ABSTRACT
Here, we present the draft genome sequence of Alteromonas gracilis strain J4, isolated from the green macroalga Caulerpa prolifera. The draft genome is 4,492,914 bp in size and contains 4,719 coding DNA sequences, 67 tRNAs, and 16 rRNA-coding genes. Strain J4 may exhibit host growth-promoting properties.
KEYWORDS: plant-microbe interactions, whole-genome sequencing, marine microbiology, growth-promoting bacteria, interspecific competition
ANNOUNCEMENT
To understand Caulerpa host-microbe interaction(1,2), we isolated bacteria from C. prolifera rhizoid tissue and conducted whole-genome sequencing. Here, we present the draft genome sequence of Alteromonas gracilis strain J4.
Strain J4 was isolated from rhizoids of hand-collected C. prolifera in the Ria Formosa lagoon (37°00′22.7″N 7°58′00.3″W, Faro, Portugal) and stored in a cooling box. Rhizoids were ground with mortar and pestle, and the lysate was plated on Difco Marine Agar 2216 and incubated in the dark at room temperature (20–25°C). After 3 days, individual colonies were replated and re-incubated. Isolate J4 was identified as an Alteromonas sp. by comparative full-length 16S rRNA gene Sanger sequencing (Applied BioSystems 3130xl Genetic Analyzer) analysis using primers 27F/1492R against the NCBI database (3). The 16S rRNA gene showed 96.80% sequence identity to A. gracilis strain 9a2 341. Genomic DNA was extracted using the peqGOLD Bacterial DNA Mini Kit (VWR). Genome sequencing was conducted on a MinION Mk1C device (Oxford Nanopore Technologies), using the Ligation Sequencing Kit (SQK-LSK110) and a Flongle Flow Cell (R9.4.1). Base-calling was performed using Guppy v6.2.7 (https://community.nanoporetech.com/downloads). 225,773 reads passed quality control with a mean Q-score of 12 and an N50 of 3.71 kb.
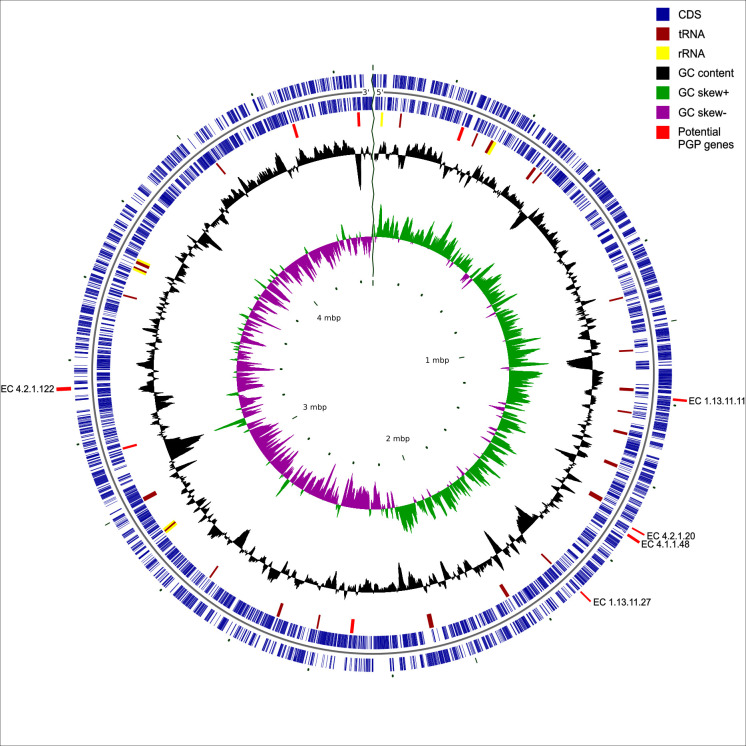
Trimming residual sequencing adaptors and splitting chimeric reads [Porechop v0.2.4 (4)] resulted in 211,623 reads. Filtlong v0.2.1 was used to remove small (<1,000 bp) and poor quality (<5%) reads (https://github.com/rrwick/Filtlong). For the remaining 172,018 reads, 12 subsamples were generated at 55× depth using Trycycler v0.5.5 (5). Three subsamples were each assembled with (i) Flye v2.9.3, (ii) Miniasm v0.3 & Minipolish v0.1.3, (iii) Raven v1.8.3, and (iv) Unicycler v0.5.0 (6–10). The consensus assembly was further generated using Trycycler v0.5.5 and polished with Homopolish v0.4.1 (11), resulting in one circular contig with a total size of 4,492,914 bp (131× coverage) and a GC content of 44.0%. The genome was reoriented using Dnaapler chromosome v0.7.0 (12) by identifying dnaA as the replication initiator gene. The genome had a completeness of 98% and contamination of 5.1% (CheckM2 v1.0.2) (13) and contained 4,719 protein-coding genes, 67 tRNA, and 16 rRNA coding genes [Prokka v1.14.6 (14); Fig. 1]. 903 genes were identified as potential pseudogenes (https://github.com/ndombrowski/j4_assembly) using Pseudofinder v1.1.0 with the UniProtKB/Swiss-Prot database as reference (15, 16). J4 likely belongs to an uncharacterized species within the genus Alteromonas. The genome exhibits 85.3% average nucleotide identity (ANI) with Alteromonas sp009811495 (GCF_016756315.1) based on a comparison with the GTDB r214 database using GTDB-Tk v2.3.2 (17, 18).
