Abstract
Peripheral mononuclear cells (PBMCs) exhibit robust changes in mitochondrial respiratory capacity in response to health and disease. While these changes do not always reflect what occurs in other tissues, such as skeletal muscle, these cells are an accessible and valuable source of viable mitochondria from human subjects. PBMCs are exposed to systemic signals that impact their bioenergetic state. Thus, expanding our tools to interrogate mitochondrial metabolism in this population will elucidate mechanisms related to disease progression. Functional assays of mitochondria are often limited to using respiratory outputs following maximal substrate, inhibitor, and uncoupler concentrations to determine the full range of respiratory capacity, which may not be achievable in vivo. The conversion of adenosine diphosphate (ADP) to adenosine triphosphate (ATP) by ATP-synthase results in a decrease in mitochondrial membrane potential (mMP) and an increase in oxygen consumption. To provide a more integrated analysis of mitochondrial dynamics, this article describes the use of high-resolution fluorespirometry to measure the simultaneous response of oxygen consumption and mitochondrial membrane potential (mMP) to physiologically relevant concentrations of ADP. This technique uses tetramethylrhodamine methylester (TMRM) to measure mMP polarization in response to ADP titrations following maximal hyperpolarization with complex I and II substrates. This technique can be used to quantify how changes in health status, such as aging and metabolic disease, affect the sensitivity of mitochondrial response to energy demand in PBMCs, T-cells, and monocytes from human subjects.
Introduction
A cell's ability to function and survive in a period of physiological stress is largely dependent on its ability to meet the energetic requirement to restore homeostasis1, 2. Energy demand rises in response to a variety of stimuli. For instance, increased muscle contraction during exercise increases the utilization of ATP and glucose by skeletal muscle, and a rise in protein synthesis following infection increases the utilization of ATP by immune cells for cytokine production and proliferation3, 4, 5, 6. A spike in energy demand triggers a series of bioenergetic processes to restore the ATP/ADP ratio. As ATP is consumed, ADP levels rise and stimulate F1F0 ATP-synthase (complex V), which requires a protonmotive force to drive its mechanical rotation and catalytic conversion of ADP to ATP within the mitochondrion7. The protonmotive force is an electrochemical gradient created by the pumping of protons during the transfer of electrons from substrates to oxygen through the electron transport system (ETS) within the inner mitochondrial membrane. The resulting difference in proton concentration (delta pH) and electrical potential (membrane potential) creates the protonmotive force that drives ATP synthesis and oxygen consumption in response to energy demand reducing the ATP/ADP ratio or raising ADP levels. The affinity of mitochondria to ADP can be determined by the calculation of the Km or EC50 of ADP-stimulated respiration of isolated mitochondria or permeabilized cells8, 9. This method has shown that permeabilized muscle fibers from older humans require a greater concentration of ADP to stimulate 50% of their maximal oxidative phosphorylation capacity than those of younger subjects9. Similarly, aging mouse skeletal muscle requires more ADP to lower the production of mitochondrial reactive oxygen species (ROS)10, 11. Additionally, ADP sensitivity is reduced in permeabilized muscle fibers of mice with diet-induced obesity relative to controls and is enhanced in the presence of insulin and following nitrate consumption12, 13. Thus, the capacity of mitochondria to respond to energy demand varies under different physiological conditions, but this has not been previously explored in the context of immune cells.
Peripheral blood mononuclear cells (PBMCs) are commonly used to investigate cellular bioenergetics in human subjects14, 15, 16, 17, 18, 19, 20. This is largely due to cells being easily obtainable from uncoagulated blood samples in clinical studies, the responsiveness of cells to metabolic perturbations, and the methods developed by various groups to interrogate mitochondrial metabolism by using inhibitors and uncouplers to determine the maximal and minimal capacity of mitochondrial respiration21, 22. These methods have led to an appreciation of the roles of bioenergetics in aging, metabolic disease, and immune function14, 20, 23, 24. Mitochondrial respiratory capacity is often reduced in skeletal muscle and PBMCs under conditions of heart failure18, 25. PBMC bioenergetics are also correlated with cardiometabolic risk factors in healthy adults17 and are responsive to treatments such as nicotinamide riboside18. PBMCs include neutrophils, lymphocytes (B-cells and T-cells), monocytes, natural killer cells, and dendritic cells, which all contribute to PBMC mitochondrial capacity26, 27, 28. In addition, cellular bioenergetics play a crucial role in immune cell activation, proliferation, and renewal23. However, a limitation of these methods is that the cells are not functioning under a physiological range of substrates. Additional methods are therefore required to interrogate mitochondrial function in substrate concentrations that are more relevant to what cells experience in vivo.
Mitochondrial membrane potential (mMP) is the major component of a protonmotive force and is essential for a variety of mitochondrial processes beyond ATP production, such as regulation of respiratory flux, reactive oxygen species production, protein and ion import, autophagy, and apoptosis. mMP can be assessed with electrochemical probes or fluorescent dyes sensitive to changes in membrane polarization like JC-1, Rhod123, DiOC6, tetramethyl rhodamine (TMRE) or methyl ester (TMRM), and safranin. The latter two are lipophilic cationic dyes that have been successfully used in high-resolution fluorespirometry of tissue homogenates, isolated mitochondria, and permeabilized tissue11, 29, 30, 31, 32, 33. In this technique, TMRM is used in quench mode, where cells are exposed to a high concentration of TMRM that accumulates in the mitochondrial matrix when polarized (high mMP and protonmotive force), resulting in the quenching of cytosolic TMRM fluorescence. When mitochondria depolarize in response to ADP or uncouplers, the dye is released from the matrix, increasing the TMRM fluorescent signal34, 35. The purpose of this method is to simultaneously measure changes in mitochondrial respiration and mMP in response to ADP titrations in human-derived PBMCs, circulating monocytes, and T-cells, and it can also be applied to mouse splenic T-cells.
Protocol
The collection of blood samples for data and methods development presented herein was approved by the Internal Review Board of the University of Washington. Representative results also include data from male C57BL/6J mice (5–7 months old) purchased from Jackson Laboratories. All animal procedures were approved by the University of Washington Office of Animal Welfare. The protocol overview is pictured in Figure 1. Reagent preparation for this protocol can be found in Supplementary File 1.
Figure 1: Overview of the protocol.
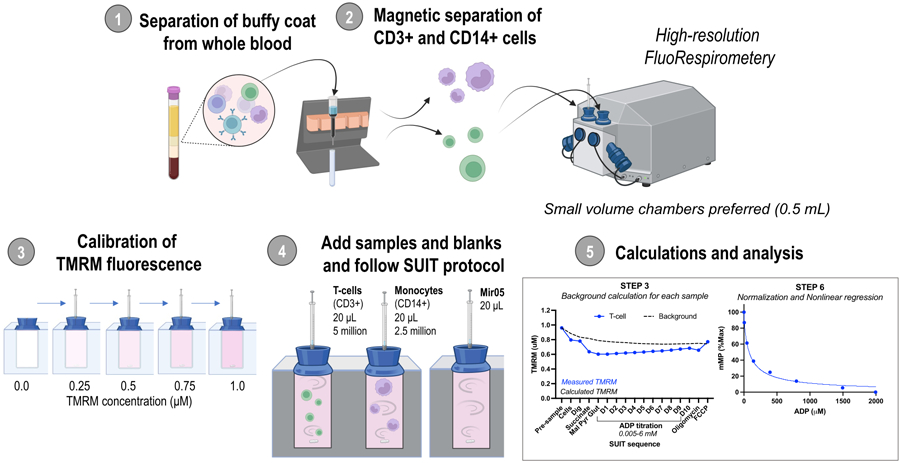
Workflow using high-resolution fluorespirometry to assess changes in mitochondrial membrane potential in isolated monocytes (CD14+ ) and T-cells (CD3+ ) from fresh human blood samples. Abbreviations: TMRM, tetramethylrhodamine methyl ester; SUIT, substrate-uncoupler-inhibitor titrations; ADP, adenosine diphosphate; Dig, digitonin; Mal, malate; Pyr, pyruvate; Glut, glutamate; D1–10, 10 consecutive ADP titrations.
