Abstract
Histone modifications are key factors in chromatin packaging, and are responsible for gene regulation during cell fate determination and development. Abnormal alterations in histone modifications potentially affect the stability of the genome and disrupt gene expression patterns, leading to many diseases, including cancer. In recent years, mounting evidence has shown that various histone modifications altered by aberrantly expressed modifier enzymes contribute to tumor development and metastasis through the induction of epigenetic, transcriptional, and phenotypic changes. In this review, we will discuss the existing histone modifications, both well-studied and rare ones, and their roles in solid tumors and hematopoietic cancers, to identify the molecular pathways involved and investigate targeted therapeutic drugs to reorganize the chromatin and enhance cancer treatment efficiency. Finally, clinical inhibitors of histone modifications are summarized to better understand the developmental stage of cancer therapy in using these drugs to inhibit the histone modification enzymes.
Keywords: Histone modification, Tumorigenesis, Histone modifier enzyme inhibitor
1. Introduction
Cancer is a complex disease, which has rising prominently to become one of the largest killers of human health together with cardiovascular diseases1. The incidence and mortality of cancer are rapidly growing in many countries, indicating that cancer is still a life-threatening disease worldwide. According to the International Agency for Research on Cancer (IARC), approximately 19.3 million new cancer cases and almost 10.0 million cancer deaths occurred in 20201. Incidence and mortality in 2040 are estimated as 30.2 million and 16.3 million, respectively (based on https://gco.iarc.fr/tomorrow/en). Thus, it is vital for scientists to investigate the regulatory mechanisms of gene expression to develop new strategies and effective drugs for cancer therapy.
Cancer is the first human disease to be correlated with epigenetic alterations2. The term ‘epigenetics’ was first defined by Conard Waddington in 19423. It is increasingly clear that epigenetics plays a key role in tumor development and metastasis through the regulation of gene expression by histone modifications, DNA methylation, histone variant incorporation, chromatin remodeling, and non-coding RNAs4. Histone modifications, established and removed by modifier enzymes called writers and erasers, respectively, alter chromatin structure and physical properties to control gene expression, including tumor suppressor genes (TSGs) and oncogenes. Evidence has revealed that histone modifications participate in almost all the DNA-based processes, such as gene transcription and replication and DNA damage repair and recombination5. Mutations within histones or chromatin remodeling complexes affect the cell phenotype, leading to various diseases, including cancer. For example, mutations within the SWI/SNF complex, which contains 15 subunits encoded by 29 genes, affect > 20% of human cancers across many tumor types6. Histone mutations within H3K27M, H3K36M, and H4G34V/R/W/L usually occur in pediatric cancers7. In particular, aberrant expression of histone modifier enzymes is associated with disruption of the histone modification machinery, leading to cancer initiation, progression, and metastasis. Epigenetic-based drugs (epidrugs) have been investigated, and several have been approved by the US Food and Drug Administration (FDA) to treat cancer with abnormal histone modifications8. However, the majority of epidrugs are still in the pre- or clinical phase, indicating that it is necessary to clarify the regulatory pattern of epigenetics, especially histone modifications, in cancer.
A wealth of evidence supports a close relationship between misregulated histone modifications and cancer. In this review, we will summarize known histone modifications, to provide a clearer view of the whole picture. In addition, gene regulation by abnormal histone modifications in solid tumors and hematopoietic cancers is discussed, and the differences are compared. Finally, clinical inhibitors of histone modifications will be summarized to better understand the developmental stage of cancer therapy using these drugs.
2. Histone modifications
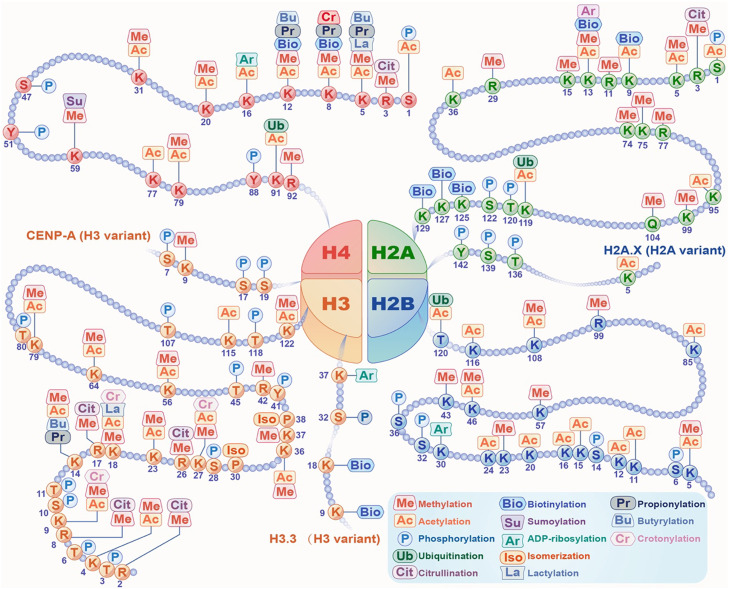
Histone modification is a covalent post-translational change to histone tails, including H2A, H2B, H3 and H4, catalyzed by proteins termed as “writers” and “erasers”. Currently, several well-studied histone modifications are involved in cancer development, such as H3K4me3 and H3K36me3, which are associated with active transcription, and H3K27me3, H3K9me2/3, and H4K20me3, which are associated with repressed genes9. A large catalogue of histone modifications has been described, but functional understanding is still lacking. Histone methylation, acetylation, and phosphorylation are the most frequent alterations in histone tails, while many other modifications have been detected, such as ubiquitination, lactylation, propionylation, crotonylation, and formylation9, 10, 11. The modified amino acid sites on histone tails are shown in Fig. 1.
Fig. 1.
Histone modifications. Various modifications could occur on tails of four core histones, H2A, H2B, H3 and H4. And variants of H2A and H3 can also be modified by different modifications, such as acetylation or phosphorylation on H2A.X, methylation or phosphorylation on CENP-A, and phosphorylation or biotinylation on H3.3. Histone methylation and acetylation are the most common modifications and usually occur on the same lysine sites of the four histones (i.e., H2AK5/13, H2BK5/46/108, H3K4/9/14/23/27/36/56/64/79/122 and H4K5/8/12/20/31/79). What's more, several amino acid sites on histone tails could be commonly modified with more than two different modifications, including H2AK13me/ac/ar/bio, H3K9me/ac/cr, H3K14me/ac/pr/bu, H3K18me/ac/la/cr, H4K5me/ac/pr/bu/la, H4K8me/ac/pr/bio/cr and H4K12me/ac/pr/bu/bio, illustrating the role that these sites play in gene regulation and cell fate determination.
2.1. Histone methylation
Histone methylation involves the transfer of methyl groups from S-adenosyl methionine (SAM) to lysine (K) and arginine (R) residues of H3 or H4 tails with indicated lysine methyltransferases (KMTs) or arginine methyltransferases (PRMTs), respectively. Lysine demethylases (KDMs) are responsible for removing methyl groups from histone lysine residues12. Histone methylation predominantly occurs via recruitment of histone-binding proteins or inhibition of recruitment. For example, H3K4me3 recruits the activating proteins such as transcription factors (TFs) to gene promoters, whereas the recruitment of repressors such as nucleosome remodeling and deacetylase (NuRD) complex is inhibited by H3K4me311,13. However, H3K9me2/3 specifically binds chromodomain proteins, such as the heterochromatin protein 1 (HP1) family, to form a higher-order architecture of heterochromatin, leading to gene repression11,13.
Mutations within histone modifier enzymes and histone genes are usually observed in cancer cells, resulting in changes in chromatin methylation patterns, which leads to tumor development and metastasis. Analysis of The Cancer Genome Atlas (TCGA) databases revealed that mutations occur in various enzymes involved in histone methylation14,15. Gain or loss of function of H3K27me3 is one of the well-known disordered histone modifications leading to aberrant gene expression and genome stability in cancer, which is usually caused by mutations in the gene encoding enhancer of zeste homologue 2 (EZH2), a histone methyltransferase5. Moreover, the enzymatic activity of EZH2 can also be influenced by mutant histones with tumor-promoting features, including H3K27M/I16. Despite the obtained advances, a better understanding of the abnormal histone methylation patterns in malignancies is still necessary to elucidate the molecular mechanisms of tumorigenesis and develop novel targeted therapeutics or combination treatments.
2.2. Histone acetylation
Histone acetylation and deacetylation are processes by which the lysine residues of tails protruding from the histone core of the nucleosome are acetylated and deacetylated by the addition or removal of acetyl groups, which is associated with many major cellular functions such as DNA replication, DNA damage repair, and RNA transcription10,17. These reactions are typically catalyzed by enzymes with histone acetyltransferase (HAT) or histone deacetylase (HDAC) activity18. In terms of dynamics, histone acetylation is one of the fastest post-translational modifications (PTMs), faster than methylation but slower than phosphorylation19.
Numerous studies have noted that changes in histone acetylation can lead to cancer development. Overexpression and enhanced activity of HDACs have been identified as drivers of tumor development and metastasis by altering histone acetylation and regulating expression of oncogenes such as p300 and CBP20,21. However, p300 and CBP have also been shown to be tumor suppressors in hematological malignancies and several solid tumors20. These findings suggest that the roles of p300 and CBP, which are also HATs, in cancer, require further investigation. Additionally, HDAC1, a component of the NuRD complex, can catalyze deacetylation of H3K27 at the STAT1 gene promoter, creating an immunosuppressive environment that promotes the progression of glioma stem-like cells (GSCs)22. Currently, four HDAC inhibitors (vorinostat, istodax and beleodap for the treatment of T cell lymphoma, and panobinostat for the treatment of multiple myeloma) have been approved by the US FDA for cancer treatment23. Considering the reversible characteristics of histone acetylation, developing new methods or combination strategies is necessary to transform the histone acetylation status in cancer cells to a normal state.
2.3. Histone phosphorylation
Another histone modification, phosphorylation, changes the chromatin structure by adding a negative charge to mainly the serine (S), threonine (T), and tyrosine (Y) of histone tails, allowing the interaction with TFs to regulate gene expression associated with cell cycle and proliferation24,25. Similar to histone methylation and acetylation, aberrant histone phosphorylation can also mediate tumor development and metastasis. For example, H3S10P, a well-known modification mediated by several kinases, is correlated with positive regulation of transcription and is thought to be a cancer biomarker (reviewed in REF.26). Deletion of N-α-acetyltransferase D (NatD) inhibits the epithelial mesenchymal transition (EMT) in lung cancer by enhancing H4S1 phosphorylation to downregulate Slug expression27. Phosphorylation of H3.3 at serine 31 can enhance the activity of p300 and histone acetylation in mouse embryonic stem cells (mESCs)28. This suggests that histone phosphorylation participates in many crosstalk events with other histone modifications, providing new insights into cancer drug discovery.
2.4. Other histone modifications
Histone ubiquitination is also a common modification that usually occurs in histones H2A and H2B29. Interestingly, ubiquitination of H2AK119 is always accompanied by H3K27me3 mediated by the polycomb repressive complex 2 (PRC2) complex30. During development, PRC1-mediated H2AK119ub and PRC2-associated H3K27me3 initially accumulate at large intergenic domains that can then spread into genes only under conditions of histone deacetylation and gene silencing31. The cullin4B-ring E3 ligase complex (CRL4B) complex can also catalyze ubiquitination of H2AK119 and cooperate with the PRC2 complex to promote tumorigenesis32. The BAP1 deubiquitinase complex can remove the ubiquitination of H2AK11933. Another ubiquitination site on H2BK120 is mediated by RNF20/40, which activates gene transcription in human cells34. This modification can also be catalyzed by UBR7 to suppress the development and metastasis of triple-negative breast cancer (TNBC)35. Crosstalk between different histone modifications or interaction between modifier enzymes represents a promising mechanism for better understanding the gene regulation patterns in cancer. For instance, ubiquitination of H2B is a prerequisite for the methylation of H3K4 and H3K79 mediated by COMPASS and DOT1L, respectively36,37. The recruitment and activity of UTX, a key component of MLL3/COMPAS and H3K27 demethylase, are dependent on BAP1, a deubiquitinase of H2AK119ub38.
In addition to the four well-known histone modifications mentioned above, there are many other types of histone modifications, such as neddylation, biotinylation, crotonylation, and lactylation9, 10, 11. In recent years, non-acetyl histone lysine acylations, such as crotonylation, butyrylation and propionylation, have been identified and always share the same “readers”, “writers”, and “erasers”39,40. For example, histone crotonylation is read by Yaf9, ENL, AF9, and Sas5 domain proteins and double plant homeodomain (PHD) fingers, catalyzed by p300, and erased by several histone deacetylases39. Fang et al. discovered that histone crotonylation increased during meso/endodermal differentiation of human ESCs, enhancing the expression of meso/endodermal genes and promoting meso/endoderm commitment39. This study suggests that histone crotonylation may be correlated with cancer stem cells (CSCs) and is involved in tumorigenesis. Histone butyrylation stimulates gene transcription by competing with H4K5ac41. However, the role of histone butyrylation in cancer development and metastasis remains poorly understood. Recently, an isomer of histone butyrylation, termed histone isobutyrylation, was identified and found to be mediated by p300 and HAT142. Like crotonylation and butyrylation, histone propionylation can also be catalyzed by acetyltransferases such as KAT6, and deficiency of this modification by KAT6-BRPF1 complexes has been shown to be related with neurodevelopmental disorders and cancer43. Lysine succinylation was first identified by Zhao lab in 2010 as a novel PTM44. HAT1, CPT1A and KAT2A are demonstrated to be the lysine succinyltransferases (KSTases) to catalyze succinylation45,46. Interestingly, the activity of carnitine palmitoyltransferase (CPTase) and KSTase of CPT1A can be separated by G710E mutation, revealing that tumor cells can acquire new mutations to alter different histone modifications to avoid targeted therapies. Other types of acylation reaction on histone tails are glutarylation (catalyzed by KAT2A and erased by SIRT7), benzoylation (erased by SIRT2), lactylation (catalyzed by p300), S-palmitoylation, and O-palmitoylation (catalyzed by LPCAT1)10,47. Although the function and mechanism of histone lactylation, a recently identified histone modification, remains largely unknown in cancer, it has been shown to link the metabolic regulation and epigenetically regulated gene expression, implying the important role of this modification in cancer48,49.
