ABSTRACT
We present the complete mitochondrial genome of Carausius morosus from Salinas, CA. The mitochondrial genome of C. morosus is circular, AT rich (78.1%), and 16,671 bp in length. It consists of 13 protein-coding, 22 transfer RNA, and 2 ribosomal RNA genes and is identical in gene content to Carausius sp.
KEYWORDS: mitogenome, non-native, Phasmatodea
ANNOUNCEMENT
Carausius morosus Brunner Von Wattenwyl 1907, the Indian walking stick, is a large polyphagous species originally described from specimens from Shembagonor and Trichinopoly, Madura Province, southern India (1, 2). It was introduced to Australia, Azores, Madagascar, Madeira, South Africa, United Kingdom, and United States (3), likely due to the escaping of captive C. morosus and the accidental discarding of eggs (4). Carausius morosus was first reported from San Diego County, CA, in 1991 and has since spread to 10 counties where it is a pest on ornamental plants (4, 5). According to the California Department of Food and Agriculture, C. morosus has a high environmental impact score but a low overall pest rating (6). To contribute to the phylogenomics of Phasmatodea and the bioinformatics of C. morosus, the complete mitochondrial genome of C. morosus from California was assembled and characterized.
The specimen of C. morosus analyzed here (Fig. 1A) was collected from a coffee house stockroom in Salinas, Monterey County, CA (36.7000 N 121.6201 W), voucher number Hartnell College Collection #272 (Salinas, CA). The DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen) following a previously published protocol (7). The 150-bp paired-end library was constructed with the KAPA HyperPlus Kit (Roche) and sequenced on an Illumina NovaSeq 6000 (Illumina Inc.). The sequencing generated 12,826,468 reads that were filtered using the default BBDuk 1.0 (8) settings in Geneious Prime 2019.1.3 (Biomatters Limited). The mitochondrial genome was assembled de novo using a kmer ≥ 79 with MEGAHIT 1.2.9 (9). The assembly yielded 97,733 contigs with an N50 of 469 and GC content of 37.5%. A single C. morosus mitochondrial contig with 119-bp overlapping ends and 49.5× coverage was identified by a two-way Nucleotide BLAST 2.15.0+ search using the default settings (10). The overlapping ends were removed manually, and the mitochondrial genome start position was adjusted to correspond with Carausius sp. (GenBank accession number OQ682524). The annotation was performed in Geneious Prime designating the Carausius sp. mitochondrial genome as the reference with a similarity index of 65%. Gene start and stop positions were confirmed by comparison to 56 complete mitochondrial genomes of Phasmatodea in GenBank and using standard, universally accepted initiation and termination codons as defined in the invertebrate mitochondrial genetic code (11, 12). Nucleotide identities were calculated by BLAST search using the default settings.
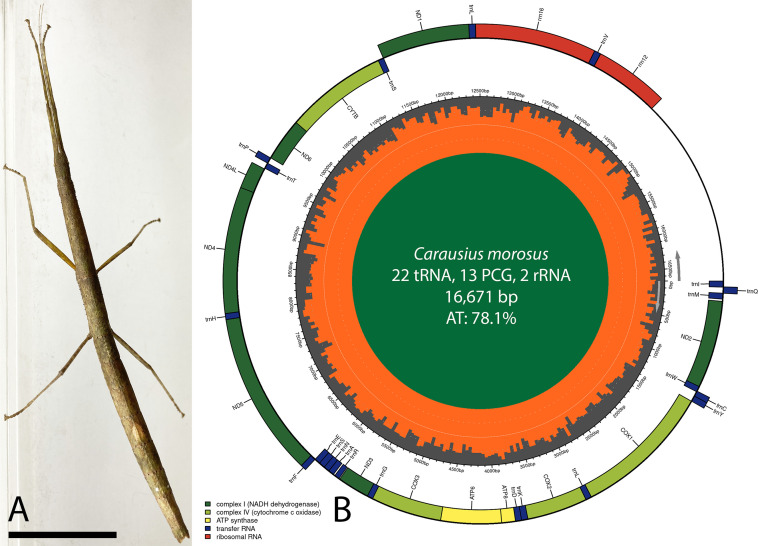
Fig 1.
Carausius morosus analyzed in this study (A) and the complete mitochondrial genome map of C. morosus (B). (A) Specimen of C. morosus genome sequenced. Scale bar = 2 cm. (B) The genome was annotated using Geneious Prime and mapped with Chloroplot 0.2.4 (13). The innermost ring displays the AT content in Saffron color and the direction of transcription, as indicated by the arrows. The final ring displays the genes. Genes transcribed clockwise are on the inside, while counterclockwise transcriptions are positioned on the outside. The color coding corresponds to genes of different groups as listed in the key in the bottom left. PCG, protein-coding genes.