1. Separation of buffy coat from whole blood
NOTE: Cell isolation is modified from Kramer et al.27.
Allow RPMI, density gradient, and centrifuge to reach room temperature. Sterilize the biosafety cabinet and materials before starting.
Collect venous blood into three 10 mL K2EDTA tubes. Invert the tubes at least 3 times.
Centrifuge the tubes at 500 x g 10 min (22 °C, 9 acceleration [acc], 2 deceleration [dec]).
Remove 1 mL of plasma from each tube and store at −80 °C for future analyses.
Transfer the plasma and half the red blood cell layer from each tube into a single 50-mL conical tube. Add RPMI up to the 40 mL mark. Invert at least 3 times.
Slowly layer 10 mL of the plasma solution into four 15 mL conical tubes containing 3 mL of the density gradient.
Centrifuge at 700 x g for 30 min (22 °C, 5 acc, 2 dec).
Collect all the plasma and the buffy coat containing the peripheral mononuclear cells (PBMCs) without disrupting the red blood cells.
Centrifuge at 500 x g for 10 min (22 °C, 5 acc, 5 dec) and aspirate the supernatant.
Wash the PBMC pellet 1x-2x by resuspending it in 10 mL of RPMI and centrifuging at 500 x g for 10 min (22 °C, 5 acc, 5 dec).
2. Magnetic separation of CD14+ and CD3+ cells
Place a column in the magnetic field of a magnetic cell separator (see the Table of Materials). Wash the column with 3 mL of RP-5.
Resuspend the PBMC pellet in 80 µL of RP-5 and 20 µL of anti-CD14 microbeads (see the Table of Materials). Incubate for 15 min at 4 °C.
Resuspend the cells with 1 mL of RP-5 and load the suspension onto the column. Collect unlabeled cells that flow through into a 15 mL conical tube labeled "Flow-through 1". Wait until all cell suspension has gone through the column, then continue washing with 3 mL of RP-5 3x, collecting all flow-through.
Carefully remove the column from the magnetic field and place it on a new 15 mL conical tube. Add 5 mL of RP-5 and immediately use the plunger to purge the column contents into a collection tube labeled "CD14+".
Centrifuge "Flow-through 1" at 500 x g for 10 min (22 °C, 5 acc, 5 dec) and aspirate the supernatant.
Using cells from Flow-through 1, repeat steps 2.2–2.5 using anti-CD3 microbeads (see the Table of Materials) to isolate T-cells.
Centrifuge tubes containing T-cells (CD3+ ) and monocytes (CD14+ ) at 300 x g for 5 min. Aspirate the supernatant and resuspend the pellet in 1 mL of RP-5.
-
Determine cell concentration using a hemocytometer or automatic cell counter.
NOTE: Cells can be counted by adding 10 µL of a cell dilution of 1:10 or 1:20 to a hemocytometer. One may refer to previously published protocols to count cells using a hemocytometer36.
-
Pipette 2.5 million monocytes or 5 million T-cells into a new centrifuge tube. Centrifuge for 30 s at 2000 x g, aspirate the supernatant and resuspend the cells in MiR05 for a total volume of 20 µL and final concentration of 125 million monocytes or 250 million T-cells per mL.
NOTE: The final concentration was selected to inject 2.5 million monocytes or 5 million T-cells in a volume of 20 µL. Mouse T-cells isolated from the spleen have also been tested using this method. The procedure is found in Supplementary File 1.
Table of Materials
| Procedure | Consumables | Source | Identifier |
|---|---|---|---|
| Cell isolation | K2EDTA blood collection tubes | BD Vacutainer | 366643 |
| RPMI Buffer | Corning | 17-105-CV | |
| Bovine Serum Albumin | Sigma-Aldrich | A6003 | |
| Histopaque 1077 | Sigma-Aldrich | 10771 | |
| CD14 Microbeads, human | Miltenyi Biotec | 130-050-201 | |
| CD3 Microbeads, human | Miltenyi Biotec | 130-050-101 | |
| LS Columns | Miltenyi Biotec | 130-042-401 | |
| Multi-MACS stand and MidiMACS Separator | Miltenyi Biotec | 130-042-301 | |
| Mir05 buffer | Ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) | Sigma-Aldrich | E4378 |
| Magnesium Chloride (MgCl2) | Sigma-Aldrich | M9272 | |
| Potassium dihydrogen phosphate (KH2PO4) | Sigma-Aldrich | P0662 | |
| HEPES sodium salt | Sigma-Aldrich | H7523 | |
| D-Sucrose | Sigma-Aldrich | 84097 | |
| Lactobionic acid | Sigma-Aldrich | 153516 | |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | A6003 | |
| Taurine | Sigma-Aldrich | T0625 | |
| Potassium Hydroxide (KOH) | Sigma-Aldrich | 221473 | |
| Fluorespirometry | Tetramethyrhodamine methyl ester perchlorite | Sigma-Aldrich | T5428 |
| Digitonin | Sigma-Aldrich | D141 | |
| L-Malic Acid | Sigma-Aldrich | M1000 | |
| Sodium Pyruvate | Sigma-Aldrich | P2256 | |
| L-Glutamic acid | Sigma-Aldrich | G1626 | |
| Succinate disodium salt | Sigma-Aldrich | S2378 | |
| Adenosine Diphosphate | Sigma-Aldrich | A5285 | |
| Oligomycin | Sigma-Aldrich | 04876 | |
| Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone | Sigma-Aldrich | C2920 | |
| Rotenone | Sigma-Aldrich | R8875 | |
| Antimycin A | Sigma-Aldrich | A8674 | |
| Equipment | O2k-FluoRespirometer series J | Oroboros | 10201-03 |
| O2k-sV-Module (0.5 chamber) | Oroboros | 11200-01 | |
| O2k-Fluo Smart-Module | Oroboros | 12100-03 | |
| Filter Set AmR | Oroboros | 44321-01 | |
| Software | DatLab | Oroboros | Version 8 |
| Prism | GraphPad | Version 10 |
3. High-resolution fluorespirometry - Oxygen and TMRM fluorescence calibration
NOTE: This method was adapted from previous work done on permeabilized fibers by Pharaoh et al.11. A high, un-inhibitory concentration of TMRM is used for quench mode, where the relationship of mMP and TMRM concentration in the matrix is inverted. Thus, a decrease in mMP leads to the release of TMRM dye from the matrix and an increase in fluorescence32.
Install 0.5 mL chambers in the O2K respirometer according to the manufacturer's instructions (see Table of Materials). Turn on the instrument and connect it to the software provided by the manufacturer for data acquisition.
Adjust the temperature to 37 °C and stir speed to 750 rpm.
Wash the chambers with distilled water 3x. Replace water with 0.54 mL of Mir05, close the stoppers fully, and remove excess buffer with the integrated suction system (ISS). Raise stoppers to allow room oxygen to equilibrate with chamber oxygen using a stopper-spacer.
-
Once the oxygen flux is stable, perform the air oxygen calibration (R1) according to the manufacturer's instructions.
