Abstract
Objective:
To assess the cellular and molecular landscape of adenomyosis.
Design:
Single-cell analysis of genome-wide messenger RNA (mRNA) expression (single-cell RNA sequencing) of matched tissues of endometrium, adenomyosis, and myometrium using relatively large numbers of viable cells.
Setting:
Not applicable.
Patient(s):
Patients (n = 3, age range 40–44 years) undergoing hysterectomy for diffuse adenomyosis.
Main Outcome Measure(s):
Definition of the molecular landscape of matched adenomyotic, endometrial and myometrial tissues from the same uterus using single-cell RNA sequencing and comparison of distinct cell types in these tissues to identify disease-specific cell populations, abnormal gene expression and pathway activation, and mesenchymal–epithelial interactions.
Result(s):
The largest cell population in the endometrium was composed of closely clustered fibroblast groups, which comprise 36% of all cells and seem to originate from pericyte progenitors differentiating to estrogen/progesterone receptor-expressing endometrial stromal-cells. In contrast, the entire fibroblast population in adenomyosis comprised a larger (50%) portion of all cells and was not linked to any pericyte progenitors. Adenomyotic fibroblasts eventually differentiate into extracellular matrix protein-expressing fibroblasts and smooth muscle cells. Hierarchical clustering of mRNA expression revealed a unique adenomyotic fibroblast population that clustered transcriptomically with endometrial fibroblasts, suggestive of an endometrial stromal cell population serving as progenitors of adenomyosis. Four other adenomyotic fibroblast clusters with disease-specific transcriptomes were distinct from those of endometrial or myometrial fibroblasts. The mRNA levels of the natural WNT inhibitors, named, secreted frizzled-related proteins 1, 2, and 4, were higher in these 4 adenomyotic fibroblast clusters than in endometrial fibroblast clusters. Moreover, we found that multiple WNTs, which originate from fibroblasts and target ciliated and unciliated epithelial cells and endothelial cells, constitute a critical paracrine signaling network in adenomyotic tissue. Compared with endometrial tissue, unciliated and ciliated epithelial cells in adenomyosis comprised a significantly smaller portion of this tissue and exhibited molecular evidence of progesterone resistance and diminished regulation of estrogen signaling.
Conclusion(s):
We found a high degree of heterogeneity in fibroblast-like cells in the adenomyotic uterus. The WNT signaling involving differential expression of secreted frizzled-related proteins, which act as decoy receptors for WNTs, in adenomyotic fibroblasts may have a key role in the pathophysiology of this disease.
Keywords: Adenomyosis, endometrium, endometriosis, scRNA-seq, SFRP, fibroblast
Abstract
Objetivo:
evaluar el panorama celular y molecular de la adenomiosis.
Diseño:
Análisis unicelular de la expresión de ARN mensajero (ARNm) de todo el genoma (secuenciación de ARN unicelular) de tejidos emparejados de endometrio, adenomiosis y miometrio utilizando cantidades relativamente grandes de ćelulas viables.
Entorno:
No aplicable.
Paciente(s):
Pacientes (n = 3, rango de edad de 40 a 44 años) sometidas a histerectomía por adenomiosis difusa.
Principales medidas de resultado:
Definición del panorama molecular de tejidos adenomióticos, endometriales y miometriales emparejados del mismo útero utilizando secuenciación de ARN de una sola ćelula y comparación de distintos tipos de ćelulas en estos tejidos para identificar poblaciones de ćelulas específicas de la enfermedad, expresión génica anormal y la activación de la vía, y las interacciones mesenquimales-epiteliales.
Resultado(s):
La población celular más grande en el endometrio estaba compuesta por grupos de fibroblastos estrechamente agrupados, que comprenden el 36 % de todas las ćelulas y parecen originarse a partir de los progenitores de pericitos que se diferencian a ćelulas del estroma endometrial que expresan receptores de estrógeno/progesterona. Por el contrario, toda la población de fibroblastos en la adenomiosis comprendía una porción más grande (50%) de todas las ćelulas y no estaba vinculada a ningún progenitor de pericitos. Los fibroblastos adenomióticos finalmente se diferencian en fibroblastos que expresan proteínas de la matriz extracelular y ćelulas de músculo liso. El agrupamiento jerárquico de la expresión de ARNm reveló una población de fibroblastos adenomióticos única que se agrupaba transcriptómicamente con fibroblastos endometriales, lo que sugiere una población de ćelulas del estroma endometrial que actuá como progenitores de la adenomiosis. Otros cuatro grupos de fibroblastos adenomióticos con transcriptomas específicos de la enfermedad eran distintos de los de los fibroblastos endometriales o miometriales. Los niveles de ARNm de los inhibidores naturales de WNT, denominados proteínas secretadas relacionadas con frizzled 1, 2 y 4, fueron más altos en estos 4 grupos de fibroblastos adenomióticos que en los grupos de fibroblastos endometriales. Además, encontramos que múltiples WNT, que se originan a partir de fibroblastos y se dirigen a ćelulas epiteliales ciliadas y no ciliadas y ćelulas endoteliales, constituyen una red de señalización paracrina crítica en el tejido adenomiótico. En comparación con el tejido endometrial, las ćelulas epiteliales ciliadas y no ciliadas en la adenomiosis comprendían una porción significativamente menor de este tejido y exhibieron evidencia molecular de resistencia a la progesterona y disminución de la regulación de la señalización de estrógenos.