Histone sumoylation of histone H4 is catalyzed by SUMO family proteins, which mediate transcriptional repression by recruiting HDACs and HP150. In addition, H4K12 sumoylation suppresses p300-mediated histone acetylation and Set1/COMPASS-mediated histone methylation, resulting in repressed transcription51. Similar to histone acetylation and methylation, ubiquitination can be influenced by histone neddylation catalyzed by RNF168, which regulates DNA damage repair52. Furthermore, the neddylation inhibitor MLN4924 has been shown to suppress the proliferation and migration of several cancers53, 54, 55. In breast cancer cells, xenografts and patient tumors, the basal histone glycation is high and further investigation found that DJ-1 is the eraser of histone glycation associated with nucleosome stability47. ADP-ribosylation of histone H2AX is another modification involved in DNA damage repair56,57. Ten eleven translocation enzyme 2 (TET2) is found to directly interact with O-linked β-N-acetylglucosamine (O-GlcNAc) transferase (OGT), promoting histone O-GlcNAcylation during gene transcription58. This histone H2B modification can also be regulated by adenosine-monophosphate activated protein kinase (AMPK)59. Due to the modification of histone lysine sites, histone N-formylation may crosstalk with other modifications, such as methylation or acetylation, contributing to the pathophysiology of oxidative and nitrosative stress60. Also, serotonylation and dopamunylation on histone H3Q5 are transamidated by TGM2 both alone and in combination with H3K3me3, and are associated with gene transcription10. Other rare histone modifications, including isomerization, biotinylation and citrullination, are reported to be capable of influencing histone methylation. However, the role of these modifications is still not well-established in cancer9.
2.5. Histone modification and DNA methylation
DNA methylation, mediated by DNA methyltransferases (DNMTs), DNMT1, DNMT3A and DNMT3B, is also involved in histone modification-regulated gene expression by operating chromatin accessibility. Dysregulation of DNMTs is also involved in cancer initiation and development. Evidence suggests that DNMT3A and DNMT3B are “readers” of methylation on H3K36 site through the PWWP domain5. DNMT3A is more preferentially recruited by H3K36me2 than H3K36me3, whereas DNMT3B is recruited by H3K36me361, 62, 63, 64. In gastric cancer, DNMT3A isoform b contributes to EMT-mediated metastasis by repressing E-cadherin expression through cooperating with H3K9me2 and H3K27me365. Dysregulated histone modification is always correlated with high expression of oncogenes in cancer, which may be regulated by DNA methylation. For example, HDAC1 is involved in PAX2 upstream regulatory region, while hypomethylation of the PAX2 promoter is detected in endometrial carcinoma. These results suggest that histone acetylation may be involved in PAX2 regulation, resulting in tamoxifen-stimulated carcinogenesis66. However, the DNMT, that mediates methylation of PAX2 promoter, still needs to be further investigated. The expression of DNMT3A can also be regulated by histone methylation mediated by KMT2C in small cell lung cancer (SCLC)67. Low level of H3K4 methylation, mediated by KMT2C, repressed the expression of DNMT3A, resulting in elevated expression of MEIS2 and SCLC metastasis. Interestingly, treatment with SAM, an approved drug for liver cirrhosis, depressive disorder, osteoarthritis and other syndromes, reversed this process, resulting in hypermethylation of H3K4 and MEIS2 promoter, which leads to retrained SCLC metastasis. These results suggest that SAM may be used for the treatment of histone and DNA hypomethylation-associated malignancy, providing a potential epigenetic therapeutic vulnerability.
3. Histone modifications in solid tumors
Disorders in histone modification are accompanied by various diseases, including cancer. According to data from Cancer Statistics 2020, the most frequently occurring solid tumor is breast cancer, followed by lung, colorectal, prostate, stomach, and liver cancers1. Reordering aberrant histone modifications is considered to represent a powerful strategy for the development of cancer therapeutic drugs. Therefore, an understanding of the role of histone modifications in gene regulation in different solid tumors is urgently required.
3.1. Breast cancer
Since 2020, breast cancer is the highest occurring cancer worldwide, based on data from the IARC1. Various genetic mutations have been identified in subclonal tumor cell populations within the tumor microenvironment, resulting in different phenotypes, including drug resistance, metastatic potential, and stem cell properties. However, the genetic mechanisms driving these phenotypes are sometimes unclear. The major epigenetic alteration that regulates gene expression is the modulation of chromatin structure by histone modification. Data from a single-cell chromatin immunoprecipitation followed by sequencing (scChIP-seq) experiment revealed that H3K27me3, a stable regulator of the transcriptional repression of genes responsible for treatment resistance, was lost in breast cancer cells from drug-resistant tumors68. CSCs within the tumor microenvironment always contributes to therapeutic failure, due to the protection of various surrounding cells and overexpression of immunosuppressive markers. It has been reported that PD-L1, an immune checkpoint molecule, is overexpressed in CSCs and contributes to immune evasion69. Furthermore, the repressive histone modifications H3K9me3 and H3K27me3 are enriched in the promoter region of PD-L1 in breast cancer cells, but are weaker in breast CSCs, leading to upregulated expression of PD-L1 in CSCs70. Even though the mortality of breast cancer has decreased owing to improved screening approaches and better therapeutics, approximately 25-40% of patients develop metastasis and even die71. Lymph nodes (LNs) are considered the most common organ of initial spread in most cancers. Then tumor cells spread to distant sites through draining LN to blood vessels. This process is highly efficient and dynamic, making cancer metastasis a more complex mechanism in tumorigenesis. HDAC11 plays an important role in regulating LN metastasis72. HDAC11 expression was upregulated in LN metastasis tumors compared to primary and lung metastasis tumors. Histone acetyl groups were removed by HDAC11, leading to downregulated expression of cell cycle-associated genes such as RRM2 and E2F8, resulting in promoted tumor growth within LNs. However, metastasis from LNs to distant organs increased when HDAC11 was blocked using HDAC inhibitor (HDACi) or shRNA. These results strongly suggest the risk of using a single HDACi to treat cancer patients, and recommend that a combination of multiple therapeutic drugs may be the most appropriate strategy. Breast cancer is one of the most common cancer types associated with bone metastasis that correlates with histone modification73. However, therapy targeting bone metastasis is still under preclinical studies, including cell culture experiments and mouse models. Thus, it is still urgently needed to investigate the histone modification patterns involved in bone metastasis. Accumulating evidence suggests a strong correlation between breast cancer and glucose metabolism. PDK1, a key enzyme in glucose metabolism, is proved to be regulated by miR-148a whose expression can be inhibited by HDAC2 and EZH2-mediated histone modification, resulting in breast cancer progression and Adriamycin resistance74. This provides new ideas and directions to counter chemotherapy resistance in breast cancer.
3.2. Lung cancer
Before 2020, lung cancer was the most common type of solid tumor worldwide and is now still the leading cause of cancer-related death, with a 5-year survival rate of only 15%. TSGs, regulated by histone modifier factors, play a key role in the initiation, progression, and metastasis of lung cancer. The histone methyltransferase G9a mediated H3K9me2 recruits transcription factors HP1 and DNMT1 to inhibit the expression of TSGs such as APC2 and WIF1, leading to Wnt activation and cancer progression in non-small cell lung cancer (NSCLC). Targeting G9a reversed this cancer-promoting effect, revealing that G9a could serve as a therapeutic target in the treatment of lung cancer75. In another study, loss of G9a or H3K9me2 reduction permitted the expression of MMP10, KRAS, and ECM genes to promote lung adenocarcinoma progression76. This finding demonstrates the controversial role of G9a in lung cancer. A CRISPR/Cas9 screening system found that histone demethylase UTX could serve as a TSG in lung tumorigenesis by regulating the homeostasis of H3K27me3 mediated by EZH2. Knockout of UTX increased the status of H3K27me3 and downregulated the expression of CDKN2a and CDKN2b to promote lung tumor proliferation in vivo77. EZH2 is also overexpressed in SCLC and contributes to immune evasion and drug resistance78. Chromodomain Y-like (CDYL) enhances the chemoresistance in patients with SCLC by decreasing CDKN1C expression through H3K27me3 mediated by EZH279. Due to the important role of EZH2 in tumorigenesis, EZH2 inhibitors are used in lung cancer therapy and acquire drug sensitivity in tumors with mutated histone modifier genes, such as histone demethylase KDM6 and H2AK119ub deubiquitinase BAP180,81. These findings imply that EZH2 inhibitors are attractive cancer drugs and may have better therapeutic efficacy when combined with epidrugs based on other types of histone modification. LSD1 (KDM1A), a flavin adenine dinucleotide (FAD)-dependent demethylase, is responsible for demethylating H3K4me1/2, and is another potential target for lung cancer. LSD1 is a component of several large chromatin-repressive complexes, such as CoREST and NuRD, exerting its function together with histone acetylation and methylation to regulate gene transcription82. ORY-1001 and GSK2879552 are LSD1 inhibitors that have been investigated in preclinical and clinical studies and show potential antitumor effects83,84. Lung is one of the most common sites of neuroendocrine tumors (NETs) with a worse 5-year survival rate (19-38%) in metastatic disease. Multiple clinical trials are ongoing using drugs interfering with epigenetic pathways, including histone modification, or combination therapies with immune-checkpoint inhibitors to NETs treatment, indicating potential usage of histone modifier inhibitors and immunotherapy as combination therapy in cancer treatment85.
3.3. Colorectal cancer
Colorectal cancer (CRC) remains a life-threatening cancer all over the world. In 2020, the incidence and mortality of CRC were 10% and 9.4%, respectively, making CRC the third most frequent cancer and the second leading cause of cancer-related deaths worldwide1. The etiology of CRC is not well established; therefore, there is an urgently need to identify potential biomarkers for the early diagnosis of CRC. Recently, the contribution of epigenetic alterations, especially histone modifications, to CRC malignancy has gained considerable attention. The spatial features of histone modifications may have prognostic potential and benefit CRC therapy. In a comparative study, H3K9me, H3K27ac, and H4K12ac levels were higher in CRC tissues than in normal colonic mucosa86, 87, 88. Histone modification markers are completely different from the primary site in metastatic organs, such as the liver. H3K4me2 and H3K9me2 have been shown to be correlated with the clinicopathological stage and may be prognostic markers for liver metastasis89,90. Furthermore, combined histone modifications are thought to be a more powerful method for detecting CRC to enhance the credibility of prognostic biomarkers. For instance, high H3K9me and H4K20me3 and low H3K4me3 in the nucleus are together associated with improved clinical prognosis, with hazard ratios (HR) of 3.81 (disease-free survival), 2.86 (locoregional recurrence-free survival) and 2.94 (distant recurrence-free survival)91. Compared with colonoscopy, liquid biopsy (serum or plasma) is considered the most efficient method for detecting CRC without resected lesions. Interestingly, H3K9me3 and H4K20me3 levels were both decreased in the circulating nucleosomes of CRC patients92,93. However, further studies are required to confirm the utility of these two biomarkers for diagnostic detection. Owing to the prognostic potential of histone modifications, several inhibitors of modifier enzymes, including HDAC inhibitors, histone methyltransferase (HMT) inhibitors, and histone demethylase (HDM) inhibitors, have been applied into preclinical and clinical studies combined with chemotherapeutic drugs94.
3.4. Prostate cancer
Prostate cancer (PCa) is the second most common cancer occurring in men, and over 1.4 million new cases of PCa were detected worldwide in 20201. The androgen receptor (AR) is widely accepted to play an essential role in the proliferation and maintenance of PCa. Although androgen deprivation therapy (ADT), such as enzalutamide (an AR antagonist), is considered the most effective treatment for PCa, the emergence of castration-resistant prostate cancer (CRPC) and enzalutamide-resistant prostate cancer (ERPC) has presented obstacles for PCa therapy95. AR can be regulated by histone modifications, such as histone methylation and phosphorylation, mediated by the correlated modifier enzymes, such as EZH2, JMJD1A, ACK1 and LSD196. Thus, these enzymes could serve as therapeutic targets for PCa treatment. The lysine methyltransferase KMT9A controls the proliferation of PCa cells by monomethylating H4K12, and inhibition of KMT9A significantly attenuates the growth of xenograft tumors97. Another histone methyltransferase, DOT1L, together with AR, improves the expression of Myc through binding to the enhancer by catalyzing H3K79me2, which inhibits the expression of E3 ubiquitin ligases HECTD4 and MYCBP2 to promote PCa. Blockade of DOT1L with its inhibitor EPZ004777 disrupts this process and enhances the degradation of Myc and AR by upregulating HECTD4 and MYCBP298. Histone demethylases, such as JMJD1A and LSD1, can also serve as coactivators of AR by epigenetic regulation of H3K9 or H3K4 methylation99,100. Other histone modifications such as ubiquitination and phosphorylation may be involved in the epigenetic process of AR activation99,101. RNF8, a RING finger E3 ligase, binds to MYC and enhances AR transcription by catalyzing ubiquitination of H2A/H2B and acetylation of H3/H4. Elevated AR/ARV7 level interact with RNF8 to form a complex that activates the expression of AR target genes, including PSA, FASN, and ALDH1A3. In this process, reduced H3K27me3 is also involved as a repressive histone marker of AR target genes101. In addition to EZH2, methylation of H3K27 can also be mediated by NSD3, a histone methyltransferase for H3K36102,103. Belonging to the same protein family, NSD2 enhances AR-mediated transcription104. However, the role of NSD3 in PCa malignancy is still a mystery.
3.5. Gastric cancer
Gastric cancer (GC) is the third most common cancer (10.5%) and the third leading cause of cancer-related deaths (12.4%) in both sexes in China, based on IARC data from 2020. Among the risk factors, such as smoking and obesity, Helicobacter pylori (Hp) is an important cause of gastric carcinoma and is responsible for 75% of cases105,106. Hp causes alterations in histone modifications in epithelial cells and macrophages within the stomach, leading to GC development. Yang et al. demonstrated that Hp infection induces the phosphorylation of H3S10 to facilitate gastric carcinogenesis107. Phosphorylation of H3S10 induced by Hp in macrophages increased the expression of IL-6 by binding to the promoter region of this gene, resulting in Hp-induced gastritis108. In contrast, Hp decreased the phosphorylation of H3S10 and H3T3 in a type IV secretion system (T4SS)-dependent manner in gastric epithelial cell lines109. Hp also affects histone acetylation. For example, Hp increases the expression of p21WAP1/CIP1 by promoting acetylation of histone H4 in the promoter region110. However, p21WAP1/CIP1 is a TSG in GC and acquires H3 hypoacetylation on its promoter region. The HDACi TSA could restore the H3 hyperacetylation to induce the p21WAP1/CIP1 expression111. These studies reveal that the exact mechanism of Hp in histone acetylation of GC needs to be further elucidated. Hp upregulates the JMJD2B expression to promote tumorigenesis in GC. JMJD2B then cooperates with NF-κB to enhance the expression of COX-2 on the promoter with decreased H3K9me3, a histone marker that is correlated with tumor stage, invasion, and recurrence112,113. Cytotoxin-associated gene A (CagA), an Hp virulence factor, is thought to be a powerful factor that has carcinogenic potential114. CagA upregulates the expression of Myc, DNMT3B, and EZH2, and increases H3K27me3 and DNA methylation on the let-7 promoter. Finally, the expression of Ras oncoprotein is upregulated in the stomach without inflammation115. DNA methylation and histone modifications always co-occur in GC, illustrating that epigenetic combination modalities may be a better method for GC therapy.