NOTE: It may take >30 min for oxygen flux to stabilize. Oxygen sensors require the determination of zero-point oxygen (R0) and background oxygen flux from 50–200 µM from separate experiments using dithionite titrations. Specific methods can be found in the manufacturer's manual.
-
Seal the chamber by closing the stoppers.
NOTE: Unlike experiments using permeabilized fibers, the chambers do not require hyperoxygenation for PBMCs. Sealing the chamber after R1 calibration provides sufficient oxygen for the experiment. The oxygen levels should be maintained between 50–250 µM. If oxygen concentration falls below the threshold, the chamber can be partially opened so that chamber oxygen can equilibrate with room air oxygen.
- TMRM calibration
- Use the Green LED fluo-sensors (ex. 525 nm) with the AmR filter set (see Table of Materials). Set fluorometer Gain to 1000 and Intensity to 1000. Turn on the fluo-sensors and begin recording the baseline.
- Inject 2.5 µL of 0.05 mM TMRM and allow the signal to stabilize (~2 min) before the next 2.5 µL injection until a total of 4 injections are performed for a total TMRM concentration of 1 µM TMRM in the chamber. Use a Hamilton syringe for all injections.
- Calibrate the fluo-sensor by selecting the fluorescent signal (voltage) for each injection representing 0, 0.25, 0.5, 0.75, and 1.0 µM of TMRM for a five-point calibration.
4. Substrate-uncoupler-inhibitor titration (SUIT) protocol
NOTE: Run blank experiments where 20 µL of Mir05 is injected into the chamber instead of 20 µL of cell suspension, as the TMRM signal will change in response to injections alone (discussed in representative results). Allow for the oxygen flux signal to stabilize (about 2–3 min) before the next injection for both blank and sample experiments. The following titration protocol and expected observations are in Table 1.
Table 1:
Example SUIT protocol to assess mitochondrial membrane potential in freshly isolated T-cells and monocytes using the 0.5 mL chambers
| SUIT labels | Injection | Injection volume (μL) | Final concentration in 0.5mL chamber | Comments/Expected observations |
|---|---|---|---|---|
| TMRM calibration | 0.05 mM TMRM | 2.5 X 4 | 0.25–1.0 μM | Increase in fluorescence per injection |
| Pre-Cell | TMRM reading just prior to cell addition | |||
| Cell | Cells suspended in Mir05 | 20 | Mono: 5 million / mL | Read for 10 minutes. Decline in fluorescence as TMRM is taken in. |
| T-cell: 10 million / mL | ||||
| DIG | 1 mg/mL digitonin | 2 | 4 μL/mL | Read for 20 minutes. Stabilization of fluorescence. |
| SUCC | 1 M succinate | 2.5 | 5 mM | Sharp decline in TMRM fluorescence (increased mMP) |
| MPG | 0.1 M malate | 5 | 1.0 mM | Simultaneous injections. Small decline in TMRM fluorescence (increased mMP) |
| 1.0 M glutamate | 5 | 10 mM | ||
| 0.5 M pyruvate | 5 | 5 mM | ||
| D1-D10 | ADP (0.5–500mM) | titrat. | 0.01– 4.0 mM | Notice ADP-driven rise in TMRM fluorescence (decrease in mMP) |
| OMY | 1 mg/mL oligomycin | 1 | 2 μg/ml | Small decrease in fluorescence (increase in mMP) |
| FCCP | 0.25 mM FCCP | 1 X 3 | 0.5–1.5 μM | Sharp increase in fluorescence (depletion of mMP) |
| ROT | 0.25 mM Rotenone | 1 | 0.5 μM | Decline in respiration. mMP values are not relevant after FCCP addition. |
| AMA | 1.25 mM Antimycin A | 1 | 2.5 μM | Inhibition of mitochondrial respiration |
Once oxygen flux is stable, select and label both the oxygen flux and TMRM signal "pre-cell".
Inject the cell suspension containing either 5 million T-cells or 2.5 million monocytes in ~20 µL and measure for about 10 min. Select and label both the oxygen flux and TMRM signal as "Cell".
-
Permeabilize cells by injecting 2 µL of 1 mg/mL digitonin (final concentration: 4 µg/mL). Wait for 20 min. Select and label both the oxygen flux and TMRM signal as "Dig".
NOTE: It is suggested to optimize the digitonin concentration in separate experiments.
Add 2.5 µL of 1 M succinate (final concentration: 5 mM). Select and label both the oxygen flux and TMRM signal as "SUCC".
Once oxygen flux is stable, add 5 µL of 100 mM malate (final concentration: 1.0 mM), 5 µL of 1 M glutamate (final concentration: 10 mM), and 5 µL of 500 mM pyruvate (final concentration: 5 mM). Select and label both the oxygen flux and TMRM signal as "MPG".
Once oxygen flux is stable, titrate ADP. Select rates for each titration and label them "D" sequentially 1 through 10, depending on the number of titrations. Use the titration scheme in Table 2.
-
Once oxygen flux is stable, perform a series of 1 µL titrations of 0.25 mM carbonyl cyanide p-(tri-fluromethoxy) phenyl-hydrazone (FCCP) until the fluorescent signal reaches its maximum. Select and label both the oxygen flux and TMRM signal representing minimum membrane potential and label it "FCCP".
NOTE: FCCP concentration of 0.5–1.0 µM is usually required to deplete mitochondrial membrane potential.
CAUTION: FCCP is toxic. Refer to the safety data sheet (SDS) for proper handling.
-
OPTIONAL: Once oxygen flux is stable, inject 1 µL of 0.25 mM rotenone to inhibit complex I and determine respiratory capacity through complex II.
NOTE: Changes in membrane potential are no longer relevant after titrating with uncoupler.
CAUTION: Rotenone is toxic. Refer to SDS for proper handling.
-
Once oxygen flux is stable, inject 1 µL of 1.25 mM (final concentration: 2.5 µM) of Antimycin A to inhibit mitochondrial respiration.
CAUTION: Antimycin A is toxic. Refer to SDS for proper handling.
Table 2:
Recommended ADP titration for 0.5 mL chamber
| SUIT labels | ADP stock (mM) | Injection volume (μL) | Final concentration (μM) |
|---|---|---|---|
| D1 | 0.5 mM ADP | 1 μL | 1 μM |
| D2 | 0.5 mM ADP | 4 μL | 5 μM |
| D3 | 0.5 mM ADP | 5 μL | 10 μM |
| D4 | 5 mM ADP | 1 μL | 20 μM |
| D5 | 5 mM ADP | 4 μL | 60 μM |
| D6 | 5 mM ADP | 4 μL | 100 μM |
| D7 | 50 mM ADP | 1 μL | 200 μM |
| D8 | 50 mM ADP | 4 μL | 600 μM |
| D9 | 50 mM ADP | 4 μL | 1000 μM |
| D10 | 500 mM ADP | 3 μL | 4000 μM |
5. Calculation of mitochondrial membrane potential and analysis
Using blank experiments, record the calibrated TMRM values (micromolar TMRM) before the injection of the blank sample ("pre-sample") and for each of the injections. See Figure 2.
-
For each blank experiment, calculate the background ratio by setting the "pre-sample" TMRM concentration to 1.0. Calculate the subsequent proportional decrease in TMRM. Calculate the average background ratio from all blank experiments.
NOTE: The number of blank experiments to include may depend on the instrumenťs precision. See the calculation example in Table 3 from five different blank experiments, where the standard deviation of the average background ratio fell between 0 and 0.016 for each titration.
Background calculation: Calculate the background for each sample experiment by multiplying "pre-sample" TMRM of the sample experiment times the average background ratio for each injection. See the calculation example in Table 3.
