ABSTRACT
Bacillus safensis strain WOB7 is a linamarin-utilizing bacterium (LUB) that was isolated from cassava wastewater obtained from a processing factory. We present here the draft genome sequence of the strain (WOB7). These data provide valuable information on the prospects of the linamarase and other genes of importance associated with cyanogen detoxification.
KEYWORDS: Bacillus safensis, linamarin-utilizing bacterium, whole-genome sequence, cassava wastewater, cyanogen, detoxify
ANNOUNCEMENT
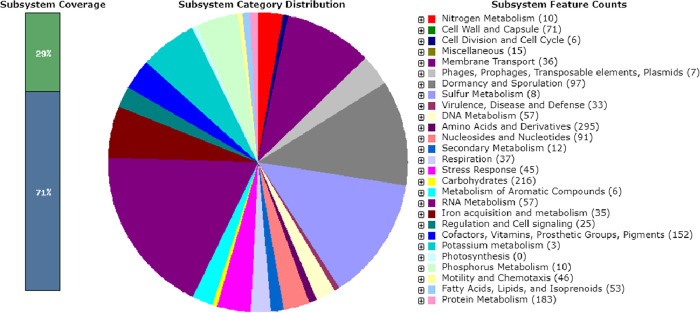
Cassava production has globally increased by 240 million metric tons. By 2025, 62% of global cassava production is predicted to come from Sub-Saharan Africa (1). Nigeria, as the world’s largest producer, generates a significant amount of cassava waste (1). Cassava mill wastewater is the liquid waste generated during cassava processing. It contains high organic content, linamarin (a cyanogenic glucoside that produces hydrocyanic acid (HCN)), minerals (2 3), and has an acidic pH (4). Therefore, it poses a significant threat to the environment and the health of communities living near cassava mills (5). Here, we report the draft genome of strain WOB7, a type strain of Bacillus safensis, a member of the Bacillaceae isolated from cassava wastewater obtained from a cassava processing factory in Odogunyan, Ikorodu, Lagos State, Nigeria (6°39′39.40236″E3°30′33.19883″). The CWW sample was mixed, serially diluted with sterile water, and then cultured on nutrient agar (NA) plate. After aerobic incubation at 37°C for 24 h, small and white colonies were picked out and then restreaked on the NA plate until pure cultures were obtained. Strain WOB7 was identified as B. safensis KX774196, using the universal primers 27F and 1492R by 16S rRNA gene sequencing (6). The strain was grown on nutrient broth medium for 24 h at 37 °C for DNA extraction. A single colony was inoculated in 5 mL of nutrient broth medium and incubated at the same condition (24 h at 37°C) to obtain cell biomass. Afterward, 2 mL of the culture was centrifuged (16,000 × g, 4 min), and the pellet was extracted for genomic DNA with the Zymo Research Fungal/Bacterial DNA MiniPrep Kit (catalog no. D4300; Zymo Research Corp., Irvine, CA, USA), according to the manufacturer’s instructions. Then xGen DNA library preparation kit was used for 2 × 150 bp paired-end sequencing with the Illumina HiSequencing platform (HiSeq4000X). The obtained raw reads from each run were quality-checked using FastQC (v.1.0.0, BaseSpace Illumina) and subsequently trimmed using FastXToolkit (v.2.2.5, BaseSpace Illumina) (7). Genome de novo assembly and assembly quality were performed using SPAdes (v.3.9.0, BaseSpace Illumina) (8, 9). The draft genome sequences were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (v.6.6), with default parameters (10). Also, the genome annotation and metabolic pathway identification were carried out by RAST (v.2.0) server (11). Default parameters were used except where otherwise noted. Table 1 shows the genomic features of Bacillus safensis WOB7. The genome of strain WOB7 consists of 15,185,963 reads and 75 contigs, with GC content of 41.5%. A total of 3.7 × 106 bp (3.7 Mb) genomic size was generated, corresponding to 569.5X genome coverage. Figure 1 shows the RAST analysis-based subsystem distribution of the whole-genome sequence of strain WOB7. These genomic data add information to clarify the B. safensis metabolic strategies to detoxify cassava wastewater.
TABLE 1.