3.6. Liver cancer
The mortality rate of liver cancer is 8.3% in both sexes worldwide, ranking the third in 20201. Although the incidence of liver cancer (9%) ranks fifth in China, the death rate is 13%, which is only lower than that of lung cancer. Abnormal epigenetic regulation is a common feature of human hepatocellular carcinoma (HCC). HCC has been shown to develop from liver disease induced by hepatitis C virus (HCV) infection. Most HCV-positive patients can still develop HCC after antiviral agents treatment due to persistent epigenetic changes such as acetylation of H3K27116. In hepatitis B virus (HBV)-positive HCC patients, overexpression of the long non-coding RNA PVT1 impairs the recruitment of EZH2 to the myc promoter region, leading to elevated Myc expression by reducing the status of H3K27me3117. This study also supports the notion that histone methylation is linked to HCC proliferation and metastasis118. H3K9me2, mediated by G9a, which is overexpressed in HCC, represses the expression of RARRES3, a TSG, to promote tumor development119. Several other histone methylation modifier enzymes, including JARID1B120, KDM4B121, KDM5C122, and SETDB1123,124, are overexpressed in HCC. HDAC3 is a member of class I HDACs and is considered a strong contributor to hepatocarcinogenesis owing to its role in cell cycle regulation and transcriptional reprogramming. HDAC3 is selectively expressed in liver cancer stem cells and contributes to their self-renewal via histone modifications125. Furthermore, HDAC deficiency increases H3K56ac level and decreases the status of H3K27me3 in liver cancer stem cells125. Interestingly, targeting mTORC2/HDAC3 signaling inhibits the stemness of HCC cells and is correlated with metabolic reprogramming126. The presence of HDAC3 increased the level of H3K9me3 mediated by histone lysine N-methyltransferase SUV39H1. H3K9me3 cooperates with the DNA damage response (DDR) complex to accumulate damaged DNA, resulting in HCC progression127. However, in liver-specific deficient mice, HDAC3 ablation enhances the H3K9ac level, activating numerous oncogenes such as KRAS, FOS, and CDK6, leading to HCC development127. Thus, the mechanism of action of HDAC inhibitors, especially those targeting class I HDACs, require further investigation in the treatment of HCC.
4. Histone modifications in hematopoietic cancers
Hematopoietic malignancies can occur at any stage of blood cell development and influence the production and function of blood cells, leading to a diminished ability to fight infection and susceptibility to uncontrolled bleeding. Aberrant regulation of gene transcription by histone modifications is an important mechanism in oncogenesis and the development of hematopoietic malignancies, including three main types: leukemia, lymphoma, and multiple myeloma128.
4.1. Leukemia
According to IARC, there were approximately 313,594 deaths caused by leukemia worldwide. Leukemia, originally developed from bone marrow with production of large amounts of abnormal white blood cells, is the most common type of cancer in children and can be divided into four main subtypes: acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic lymphocytic leukemia (CLL), and chronic myeloid leukemia (CML). However, over 90% of leukemia cases are diagnosed in adults, of which CLL and AML are the most common. A study conducted using a reverse phase protein array (RPPA) demonstrated that multiple histone-modifying proteins are associated with the survival of newly diagnosed AML patients129. For instance, KDM6A expression is upregulated in AML and contributes to poor prognosis, while inhibition of KDM6A by GSK-J4 inhibitor reduces the progression and proliferation rate of both primary and immortalized AML cells with increased H3K27me3 level130. H3K27me3 is catalyzed by EZH2, and its loss induces poor outcomes in patients with AML131. Moreover, loss of EZH2 contributes to resistance to multiple drugs in AML132. EZH2 is an oncoprotein overexpressed in CLL133. Inhibition of EZH2 enzymatic activity by drugs induces downregulation of H3K27me3, leading to increased cell apoptosis134. As reviewed elsewhere, disruption of the balance of histone acetylation is also a major factor in AML135.
In recent years, no histone modification-based drug has been approved by the FDA for leukemia. Thus, more progress is still required in the research area of leukemia therapy, and the detailed mechanism needs to be fully elucidated for different leukemia subtypes. Drug discovery may focus on the direction of individualized therapy, according to the regulatory mechanism of histone modifications.
4.2. Lymphoma
Lymphoma is a group of blood and lymph tumors that develop from lymphocytes. The two main subtypes of lymphoma are non-Hodgkin lymphoma (NHL) and Hodgkin lymphoma (HL). The most frequent lymphoma is NHL that accounts for 90% of all cases. Based on IARC data, NHL caused 259,793 deaths worldwide in 2020, while HL caused only 23,376 deaths. Frequent mutation analysis demonstrated that genes associated with histone modification are common targets of somatic mutations in B-cell NHL136. MLL2, also known as histone lysine N-methyltransferase (KMT2D), was the most frequent gene with the largest number of single nucleotide variants (SNVs) distributed across the whole MLL2 gene sequence. MLL2 is a major mammalian histone methyltransferase that mono-methylates H3K4 and is a TSG in NHL137,138. Another gene acquiring mutations in NHL is MEF2B, a TF linked to histone acetylation and methylation in a calcium-regulated manner139. The most common mutation in MEF2B leads to amino acid changes in D83V140. This mutation cannot alter DNA interactions, but disrupts the interaction with some repressive complexes, including the HUCA complex and HDAC class IIa members, resulting in abnormal expression of histone markers at H3K27139,141. Therefore, targeting the various mutated TFs associated with anomalous histone modifications is an attractive strategy for lymphoma treatment.
These studies reveal that mutations in histone modifier genes or correlated TFs alter the normal status of histone modifications, resulting in lymphomagenesis. A better understanding of the associated mechanisms will aid in the design and discovery of targeted drugs. Three HDAC inhibitors (vorinostat, romidepsin and beleodap) have already been approved by the FDA for treatment of T-cell lymphoma142.
4.3. Multiple myeloma
In 2020, multiple myeloma (MM) caused 117,077 deaths in both sexes worldwide, and is the second most common type of blood cancer in high-income countries and is characterized by the uncontrolled proliferation of plasma cells in the bone marrow. Despite genetic regulation, accumulating evidence has revealed that alteration of histone modifications also plays a central role in supporting MM. NSD2, also known as MMSET/WHSC1, is the most well-studied histone methylase for H3K36me2 in MM and is overexpressed in all MM cases with t(4;14), one of the most common translocations143,144. Elevated levels of H3K36me2 are associated with active chromatin, which drives oncogene expression, leading to MM progression145,146. NSD2 can also act as a component of the repressor complex through its association with HDAC1/2 and LSD1, resulting in increased H4K20me3 and decreased histone acetylation147. Furthermore, NSD2, which interacts with KAP1 and HDAC1, induces H3K9me3 and represses H3ac, resulting in the inhibited expression of miRNA-126* and enhanced c-Myc expression, leading to MM pathogenesis148. Thus, NSD2 may be a regulatory center that affects overall histone methylation in MM. Other well-known aberrant histone modifiers, such as EZH2, PRMT5, KDM6B, and KDM3A, are also involved in disrupting histone modifications that contribute to MM development149. Except the functions of HDAC1/2 mentioned above, HDAC3 is much more important in MM. Inhibition of HDAC3 induced a stronger decrease in MM cell growth than inhibition of HDAC1 and HDAC2, indicating that HDAC3 is a more attractive target in MM150. Therefore, MS-275, an HDAC1/2/3 inhibitor, was more toxic to MM cells than Merck60, an HDAC1/2 inhibitor. HDAC3 also inhibits the acetylation of c-Myc and DNMT1 to maintain their stability and promote MM cell survival151. To date, multiple therapeutic approaches have been investigated for the treatment of MM, including alkylating drugs, steroids, anthracyclines, proteasome inhibitors, immunomodulatory drugs, HDAC inhibitors, monoclonal antibodies, antibody-drug conjugates, and nuclear export inhibitors152. However, only one HDAC inhibitor, panobinostat, has been approved by the FDA for relapsed MM therapy153. More efforts are required to understand the histone modification pattern in MM for targeted drug development.
5. Clinical inhibitors of histone modifier enzymes
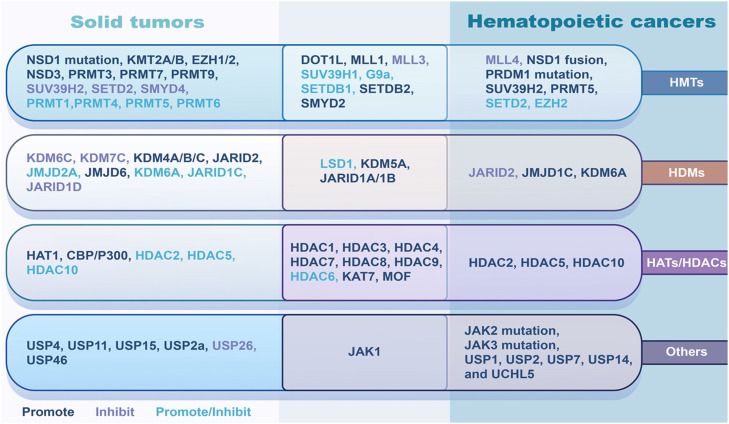
Dysregulation of histone modification enzymes plays an important role in tumorigenesis. The high expression of HMTs, including SMYD2, SETDB2, or MLL1 mutations, and the loss of function of MLL3 are associated with tumor growth, invasion, metastasis, and poor prognosis in various types of cancers154,155. Histone demethylases JARID1A/1B and histone acetyltransferases KAT7156 and MOF, and histone deacetylases HDAC1, HDAC3, HDAC4, HDAC7, HDAC8, and HDAC9 are positively associated with the aggressive progression of solid tumors and hematologic malignancies157,158. SUV39H1, G9a, SETDB1, histone demethylase HDM LSD1, and histone deacetylase HDAC6 mainly act as tumor promoters but sometimes play different roles in the tumorigenesis of solid tumors and hematologic malignancies159, 160, 161. A large number of histone modification enzymes play a different role in some specific tumors. For example, some histone modification enzymes such as EZH2, SETD2, DOT1L, KDM6A, JARID2, HDAC2, HDAC5, and HDAC10, play different roles in solid tumors and hematologic malignancies162,163. What's more, some tumor-specific enzymes exist and function in different cancers. Here, we briefly list some dysregulation of the histone modification enzymes and their roles in cancer (Fig. 2).
Fig. 2.
Main roles of histone modifier enzymes alteration in solid tumors and hematopoietic cancers. The alterations of histone modifier enzymes could induce various diseases, including cancer. Some enzymes such as DOT1L, SMYD2, JARID1 and several HDACs play similar role in tumorigenesis, promoting tumor development in both solid and hematopoietic cancers. Some of them play different, even opposite, role between solid and hematopoietic cancers. For instance, PRMT1, PRMT5, KDM6A, HDAC2, HDAC5 and HDAC10 can promote the development of hematopoietic cancers. However, the exact function of these molecules in solid tumors is controversial, which may depend on the individual situation. For example, SUV39H2, a HMT, functions as a tumor suppressor in solid tumors, but promotes the tumorigenesis of hematopoietic cancers. Similarly, JARID2 is an oncoprotein in solid tumors, but a tumor suppressor in hematopoietic cancers. HAT, histone acetyltransferase; HDAC, histone deacetylases; HDM, histone demethylase; HMT, histone methyltransferase.
5.1. Histone acetyltransferase and deacetylase inhibitors
Acetylation or deacetylation of histone proteins regulates gene expression. A small number of cancer patients have reported abnormal HATs. Tip60 (KAT5) and p300 are typical HATs that have participated in multiple physiopathological processes including DNA damage and repair, transcriptional regulation, and cell signaling. Histone acetyltransferase inhibitors (HATi) of Tip60 and p300 have been developed in clinical trials for cancer treatment164,165. Inhibitors of histone deacetylation enzymes are considered one of the most promising anticancer targets. Relaxation of the DNA wrapped around histone proteins promotes the binding of DNA to gene transcription factors166. The HDAC family of histone demethylases can be divided into four groups according to their sequence homology to yeast. Class I consists of HDAC1, 2, 3, and 8, class II consists of HDAC IIA (HDAC4, 5, 7, and 9) and IIB subgroups (HDAC6 and 10), class III comprises seven Sir2-like enzymes (SIRT1-7), and class IV has only one member, HDAC11. HDACs are involved in regulating a variety of cellular biological processes, including cellular metabolism, growth, metastasis, and aging. To a variable extent, HDACis induce the expression of proteins related to cell differentiation, cell growth arrest, and apoptosis, thereby inhibiting cancer progression167. The identified HDACis are classified as short-chain fatty acids (sodium butyrate, valproic acid, pivanex, AR-42, and phenylbutyrate), hydroxamic acids (TSA, oxamflatin, SAHA, hexamethylene bisacetamide (HMBA), pyroxamide, CHAPs), depsipeptide (FK-228), cyclic tetrapeptides (trapoxin and apicidin), and benzamides (MS-275, MGCD0103, CI-994), some of which have already entered the different stages of clinical trials168,169 (Table 1). The anticancer agents vorinostat, romidepsin, belinostat, and panobinostat (LBH-589) are four granted FDA-approval HDACi that have been shown to inhibit histone deacetylation in clinical trials170,171 (Table 1).
Table 1.