Conclusión(es):
Encontramos un alto grado de heterogeneidad en las ćelulas similares a fibroblastos en el útero adenomiótico. La señalización de WNT que implica la expresión diferencial de proteínas secretadas relacionadas con frizzled, que actuán como receptores señuelo para WNT, en fibroblastos adenomióticos puede tener un papel clave en la fisiopatología de esta enfermedad.
Adenomyosis is an estrogen-dependent gynecologic disorder in which endometrial epithelial cells and stromal fibroblasts appear in the myometrium as islands surrounded by hypertrophic smooth muscle cells (1–3). It is associated with heavy menstrual bleeding, dysmenorrhea, chronic pelvic pain, and infertility (3, 4). The etiology and underlying mechanisms of adenomyosis have remained mostly unclear, although 2 main explanations have been proposed: invagination of the basal layer of the endometrium = and = metaplasia = of = Müllerian = remnant progenitor cells inside the myometrium (1, 5). Another explanation, proposed by Leyendecker, describes a “tissue injury and repair” mechanism that is activated by hyperestrogenism and hyperperistalsis leading to uterine autotraumatization (6, 7). According to the tissue injury and repair theory, adenomyosis arises from a vicious cycle involving hyperestrogen-initiated local injury that leads to increased production of prostaglandin E2, hyperperistalsis, and trauma at the archimetria (endometrial glands, endometrial stroma, and subendometrial myometrium), endometrial invagination, and chronic inflammation, which leads to further local hyperestrogenism (1, 7–9).
In its advanced stages, the disease imitates malignancy, not in terms of histology but by its clinical course. The unclear etiology and lack of effective medical therapy make adenomyosis a challenging clinical disorder. It is clear, however, that estrogen is essential for the growth of ectopic endometrium outside the uterus (1, 5). Therefore, medical strategies blocking ovarian steroidogenesis and local estrogen synthesis by the aromatase enzyme represent an indispensable option for treatment (10). However, the majority of patients require hysterectomy (11, 12), which is a radical treatment but not an appropriate option for women who desire pregnancy.
Adenomyosis once was seen as a disease of parous women that could be diagnosed only by surgical resection and pathological examination (13). However, advancements in high-resolution imaging now allow for noninvasive diagnosis by magnetic resonance imaging or transvaginal ultrasound (14, 15). Ultrasound features of the adenomyotic uterus include an enlarged uterus, asymmetry of the myometrial thickness, a poorly defined endomyometrial junction, and heterogeneously hypoechoic myometrium (11). Development of noninvasive diagnostic techniques also revealed a higher prevalence of adenomyosis in infertile women (16, 17); suggested mechanisms underlying uterine factor infertility include abnormal decidualization resulting in impaired endometrial receptivity, altered peristalsis of the uterus, and disruption of the endomyometrial junction (18, 19). Alterations in the immune system linked to sex steroid hormone changes and epithelial mesenchymal transition in women with adenomyosis may promote inflammation and impair fertility (20). Some studies have shown that adenomyosis is associated not only with infertility but also increased risk of miscarriage (21, 22).
Recent research has led to new insights into the molecular pathophysiology of adenomyosis. Evidence from next-generation screening-based studies identified KRAS mutations in endometrial epithelial cell clones and adjacent adenomyotic tissue (5, 23). These activating KRAS mutations, which also have been found in endometriosis, are one of the most frequent driver mutations in malignancies, such as lung, pancreatic, and colorectal cancers (24). KRAS mutations in adenomyosis and endometriosis are confined to endometrial epithelial cells and associated with specific pathways that increase cell survival and proliferation (25). These mutations also have been associated with progesterone resistance in adenomyosis (23). The findings of next-generation screening-based studies favor mechanisms involving invagination of mutated cells rather than metaplasia or differentiation of progenitor cells; however, more work is needed to understand gene expression in the extremely heterogeneous adenomyotic cell populations and cross talk with surrounding tissues.