Genome features of Bacillus safensis strain WOB7a
| Attribute | Value |
|---|---|
| Total reads | 15,185,963 |
| Genome coverage (X) | 569.5 |
| Genome size (Mb) | 3.7 |
| CDSs (total) | 3,744 |
| CDSs (with protein) | 3,711 |
| CDSs (without protein) | 33 |
| Genes (total) | 3,836 |
| Number of contigs | 75 |
| Contig N50 (kb) | 149.5 |
| Contig L50 | 8 |
| GC content (%) | 41.5 |
| tRNA | 71 |
| rRNA | 4,10, 2 |
| Genes (RNA) | 92 |
| ncRNA | 5 |
| Accession number | JAYSGV010000000 |
CDS, coding sequence; GC, guanine-cytosine content; ncRNA, non-coding RNA; rRNA, ribosomal RNA; tRNA, transfer RNA.
Fig 1.
The RAST analysis-based subsystem distribution of whole-genome sequence of Bacillus safensis strain WOB7. Each colour in the pi graph represents a particular group of genes mentioned on the right site of the graph.
ACKNOWLEDGMENTS
The authors thank the Laragen Biotechnology Laboratory, Culver City, Virginia, USA, for their technical assistance in the course of the project.
O.A.K.: conceptualization, investigation, methodology, project administration, writing (original draft); B.M.O., A.O.O., and O.B.C.: investigation and writing (review and editing); D.W.P.: investigation, data curation, and writing (review and editing); S.T.A., I.M.O., and A.O.O.: supervision, writing (review and editing).
Contributor Information
Adewale K. Ogunyemi, Email: waleogunyemi2002@yahoo.com.
Kenneth M. Stedman, Portland State University, Portland, Oregon, USA
DATA AVAILABILITY
A whole-genome sequence of Bacillus safensis strain WOB7 has been deposited in GenBank under accession number JAYSGV010000000, BioProject number PRJNA1057587, BioSample number SAMN38321375, and Sequence Read Archive accession number SRX24013597.
REFERENCES
- 1. FAOSTAT . 2020. Food and Agriculture organization of United Nations, Rome, Italy. Edited by Glicksman M., Staches M., and Glicksman M.. Gum Technology in the Food Industry, New York: Academic Press. Available from: https://www.fao.org/faostat/en/#data [Google Scholar]
- 2. Duarte A de S, Rolim MM, Silva ÊF de F e, Pedrosa EMR, Albuquerque F da S, Magalhães AG. 2013. Alterações dos atributos físicos e químicos de um Neossolo após aplicação de doses de manipueira. Rev bras eng agríc ambient 17:938–946. doi: 10.1590/S1415-43662013000900005 [DOI] [Google Scholar]
- 3. Osunbitan JA. 2012. Short-term effects of cassava processing wastewater on some chemical properties of loamy sand soil in Nigeria. J Soil Sci Environ Manage 96:164–171. doi: 10.11648/j.be.20180201.13 [DOI] [Google Scholar]
- 4. Izonfuo WAL, Bariweni PA, George DMC. 2013. Soil contamination from cassava wastewater discharges in a rural community in the Niger Delta, Nigeria. J Appl Sci Environ Mgt 17:105–110. https://hdl.handle.net/1807/51957. [Google Scholar]
- 5. Vasconcellos SP, Cereda MP, Cagnon JR, Foglio MA, Rodrigues RA, Manfio GP, Oliveira VM. 2009. In vitro degradation of linamarin by microorganisms isolated from cassava wastewater treatment lagoon. Braz J Microbiol 40:879–883. doi: 10.1590/S1517-83822009000400019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Timm CM, Loomis K, Stone W, Mehoke T, Brensinger B, Pellicore M, Staniczenko PPA, Charles C, Nayak S, Karig DK. 2020. Isolation and characterization of diverse microbial representatives from the human skin microbiome. Microbiome 8:58. doi: 10.1186/s40168-020-00831-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hannon G. 2010. FASTX-toolkit package. Available online at. Available from: http://hannonlab.cshl.edu/fastx_toolkit/
- 8. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Y, Cui Z, Liu F, Chen N. 2021. Mitochondrial genome and phylogenomic analysis of pseudonitzschia micropora (Bacillariophyceae, Bacillariophyta). Mitochondrial DNA B Resour 6:2035–2037. doi: 10.1080/23802359.2021.1923426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A whole-genome sequence of Bacillus safensis strain WOB7 has been deposited in GenBank under accession number JAYSGV010000000, BioProject number PRJNA1057587, BioSample number SAMN38321375, and Sequence Read Archive accession number SRX24013597.