Histone acetyltransferase inhibitors and histone deacetylase inhibitors.
| Chemical class | Compound | Clinical stage | Cancer specificity | Reference/NCT number |
|---|---|---|---|---|
| HATi | TH1834 | Preclinical | Breast cancer | 172 |
| C646 | Preclinical | Gastric cancer | 164 | |
| HDACi: Cyclic tetrapeptides | FK-228 (Romidepsin) | FDA approved (2009) | Cutaneous/peripheral T cell lymphoma | NCT00106431 |
| HDACi: Hydroxamic acid | SAHA (Vorinostat) | FDA approved (2006) | Cutaneous T cell lymphoma | NCT00918489 |
| PXD-101 (Belinostat) | FDA approved (2014) | Peripheral T cell lymphoma | NCT00413075 | |
| LBH-589 (Panobinostat) | FDA approved (2015) | Multiple myeloma | NCT00739414 | |
| Pyroxamide | Phase I and II | Solid and hematological tumors | NCT00042900 | |
| Resminostat | Phase II | Colorectal, hepatocellular carcinoma, lymphoma | NCT01037478 | |
| Givinostat | Phase II | Lymphoma, Hodgkin lymphoma, myeloma | NCT01761968 | |
| Pracinostat | Phase II | Acute myeloid leukemia | NCT03151304 | |
| CHAPs | Phase II | Hematological tumors | NCT03986970 | |
| TSA | Phase I | Solid tumors | NCT02959905 | |
| CBHA | Preclinical | Solid and hematological tumors | 173 | |
| Oxamflatin | Preclinical | Solid and hematological tumors | 174 | |
| Abexinostat | Phase I | Solid and hematological tumors | NCT03939182 | |
| Quisinostat | Phase I and II | Solid tumor, cutaneous T cell lymphoma | NCT01486277 | |
| MPT0E028 | Phase I | Solid tumor, B-cell lymphoma | NCT02350868 | |
| CHR 3996 | Phase I | Solid tumor | NCT00697879 | |
| CUDC 101 | Phase I | Solid tumor | NCT01171924 | |
| HDACi: Benzamides | Entinostat (MS-275) | Phase I and II | Solid and hematological malignancies | NCT04708470 |
| Chidamide | Phase II and II | Breast cancer; non-small-cell lung cancer | NCT04582955 | |
| Ricolinostat | Phase I and II | Multiple myeloma, lymphoma | NCT02091063 | |
| Tacedinaline | Phase II and III | Lung and pancreatic cancer; myeloma | NCT00005093 | |
| Mocetinostat | Phase I and II | Solid and hematological malignancies | NCT02282358 | |
| HDACi: Short-chain fatty acids | Valproic acid | Phase I and II | Solid and hematological malignancies | NCT01861990 |
| Sodium butyrate | Phase I | Colorectal | NCT05456763 | |
| AR-42 | Phase I | Acute myeloid leukemia | NCT01798901 | |
| Phenylbutyrate | Phase II | Solid and hematological tumors | NCT00005639 | |
| Pivanex | Phase II | Non-small cell lung cancer, myeloma, leukemia | NCT00073385 |
Abbreviations: HATi, histone acetyltransferase inhibitor; HDACi, histone deacetylase inhibitor.
5.2. Histone methyltransferase and demethylase inhibitors
HMTs can methylate specific residues on histone proteins, leading to the alteration of chromatin structure, which plays an important role in tumorigenesis. Therefore, inhibition of abnormal HMTs is crucial for blocking tumor growth and development. High level of histone methyltransferase enzymes usually correlates with aggressive cancer progression. Histone lysine methyltransferase EZH2 catalyzing the methylation of H3K27 plays an important role in cancer development, and is frequently overexpressed in breast cancer and other human cancers including melanoma, prostate, gastric, bladder, and colon cancer. High expression of EZH2 is associated with aggressive cancer progression175. The emergence of different EZH2 inhibitors provides new insights into cancer therapy. The classification and details of the EZH2 inhibitors are shown in Table 2. Based on structural differences, EZH2 inhibitors can be classified into pyridone-indazole scaffolds (e.g., EPZ005687, UNC1999, GSK343), pyridone-indole scaffolds (e.g., CPI-1205, GSK126, EI1), and pyridone-phenyl scaffolds (e.g., EPZ6438, EPZ006088). In addition, the histone lysine methyltransferase G9a catalyzes the methylation of H3K9 and plays an essential role in cancer proliferation, invasion, and metastasis. Many G9a inhibitors have been reported and these can be divided into several groups, including substrate competitive inhibitors (BIX01294, UNC0638, and A-366) and S-adenosyl-methionine cofactor competitive inhibitors (BRD4770 and BRD9539)176. G9a inhibitors can induce cell cycle arrest and impede cancer development177. SMYD2 is a lysine methyltransferase that catalyzes the methylation of H3K36. The activity of SMYD2 is associated with normal organismal development and a series of pathophysiological processes. The aberrant expression of SMYD2 usually leads to multiple diseases, including cancer. AZ-505 and LLY-507 are well-studied SMYD2 inhibitors that prevent the growth and progression of various cancers178,179. DOT1L methyltransferase calculates the methylation of H3K79 and plays an important role in embryogenesis and leukemia tumorigenesis. EPZ004777, EPZ5676, and SYC-522 are selective inhibitors of DOT1L in cancer therapy98,180,181.
Table 2.
Histone methyltransferase/demethylase inhibitors.
| Chemical class | Compound | Clinical stage | Cancer specificity | Reference/NCT number |
|---|---|---|---|---|
| G9a substrate competitive inhibitor | BIX01294 | Preclinical | Breast cancer, myeloid leukemia | 197, 198 |
| UNC0638 | Preclinical | Breast cancer | 199 | |
| A-366 | Preclinical | Leukemia | 200 | |
| G9a inhibitor | BRD4770 | Preclinical | Pancreatic cancer | 201 |
| SMYD2 inhibitor | AZ505 | Preclinical | Polycystic kidney | 202 |
| LLY-507 | Preclinical | Esophageal, liver | 179 | |
| DOT1L inhibitor | EPZ-5676 | Phase I | Leukemia | NCT02141828 |
| SYC-552 | Preclinical | Leukemia | 181 | |
| EZH2 inhibitor | EPZ005687 | Preclinical | Lymphoma | 203 |
| UNC1999 | Preclinical | Lymphoma | 204 | |
| GSK343 | Preclinical | Breast cancer, prostate cancer | 205 | |
| CPI-1205 | Phase I and II | B cell lymphoma, solid tumor | NCT02395601 | |
| GSK2816126 | Phase I | Cancer, neoplasm | NCT02082977 | |
| EI1 | Preclinical | Lymphoma | 206 | |
| Tazemetostat | FDA approved (2020) | Epithelioid sarcoma | NCT02875548 | |
| EPZ6438 | Phase I and II | Solid and hematological tumors | NCT01897571 | |
| DZNeo | Preclinical | Solid tumor, glioblastoma | 207 | |
| DS-3201 | Phase I and II | Leukemia, small cell lung carcinoma, lymphoma | NCT04388852 | |
| PF-06821497 | Phase I | small cell lung carcinoma | NCT03460977 | |
| MAK683 | Phase I and II | Lymphoma, advanced solid tumor | NCT02900651 | |
| SHR2554 | Phase I | Prostate cancer, lymphoid neoplasm | NCT04407741 | |
| PRMT1 inhibitor | DB75 | Phase I and II | Leukemia | NCT00408369 |
| PRMT4 inhibitor | TP-064 | Preclinical | Multiple myeloma | 208 |
| PRMT5 inhibitor | GSK3326595 | Phase I and II | Solid tumor, lymphoma | NCT03614728 |
| JNJ-64619178 | Phase I | Solid and hematological tumors | NCT03573310 | |
| PF-06939999 | Phase I | Advanced or metastatic solid tumor | NCT03854227 | |
| LSD1 inhibitor | Tranylcypromine | Phase I and II | Solid and hematological tumors | NCT02273102 |
| Bizine | Preclinical | Lung cancer, prostate cancer | 209 | |
| PG11144 | Preclinical | Breast cancer | 210 | |
| Namoline | Preclinical | Prostate cancer | 211 | |
| ORY-1001 | Preclinical | Acute leukemia | 212 | |
| GSK2879552 | Phase I and II | Small cell lung cancer, acute myeloid leukemia | NCT02177812 | |
| CC-90011 | Phase I and II | Solid tumor and lymphoma | NCT04748848 | |
| INCB059872 | Phase I and II | Solid and hematological tumors | NCT02712905 | |
| IMG-7289 | Phase I and II | Acute myeloid leukemia | NCT02842827 | |
| KDM inhibitor | JIB-04 | Preclinical | Breast cancer | 213 |
| IOX1 | Preclinical | Esophageal squamous cell carcinoma | 214 |
Enzymes of the PRMT family catalyze the methylation of histone arginine residues. Dysregulation of PRMTs has been observed in various cancers. Many selective PRMT inhibitors (PRMTi) have been developed for cancer therapy182. PRMT1 regulates the methylation of H4R3 and numerous non-histone substrates. PRMT1 plays an essential role in a large number of biological and pathology processes including DNA repair, signal transduction, and tumorigenesis. DB75 is a selective inhibitor of PRMT1183. The protein arginine methyltransferase 3 catalyzes asymmetric dimethylarginine, and plays an essential role in tumor cell proliferation and metastasis. SGC707 is a selective inhibitor of PRMT3184. PRMT4 (CARM1) is a type I PRMT that catalyzes the asymmetrically dimethylation of protein arginine residues. TP-064 and EZM2302 are selective inhibitors of CARM1 for MM treatment185,186. The PRMT5 usually catalyzes the dimethylation of mono- and symmetric arginine residues. PRMT5 plays an important role in physiological and pathological processes by regulating the cell cycle, cell proliferation and differentiation. GSK3326595, JNJ-64619178, LLY-283, and PF-06939999 are potent inhibitors of PRMT5 in tumor therapy187, 188, 189, 190, while EPZ020411 is a selective PRMT6 specific inhibitor191. Table 2 highlights some HMT inhibitors of EZH2, G9a, SMYD2, DOT1L, LSD1 and PRMT1/4/5.
Histone demethylases catalyze the removal of lysine or arginine methyl groups, which regulate the dynamic balance of the lysine or arginine residues methylation192. Abnormal expression of the histone demethylase LSD1 plays an essential role in tumorigenesis, and targeting LSD1 is an emerging option for cancer therapy193. Numerous LSD1 inhibitors have been discovered, including TCP, PCPA, phenelzine, pargyline, ORY-1001, GSK2879552, INCB059872, IMG-7289, and CC-90011, in various cancer therapy194. In addition, some natural products, such as cyclic peptides, flavonoids, protoberberine alkaloids, melatonin, stilbene, and diarylheptanoids, have been identified as LSD1 inhibitors that restrict tumor growth and progression195. KDM2A is a histone demethylase containing the JmjC domain, which most inhibitors are designed to target. KDM2A is usually overexpressed in various cancers such as lung cancer and breast cancer, thus inhibiting KDM2A could decrease the growth and metastasis of tumors. JmjC KDM inhibitors include the 8-hydroxyquinoline analogs IOX1 and JIB-04196. Several other molecules that inhibit the activity of KDMs have also been identified (not listed here). Table 2 highlights some of the developed KDM inhibitor compounds.
5.3. Other histone modification enzyme inhibitors
Abnormal regulation of histone modifier enzymes plays an essential role in tumor growth and development. In addition to the above-mentioned histone-modifying enzymes, histone phosphorylases and ubiquitinases also have important functions in cancer treatment. Aurora kinase A (AURKA), Aurora kinase B (AURKB) and aurora kinase C (AURKC) belong to the aurora kinase family that are mitotic serine/threonine protein kinases. The expression level of these protein kinases is frequently linked to tumor cell proliferation, invasion and drug resistance215. The alteration of cyclin-dependent kinase (CDK) activity is associated with tumor cell cycle defects. Some CDK inhibitors including CDK4/6 or CDK8 inhibitors, have been developed as potential anti-cancer drugs216,217. PIM serine/threonine kinases behave in 3 isoforms: PIM1, PIM2, and PIM3. These PIM kinases are engaged in various scopes of the biological process including cell proliferation, drug resistance, apoptosis, and immune response218. Targeting PIM kinases and signaling pathways has been recognized as potential therapeutics. Some small molecules, including barasertib, BI-847325, alisertib, flavopiridol, SEL120, SEL24/MEN1703, and PIM447, target different histone phosphorylases219. Non-receptor tyrosine kinases are involved in autoimmune diseases and cancers. Janus kinase 1 (JAK1) is one of the Janus kinase family proteins. JAK1 plays a critical role in inflammatory cytokine signaling, cancer progression, and oncogenic signaling activation221. JAK2 is frequently mutated in cancers, mediating the activation of downstream signaling responses to cytokines and growth factors220. Ruxolitinib is a JAK2 inhibitor used for the treatment of lymphoma and itacitinib is a JAK1 inhibitor that inhibits tumor growth in solid tumors, leukemia, and lymphoma222. Deubiquitinases (DUBs) could regulate the deubiquitination of substrate proteins to control the modification of proteins. The aberration of ubiquitination caused by the abnormal function of DUBs is frequently associated with various diseases, especially the development and progression of cancer. Ubiquitin-specific peptidase 1 (USP1), USP2, USP7, and USP14 are members of the DUBs family. Inhibition of these ubiquitin-specific peptidases can inhibit tumor cell proliferation, metastasis and stemness, and promote tumor cell senescence to varying degrees223. ML323, ML364, b-AP15, and P5091 are selective inhibitors of histone ubiquitinases in cancer224. However, inhibitors of histone phosphorylase and histone deubiquitinase are still very limited in clinical trials, and require further study. Table 3 highlights some other histone modification enzyme inhibitors that have been developed.
Table 3.
Other histone modification enzyme inhibitors.
| Chemical class | Compound | Clinical stage | Cancer specificity | Reference/NCT number |
|---|---|---|---|---|
| AURKB/A inhibitor | Barasertib | Phase I and II | Acute myeloid leukemia, solid tumor | NCT03217838 |
| AURKA/B/C inhibitor | BI-847325 | Phase I | Solid tumor | NCT01324830 |
| AURKA inhibitor | Alisertib | Phase II | Solid tumor | NCT01898078 |
| CDKs inhibitor | Flavopiridol | Phase I and II | Myelodysplastic syndrome, acute myeloid leukemia | NCT00058240 |
| CDK8 inhibitor | SEL120 | Phase I | High-risk myelodysplastic syndrome, acute myeloid leukemia | NCT04021368 |
| PIM kinases inhibitor | SEL24/MEN1703 | Phase I and II | Acute myeloid leukemia | NCT03008187 |
| PIM447 | Phase I | Myelofibrosis | NCT02160951 | |
| JAK2 inhibitor | Ruxolitinib | Phase II | B cell lymphoma, T cell lymphoma | NCT02912754 |
| JAK1 inhibitor | Itacibinib | Preclinical | Leukemia, lymphoma | 225 |
| USP1 inhibitor | ML323 | Preclinical | Solid tumor, leukemia | 226 |
| USP2 inhibitor | ML364 | Preclinical | Colorectal cancer, mantle cell lymphoma | 227 |
| USP14/UCHL5 inhibitor | b-AP15 | Preclinical | Acute myeloid leukemia, multiple myeloma | 228 |
| USP7 inhibitor | P5091 | Preclinical | Multiple myeloma | 229 |
Abbreviation: CDKs, cyclin-dependent kinases.