The histologically complex nature of adenomyosis, with island-like structures inside the myometrium that also contain endometrial and myometrial cells, makes it difficult to identify pathophysiologic mechanisms based on the average gene expression of the whole tissue because of the background noise or dilution issues. Single-cell RNA sequencing (scRNA-seq) has opened a new era in research that enables investigators to reveal the heterogeneity in complex tissues and understand how distinct cell types contribute to disease (26). In a recent study, scRNA-seq of endometrial tissues collected throughout the menstrual cycle from healthy women revealed 7 cell types based on the transcriptomic profile of epithelial cells and stromal fibroblasts during 4 major phases of endometrial transformation (27). Here, we applied scRNA-seq to analyze the cellular composition and transcriptomic profiles of matched tissues of endometrium, normal myometrium, and adenomyosis from the same hysterectomy specimens collected from 3 women, with the goal of identifying specific signaling pathways and cell-cell interactions in the adenomyotic uterus that contribute to this complex disease.
MATERIALS AND METHODS
Tissue Collection and Patient Characteristics
Northwestern University’s institutional review board approved the use of human tissue for this study. After obtaining written informed consent, adenomyotic tissue and adjacent normal endometrium and normal myometrium were obtained from premenopausal women undergoing hysterectomy for adenomyosis. Three samples were collected from each patient, for a total of 9 samples from 3 patients. Briefly, after hysterectomy, 3 samples from each hysterectomy specimen were dissected by a senior pathologist immediately and confirmed histologically using hematoxylin and eosin staining. Normal-appearing endometrial and myometrial samples were obtained at 1 cm from adenomyotic tissue. For normal-appearing endometrium we obtained a full thickness section of endometrial tissue that included functionalis and basalis layers. After tissue dissection and digestion as described below, single-cell suspensions were submitted for scRNA-seq in the Northwestern University NUSeq Core (Fig. 1A). The age range of the patients was 40–44 years and each patient had at least 1 previous delivery. No patient had been receiving hormonal treatment at the time of tissue collection for a period of at least 3 months preoperatively. All tissues were collected during the follicular phase. The cycle phase was determined based on last menstrual period and histology. All 3 patients had diagnoses of diffuse adenomyosis confirmed by pathology reports.
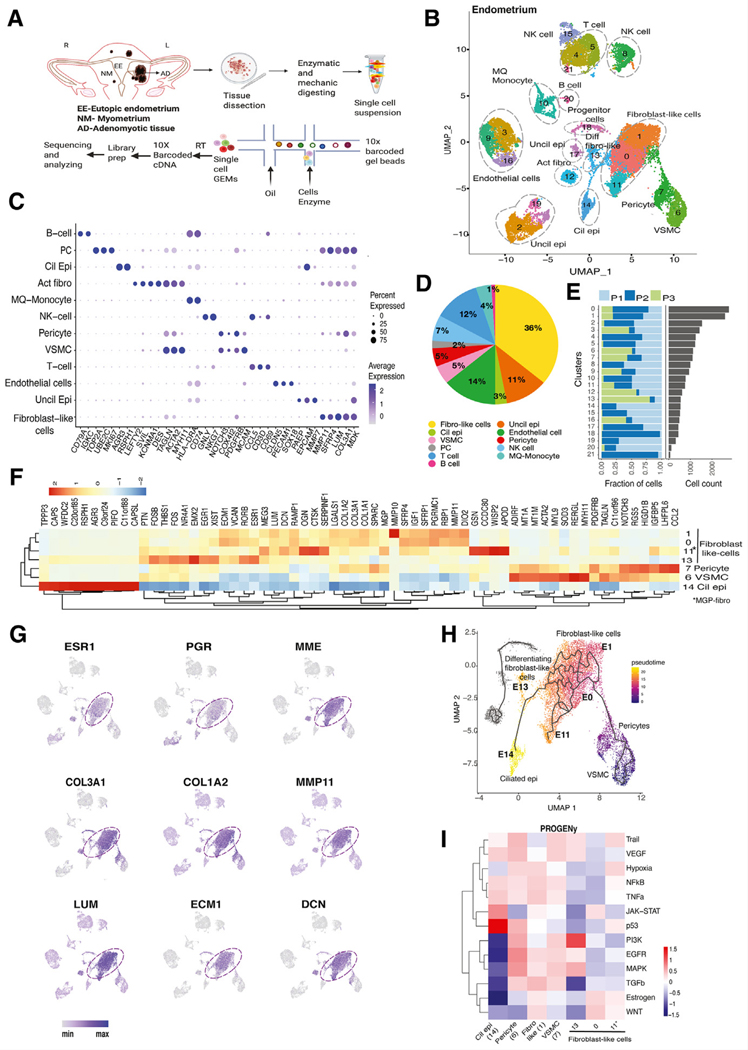
FIGURE 1.