The complete circular mitochondrial genome of C. morosus is 16,671 bp in length and has a AT bias of 78.1%. Genome content and structure are identical to Carausius sp. (14). The genome contains 37 genes including 13 protein-coding, 22 tRNA, and 2 rRNA genes (Fig. 1B). Five of the protein-coding genes initiate with ATA (ATP6, ATP8, COX2, ND2, and ND4L), three with ATT (ND3, ND5, and ND6) and ATG (COX1, COX3, and CYTB), and one with GTG (ND4) and TTG (ND1) (Table 1). The TAA termination codon is found in all genes except CYTB (TAG) and COX2 [T, stop codon is completed by the addition of 3′ A residues to the mRNA, as is common in animal mitochondrial genomes (15, 16)]. The entire mitochondrial genome sequence of C. morosus is 80.85% similar to the genome of Carausius sp. from China and 100% and 99.3% similar to the gene sequences COX1/COX2 and 16S rRNA, respectively, of C. morosus from India.
TABLE 1.
Mitochondrial genome content, organization, and codon information of Carausius morosus
| Gene | Type | Minimum nucleotide position | Maximum nucleotide position | Length | Start codon | Stop codon | Direction |
|---|---|---|---|---|---|---|---|
| tRNA-Ile | tRNA | 1 | 67 | 67 | –a | – | Forward |
| tRNA-Gln | tRNA | 65 | 134 | 70 | – | – | Reverse |
| tRNA-Met | tRNA | 134 | 198 | 65 | – | – | Forward |
| ND2 | CDS | 223 | 1,194 | 985 | ATA | TAA | Forward |
| tRNA-Trp | tRNA | 1,193 | 1,256 | 64 | – | – | Forward |
| tRNA-Cys | tRNA | 1,249 | 1,310 | 62 | – | – | Reverse |
| tRNA-Tyr | tRNA | 1,310 | 1,373 | 64 | – | – | Reverse |
| COX1 | CDS | 1,375 | 2,908 | 1534 | ATG | TAA | Forward |
| tRNA-Leu | tRNA | 2,909 | 2,973 | 65 | – | – | Forward |
| COX2 | CDS | 2,974 | 3,643 | 670 | ATA | T | Forward |
| tRNA-Lys | tRNA | 3,644 | 3,714 | 71 | – | – | Forward |
| tRNA-Asp | tRNA | 3,714 | 3,780 | 67 | – | – | Forward |
| ATP8 | CDS | 3,781 | 3,936 | 156 | ATA | TAA | Forward |
| ATP6 | CDS | 3,933 | 4,604 | 672 | ATA | TAA | Forward |
| COX3 | CDS | 4,604 | 5,386 | 783 | ATG | TAA | Forward |
| tRNA-Gly | tRNA | 5,386 | 5,449 | 64 | – | – | Forward |
| ND3 | CDS | 5,450 | 5,803 | 354 | ATT | TAA | Forward |
| tRNA-Arg | tRNA | 5,814 | 5,869 | 56 | – | – | Forward |
| tRNA-Ala | tRNA | 5,889 | 5,952 | 64 | – | – | Forward |
| tRNA-Asn | tRNA | 5,953 | 6,018 | 66 | – | – | Forward |
| tRNA-Ser | tRNA | 6,017 | 6,086 | 70 | – | – | Forward |
| tRNA-Glu | tRNA | 6,086 | 6,148 | 63 | – | – | Forward |
| tRNA-Phe | tRNA | 6,149 | 6,209 | 61 | – | – | Reverse |
| ND5 | CDS | 6,210 | 7,934 | 1721 | ATT | TAA | Reverse |
| tRNA-His | tRNA | 7,935 | 7,997 | 63 | – | – | Reverse |
| ND4 | CDS | 7,998 | 9,329 | 1332 | GTG | TAA | Reverse |
| ND4L | CDS | 9,323 | 9,607 | 285 | ATA | TAA | Reverse |
| tRNA-Thr | tRNA | 9,616 | 9,678 | 63 | – | – | Forward |
| tRNA-Pro | tRNA | 9,679 | 9,744 | 66 | – | – | Reverse |
| ND6 | CDS | 9,746 | 10,222 | 478 | ATT | TAA | Forward |
| CYTB | CDS | 10,222 | 11,355 | 1134 | ATG | TAG | Forward |
| tRNA-Ser | tRNA | 11,354 | 11,417 | 64 | – | – | Forward |
| ND1 | CDS | 11,416 | 12,384 | 969 | TTG | TAA | Reverse |
| tRNA-Leu | tRNA | 12,385 | 12,453 | 69 | – | – | Reverse |
| 16S rRNA | rRNA | 12,455 | 13,726 | 1272 | – | – | Reverse |
| tRNA-Val | tRNA | 13,727 | 13,794 | 68 | – | – | Reverse |
| 12S rRNA | rRNA | 13796 | 14578 | 783 | – | – | Reverse |
–, not applicable.