5.4. Combination of histone modification enzyme inhibitors with other inhibitors
In addition to the above-mentioned histone modifying enzyme inhibitors, the combination of multiple histone inhibitors or in combination with chemotherapy and immunotherapy have been widely used in clinical trials for cancer treatments and overcoming drug resistance. A large number of HDACi have been used in combination therapy for various cancer treatment. For example, HDACi vorinostat combined with Olaparib (PARP inhibitor) or panobinostat co-treatment displayed powerful anti-cancer activity in leukemia and colon adenocarcinoma, and breast cancer by causing enhanced apoptosis of cancer cells230. Synergistic effects of histone inhibitors such as EZH2 inhibitors tazemetostat with doxorubicin plus placebo or plus the combination of rituximab, vincristine, cyclophosphamide, prednisolone, doxorubicin, and so on have been under evaluation in clinical trials for cancer treatment231. DNMT inhibitors combined with Cytarabine or Talacotuzumab or Chelated Zinc, are widely implicated in the treatment of hematologic malignancies leading to cell cycle arrest, growth inhibition, and apoptosis232. The combination of epi-drugs or with other inhibitors have displayed favorable outcomes in clinical trials for cancer therapy. Here, we briefly summarized part of the epi-drugs combination currently in clinical trials in Table 4.
Table 4.
Combination of histone modification enzyme inhibitors.
| Epi-drug | Combination | Clinical stage | Cancer specificity | NCT number |
|---|---|---|---|---|
| EZH2 inhibitor tazemetostat | Cyclophosphamide/doxorubicin/ oncovin/prednisone | Phase III | EZH2 mutant DLBCL | NCT04204941 |
| HDAC inhibitor vorinostat | Olaparid | Phase I | Breast cancer | NCT03742245 |
| HDAC inhibitor mocetinostat | Gemcitabine | Phase II | Metastatic leiomyosarcoma | NCT02303262 |
| HDAC inhibitor depsipeptide/FK228 | Alisertib/pralatrexate/gemcitabine | Phase III | Relapsed PTCL | NCT01482962 |
| HDAC inhibitor resminostat | Sorafenib | Phase I and II | Hepatocellular carcinoma | NCT02400788 |
| DNMT inhibitor disulfiram | Chelated zinc | Phase II | Melamoma | NCT02101008 |
| DNMT inhibitor azacytidine | Cytarabine | Phase III | Acute myeloid leukemia | NCT01839240 |
Abbreviations: DLBCL, diffuse large B-cell lymphoma; DNMT, DNA methyltransferases; HDAC, histone deacetylase; PTCL, peripheral T-cell lymphoma.
6. Conclusions
Histone modification is a key step in gene regulation that determines cell fate. Abnormal modifications of histone tails contribute to the development of various diseases including cancer, making targeting or reshaping aberrant histone modifications an attractive approach in cancer therapy. However, histone tail-modifying enzymes play different roles in solid tumors and hematopoietic cancers. There are four FDA-approved drugs based on histone modifier enzymes to treat hematopoietic cancers142,153, whereas only tazemetostat, an EZH2 inhibitor, was approved by the FDA for treatment of epithelioid sarcoma233. Thus, further investigations should focus on discrete histone modification patterns in solid tumors and hematopoietic cancers. In addition to the mentioned histone modification enzyme inhibitors, many other histone modification enzyme inhibitors are entering clinical trials for therapy of both solid tumors and hematological malignancies. Furthermore, the combination of these inhibitors or their combination with other drugs in clinical trials for cancer is worth further investigation. Therefore, further studies are required to reveal the functions and crosstalk of these histone modification enzymes, which may facilitate the development of more efficient cancer therapy methods with histone modification enzyme inhibitors.
Cancer/tumor is a complex and systemic disease instead of a single organ/tissue failure. Therefore, a single drug cannot cure the tumor completely resulting in tumor recurrence or resistance. Recently, a concept “network medicine” provides a potential improvement in the diagnosis, prognosis, and treatment of cancer using network science approaches and computational biology. There are several network medicine approaches in cancer, including functional epigenetic modules (FEM), oxidative bisulfite and bisulfite (OxyBS), whole-genome bisulfite sequencing (WGBS) combined with whole-genome sequencing (WGS) and weighted correlation network analysis (WGCNA)234. Network medicine integrates multiple datasets, including epigenetics, illustrating the molecular interactome to develop drugs235,236. Traditional therapeutic approaches cannot fulfill requirement of every patient because every single cancer patient has a unique tumor microenvironment. Precision medicine may be the potential strategy to fill the gap through clinical image-based deep learning architectures237. Precision medicine can not only truly reflect the clinical problems of cancer patients, but also propose unique treatment plans for specific patients. It's a very promising direction in cancer therapy.
Declaration of competing interest
The authors declare that they have no conflict of interests.
Acknowledgments
This study was supported by grants from National Natural Science Foundation of China (grant number: 42125707, 41931291), Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (grant number: 2019PT310027), Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (grant number: 2021-RC310-006, 2021-I2M-1-018, 2021-RC310-018, 2020-RC310-008), China Postdoctoral Science Foundation (grant number: 2022M710454).
Author contributions
Y.W. conceived this study. Y.K. and M.Z. wrote the original draft and prepared the figures and tables. Y.W. supervised the whole process and revised the manuscript.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;0:1–41. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 3.Waddington CH. The epigenotype. Int J Epidemiol. 2012;41(1):10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- 4.Lee JE, Kim MY. Cancer epigenetics: past, present and future. Semin Cancer Biol. 2022;83:4–14. doi: 10.1016/j.semcancer.2021.03.025. [DOI] [PubMed] [Google Scholar]
- 5.Zhao S, Allis CD, Wang GG. The language of chromatin modification in human cancers. Nat Rev Cancer. 2021;21(7):413–430. doi: 10.1038/s41568-021-00357-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribeiro-Silva C, Vermeulen W, Lans H. SWI/SNF: complex complexes in genome stability and cancer. DNA Repair. 2019;77:87–95. doi: 10.1016/j.dnarep.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Lowe BR, Maxham LA, Hamey JJ, et al. Histone H3 mutations: an updated view of their role in chromatin deregulation and cancer. Cancers (Basel) 2019;11(5):660. doi: 10.3390/cancers11050660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Wang Y. Role of epigenetic regulation in plasticity of tumor immune microenvironment. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.640369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Z, Shilatifard A. Epigenetic modifications of histones in cancer. Genome Biol. 2019;20(1):1–16. doi: 10.1186/s13059-019-1870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan JC, Maze I. Nothing is yet set in (hi)stone: novel post-translational modifications regulating chromatin function. Trends Biochem Sci. 2020;45(10):829–844. doi: 10.1016/j.tibs.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saleh R, Toor SM, Sasidharan Nair V, et al. Role of epigenetic modifications in inhibitory immune checkpoints in cancer development and progression. Front Immunol. 2020;11:1469. doi: 10.3389/fimmu.2020.01469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xhemalce B, Dawson MA, Bannister AJ. In: In Reviews in Cell Biology and Molecular Medicine. Meyers R.A., editor. 2011. Histone modifications. [DOI] [Google Scholar]
- 14.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Sharma A, Liu H, Herwig-Carl MC, et al. Epigenetic regulatory enzymes: mutation prevalence and coexistence in cancers. Cancer Investig. 2021;39(3):257–273. doi: 10.1080/07357907.2021.1872593. [DOI] [PubMed] [Google Scholar]
- 16.Mohammad F, Helin K. Oncohistones: drivers of pediatric cancers. Genes Dev. 2017;31(23-24):2313–2324. doi: 10.1101/gad.309013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 18.Martin BJE, Brind'Amour J, Kuzmin A, et al. Transcription shapes genome-wide histone acetylation patterns. Nat Commun. 2021;12(1):210. doi: 10.1038/s41467-020-20543-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kori Y, Sidoli S, Yuan ZF, et al. Proteome-wide acetylation dynamics in human cells. Sci Rep. 2017;7(1):10296. doi: 10.1038/s41598-017-09918-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen I, Poreba E, Kamieniarz K, et al. Histone modifiers in cancer: friends or foes? Genes Cancer. 2011;2(6):631–647. doi: 10.1177/1947601911417176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugiura M, Sato H, Kanesaka M, et al. Epigenetic modifications in prostate cancer. Int J Urol. 2021;28:140–149. doi: 10.1111/iju.14406. [DOI] [PubMed] [Google Scholar]
- 22.Zhan X, Guo S, Li Y, et al. Glioma stem-like cells evade interferon suppression through MBD3/NuRD complex-mediated STAT1 downregulation. J Exp Med. 2020;4(5) doi: 10.1084/jem.20191340. 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17(10):630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 24.Villanueva L, Álvarez-Errico D, Esteller M. The contribution of epigenetics to cancer immunotherapy. Trends Immunol. 2020;41(8):676–691. doi: 10.1016/j.it.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Surapaneni SK, Bhat ZR, Tikoo K. MicroRNA-941 regulates the proliferation of breast cancer cells by altering histone H3 Ser 10 phosphorylation. Sci Rep. 2020;10(1):17954. doi: 10.1038/s41598-020-74847-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komar D, Juszczynski P. Rebelled epigenome: histone H3S10 phosphorylation and H3S10 kinases in cancer biology and therapy. Clin Epigenetics. 2020;12(1):147. doi: 10.1186/s13148-020-00941-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ju J, Chen A, Deng Y, et al. NatD promotes lung cancer progression by preventing histone H4 serine phosphorylation to activate Slug expression. Nat Commun. 2017;8(1):928. doi: 10.1038/s41467-017-00988-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martire S, Gogate AA, Whitmill A, et al. Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. Nat Genet. 2019;51(6):941–946. doi: 10.1038/s41588-019-0428-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattiroli F, Penengo L. Histone ubiquitination: an integrative signaling platform in genome stability. Trends Genet. 2021;37(6):566–581. doi: 10.1016/j.tig.2020.12.005. [DOI] [PubMed] [Google Scholar]
- 30.van Mierlo G, Veenstra GJC, Vermeulen M, et al. The complexity of PRC2 subcomplexes. Trends Cell Biol. 2019;29(8):660–671. doi: 10.1016/j.tcb.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Zylicz JJ, Bousard A, Zumer K, et al. The implication of early chromatin changes in X chromosome inactivation. Cell. 2019;176(1-2):182–197. doi: 10.1016/j.cell.2018.11.041. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu H, Yang Y, Ji Q, et al. CRL4B catalyzes H2AK119 monoubiquitination and coordinates with PRC2 to promote tumorigenesis. Cancer Cell. 2012;22(6):781–795. doi: 10.1016/j.ccr.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 33.Tamburri S, Conway E, Pasini D. Polycomb-dependent histone H2A ubiquitination links developmental disorders with cancer. Trends Genet. 2022;38(4):333–352. doi: 10.1016/j.tig.2021.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Kim J, Guermah M, McGinty RK, et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137(3):459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adhikary S, Chakravarti D, Terranova C, et al. Atypical plant homeodomain of UBR7 functions as an H2BK120Ub ligase and breast tumor suppressor. Nat Commun. 2019;10(1):1398. doi: 10.1038/s41467-019-08986-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 37.Serrano-Quilez J, Roig-Soucase S, Rodriguez-Navarro S. Sharing marks: H3K4 methylation and H2B ubiquitination as features of meiotic recombination and transcription. Int J Mol Sci. 2020;21(12):4510. doi: 10.3390/ijms21124510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Zhao Z, Ozark PA, et al. Resetting the epigenetic balance of Polycomb and COMPASS function at enhancers for cancer therapy. Nat Med. 2018;24(6):758–769. doi: 10.1038/s41591-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang Y, Xu X, Ding J, et al. Histone crotonylation promotes mesoendodermal commitment of human embryonic stem cells. Cell Stem Cell. 2021;28(4):748–763. doi: 10.1016/j.stem.2020.12.009. e747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Sabari BR, Panchenko T, et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol Cell. 2016;62(2):181–193. doi: 10.1016/j.molcel.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goudarzi A, Zhang D, Huang H, et al. Dynamic competing histone H4 K5K8 acetylation and butyrylation are hallmarks of highly active gene promoters. Mol Cell. 2016;62(2):169–180. doi: 10.1016/j.molcel.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Z, Han Z, Halabelian L, et al. Identification of lysine isobutyrylation as a new histone modification mark. Nucleic Acids Res. 2021;49(1):177–189. doi: 10.1093/nar/gkaa1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan K, Rousseau J, Machol K, et al. Deficient histone H3 propionylation by BRPF1-KAT6 complexes in neurodevelopmental disorders and cancer. Sci Adv. 2020;6(4):eaax0021. doi: 10.1126/sciadv.aax0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z, Tan M, Xie Z, et al. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7(1):58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sreedhar A, Wiese EK, Hitosugi T. Enzymatic and metabolic regulation of lysine succinylation. Genes Dis. 2020;7(2):166–171. doi: 10.1016/j.gendis.2019.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X, Hou C, Yang G. Highlighted multi-modifications of enzymes: a novel succinylation mediated by histone acetyltransferase 1 in tumors. Cancer Biol Med. 2021;19(2):133–135. doi: 10.20892/j.issn.2095-3941.2021.0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jing Y, Li X, Liu Z, et al. Roles of negatively charged histone lysine acylations in regulating nucleosome structure and dynamics. Front Mol Biosci. 2022;9 doi: 10.3389/fmolb.2022.899013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liberti MV, Locasale JW. Histone lactylation: a new role for glucose metabolism. Trends Biochem Sci. 2020;45(3):179–182. doi: 10.1016/j.tibs.2019.12.004. [DOI] [PubMed] [Google Scholar]