Background correction: Subtract the experimenťs background to the measured TMRM values of the sample. See the calculation example in Table 4.
FCCP correction: Subtract the FCCP background-corrected mMP from each injection. See the calculation example in Table 4.
-
ADP sensitivity curve: Normalize the ADP-driven decrease in mMP by setting the highest and lowest membrane potential as 100% and 0%, respectively, using the mMP values collected throughout the ADP titration. Fit the data into a non-linear fit regression model using the preferred statistical software to calculate the half-maximal inhibitory concentration (IC50) of ADP on mMP.
NOTE: The curve fits the [Inhibitor] vs. normalized response - Variable slope in Prism.
Figure 2: Calculating mitochondrial membrane potential (mMP) and ADP sensitivity from TMRM fluorescence.
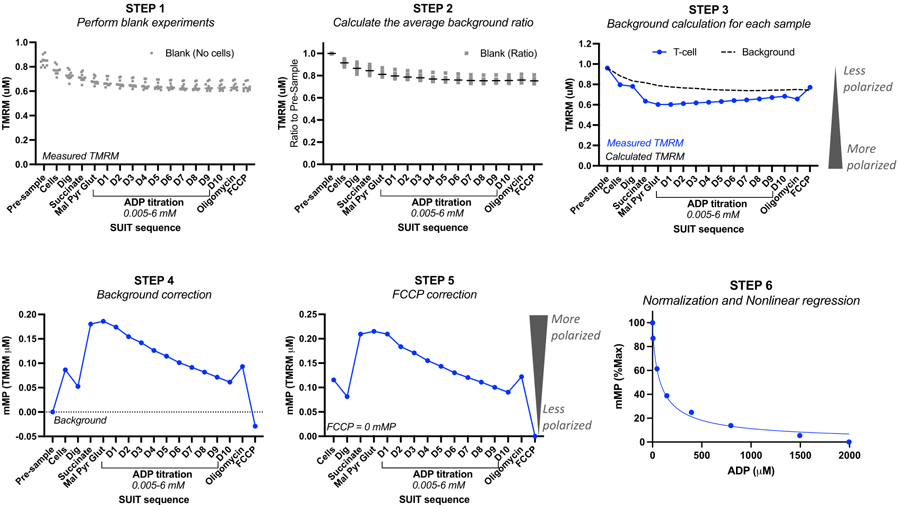
Steps for calculating mitochondrial membrane potential (mMP) and ADP sensitivity from measurements of TMRM fluorescence by high-resolution fluorespirometry of one sample of T-cells (n = 1). Step 1: TMRM fluorescence is measured in blank samples as done in the biological sample. Step 2: Determine the ratio in the TMRM signal with each titration relative to the signal prior to the sample for each blank experiment. Calculate the average for each titration of all blank experiments. Step 3: Calculate the background for each sample experiment by multiplying the "pre-sample" fluorescence by the average background ratio for each titration. Step 4: Calculate the difference between background and sample TMRM fluorescence for each titration to express data as mMP or mitochondrial TMRM uptake. Step 5: Correct mMP so that full uncoupling with FCCP reflects zero mMP. Step 6: Perform non-linear regression to graph changes in mMP with increasing ADP concentrations. Measurements were performed in 0.5 mL chambers, one containing 5 million T-cells from a healthy volunteer. Averaged data are expressed as mean ± SEM. Single data points of a single replicate are expressed without error bars.
Table 3:
Calculating the average background ratio using five independent blank experiments
| SUIT protocol | TMRM (μM) | Background ratio | Average background ratio | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| From O2K blank experiments | Divide by pre-sample value for each experiment | ||||||||||
| Blank experiment | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| Pre-sample | 0.916 | 0.851 | 0.903 | 0.842 | 0.851 | 1 | 1 | 1 | 1 | 1 | 1 |
| CELL | 0.834 | 0.77 | 0.82 | 0.777 | 0.77 | 0.91 | 0.904 | 0.908 | 0.923 | 0.904 | 0.91 |
| DIG | 0.789 | 0.727 | 0.776 | 0.733 | 0.727 | 0.861 | 0.854 | 0.86 | 0.87 | 0.854 | 0.86 |
| SUCC | 0.769 | 0.711 | 0.757 | 0.712 | 0.711 | 0.839 | 0.835 | 0.838 | 0.846 | 0.835 | 0.839 |
| MPG | 0.735 | 0.676 | 0.725 | 0.68 | 0.676 | 0.802 | 0.794 | 0.803 | 0.808 | 0.794 | 0.8 |
| D1 | 0.724 | 0.661 | 0.724 | 0.661 | 0.661 | 0.79 | 0.777 | 0.802 | 0.785 | 0.777 | 0.786 |
| D2 | 0.713 | 0.653 | 0.718 | 0.653 | 0.653 | 0.778 | 0.767 | 0.795 | 0.775 | 0.767 | 0.776 |
| D3 | 0.708 | 0.647 | 0.712 | 0.643 | 0.647 | 0.772 | 0.76 | 0.789 | 0.763 | 0.76 | 0.769 |
| D4 | 0.701 | 0.638 | 0.702 | 0.631 | 0.638 | 0.765 | 0.75 | 0.777 | 0.749 | 0.75 | 0.758 |
| D5 | 0.696 | 0.634 | 0.695 | 0.627 | 0.634 | 0.759 | 0.745 | 0.77 | 0.745 | 0.745 | 0.753 |
| D6 | 0.689 | 0.629 | 0.692 | 0.624 | 0.629 | 0.752 | 0.739 | 0.766 | 0.741 | 0.739 | 0.747 |
| D7 | 0.684 | 0.622 | 0.688 | 0.619 | 0.622 | 0.746 | 0.731 | 0.762 | 0.735 | 0.731 | 0.741 |
| D8 | 0.682 | 0.621 | 0.686 | 0.62 | 0.621 | 0.744 | 0.729 | 0.76 | 0.736 | 0.729 | 0.74 |
| D9 | 0.684 | 0.624 | 0.692 | 0.62 | 0.624 | 0.746 | 0.732 | 0.766 | 0.737 | 0.732 | 0.743 |
| D10 | 0.684 | 0.623 | 0.691 | 0.618 | 0.623 | 0.746 | 0.732 | 0.765 | 0.734 | 0.732 | 0.742 |
| OMY | 0.688 | 0.627 | 0.695 | 0.619 | 0.627 | 0.751 | 0.736 | 0.769 | 0.735 | 0.736 | 0.745 |
| FCCP | 0.684 | 0.622 | 0.69 | 0.612 | 0.622 | 0.747 | 0.731 | 0.764 | 0.727 | 0.731 | 0.74 |
Table 4:
Calculating mitochondrial membrane potential (mMP) from sample experiment
| Sample TMRM (μM) | Average background ratio | Experiment background (μM) | Background corrected mMP | FCCP corrected mMP | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Source/Calculation | From O2K Experiment | Calculated from blank experiments | Multiply ”Pre-sample" by the background ratio | Subtract TMRM background - sample | Subtract by FCCP mMP value | |||