Fig 1.
The genome map of Alteromonas gracilis strain J4. Each circle from inner to outer indicates potential plant growth-promoting (PGP) genes, coding sequences (CDS) in the leading strand, CDS in the lagging strand, tRNA and rRNA, GC content, GC skew+, and GC skew−. Potential PGP genes are indicated in red and labeled with EC numbers. If not labeled, tryptophan halogenases are represented.
The annotation analysis identified two genes encoding 4-hydroxyphenylpyruvate dioxygenase (EC 1.13.11.27), pivotal in melanin catalysis (19). Melanin plays a role in ensuring survival during symbiotic interactions (20). Protein genes cspD, dnaJ, dnaK, and grpE, described to protect against cold/heat and oxidative stress, were detected (21), as well as 12 genes related to sulfur metabolism (22). Four indole-3-glycerol phosphate synthases (EC 4.1.1.48) and two tryptophan 2,3-dioxygenases (EC 1.13.11.11), both key precursors in indole-3-acetic acid biosynthesis were found. In all, 24 putative genes encoding tryptophan halogenases and six tryptophan synthases were found, suggesting potential growth-promoting properties in strain J4 with biotechnological applications (23, 24).
ACKNOWLEDGMENTS
The research leading to the results presented in this publication was financially supported by CCMAR under reference number CCMAR/BD/07/2022 for H.D., CEECINST/00114/2018 for AE, and funded by The BlueForests project of EEA Grants (PT-INNOVATION-0081). T.A. was supported by the fellowship reference—SFRH/BPD/116774/2016) from FCT and M.C. was supported by FCT (DivRestore/0013/2020). G.M. and P.K. were supported by the 2020–2021 Biodiversa+ and water JPI joint call for research projects, under the BiodivRestore ERA-NET Cofund (GA N°101003777) with the EU and the Dutch Ministry of Agriculture, Nature and Food Quality. This study received Portuguese national funds from FCT—Foundation for Science and Technology through projects UIDB/04326/2020 (DOI:10.54499/UIDB/04326/2020), UIDP/04326/2020 (DOI:10.54499/UIDP/04326/2020), and LA/P/0101/2020 (DOI:10.54499/LA/P/0101/2020).
Contributor Information
Hannah J. van Duijnhoven, Email: juliavanduijnhoven@msn.com.
Frank J. Stewart, Montana State University, Bozeman, Montana, USA
DATA AVAILABILITY
The 16S rRNA sequence data, raw Nanopore sequence reads, and the assembled genome sequence have been deposited in GenBank under BioProject number PRJNA1077798, with BioSample accession numbers PP541516, SAMN40604929, and SAMN39982826, respectively, and the reported genome is the second version, CP145482.2.