- 49.Zhang D, Tang Z, Huang H, et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryu HY, Hochstrasser M. Histone sumoylation and chromatin dynamics. Nucleic Acids Res. 2021;49(11):6043–6052. doi: 10.1093/nar/gkab280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leonen CJA, Shimada M, Weller CE, et al. Sumoylation of the human histone H4 tail inhibits p300-mediated transcription by RNA polymerase II in cellular extracts. Elife. 2021;10:e67952. doi: 10.7554/eLife.67952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng YC, Guo YJ, Wang B, et al. Targeting neddylation E2s: a novel therapeutic strategy in cancer. J Hematol Oncol. 2021;14(1):57. doi: 10.1186/s13045-021-01070-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi CS, Kuo KL, Lin WC, et al. Neddylation inhibitor, MLN4924 suppresses angiogenesis in huvecs and solid cancers: in vitro and in vivo study. Am J Cancer Res. 2020;10(3):953–964. [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y, Du M, Yusuying S, et al. Nedd8-activating enzyme inhibitor MLN4924 (Pevonedistat), inhibits miR-1303 to suppress human breast cancer cell proliferation via targeting p27(Kip1) Exp Cell Res. 2020;392(2) doi: 10.1016/j.yexcr.2020.112038. [DOI] [PubMed] [Google Scholar]
- 55.Ferris J, Espona-Fiedler M, Hamilton C, et al. Pevonedistat (MLN4924): mechanism of cell death induction and therapeutic potential in colorectal cancer. Cell Death Discov. 2020;6:61. doi: 10.1038/s41420-020-00296-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Messner S, Hottiger MO. Histone ADP-ribosylation in DNA repair, replication and transcription. Trends Cell Biol. 2011;21(9):534–542. doi: 10.1016/j.tcb.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 57.Chen Q, Bian C, Wang X, et al. ADP-ribosylation of histone variant H2AX promotes base excision repair. EMBO J. 2021;40(2) doi: 10.15252/embj.2020104542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Q, Chen Y, Bian C, et al. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493(7433):561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu Q, Yang C, Du Y, et al. AMPK regulates histone H2B O-GlcNAcylation. Nucleic Acids Res. 2014;42(9):5594–5604. doi: 10.1093/nar/gku236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang T, Zhou X, Taghizadeh K, et al. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc Natl Acad Sci U S A. 2007;104(1):60–65. doi: 10.1073/pnas.0606775103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinberg DN, Papillon-Cavanagh S, Chen H, et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019;573(7773):281–286. doi: 10.1038/s41586-019-1534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu W, Li J, Rong B, et al. DNMT3A reads and connects histone H3K36me2 to DNA methylation. Protein Cell. 2020;11(2):150–154. doi: 10.1007/s13238-019-00672-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dukatz M, Holzer K, Choudalakis M, et al. H3K36me2/3 binding and DNA binding of the DNA methyltransferase DNMT3A PWWP domain both contribute to its chromatin interaction. J Mol Biol. 2019;431(24):5063–5074. doi: 10.1016/j.jmb.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 64.Baubec T, Colombo DF, Wirbelauer C, et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520(7546):243–247. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- 65.Cui H, Hu Y, Guo D, et al. DNA methyltransferase 3A isoform b contributes to repressing E-cadherin through cooperation of DNA methylation and H3K27/H3K9 methylation in EMT-related metastasis of gastric cancer. Oncogene. 2018;37(32):4358–4371. doi: 10.1038/s41388-018-0285-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu H, Chen Y, Liang J, et al. Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature. 2005;438(7070):981–987. doi: 10.1038/nature04225. [DOI] [PubMed] [Google Scholar]
- 67.Na F, Pan X, Chen J, et al. KMT2C deficiency promotes small cell lung cancer metastasis through DNMT3A-mediated epigenetic reprogramming. Nat Cancer. 2022;3(6):753–767. doi: 10.1038/s43018-022-00361-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grosselin K, Durand A, Marsolier J, et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat Genet. 2019;51(6):1060–1066. doi: 10.1038/s41588-019-0424-9. [DOI] [PubMed] [Google Scholar]
- 69.Hsu JM, Xia W, Hsu YH, et al. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat Commun. 2018;9(1):1908. doi: 10.1038/s41467-018-04313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darvin P, Sasidharan Nair V, Elkord E. PD-L1 expression in human breast cancer stem cells is epigenetically regulated through posttranslational histone modifications. J Oncol. 2019;2019 doi: 10.1155/2019/3958908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 72.Leslie PL, Chao YL, Tsai YH, et al. Histone deacetylase 11 inhibition promotes breast cancer metastasis from lymph nodes. Nat Commun. 2019;10(1):4192. doi: 10.1038/s41467-019-12222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Nigris F, Ruosi C, Napoli C. Clinical efficiency of epigenetic drugs therapy in bone malignancies. Bone. 2021;143 doi: 10.1016/j.bone.2020.115605. [DOI] [PubMed] [Google Scholar]
- 74.Xie Y, Shi Z, Qian Y, et al. HDAC2- and EZH2-mediated histone modifications induce PDK1 expression through miR-148a downregulation in breast cancer progression and adriamycin resistance. Cancers (Basel) 2022;14(15):3600. doi: 10.3390/cancers14153600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang K, Wang J, Yang L, et al. Targeting histone methyltransferase G9a inhibits growth and Wnt signaling pathway by epigenetically regulating HP1alpha and APC2 gene expression in non-small cell lung cancer. Mol Cancer. 2018;17(1):153. doi: 10.1186/s12943-018-0896-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rowbotham SP, Li F, Dost AFM, et al. H3K9 methyltransferases and demethylases control lung tumor-propagating cells and lung cancer progression. Nat Commun. 2018;9(1):4559. doi: 10.1038/s41467-018-07077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu Q, Tian Y, Zhang J, et al. In vivo CRISPR screening unveils histone demethylase UTX as an important epigenetic regulator in lung tumorigenesis. Proc Natl Acad Sci U S A. 2018;115(17):E3978–E3986. doi: 10.1073/pnas.1716589115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burr ML, Sparbier CE, Chan KL, et al. An evolutionarily conserved function of polycomb silences the MHC Class I antigen presentation pathway and enables immune evasion in cancer. Cancer Cell. 2019;36(4):385–401. doi: 10.1016/j.ccell.2019.08.008. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qiu Z, Zhu W, Meng H, et al. CDYL promotes the chemoresistance of small cell lung cancer by regulating H3K27 trimethylation at the CDKN1C promoter. Theranostics. 2019;9(16):4717–4729. doi: 10.7150/thno.33680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.LaFave LM, Beguelin W, Koche R, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med. 2015;21(11):1344–1349. doi: 10.1038/nm.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ezponda T, Dupere-Richer D, Will CM, et al. UTX/KDM6A loss enhances the malignant phenotype of multiple myeloma and sensitizes cells to EZH2 inhibition. Cell Rep. 2017;21(3):628–640. doi: 10.1016/j.celrep.2017.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y, Zhang H, Chen Y, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138(4):660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 83.Lu Z, Guo Y, Zhang X, et al. ORY-1001 suppresses cell growth and induces apoptosis in lung cancer through triggering HK2 mediated Warburg effect. Front Pharmacol. 2018;9:1411. doi: 10.3389/fphar.2018.01411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bauer TM, Besse B, Martinez-Marti A, et al. Phase I, open-label, dose-escalation study of the safety, pharmacokinetics, pharmacodynamics, and efficacy of GSK2879552 in relapsed/refractory SCLC. J Thorac Oncol. 2019;14(10):1828–1838. doi: 10.1016/j.jtho.2019.06.021. [DOI] [PubMed] [Google Scholar]
- 85.Colao A, de Nigris F, Modica R, et al. Clinical epigenetics of neuroendocrine tumors: the road ahead. Front Endocrinol. 2020;11 doi: 10.3389/fendo.2020.604341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ashktorab H, Belgrave K, Hosseinkhah F, et al. Global histone H4 acetylation and HDAC2 expression in colon adenoma and carcinoma. Dig Dis Sci. 2009;54(10):2109–2117. doi: 10.1007/s10620-008-0601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakazawa T, Kondo T, Ma D, et al. Global histone modification of histone H3 in colorectal cancer and its precursor lesions. Hum Pathol. 2012;43(6):834–842. doi: 10.1016/j.humpath.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 88.Karczmarski J, Rubel T, Paziewska A, et al. Hitone H3 lysine 27 acetylation is altered in colon cancer. Clin Proteom. 2014;11(1):24. doi: 10.1186/1559-0275-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tamagawa H, Oshima T, Shiozawa M, et al. The global histone modification pattern correlates with overall survival in metachronous liver metastasis of colorectal cancer. Oncol Rep. 2012;27(3):637–642. doi: 10.3892/or.2011.1547. [DOI] [PubMed] [Google Scholar]
- 90.Yokoyama Y, Hieda M, Nishioka Y, et al. Cancer-associated upregulation of histone H3 lysine 9 trimethylation promotes cell motility in vitro and drives tumor formation in vivo. Cancer Sci. 2013;104(7):889–895. doi: 10.1111/cas.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Benard A, Goossens-Beumer IJ, Hoesel AQ, et al. Histone trimethylation at H3K4, H3K9 and H4K20 correlates with patient survival and tumor recurrence in early-stage colon cancer. BMC Cancer. 2014;14:531. doi: 10.1186/1471-2407-14-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gezer U, Ustek D, Yoruker EE, et al. Characterization of H3K9me3- and H4K20me3-associated circulating nucleosomal DNA by high-throughput sequencing in colorectal cancer. Tumour Biol. 2013;34(1):329–336. doi: 10.1007/s13277-012-0554-5. [DOI] [PubMed] [Google Scholar]
- 93.Gezer U, Yoruker EE, Keskin M, et al. Histone methylation marks on circulating nucleosomes as novel blood-based biomarker in colorectal cancer. Int J Mol Sci. 2015;16(12):29654–29662. doi: 10.3390/ijms161226180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jung G, Hernandez-Illan E, Moreira L, et al. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020;17(2):111–130. doi: 10.1038/s41575-019-0230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Monga J, Subramani D, Bharathan A, et al. Pharmacological and genetic targeting of 5-lipoxygenase interrupts c-Myc oncogenic signaling and kills enzalutamide-resistant prostate cancer cells via apoptosis. Sci Rep. 2020;10(1):6649. doi: 10.1038/s41598-020-62845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ge R, Wang Z, Montironi R, et al. Epigenetic modulations and lineage plasticity in advanced prostate cancer. Ann Oncol. 2020;31(4):470–479. doi: 10.1016/j.annonc.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 97.Metzger E, Wang S, Urban S, et al. KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells. Nat Struct Mol Biol. 2019;26(5):361–371. doi: 10.1038/s41594-019-0219-9. [DOI] [PubMed] [Google Scholar]
- 98.Vatapalli R, Sagar V, Rodriguez Y, et al. Histone methyltransferase DOT1L coordinates AR and MYC stability in prostate cancer. Nat Commun. 2020;11(1):4153. doi: 10.1038/s41467-020-18013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cai C, He HH, Gao S, et al. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep. 2014;9(5):1618–1627. doi: 10.1016/j.celrep.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fan L, Zhang F, Xu S, et al. Histone demethylase JMJD1A promotes alternative splicing of AR variant 7 (AR-V7) in prostate cancer cells. Proc Natl Acad Sci U S A. 2018;115(20):E4584–E4593. doi: 10.1073/pnas.1802415115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou T, Wang S, Song X, et al. RNF8 up-regulates AR/ARV7 action to contribute to advanced prostate cancer progression. Cell Death Dis. 2022;13(4):352. doi: 10.1038/s41419-022-04787-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morishita M, Mevius D, Luccio E. In vitro histone lysine methylation by NSD1, NSD2/MMSET/WHSC1 and NSD3/WHSC1L. BMC Struct Biol. 2014;14:25. doi: 10.1186/s12900-014-0025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim SM, Kee HJ, Eom GH, et al. Characterization of a novel WHSC1-associated SET domain protein with H3K4 and H3K27 methyltransferase activity. Biochem Biophys Res Commun. 2006;345(1):318–323. doi: 10.1016/j.bbrc.2006.04.095. [DOI] [PubMed] [Google Scholar]
- 104.Kang HB, Choi Y, Lee JM, et al. The histone methyltransferase, NSD2, enhances androgen receptor-mediated transcription. FEBS Lett. 2009;583(12):1880–1886. doi: 10.1016/j.febslet.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 105.Yousefi B, Mohammadlou M, Abdollahi M, et al. Epigenetic changes in gastric cancer induction by Helicobacter pylori. J Cell Physiol. 2019;234(12):21770–21784. doi: 10.1002/jcp.28925. [DOI] [PubMed] [Google Scholar]
- 106.Rivas-Ortiz CI, Lopez-Vidal Y, Arredondo-Hernandez LJR, et al. Genetic alterations in gastric cancer associated with helicobacter pylori infection. Front Med. 2017;4:47. doi: 10.3389/fmed.2017.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yang TT, Cao N, Zhang HH, et al. Helicobacter pylori infection-induced H3Ser10 phosphorylation in stepwise gastric carcinogenesis and its clinical implications. Helicobacter. 2018;23(3):e12486. doi: 10.1111/hel.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pathak SK, Basu S, Bhattacharyya A, et al. TLR4-dependent NF-kappaB activation and mitogen- and stress-activated protein kinase 1-triggered phosphorylation events are central to Helicobacter pylori peptidyl prolyl cis-, trans-isomerase (HP0175)-mediated induction of IL-6 release from macrophages. J Immunol. 2006;177(11):7950–7958. doi: 10.4049/jimmunol.177.11.7950. [DOI] [PubMed] [Google Scholar]
- 109.Fehri LF, Rechner C, Janssen S, et al. Helicobacter pylori-induced modification of the histone H3 phosphorylation status in gastric epithelial cells reflects its impact on cell cycle regulation. Epigenetics. 2009;4(8):577–586. doi: 10.4161/epi.4.8.10217. [DOI] [PubMed] [Google Scholar]
- 110.Xia G, Schneider-Stock R, Diestel A, et al. Helicobacter pylori regulates p21(WAF1) by histone H4 acetylation. Biochem Biophys Res Commun. 2008;369(2):526–531. doi: 10.1016/j.bbrc.2008.02.073. [DOI] [PubMed] [Google Scholar]