| Pre-sample | 0.859 | 1 | x 0.859 = | 0.859 | − 0.859 = | 0 | ||
| CELL | 0.709 | 0.91 | x 0.859 = | 0.782 | − 0.709 = | 0.073 | − (−0.102) = | 0.175 |
| DIG | 0.711 | 0.86 | x 0.859 = | 0.739 | − 0.711 = | 0.028 | − (−0.102) = | 0.13 |
| SUCC | 0.552 | 0.838 | x 0.859 = | 0.72 | − 0.552 = | 0.168 | − (−0.102) = | 0.27 |
| MPG | 0.51 | 0.8 | x 0.859 = | 0.687 | − 0.510 = | 0.177 | − (−0.102) = | 0.279 |
| D1 | 0.505 | 0.786 | x 0.859 = | 0.675 | − 0.505 = | 0.17 | − (−0.102) = | 0.272 |
| D2 | 0.501 | 0.776 | x 0.859 = | 0.666 | − 0.501 = | 0.165 | − (−0.102) = | 0.267 |
| D3 | 0.504 | 0.769 | x 0.859 = | 0.66 | − 0.504 = | 0.157 | − (−0.102) = | 0.259 |
| D4 | 0.51 | 0.758 | x 0.859 = | 0.651 | − 0.510= | 0.141 | − (−0.102) = | 0.243 |
| D5 | 0.52 | 0.753 | x 0.859 = | 0.647 | − 0.520 = | 0.127 | − (−0.102) = | 0.229 |
| D6 | 0.534 | 0.747 | x 0.859 = | 0.642 | − 0.534 = | 0.108 | − (−0.102) = | 0.21 |
| D7 | 0.543 | 0.741 | x 0.859 = | 0.636 | − 0.543 = | 0.093 | − (−0.102) = | 0.195 |
| D8 | 0.557 | 0.739 | x 0.859 = | 0.635 | − 0.557 = | 0.077 | − (−0.102) = | 0.179 |
| D9 | 0.575 | 0.743 | x 0.859 = | 0.638 | − 0.575 = | 0.063 | − (−0.102) = | 0.165 |
| D10 | 0.592 | 0.742 | x 0.859 = | 0.637 | − 0.592 = | 0.046 | − (−0.102) = | 0.148 |
| OMY | 0.59 | 0.746 | x 0.859 = | 0.641 | − 0.590 = | 0.05 | − (−0.102) = | 0.152 |
| FCCP | 0.738 | 0.74 | x 0.859 = | 0.636 | − 0.738 = | −0.102 | − (−0.102) = | 0 |
Representative Results
To illustrate the differences in optimal cell concentration for the assay, 5 million T-cells were loaded into one 0.5 mL chamber (10 million cells/mL), and 1.25 million cells were loaded into another chamber (2.5 million cells/mL) containing 1 µM TMRM (Figure 3A–G). Three blank experiments were also included to calculate the TMRM background. We found that a higher concentration of T-cells resulted in a more distinguishable change in TMRM fluorescence relative to the background (Figure 3B,D). In addition, a higher cell concentration allowed us to detect the expected increase in oxygen consumption and simultaneous depletion of the mMP in response to the addition of FCCP (Figure 3E,F). Using a low concentration of cells yielded a weak change in fluorescence that paralleled the background. Since the calculation of mMP subtracts the background from the signal, a low cell concentration does not allow for the determination of changes in mMP in response to substrates and uncouplers. In addition to using the higher concentrations of cells in this assay, we recommend keeping the cell concentration constant for each cell type between experiments.
Figure 3: High-resolution fluorespirometry experiments.
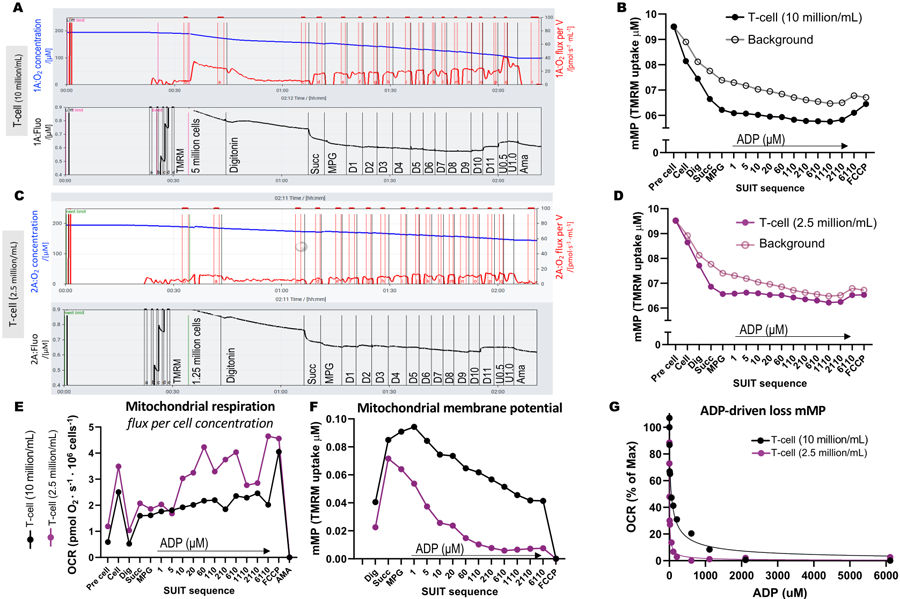
(A-D) Trace of high-resolution fluorespirometry experiments using T-cell concentrations of 10 million cells/mL and 2.5 million cells/mL in 0.5 mL chambers. (A) 10 million cells/mL in 0.5 mL chambers. (C) 2.5 million cells/mL in 0.5 mL chambers. Oxygen flux (pmol/s/mL) is shown in the top panel (red), and the calibrated TMRM signal is shown in the bottom panel (black). Changes in TMRM throughout the SUIT for the sample and its calculated background were plotted for the chambers containing (B) 10 million cells/mL and (D) 2.5 million cells/mL. (E) For each cell concentration, oxygen flux (pmol/s/million cells) and (F) mitochondrial membrane potential were calculated. (G) ADP sensitivity curve was plotted and fit to a non-linear regression model (solid lines). Abbreviations: mMP, mitochondrial membrane potential; TMRM, tetramethylrhodamine methyl ester; SUIT, substrate-uncoupler-inhibitor titrations; ADP, adenosine diphosphate; Dig, digitonin; Mal, malate; Pyr, pyruvate; Glut, glutamate; D1–11, 11 consecutive ADP titrations; U, uncoupler FCCP of 0.5 and 1.0 μM; AMA, antimycin A.
To validate the influence of ATP-synthase in the dissipation of mMP with ADP titrations, we ran parallel experiments on PBMCs and T-cells where one chamber received oligomycin before ADP titration (Figure 4). We found no dissipation of mMP in response to ADP in cells treated with oligomycin, suggesting that the gradual decrease in mMP with ADP is a result of proton flux through ATP-synthase (Figure 4A–F). We also compared ADP sensitivity between T-cells and PBMCs of the same participant and found ADP sensitivity to be lower (higher EC50) in the T-cell fraction (Figure 4G,H).
Figure 4: ATP-synthase drives ADP-driven decrease in membrane potential in T-cells and PBMCs.