Single-cell transcriptomic analysis of endometrium. (A) Experimental design of the study. (B) UMAP presentation of endometrial cell clusters. Although the fibroblast-like population contained the highest numbers of cells, there also are prominent groups of T cells, NK cells, endothelial cells and unciliated and ciliated epithelial cells. (C) Dot plot of identified markers of each cell type. (D) Pie chart showing percentages of identified cell types. (E) Bar graph showing the number of cells from each patient (n = 3) in each cluster of integrated data. Each patient sample contributed data to each cluster. (F) Heatmap of highly expressed genes compared between progenitor cells, fibroblast-like cells, SMCs, and matrix Gla protein (MGP)-expressing fibroblasts. Ciliated epithelial cells highly expressed cilia-associated genes such as TPPP3, PIFO, and RSPH1. (G) Feature plots showing the distribution of key messenger RNAs s such as estrogen and progesterone receptors and select ECM-associated genes in fibroblast-like cells. (H) Pseudotime trajectory that was started from pericytes. It appeared that the pericytes might differentiate to the fibroblast-like group, including E0/E1/E11/E13, a process terminated in ciliated epithelial cells, which suggested mesenchymal–epithelial transition. (I) Heatmap of the PROGENy-predicted pathway activity scores of selected clusters (ciliated epithelial cells, pericytes, fibroblast-like cells, VSMCs). Interestingly, estrogenic and WNT pathways were upregulated in most fibroblast-like cells and downregulated in ciliated epithelial cells. UMAP, uniform manifold approximation and projection; SMCs, smooth muscle cells; VSMCs, vascular smooth muscle cells.
Single-Cell Suspension Protocol
Adenomyotic, endometrial, and myometrial tissues were dissociated and cells were isolated using a previously described protocol (28). Briefly, tissues were digested or frozen within 12 hours after hysterectomy. Samples from 2 patients (6 samples) were processed fresh. The remaining patient’s samples (3 samples) were frozen due to lack of available appointments in the Northwestern University NUSeq Core. For the fresh samples, tissues were kept in HypoThermosol FRS Solution (BioLife Solutions, Bothell, WA) for up to 12 hours. Tissues were minced into very small pieces in a petri dish with Hank’s balanced salt solution, then placed in a 50 mL Falcon tube with digestion enzyme mixture (collagenase, protease, DNase, hyaluronidase, and calcium chloride) in 10 mL Hank’s balanced salt solution. Tissues were incubated for 2 hours in a 37°C bead bad incubator with 5 minutes of gentle shaking every 30 minutes. After enzymatic and mechanical digestion, the tissue fraction was allowed to sediment out (approximately 2–5 minutes). The stromal cells and smooth muscle cells were collected by passing the digested supernatant through 70-μm cell sieves and debris was left in the filter. Red blood cell lysis was performed for the stromal cell fraction. For adenomyotic and endometrial tissues, the epithelial cell pellets were backwashed by inverting the cell strainer and centrifugation. The supernatant was removed, and epithelial cells were digested further with 1 mL TrypLE express enzyme (Gibco, Billings, MNT) at 37°C for 10 minutes, followed by passing through 70-μm cell sieves to obtain single epithelial cell suspensions. Epithelial and stroma cell suspensions were centrifuged at 300 relative centrifugal force for 5 minutes at room temperature. After removing the supernatant without disrupting the pellet, stromal and epithelial cells were suspended and mixed together in ×1 phosphate-buffered saline with 0.04% bovine serum albumin (400 μg/mL) to achieve the target cell concentration (<5 million cells per tube in 1 mL washing solution in 2 Ml DNA LoBind Tubes). Epithelial cell digestion steps were skipped for normal myometrium tissue because significant numbers of epithelial cells were expected in the sample. For all 3 tissue types, to remove the remaining cell debris and large clumps, the cell suspension was pipetted gently and filtered with Scienceware 40 μm Flowm Tip strainers (VWR, Radnor, PA). For frozen samples, the tissue was minced into very small pieces immediately after collection from the pathology core and split into cryovials containing Cryostor CS10 Freeze media (BioLife Solutions). Before scRNA-seq, frozen samples were thawed in a 37°C bath for 1 minute and digested to obtain single-cell suspensions, as described above.