- 111.Mitani Y, Oue N, Hamai Y, et al. Histone H3 acetylation is associated with reduced p21(WAF1/CIP1) expression by gastric carcinoma. J Pathol. 2005;205(1):65–73. doi: 10.1002/path.1684. [DOI] [PubMed] [Google Scholar]
- 112.Han F, Ren J, Zhang J, et al. JMJD2B is required for Helicobacter pylori-induced gastric carcinogenesis via regulating COX-2 expression. Oncotarget. 2016;7(25):38626–38637. doi: 10.18632/oncotarget.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Park YS, Jin MY, Kim YJ, et al. The global histone modification pattern correlates with cancer recurrence and overall survival in gastric adenocarcinoma. Ann Surg Oncol. 2008;15(7):1968–1976. doi: 10.1245/s10434-008-9927-9. [DOI] [PubMed] [Google Scholar]
- 114.Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4(9):688–694. doi: 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- 115.Hayashi Y, Tsujii M, Wang J, et al. CagA mediates epigenetic regulation to attenuate let-7 expression in Helicobacter pylori-related carcinogenesis. Gut. 2013;62(11):1536–1546. doi: 10.1136/gutjnl-2011-301625. [DOI] [PubMed] [Google Scholar]
- 116.Hamdane N, Juhling F, Crouchet E, et al. HCV-induced epigenetic changes associated with liver cancer risk persist after sustained virologic response. Gastroenterology. 2019;156(8):2313–2329. doi: 10.1053/j.gastro.2019.02.038. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jiang B, Yang B, Wang Q, et al. lncRNA PVT1 promotes hepatitis B viruspositive liver cancer progression by disturbing histone methylation on the cMyc promoter. Oncol Rep. 2020;43(2):718–726. doi: 10.3892/or.2019.7444. [DOI] [PubMed] [Google Scholar]
- 118.Han TS, Ban HS, Hur K, et al. The epigenetic regulation of HCC metastasis. Int J Mol Sci. 2018;19(12):3978. doi: 10.3390/ijms19123978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wei L, Chiu DK, Tsang FH, et al. Histone methyltransferase G9a promotes liver cancer development by epigenetic silencing of tumor suppressor gene RARRES3. J Hepatol. 2017;67(4):758–769. doi: 10.1016/j.jhep.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 120.Tang B, Qi G, Tang F, et al. JARID1B promotes metastasis and epithelial-mesenchymal transsition via PTEN/AKT signaling in hepatocellular carcinoma cells. Oncotarget. 2015;6(14):12723–12739. doi: 10.18632/oncotarget.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen Z, Wang X, Liu R, et al. KDM4B-mediated epigenetic silencing of miRNA-615-5p augments RAB24 to facilitate malignancy of hepatoma cells. Oncotarget. 2017;8(11):17712–17725. doi: 10.18632/oncotarget.10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ji X, Jin S, Qu X, et al. Lysine-specific demethylase 5C promotes hepatocellular carcinoma cell invasion through inhibition BMP7 expression. BMC Cancer. 2015;15:801. doi: 10.1186/s12885-015-1798-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang Y, Huang J, Li Q, et al. Histone methyltransferase SETDB1 promotes cells proliferation and migration by interacting withTiam1 in hepatocellular carcinoma. BMC Cancer. 2018;18(1):539. doi: 10.1186/s12885-018-4464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wong CM, Wei L, Law CT, et al. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology. 2016;63(2):474–487. doi: 10.1002/hep.28304. [DOI] [PubMed] [Google Scholar]
- 125.Liu C, Liu L, Shan J, et al. Histone deacetylase 3 participates in self-renewal of liver cancer stem cells through histone modification. Cancer Lett. 2013;339(1):60–69. doi: 10.1016/j.canlet.2013.07.022. [DOI] [PubMed] [Google Scholar]
- 126.Zhang HL, Chen P, Yan HX, et al. Targeting mTORC2/HDAC3 inhibits stemness of liver cancer cells against glutamine starvation. Adv Sci (Weinh) 2022;9(20) doi: 10.1002/advs.202103887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ji H, Zhou Y, Zhuang X, et al. HDAC3 deficiency promotes liver cancer through a defect in H3K9ac/H3K9me3 transition. Cancer Res. 2019;79(14):3676–3688. doi: 10.1158/0008-5472.CAN-18-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hu D, Shilatifard A. Epigenetics of hematopoiesis and hematological malignancies. Genes Dev. 2016;30(18):2021–2041. doi: 10.1101/gad.284109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.van Dijk AD, Hu CW, de Bont E, et al. Histone modification patterns using RPPA-based profiling predict outcome in acute myeloid leukemia patients. Proteomics. 2018;18(8) doi: 10.1002/pmic.201700379. [DOI] [PubMed] [Google Scholar]
- 130.Li Y, Zhang M, Sheng M, et al. Therapeutic potential of GSK-J4, a histone demethylase KDM6B/JMJD3 inhibitor, for acute myeloid leukemia. J Cancer Res Clin Oncol. 2018;144(6):1065–1077. doi: 10.1007/s00432-018-2631-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.van Dijk AD, Hoff FW, Qiu Y, et al. Loss of H3K27 methylation identifies poor outcome in adult-onset acute myeloid leukemia. Blood. 2020;136(Supplement 1):24. doi: 10.1182/blood-2020-139387. -24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gollner S, Oellerich T, Agrawal-Singh S, et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med. 2017;23(1):69–78. doi: 10.1038/nm.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rabello Ddo A, Lucena-Araujo AR, Alves-Silva JC, et al. Overexpression of EZH2 associates with a poor prognosis in chronic lymphocytic leukemia. Blood Cells Mol Dis. 2015;54(1):97–102. doi: 10.1016/j.bcmd.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 134.Chartomatsidou E, Ntoufa S, Kotta K, et al. Inhibition of EZH2 and immune signaling exerts synergistic antitumor effects in chronic lymphocytic leukemia. Blood Adv. 2019;3(12):1891–1896. doi: 10.1182/bloodadvances.2018030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dhall A, Zee BM, Yan F, et al. Intersection of epigenetic and metabolic regulation of histone modifications in acute myeloid leukemia. Front Oncol. 2019;9:432. doi: 10.3389/fonc.2019.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huang YH, Cai K, Xu PP, et al. CREBBP/EP300 mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-associated macrophage polarization via FBXW7-NOTCH-CCL2/CSF1 axis. Signal Transduct Target Ther. 2021;6(1):10. doi: 10.1038/s41392-020-00437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ortega-Molina A, Boss IW, Canela A, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21(10):1199–1208. doi: 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang J, Dominguez-Sola D, Hussein S, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21(10):1190–1198. doi: 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ying CY, Dominguez-Sola D, Fabi M, et al. MEF2B mutations lead to deregulated expression of the oncogene BCL6 in diffuse large B cell lymphoma. Nat Immunol. 2013;14(10):1084–1092. doi: 10.1038/ni.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Brescia P, Schneider C, Holmes AB, et al. MEF2B instructs germinal center development and acts as an oncogene in B cell lymphomagenesis. Cancer Cell. 2018;34(3):453–465. doi: 10.1016/j.ccell.2018.08.006. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang Q, Wang S, Chen J, et al. Histone deacetylases (HDACs) guided novel therapies for T-cell lymphomas. Int J Med Sci. 2019;16(3):424–442. doi: 10.7150/ijms.30154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Keats JJ, Reiman T, Maxwell CA, et al. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood. 2003;101(4):1520–1529. doi: 10.1182/blood-2002-06-1675. [DOI] [PubMed] [Google Scholar]
- 144.Santra M, Zhan F, Tian E, et al. A subset of multiple myeloma harboring the t(4;14)(p16;q32) translocation lacks FGFR3 expression but maintains an IGH/MMSET fusion transcript. Blood. 2003;101(6):2374–2376. doi: 10.1182/blood-2002-09-2801. [DOI] [PubMed] [Google Scholar]
- 145.Martinez-Garcia E, Popovic R, Min DJ, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2011;117(1):211–220. doi: 10.1182/blood-2010-07-298349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kuo AJ, Cheung P, Chen K, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44(4):609–620. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Marango J, Shimoyama M, Nishio H, et al. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood. 2008;111(6):3145–3154. doi: 10.1182/blood-2007-06-092122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Min DJ, Ezponda T, Kim MK, et al. MMSET stimulates myeloma cell growth through microRNA-mediated modulation of c-MYC. Leukemia. 2013;27(3):686–694. doi: 10.1038/leu.2012.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ohguchi H, Hideshima T, Anderson KC. The biological significance of histone modifiers in multiple myeloma: clinical applications. Blood Cancer J. 2018;8(9):83. doi: 10.1038/s41408-018-0119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Minami J, Suzuki R, Mazitschek R, et al. Histone deacetylase 3 as a novel therapeutic target in multiple myeloma. Leukemia. 2014;28(3):680–689. doi: 10.1038/leu.2013.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Harada T, Ohguchi H, Grondin Y, et al. HDAC3 regulates DNMT1 expression in multiple myeloma: therapeutic implications. Leukemia. 2017;31(12):2670–2677. doi: 10.1038/leu.2017.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.van de Donk NWCJ, Pawlyn C, Yong KL. Multiple myeloma. Lancet. 2021;397(10272):410–427. doi: 10.1016/S0140-6736(21)00135-5. [DOI] [PubMed] [Google Scholar]
- 153.Raedler LA. Farydak (Panobinostat): first HDAC inhibitor approved for patients with relapsed multiple myeloma. Am Health Drug Benefits. 2016;9(Spec Feature):84–87. [PMC free article] [PubMed] [Google Scholar]
- 154.Fagan RJ, Dingwall AK. COMPASS ascending: emerging clues regarding the roles of MLL3/KMT2C and MLL2/KMT2D proteins in cancer. Cancer Lett. 2019;458:56–65. doi: 10.1016/j.canlet.2019.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wu L, Kou F, Ji Z, et al. SMYD2 promotes tumorigenesis and metastasis of lung adenocarcinoma through RPS7. Cell Death Dis. 2021;12(5):439. doi: 10.1038/s41419-021-03720-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Au YZ, Gu M, De Braekeleer E, et al. KAT7 is a genetic vulnerability of acute myeloid leukemias driven by MLL rearrangements. Leukemia. 2021;35(4):1012–1022. doi: 10.1038/s41375-020-1001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang P, Wang Z, Liu J. Role of HDACs in normal and malignant hematopoiesis. Mol Cancer. 2020;19(1):5. doi: 10.1186/s12943-019-1127-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Harmeyer KM, Facompre ND, Herlyn M, et al. JARID1 histone demethylases: emerging targets in cancer. Trends Cancer. 2017;3(10):713–725. doi: 10.1016/j.trecan.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Saha N, Muntean AG. Insight into the multi-faceted role of the SUV family of H3K9 methyltransferases in carcinogenesis and cancer progression. Biochim Biophys Acta Rev Cancer. 2021;1875(1) doi: 10.1016/j.bbcan.2020.188498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Strepkos D, Markouli M, Klonou A, et al. Histone methyltransferase SETDB1: a common denominator of tumorigenesis with therapeutic potential. Cancer Res. 2021;81(3):525–534. doi: 10.1158/0008-5472.Can-20-2906. [DOI] [PubMed] [Google Scholar]
- 161.Hu C, Zhang M, Moses N, et al. The USP10-HDAC6 axis confers cisplatin resistance in non-small cell lung cancer lacking wild-type p53. Cell Death Dis. 2020;11(5):328. doi: 10.1038/s41419-020-2519-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang X, Wang H, Xu B, et al. Depletion of H3K79 methyltransferase Dot1L promotes cell invasion and cancer stem-like cell property in ovarian cancer. Am J Transl Res. 2019;11(2):1145–1153. [PMC free article] [PubMed] [Google Scholar]
- 163.Hogg SJ, Beavis PA, Dawson MA, et al. Targeting the epigenetic regulation of antitumour immunity. Nat Rev Drug Discov. 2020;19(11):776–800. doi: 10.1038/s41573-020-0077-5. [DOI] [PubMed] [Google Scholar]
- 164.Wang YM, Gu ML, Meng FS, et al. Histone acetyltransferase p300/CBP inhibitor C646 blocks the survival and invasion pathways of gastric cancer cell lines. Int J Oncol. 2017;51(6):1860–1868. doi: 10.3892/ijo.2017.4176. [DOI] [PubMed] [Google Scholar]
- 165.Brown JA, Bourke E, Eriksson LA, et al. Targeting cancer using KAT inhibitors to mimic lethal knockouts. Biochem Soc Trans. 2016;44(4):979–986. doi: 10.1042/bst20160081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ye F, Huang J, Wang H, et al. Targeting epigenetic machinery: emerging novel allosteric inhibitors. Pharmacol Ther. 2019;204 doi: 10.1016/j.pharmthera.2019.107406. [DOI] [PubMed] [Google Scholar]
- 167.Ho TCS, Chan AHY, Ganesan A. Thirty years of HDAC inhibitors: 2020 insight and hindsight. J Med Chem. 2020;63(21):12460–12484. doi: 10.1021/acs.jmedchem.0c00830. [DOI] [PubMed] [Google Scholar]
- 168.Marks P, Rifkind RA, Richon VM, et al. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001;1(3):194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 169.Romero D. HDAC inhibitors tested in phase III trial. Nat Rev Clin Oncol. 2019;16(8):465. doi: 10.1038/s41571-019-0224-2. [DOI] [PubMed] [Google Scholar]
- 170.Manal M, Chandrasekar MJ, Gomathi Priya J, et al. Inhibitors of histone deacetylase as antitumor agents: a critical review. Bioorg Chem. 2016;67:18–42. doi: 10.1016/j.bioorg.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 171.Singh AK, Bishayee A, Pandey AK. Targeting histone deacetylases with natural and synthetic agents: an emerging anticancer strategy. Nutrients. 2018;10(6):731. doi: 10.3390/nu10060731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gao C, Bourke E, Scobie M, et al. Rational design and validation of a Tip60 histone acetyltransferase inhibitor. Sci Rep. 2014;4:5372. doi: 10.1038/srep05372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Coffey DC, Kutko MC, Glick RD, et al. The histone deacetylase inhibitor, CBHA, inhibits growth of human neuroblastoma xenografts in vivo, alone and synergistically with all trans retinoic acid. Cancer Res. 2001;61(9):3591–3594. PMID: 11325825. [PubMed] [Google Scholar]
- 174.Balliu M, Guandalini L, Romanelli MN, et al. HDAC-inhibitor (S)-8 disrupts HDAC6-PP1 complex prompting A375 melanoma cell growth arrest and apoptosis. J Cell Mol Med. 2015;19(1):143–154. doi: 10.1111/jcmm.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Eich ML, Athar M, Ferguson JE, 3rd, et al. EZH2-targeted therapies in cancer: hype or a reality. Cancer Res. 2020;80(24):5449–5458. doi: 10.1158/0008-5472.Can-20-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cao H, Li L, Yang D, et al. Recent progress in histone methyltransferase (G9a) inhibitors as anticancer agents. Eur J Med Chem. 2019;179:537–546. doi: 10.1016/j.ejmech.2019.06.072. [DOI] [PubMed] [Google Scholar]