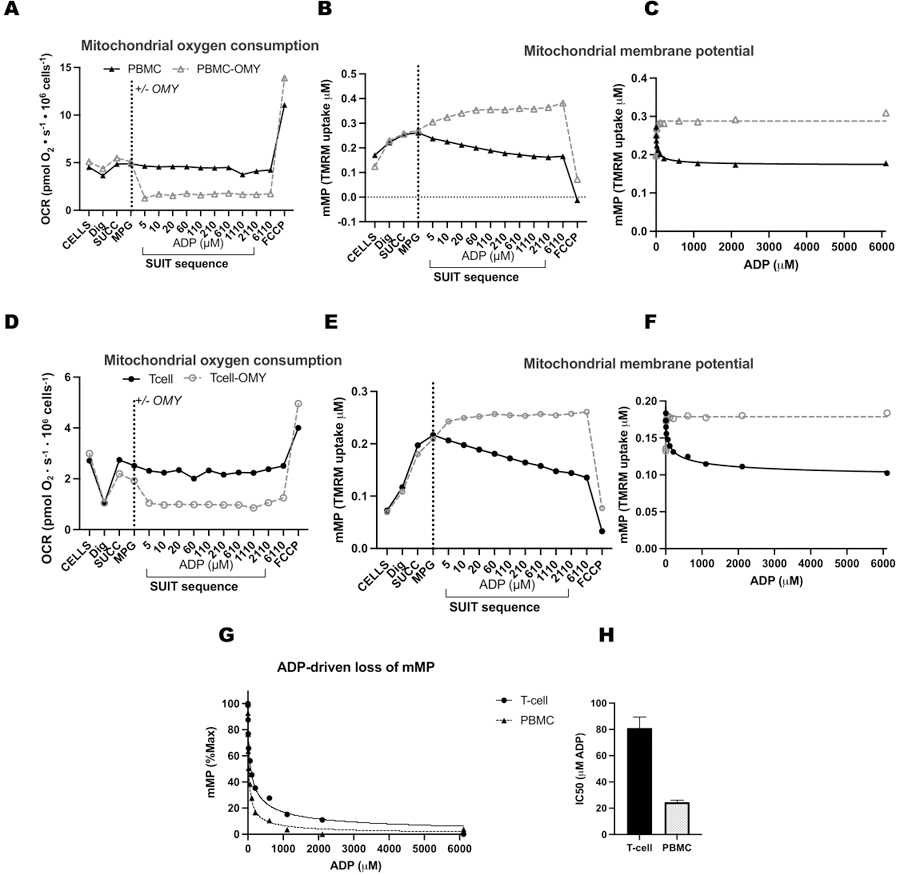
(A-H) The protocol described here was tested in PBMCs and T-cells. Two O2K chambers were injected with PBMCs, and two chambers of an additional O2K were injected with T-cells from the same participant. After injecting substrates malate, pyruvate, and glutamate in all chambers, one chamber of PBMCs and T-cells received oligomycin. Oligomycin prevented any ADP-driven rise in respiration in (A) PBMCs and (D) T-cells or decline in mitochondrial membrane potential in (B,C) PBMCs and (E,F) T-cells. (G,H) ADP sensitivity was greater in PBMCs compared to T-cells.
We conducted a series of blank experiments to determine the influence of time or the SUIT protocol on TMRM fluorescence. We found that the TMRM signal in blank experiments is mostly influenced by SUIT titrations (Figure 5A) as opposed to the timing of the titrations (Figure 5B).
Figure 5: Blank experiments show the change in TMRM fluorescence in response to time and titrations of substrates, uncouplers, and inhibitors (SUIT).
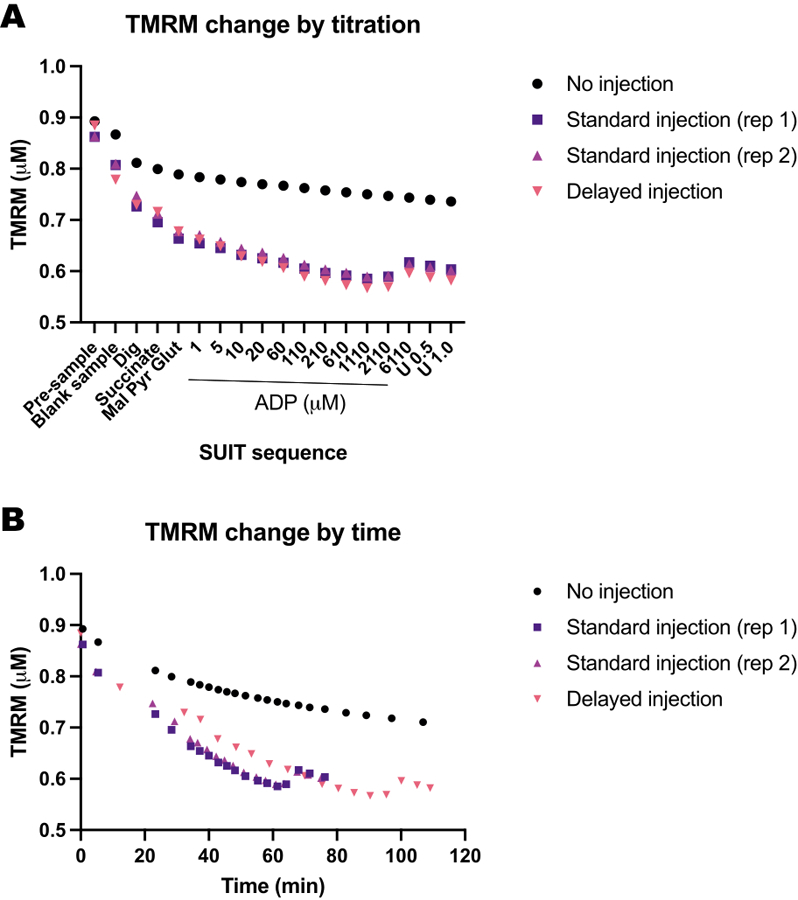
(A) Change in TMRM fluorescence in response to titration. (B) Change in TMRM fluorescence in response to time. Experiments were conducted in 0.5 mL chambers filled with Mir05 containing 1 μM TMRM. One chamber did not receive any SUIT titrations (no injection); two chambers in two different instruments received a standard suit protocol (standard injection); one chamber received the same SUIT titrations but with a delay between each injection (delayed injection).
We compared ADP-driven changes in oxygen consumption rates (OCR) and in mMP in T-cells and monocytes from 11 healthy, community-dwelling volunteers (Figure 6A–H). Similar to the results of previously published experiments using extracellular flux and enzymatic assays, monocytes exhibited a greater mitochondrial respiratory capacity than lymphocytes26, 27 (Figure 6A,H). However, we did not detect a typical dose-response increase in OCR with ADP in either cell type (Figure 6C,D), contrary to what this method shows when using highly metabolic tissues like mouse liver (Figure 7A–H). On the other hand, the use of TMRM allowed us to detect a gradual decline in mMP with ADP in human immune cells (Figure 6E–G) and in splenic T-cells from mice (Figure 7E–H). While we did not directly compare human and mouse T-cells using the same titration protocol, we did find that the IC50 of mouse T cells was lower by a factor of 10 compared with that of circulating T-cells from human subjects.
Figure 6: Differences in ADP sensitivity between T-cells and monocytes using OCR and mMP.
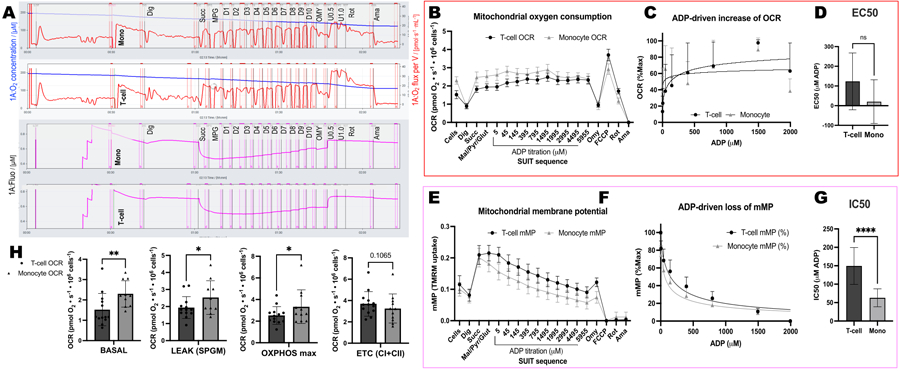
(A) Trace of high-resolution fluorespirometry experiment from a subjecťs monocyte and T-cell sample. (B) Oxygen consumption in monocytes (n= 11) and T-cells (n= 13) from the blood of healthy volunteers. (C,D) Non-linear regression fitting of the plotted rise in respiration with ADP titrations to calculate an EC50. (E) Simultaneous measurement of mitochondrial membrane potential. (F,G) Non-linear regression fitting of the plotted decline in mitochondrial membrane potential with ADP titrations to calculate an IC50. (H) Parameters of respiratory capacity of monocytes and T-cells. Data are expressed as mean ± SEM for line graphs and mean ± SD for bar graphs. Statistically significant differences following t-tests are expressed as *p < 0.05. **p < 0.01, and ****p < for 0.0001.
