21.Whitehead S S, Juhasz K, Firestone C Y, Collins P L, Murphy B R. Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J Virol. 1998;72:4467–4471. doi: 10.1128/jvi.72.5.4467-4471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]