- 177.Biswas S, Rao CM. Epigenetic tools (The Writers, The Readers and The Erasers) and their implications in cancer therapy. Eur J Pharmacol. 2018;837:8–24. doi: 10.1016/j.ejphar.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 178.Li LX, Zhou JX, Calvet JP, et al. Lysine methyltransferase SMYD2 promotes triple negative breast cancer progression. Cell Death Dis. 2018;9(3):326. doi: 10.1038/s41419-018-0347-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nguyen H, Allali-Hassani A, Antonysamy S, et al. LLY-507, a cell-active, potent, and selective inhibitor of protein-lysine methyltransferase SMYD2. J Biol Chem. 2015;290(22):13641–13653. doi: 10.1074/jbc.M114.626861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cao K, Ugarenko M, Ozark PA, et al. DOT1L-controlled cell-fate determination and transcription elongation are independent of H3K79 methylation. Proc Natl Acad Sci U S A. 2020;117(44):27365–27373. doi: 10.1073/pnas.2001075117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu W, Deng L, Song Y, et al. DOT1L inhibition sensitizes MLL-rearranged AML to chemotherapy. PLoS One. 2014;9(5):e98270. doi: 10.1371/journal.pone.0098270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kaniskan HU, Martini ML, Jin J. Inhibitors of protein methyltransferases and demethylases. Chem Rev. 2018;118(3):989–1068. doi: 10.1021/acs.chemrev.6b00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cheng D, Yadav N, King RW, et al. Small molecule regulators of protein arginine methyltransferases. J Biol Chem. 2004;279(23):23892–23899. doi: 10.1074/jbc.M401853200. [DOI] [PubMed] [Google Scholar]
- 184.Kaniskan HU, Szewczyk MM, Yu Z, et al. A potent, selective and cell-active allosteric inhibitor of protein arginine methyltransferase 3 (PRMT3) Angew Chem Int Ed Engl. 2015;54(17):5166–5170. doi: 10.1002/anie.201412154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Nakayama K, Szewczyk MM, Sena CD, et al. TP-064, a potene and selective small molecule inhibitor of PRMT4 for multiple myeloma. Oncotarget. 2018;9(26):18480–18493. doi: 10.18632/oncotarget.24883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Drew AE, Moradei O, Jacques SL, et al. Identification of a CARM1 inhibitor with potent in vitro and in vivo activity in preclinical models of multiple myeloma. Sci Rep. 2017;7(1):17993. doi: 10.1038/s41598-017-18446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bonday ZQ, Cortez GS, Grogan MJ, et al. LLY-283, a potent and selective inhibitor of arginine methyltransferase 5, PRMT5, with antitumor activity. ACS Med Chem Lett. 2018;9(7):612–617. doi: 10.1021/acsmedchemlett.8b00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.AbuHammad S, Cullinane C, Martin C, et al. Regulation of PRMT5-MDM4 axis is critical in the response to CDK4/6 inhibitors in melanoma. Proc Natl Acad Sci U S A. 2019;116(36):17990–18000. doi: 10.1073/pnas.1901323116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Brehmer D, Beke L, Wu T, et al. Discovery and pharmacological characterization of JNJ-64619178, a novel small-molecule inhibitor of PRMT5 with potent antitumor activity. Mol Cancer Ther. 2021;20(12):2317–2328. doi: 10.1158/1535-7163.MCT-21-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Jensen-Pergakes K, Tatlock J, Maegley KA, et al. SAM-Competitive PRMT5 inhibitor PF-06939999 demonstrates antitumor activity in splicing dysregulated NSCLC with decreased liability of drug resistance. Mol Cancer Ther. 2022;21(1):3–15. doi: 10.1158/1535-7163.Mct-21-0620. [DOI] [PubMed] [Google Scholar]
- 191.Mitchell LH, Drew AE, Ribich SA, et al. Aryl pyrazoles as potent inhibitors of arginine methyltransferases: identification of the first PRMT6 tool compound. ACS Med Chem Lett. 2015;6(6):655–659. doi: 10.1021/acsmedchemlett.5b00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dimitrova E, Turberfield AH, Klose RJ. Histone demethylases in chromatin biology and beyond. EMBO Rep. 2015;16(12):1620–1639. doi: 10.15252/embr.201541113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gao S, Chen S, Han D, et al. Chromatin binding of FOXA1 is promoted by LSD1-mediated demethylation in prostate cancer. Nat Genet. 2020;52(10):1011–1017. doi: 10.1038/s41588-020-0681-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Fang Y, Liao G, Yu B. LSD1/KDM1A inhibitors in clinical trials: advances and prospects. J Hematol Oncol. 2019;12(1):129. doi: 10.1186/s13045-019-0811-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.He X, Zhang H, Zhang Y, et al. Drug discovery of histone lysine demethylases (KDMs) inhibitors (progress from 2018 to present) Eur J Med Chem. 2022;231 doi: 10.1016/j.ejmech.2022.114143. [DOI] [PubMed] [Google Scholar]
- 196.Macedo-Silva C, Miranda-Gonçalves V, Lameirinhas A, et al. JmjC-KDMs KDM3A and KDM6B modulate radioresistance under hypoxic conditions in esophageal squamous cell carcinoma. Cell Death Dis. 2020;11(12):1068. doi: 10.1038/s41419-020-03279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kim K, Son MY, Jung CR, et al. EHMT2 is a metastasis regulator in breast cancer. Biochem Biophys Res Commun. 2018;496(2):758–762. doi: 10.1016/j.bbrc.2018.01.074. [DOI] [PubMed] [Google Scholar]
- 198.Vitkeviciene A, Baksiene S, Borutinskaite V, et al. Epigallocatechin-3-gallate and BIX-01294 have different impact on epigenetics and senescence modulation in acute and chronic myeloid leukemia cells. Eur J Pharmacol. 2018;838:32–40. doi: 10.1016/j.ejphar.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 199.Vedadi M, Barsyte-Lovejoy D, Liu F, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol. 2011;7(8):566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sweis RF, Pliushchev M, Brown PJ, et al. Discovery and development of potent and selective inhibitors of histone methyltransferase g9a. ACS Med Chem Lett. 2014;5(2):205–209. doi: 10.1021/ml400496h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yuan Y, Wang Q, Paulk J, et al. A small-molecule probe of the histone methyltransferase G9a induces cellular senescence in pancreatic adenocarcinoma. ACS Chem Biol. 2012;7(7):1152–1157. doi: 10.1021/cb300139y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Li LX, Fan LX, Zhou JX, et al. Lysine methyltransferase SMYD2 promotes cyst growth in autosomal dominant polycystic kidney disease. J Clin Investig. 2017;127(7):2751–2764. doi: 10.1172/jci90921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Knutson SK, Wigle TJ, Warholic NM, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8(11):890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 204.Konze KD, Ma A, Li F, et al. An orally bioavailable chemical probe of the lysine methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013;8(6):1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Verma SK, Tian X, LaFrance LV, et al. Identification of potent, selective, cell-active inhibitors of the histone lysine methyltransferase EZH2. ACS Med Chem Lett. 2012;3(12):1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Qi W, Chan H, Teng L, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A. 2012;109(52):21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hayden A, Johnson PW, Packham G, et al. S-adenosylhomocysteine hydrolase inhibition by 3-deazaneplanocin A analogues induces anti-cancer effects in breast cancer cell lines and synergy with both histone deacetylase and HER2 inhibition. Breast Cancer Res Treat. 2011;127(1):109–119. doi: 10.1007/s10549-010-0982-0. [DOI] [PubMed] [Google Scholar]
- 208.Zhang Y, Verwilligen RAF, de Boer M, et al. PRMT4 inhibitor TP-064 impacts both inflammatory and metabolic processes without changing the susceptibility for early atherosclerotic lesions in male apolipoprotein E knockout mice. Atherosclerosis. 2021;338:23–29. doi: 10.1016/j.atherosclerosis.2021.11.001. [DOI] [PubMed] [Google Scholar]
- 209.Prusevich P, Kalin JH, Ming SA, et al. A selective phenelzine analogue inhibitor of histone demethylase LSD1. ACS Chem Biol. 2014;9(6):1284–1293. doi: 10.1021/cb500018s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zhu Q, Huang Y, Marton LJ, et al. Polyamine analogs modulate gene expression by inhibiting lysine-specific demethylase 1 (LSD1) and altering chromatin structure in human breast cancer cells. Amino Acids. 2012;42(2-3):887–898. doi: 10.1007/s00726-011-1004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Willmann D, Lim S, Wetzel S, et al. Impairment of prostate cancer cell growth by a selective and reversible lysine-specific demethylase 1 inhibitor. Int J Cancer. 2012;131(11):2704–2709. doi: 10.1002/ijc.27555. [DOI] [PubMed] [Google Scholar]
- 212.Ogasawara D, Itoh Y, Tsumoto H, et al. Lysine-specific demethylase 1-selective inactivators: protein-targeted drug delivery mechanism. Angew Chem Int Ed Engl. 2013;52(33):8620–8624. doi: 10.1002/anie.201303999. [DOI] [PubMed] [Google Scholar]
- 213.Wang L, Chang J, Varghese D, et al. A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat Commun. 2013;4:2035. doi: 10.1038/ncomms3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hopkinson RJ, Tumber A, Yapp C, et al. 5-carboxy-8-hydroxyquinoline is a broad spectrum 2-oxoglutarate oxygenase inhibitor which causes iron translocation. Chem Sci. 2013;4(8):3110–3117. doi: 10.1039/c3sc51122g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tang A, Gao K, Chu L, et al. Aurora kinases: novel therapy targets in cancers. Oncotarget. 2017;8(14):23937–23954. doi: 10.18632/oncotarget.14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Yu M, Teo T, Yang Y, et al. Potent and orally bioavailable CDK8 inhibitors: design, synthesis, structure-activity relationship analysis and biological evaluation. Eur J Med Chem. 2021;214 doi: 10.1016/j.ejmech.2021.113248. [DOI] [PubMed] [Google Scholar]
- 217.Ammazzalorso A, Agamennone M, De Filippis B, et al. Development of CDK4/6 inhibitors: a five years update. Molecules. 2021;26(5):1488. doi: 10.3390/molecules26051488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wu J, Chu E, Kang Y. PIM kinases in multiple myeloma. Cancers (Basel) 2021;13(17):4304. doi: 10.3390/cancers13174304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Szydłowski M, Garbicz F, Jabłońska E, et al. Inhibition of PIM kinases in DLBCL targets MYC transcriptional program and augments the efficacy of anti-CD20 antibodies. Cancer Res. 2021;81(23):6029–6043. doi: 10.1158/0008-5472.Can-21-1023. [DOI] [PubMed] [Google Scholar]
- 220.Consortium APG AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7(8):818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wehde BL, Radler PD, Shrestha H, et al. Janus kinase 1 plays a critical role in mammary cancer progression. Cell Rep. 2018;25(8):2192–2207. doi: 10.1016/j.celrep.2018.10.063. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Beatty GL, Shahda S, Beck T, et al. A phase Ib/II study of the JAK1 inhibitor, itacitinib, plus nab-paclitaxel and gemcitabine in advanced solid tumors. Oncologist. 2019;24(1):14. doi: 10.1634/theoncologist.2017-0665. -e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Young MJ, Hsu KC, Lin TE, et al. The role of ubiquitin-specific peptidases in cancer progression. J Biomed Sci. 2019;26(1):42. doi: 10.1186/s12929-019-0522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Gutierrez-Diaz BT, Gu W, Ntziachristos P. Deubiquitinases: pro-oncogenic activity and therapeutic targeting in blood malignancies. Trends Immunol. 2020;41(4):327–340. doi: 10.1016/j.it.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Huarte E, O'Connor RS, Peel MT, et al. Itacitinib (INCB039110), a JAK1 inhibitor, reduces cytokines associated with cytokine release syndrome induced by CAR T-cell therapy. Clin Cancer Res. 2020;26(23):6299–6309. doi: 10.1158/1078-0432.Ccr-20-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Liang Q, Dexheimer TS, Zhang P, et al. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat Chem Biol. 2014;10(4):298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Davis MI, Pragani R, Fox JT, et al. Small molecule inhibition of the ubiquitin-specific protease USP2 accelerates cyclin D1 degradation and leads to cell cycle arrest in colorectal cancer and mantle cell lymphoma models. J Biol Chem. 2016;291(47):24628–24640. doi: 10.1074/jbc.M116.738567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ma YS, Wang XF, Yu F, et al. Inhibition of USP14 and UCH37 deubiquitinating activity by b-AP15 as a potential therapy for tumors with p53 deficiency. Signal Transduct Target Ther. 2020;5(1):30. doi: 10.1038/s41392-020-0143-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.He Y, Wang S, Tong J, et al. The deubiquitinase USP7 stabilizes Maf proteins to promote myeloma cell survival. J Biol Chem. 2020;295(7):2084–2096. doi: 10.1074/jbc.RA119.010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Chao OS, Goodman OB., Jr Synergistic loss of prostate cancer cell viability by co-inhibition of HDAC and PARP. Mol Cancer Res. 2014;12(12):1755–1766. doi: 10.1158/1541-7786.Mcr-14-0173. [DOI] [PubMed] [Google Scholar]
- 231.Duan R, Du W, Guo W. EZH2: a novel target for cancer treatment. J Hematol Oncol. 2020;13(1):104. doi: 10.1186/s13045-020-00937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lu Y, Chan YT, Tan HY, et al. Epigenetic regulation in human cancer: the potential role of epi-drug in cancer therapy. Mol Cancer. 2020;19(1):79. doi: 10.1186/s12943-020-01197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hoy SM. Tazemetostat: first approval. Drugs. 2020;80(5):513–521. doi: 10.1007/s40265-020-01288-x. [DOI] [PubMed] [Google Scholar]
- 234.Sarno F, Benincasa G, List M, et al. Clinical epigenetics settings for cancer and cardiovascular diseases: real-life applications of network medicine at the bedside. Clin Epigenetics. 2021;13(1) doi: 10.1186/s13148-021-01047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Silverman EK, Schmidt H, Anastasiadou E, et al. Molecular networks in network medicine: development and applications. Wiley Interdiscip Rev Syst Biol Med. 2020;12(6):e1489. doi: 10.1002/wsbm.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kurnat-Thoma E, Baranova A, Baird P, et al. Recent advances in systems and network medicine: meeting report from the first international conference in systems and network medicine. Syst Med. 2020;3(1):22–35. doi: 10.1089/sysm.2020.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Boehm KM, Khosravi P, Vanguri R, et al. Harnessing multimodal data integration to advance precision oncology. Nat Rev Cancer. 2022;22(2):114–126. doi: 10.1038/s41568-021-00408-3. [DOI] [PMC free article] [PubMed] [Google Scholar]