Figure 7: Comparing ADP response in respiration and mitochondrial membrane potential (mMP) in permeabilized mouse splenic T-cells and liver.
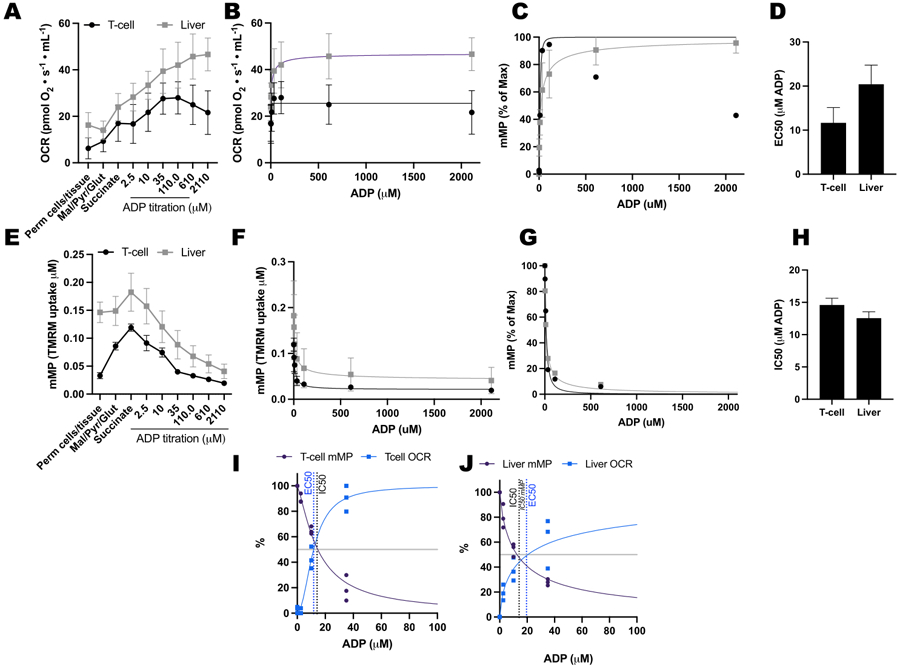
(A-D) Response in respiration in permeabilized mouse splenic T-cells and liver. (E-H) Response in mMP in permeabilized mouse splenic T-cells and liver. Fresh liver and spleen were dissected from three mice following cervical dislocation. Splenic Pan T-cells were isolated using antibody-conjugated magnetic bead separation. Both samples underwent the same SUIT protocol in the presence of 1 μM TMRM. (I,J) Comparison of EC50 calculated from the increase in oxygen consumption (OCR) and IC50 from the decrease in mMP in response to ADP. N = 3 per group. Data are expressed as mean ± SEM.
Discussion
This protocol uses high-resolution fluorespirometry to measure the sensitivity of the mitochondrial response to energy demand by measuring the dissipation of mMP in response to increasing levels of ADP in PBMCs, monocytes, and T-cells. This is done by adding complex I and II substrates to maximize the mitochondrial membrane potential and titrating ADP to gradually stimulate ATP-synthase to use the proton gradient for ATP generation.
Critical steps in the protocol include setting the gain and intensity of the fluorophore to 1000 and making sure a TMRM fluorescent signal is acquired during the TMRM titration. Because TMRM fluorescence declines following each titration (a limitation of this method), it is imperative to run background experiments using blank samples. We have also found that DMSO has an inhibitory effect on mitochondrial respiration and membrane potential and, therefore, recommend diluting the working solution of TMRM in Mir05 (Supplementary Figure 1).
Some modifications that may be used when trying this protocol are adjusting cell concentrations and using the standard 2 mL chamber. However, the 0.5 mL chamber is preferred for T-cells and monocytes because of the high concentration of cells needed for optimal response in membrane potential and oxygen flux. A lower concentration of cells may be optimal when testing cells with greater respiratory capacity, like macrophages.
Additional limitations of the method presented here include the requirement for at least 5 million T-cells and 2.5 million monocytes. We can often obtain enough cells from ~20 mL of blood from healthy participants, but these numbers can vary by health status, age, and sex26. In addition, as in most methods assessing mitochondrial capacity, the cells need to be freshly isolated. However, this method could be tried in cryopreserved cells in the future. In comparison with the yield from human blood, the T-cell yield from spleens of healthy mice is high enough to conduct this assay.
Circulating T-cells, particularly long-lived memory (TM) and regulatory (Treg) cells, rely on oxidative phosphorylation for energy37. While their energy demand and oxygen consumption are low (e.g., compared with that of resting muscle), their survival is essential for an effective immune response to reinfection and cancer38, 39, 40. A reduction in T-cell oxidative phosphorylation results in impaired proliferative capacity and promotes T-cell exhaustion and senescence5, 41. Additionally, mitochondrial hyperpolarization promotes a sustained production of cytokines (IL-4 and IL-21) by effector CD4 T-cells during activation42. Upon infection, the energy requirement for activation and proliferation of immune cells can be as high as 25%-30% of the basal metabolic rate43. Therefore, immune cells function in a wide and extreme range of energy demands, and this protocol can test mitochondrial responses within that range.
Chronic inflammation is a common feature of obesity, diabetes, and aging. Dysregulated levels of circulating hormones, lipids, and glucose have systemic impacts and can thus affect how mitochondria respond to an energetic challenge. Here, we have presented a method to assess mitochondrial ADP sensitivity in circulating PBMCs. Further studies are needed to determine how ADP sensitivity may be modulated in metabolic disease and how it impacts health status.
Supplementary Material
Supplementary File 1: Reagent preparation and protocol for isolating T-cells from mouse spleen.
Supplementary Figure 1: Effect of Mir05 and DMSO on mitochondrial respiration and membrane potential.
Acknowledgments
We would like to thank the kind volunteers who donated blood for this project. We also extend our sincere appreciation to Dr. Ellen Schur and her team for providing us with additional samples from their study. We would also like to thank Andrew Kirsh for reviewing the manuscript and editing it for legibility. This work was supported by the following funding sources: P01AG001751, R01AG078279, P30AR074990, P30DK035816, P30DK017047, R01DK089036, K01HL154761, T32AG066574.
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/66863.
Disclosures
The authors declare no conflicts of interest.