ACKNOWLEDGMENTS
This research was supported by NSF award number 1832446 to Mohammed Yahdi at Hartnell College.
We wish to thank Barbara Black for gifting the specimen analyzed in this work to Hartnell College.
Contributor Information
Jeffery R. Hughey, Email: jhughey@hartnell.edu.
Julie C. Dunning Hotopp, University of Maryland School of Medicine, Baltimore, Maryland, USA
DATA AVAILABILITY
The complete mitochondrial genome sequence of Carausius morosus is available in GenBank under accession number PP230539. The associated BioProject, SRA, and BioSample numbers are PRJNA1089237, SRS20783361, and SAMN40533979, respectively. The mitochondrial genome referenced in the text is Carausius sp. GenBank accession number OQ682524.
REFERENCES
- 1. von Wattenwyl B. 1907. Die Insektenfamilie der Phasmiden. II. Phasmidae anareolatae (Clitumnini, Lonchodini, Bacunculini), p 181–340. Wilhelm Engelmann, Leipzig. [Google Scholar]
- 2. Baker E. 2015. Carausius morosus introduced in San Diego. Figshare. doi: 10.6084/m9.figshare.1304202 [DOI] [Google Scholar]
- 3. Brock PD, Büscher T, Baker E. 2024. Phasmida species file online. Version 5.0/5.0. Available from: http://Phasmida.SpeciesFile.org. Retrieved 2 Feb 2024.
- 4. Arakelian G. 2015. Indian stick insect (Carausius morosus). Pest Bull Los Angeles Cty Dep Agric Comm. [Google Scholar]
- 5. Headrick DH, Wilen CA. 2011. Indian walking stick. Univ Calif Pest Notes. 74157 [Google Scholar]
- 6. Beucke K. 2020. California pest rating proposal: Carausius morosus (Sinéty): Indian walking stick. Available from: https://blogs.cdfa.ca.gov/Section3162/wp-content/uploads/2020/09/Carausius-morosus.pdf. Retrieved 2 Feb 2024.
- 7. Garcia AN, Ramos JH, Mendoza AG, Muhrram A, Vidauri JM, Hughey JR, Hartnell College Genomics Group . 2022. Complete chloroplast genome of topotype material of the coast live oak Quercus agrifolia Née var. agrifolia (Fagaceae) from California. Microbiol Resour Announc 11:e0000422. doi: 10.1128/mra.00004-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner. United States. Available from: https://www.osti.gov/biblio/1241166. Retrieved 16 Apr 2024.
- 9. Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033 [DOI] [PubMed] [Google Scholar]
- 10. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Osawa S, Jukes TH, Watanabe K, Muto A. 1992. Recent evidence for evolution of the genetic code. Microbiol Rev 56:229–264. doi: 10.1128/mr.56.1.229-264.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jukes TH, Osawa S. 1993. Evolutionary changes in the genetic code. Comp Biochem Physiol B 106:489–494. doi: 10.1016/0305-0491(93)90122-l [DOI] [PubMed] [Google Scholar]
- 13. Zheng S, Poczai P, Hyvönen J, Tang J, Amiryousefi A. 2020. Chloroplot: an online program for the versatile plotting of organelle genomes. Front Genet 11:576124. doi: 10.3389/fgene.2020.576124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yuan Y, Zhang L, Li K, Hong Y, Storey KB, Zhang J, Yu D. 2023. Nine mitochondrial genomes of Phasmatodea with two novel mitochondrial gene rearrangements and phylogeny. Insects 14:485. doi: 10.3390/insects14050485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature 290:470–474. doi: 10.1038/290470a0 [DOI] [PubMed] [Google Scholar]
- 16. Boore JL, Macey JR, Medina M. 2005. Sequencing and comparing whole mitochondrial genomes of animals. Methods Enzymol 395:311–348. doi: 10.1016/S0076-6879(05)95019-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete mitochondrial genome sequence of Carausius morosus is available in GenBank under accession number PP230539. The associated BioProject, SRA, and BioSample numbers are PRJNA1089237, SRS20783361, and SAMN40533979, respectively. The mitochondrial genome referenced in the text is Carausius sp. GenBank accession number OQ682524.