REFERENCES
- 1. Alexandre A, Santos R. 2020. High nitrogen and phosphorous acquisition by belowground parts of Caulerpa prolifera (Chlorophyta) contribute to the species’ rapid spread in Ria Formosa lagoon, Southern Portugal. J Phycol 56:608–617. doi: 10.1111/jpy.12988 [DOI] [PubMed] [Google Scholar]
- 2. Parreira F, Martínez-Crego B, Lourenço Afonso CM, Machado M, Oliveira F, Manuel dos Santos Gonçalves J, Santos R. 2021. Biodiversity consequences of Caulerpa prolifera takeover of a coastal lagoon. Estuar Coast Shelf Sci 255:107344. doi: 10.1016/j.ecss.2021.107344 [DOI] [Google Scholar]
- 3. Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–9. doi: 10.1093/nar/gkn201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wick R. 2017. Porechop: adapter trimmer for Oxford Nanopore reads. Available from: https://github.com/rrwick/Porechop/
- 5. Wick RR, Judd LM, Cerdeira LT, Hawkey J, Méric G, Vezina B, Wyres KL, Holt KE. 2021. Trycycler: consensus long-read assemblies for bacterial genomes. Genome Biol 22:266. doi: 10.1186/s13059-021-02483-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37:540–546. doi: 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- 7. Li H. 2016. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32:2103–2110. doi: 10.1093/bioinformatics/btw152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wick RR, Holt KE. 2019. Benchmarking of long-read assemblers for prokaryote whole genome sequencing. F1000Res 8:2138. doi: 10.12688/f1000research.21782.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vaser R, Šikić M. 2021. Time-and memory-efficient genome assembly with Raven. Nat Comput Sci 1:332–336. doi: 10.1038/s43588-021-00073-4 [DOI] [PubMed] [Google Scholar]
- 10. Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang YT, Liu PY, Shih PW. 2021. Homopolish: a method for the removal of systematic errors in nanopore sequencing by homologous polishing. Genome Biol 22:95. doi: 10.1186/s13059-021-02282-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bouras G, Grigson SR, Papudeshi B, Mallawaarachchi V, Roach MJ. 2024. Dnaapler: a tool to reorient circular microbial genomes. J Open Source Softw 9:5968. doi: 10.21105/joss.05968 [DOI] [Google Scholar]
- 13. Chklovski A, Parks DH, Woodcroft BJ, Tyson GW. 2023. CheckM2: a rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. Nat Methods 20:1203–1212. doi: 10.1038/s41592-023-01940-w [DOI] [PubMed] [Google Scholar]
- 14. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 15. Syberg-Olsen MJ, Garber AI, Keeling PJ, McCutcheon JP, Husnik F. 2022. Pseudofinder: detection of pseudogenes in prokaryotic genomes. Mol Biol Evol 39:msac153. doi: 10.1093/molbev/msac153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. UniProt Consortium . 2023. Uniprot: the universal protein knowledgebase in 2023. Nucleic Acids Res 51:D523–D531. doi: 10.1093/nar/gkac1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. 2019. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parks DH, Chuvochina M, Chaumeil PA, Rinke C, Mussig AJ, Hugenholtz P. 2020. A complete domain-to-species taxonomy for bacteria and archaea. Nat Biotechnol 38:1079–1086. doi: 10.1038/s41587-020-0501-8 [DOI] [PubMed] [Google Scholar]
- 19. Chen J, Wang X, Zhu S, Chen Y, Yang J. 2016. Complete genome sequence of Alteromonas stellipolaris LMG 21856, a budding brown pigment-producing oligotrophic bacterium isolated from the Southern Ocean. Genome Announc 4:10–1128. doi: 10.1128/genomeA.00137-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pavan ME, López NI, Pettinari MJ. 2020. Melanin biosynthesis in bacteria, regulation and production perspectives. Appl Microbiol Biotechnol 104:1357–1370. doi: 10.1007/s00253-019-10245-y [DOI] [PubMed] [Google Scholar]
- 21. Torres M, Hong KW, Chong TM, Reina JC, Chan KG, Dessaux Y, Llamas I. 2019. Genomic analyses of two Alteromonas stellipolaris strains reveal traits with potential biotechnological applications. Sci Rep 9:1215. doi: 10.1038/s41598-018-37720-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guédon E, Martin-Verstraete I. 2006. Cysteine metabolism and its regulation in bacteria, p 195–218. In Amino acid biosynthesis~ pathways, regulation and metabolic engineering. Springer, Berlin, Heidelberg. [Google Scholar]
- 23. Gago JF, Viver T, Urdiain M, Pastor S, Kämpfer P, Ferreira E, Rossello-Mora R. 2021. Description of three new Alteromonas species Alteromonas antoniana sp. nov., Alteromonas lipotrueae sp. nov. and Alteromonas lipotrueiana sp. nov. isolated from marine environments, and proposal for reclassification of the genus Salinimonas as Alteromonas. Syst Appl Microbiol 44:126226. doi: 10.1016/j.syapm.2021.126226 [DOI] [PubMed] [Google Scholar]
- 24. Navarro-Torre S, Carro L, Rodríguez-Llorente ID, Pajuelo E, Caviedes MÁ, Igual JM, Klenk H-P, Montero-Calasanz MDC. 2020. Pseudoalteromonas rhizosphaerae sp. nov., a novel plant growth-promoting bacterium with potential use in phytoremediation. Int J Syst Evol Microbiol 70:3287–3294. doi: 10.1099/ijsem.0.004167 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The 16S rRNA sequence data, raw Nanopore sequence reads, and the assembled genome sequence have been deposited in GenBank under BioProject number PRJNA1077798, with BioSample accession numbers PP541516, SAMN40604929, and SAMN39982826, respectively, and the reported genome is the second version, CP145482.2.