References
- 1.Eisner V, Picard M, Hajnóczky G Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat Cell Biol 20 (7), 755–765 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sokolova I Bioenergetics in environmental adaptation and stress tolerance of aquatic ectotherms: linking physiology and ecology in a multi-stressor landscape. J Exp Biol 224 (Pt Suppl 1), jeb.236802 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Buttgereit F, Brand MD A hierarchy of ATP-consuming processes in mammalian cells. Biochem J 312 (Pt 1), 163–167 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmid D, Burmester GR, Tripmacher R, Kuhnke A, Buttgereit F Bioenergetics of human peripheral blood mononuclear cell metabolism in quiescent, activated, and glucocorticoid-treated states. Biosci Rep 20 (4), 289–302 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Vardhana SA et al. Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat Immunol 21 (9), 1022–1033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson SR, Li A, Beck-Previs S, Kennedy GG, Warshaw DM Imaging ATP consumption in resting skeletal muscle: One molecule at a time. Biophys J 119 (6), 1050–1055 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyrat A, von Ballmoos C ATP synthesis at physiological nucleotide concentrations. Sci Rep 9 (1), 3070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gouspillou G et al. Accurate determination of the oxidative phosphorylation affinity for ADP in isolated mitochondria. PLoS One. 6 (6), e20709 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holloway GP et al. Age-associated impairments in mitochondrial ADP sensitivity contribute to redox stress in senescent human skeletal muscle. Cell Rep 22 (11), 2837–2848 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Pharaoh G, Brown J, Ranjit R, Ungvari Z, Van Remmen H Reduced adenosine diphosphate sensitivity in skeletal muscle mitochondria increases reactive oxygen species production in mouse models of aging and oxidative stress but not denervation. JCSM Rapid Commun 4 (1), 75–89 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pharaoh G et al. The mitochondrially targeted peptide elamipretide (SS-31) improves ADP sensitivity in aged mitochondria by increasing uptake through the adenine nucleotide translocator (ANT). Geroscience. 45 (6), 3529–3548 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brunetta HS, Petrick HL, Vachon B, Nunes EA, Holloway GP Insulin rapidly increases skeletal muscle mitochondrial ADP sensitivity in the absence of a high lipid environment. Biochem J 478 (13), 2539–2553 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Brunetta HS et al. Nitrate consumption preserves HFD-induced skeletal muscle mitochondrial ADP sensitivity and lysine acetylation: A potential role for SIRT1. Redox Biol 52, 102307 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyrrell DJ et al. Blood-cell bioenergetics are associated with physical function and inflammation in overweight/ obese older adults. Exp Gerontol 70, 84–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liepinsh E et al. Low-intensity exercise stimulates bioenergetics and increases fat oxidation in mitochondria of blood mononuclear cells from sedentary adults. Physiol Rep 8 (12), e14489 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedges CP et al. Peripheral blood mononuclear cells do not reflect skeletal muscle mitochondrial function or adaptation to high-intensity interval training in healthy young men. J Appl Physiol (1985). 126 (2), 454–461 (2019). [DOI] [PubMed] [Google Scholar]
- 17.DeConne TM, Muñoz ER, Sanjana F, Hobson JC, Martens CR Cardiometabolic risk factors are associated with immune cell mitochondrial respiration in humans. Am J Physiol Heart Circ Physiol 319 (2), H481–H487 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou B et al. Boosting NAD level suppresses inflammatory activation of PBMCs in heart failure. J Clin Invest 130 (11), 6054–6063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altintas MM, DiBartolo S, Tadros L, Samelko B, Wasse H Metabolic changes in peripheral blood mononuclear cells isolated from patients with end stage renal disease. Front Endocrinol (Lausanne) 12, 629239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pence BD, Yarbro JR Aging impairs mitochondrial respiratory capacity in classical monocytes. Exp Gerontol 108, 112–117 (2018). [DOI] [PubMed] [Google Scholar]
- 21.van der Windt GJW, Chang CH, Pearce EL Measuring bioenergetics in T Cells using a Seahorse extracellular flux analyzer. Curr Protoc Immunol 113, 3.16B.11–13.16B.14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chacko BK et al. Methods for defining distinct bioenergetic profiles in platelets, lymphocytes, monocytes, and neutrophils, and the oxidative burst from human blood. Lab Invest 93 (6), 690–700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buck MD et al. Mitochondrial dynamics controls T Cell fate through metabolic programming. Cell. 166 (1), 63–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinn KM et al. Metabolic characteristics of CD8. Nat Commun 11 (1), 2857 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scandalis L et al. Skeletal muscle mitochondrial respiration and exercise intolerance in patients with heart failure with preserved ejection fraction. JAMA Cardiol 8 (6), 575–584 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rausser S et al. Mitochondrial phenotypes in purified human immune cell subtypes and cell mixtures. Elife. 10, e70899 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer PA et al. Bioenergetics and the oxidative burst: protocols for the isolation and evaluation of human leukocytes and platelets. J Vis Exp 85, 51301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kramer PA, Ravi S, Chacko B, Johnson MS, Darley-Usmar VM A review of the mitochondrial and glycolytic metabolism in human platelets and leukocytes: implications for their use as bioenergetic biomarkers. Redox Biol 2, 206–210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teodoro JS, Machado IF, Castela AC, Rolo AP, Palmeira CM The evaluation of mitochondrial membrane potential using fluorescent dyes or a membrane-permeable cation (TPP+ ) electrode in isolated mitochondria and intact cells. Methods Mol Biol 2184, 197–213 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Hassan H, Zakaria F, Makpol S, Karim NA A link between mitochondrial dysregulation and idiopathic autism spectrum disorder (ASD): Alterations in mitochondrial respiratory capacity and membrane potential. Curr Issues Mol Biol 43 (3), 2238–2252 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams AS et al. Disruption of Acetyl-Lysine turnover in muscle mitochondria promotes insulin resistance and redox stress without overt respiratory dysfunction. Cell Metab 31 (1), 131–147.e111 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krumschnabel G, Eigentler A, Fasching M, Gnaiger E Use of safranin for the assessment of mitochondrial membrane potential by high-resolution respirometry and fluorometry. Methods Enzymol 542, 163–181 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Chowdhury SR, Djordjevic J, Albensi BC, Fernyhough P Simultaneous evaluation of substrate-dependent oxygen consumption rates and mitochondrial membrane potential by TMRM and safranin in cortical mitochondria. Biosci Rep 36 (1), e00286 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vianello C et al. High-throughput microscopy analysis of mitochondrial membrane potential in 2D and 3D models. Cells. 12 (7) (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perry SW, Norman JP, Barbieri J, Brown EB, Gelbard HA Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. Biotechniques. 50 (2), 98–115 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.JoVE Science Education Database. Basic Methods in Cellular and Molecular Biology. Using a Hemacytometer to Count Cells. JoVE, Cambridge, MA: (2023). [Google Scholar]
- 37.Geltink RIK, Kyle RL, & Pearce EL Unraveling the complex interplay between T cell metabolism and function. Annu Rev Immunol 36, 461–488 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sukumar M et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J Clin Invest 123 (10), 4479–4488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerriets VA et al. Foxp3 and Toll-like receptor signaling balance T. Nat Immunol 17 (12), 1459–1466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacIver NJ, Michalek RD, Rathmell JC Metabolic regulation of T lymphocytes. Annu Rev Immunol 31, 259–283 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desdín-Micó G et al. T cells with dysfunctional mitochondria induce multimorbidity and premature senescence. Science. 368 (6497), 1371–1376 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang R et al. Mitochondrial Ca2+ and membrane potential, an alternative pathway for Interleukin 6 to regulate CD4 cell effector function. Elife. 4, e06376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Straub RH, Cutolo M, Buttgereit F, Pongratz G Energy regulation and neuroendocrine-immune control in chronic inflammatory diseases. J Intern Med 267 (6), 543–560 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1: Reagent preparation and protocol for isolating T-cells from mouse spleen.
Supplementary Figure 1: Effect of Mir05 and DMSO on mitochondrial respiration and membrane potential.