Single-Cell RNA Sequencing
Single-cell suspensions of each tissue in DNA LoBind tubes were placed on ice and submitted to the NUSeq Core immediately to check the viability and cell number. Cell yield and viability of the frozen and thawed samples and fresh samples were comparable and >75%, as determined by the trypan blue cell viability assay. From the 9 samples, a total of approximately 90,000 cells were collected. After checking for sample quality and filtering out dead cells, the final total cell number was 66,000. Overall, the contribution from each of the 3 patients to cell clusters was comparable in terms of total cell numbers. However, we noted that each patient sample contributed variably to 3 cell clusters in endometrial and adenomyotic issues. The contribution came primarily from a single patient in 3 extremely small clusters: 2 endometrial clusters (progenitor and T-cell) and 1 adenomyotic cluster (neuron-like). Data from these 3 clusters were excluded from our analyses. Due to high cell yield, suspensions were diluted before sequencing to obtain the target cell concentration. Single-cell suspensions from adenomyotic uteri with approximately 80% cell viability was used as input material with an optimal cell concentration of 700–1200 cells/μL. Samples then were processed for construction of 10× libraries for sequencing using the 10× Genomics Single-Cell 3′ reagent kit protocol. Samples were loaded onto the 10× Genomics Chromium platform to generate single-cell gel beads in emulsion. Cells underwent reverse transcription reactions to generate barcoded cDNA, which shares a 10× barcode with all cDNA from its individual cell of origin. Subsequently, the gel beads in emulsion were broken, and all uniquely barcoded cDNAs were pooled and amplified by polymerase chain reaction to generate material for sequencing on the Illumina HiSeq 4000 at a depth of average 37,000 reads per cell.
Single-Cell RNA-Sequencing Data Analysis
Pre-processing and integration.
Processed data from the Cell Ranger pipeline was analyzed using the R package Seurat (v3.2) as described previously (29). Quality control analysis was performed before data integration. Cells with ≤300 gene counts and cells with >15% mitochondrial RNA content were filtered out to remove low-quality cells. After log normalization of the filtered data from the 3 patients, data were aligned using the FindIntegrationAnchors and IntegrateData functions to remove batch effects. This process was performed for data from adenomyosis, endometrium, and myometrium samples separately to identify and compare tissue-specific cell types.
Cell clustering, visualization, and cell-type annotation.
After scaling with a coefficient and regressing out the effects of mitochondrial genes and the cell cycle effect (S Scores and iG2 Scores), principal component analysis was performed with 30 principal components and a default resolution parameter using the FindCluster function. Identified cell clusters were visualized by a nonlinear dimensionality reduction algorithm, UMAP. To identify the cell type of each cluster, canonical cell marker gene information was collected from the published literature, then marker genes for every cluster were identified using the Wilcoxon rank-sum test option of the FindAll-Markers function, and clusters were annotated based on the presence of reference marker genes in these clusters.
Pseudotime, DoRothEA, PROGENy, and gene enrichment analyses.
After transferring Seurat-generated cluster embeddings into a monocle object, pseudotime analysis was performed on the fibroblast-like cell clusters using monocle3 (v0.2.1) (30). Transcription factor (TF) enrichment analysis was performed on fibroblast-like cell clusters of all tissues using the discriminant regulon expression analysis tool, DoRothEA (v2), which is a curated resource of TFs and their transcriptional targets (31). First, regulons with A-C confidence level were selected for the analysis, then the 20 most variable TFs in selected clusters were visualized. Pathway activities of selected clusters were estimated by PROGENy, which infers pathway activity from perturbation experiments (32). Gene ontology terms for differentially upregulated genes in unciliated epithelial cells in adenomyosis and endometrium samples were identified using cluster Profiler on Gene Set Enrichment Analysis using the gseGO function for all ontologies. Normalized enrichment scores were calculated for enrichment after correcting for multiple testing with the Benjamini-Hochberg method. Dot plots were generated to show significant GO terms based on their p-adjusted value and Gene Ratio, derived by dividing the total number of genes in the gene set by the number of overlapping genes. In these pathway analyses, we excluded the clusters that originated primarily from the tissue of from a single patient; i.e., E18 and E21 from endometrium and A24 from adenomyosis (see Figs. 1E, 2C).
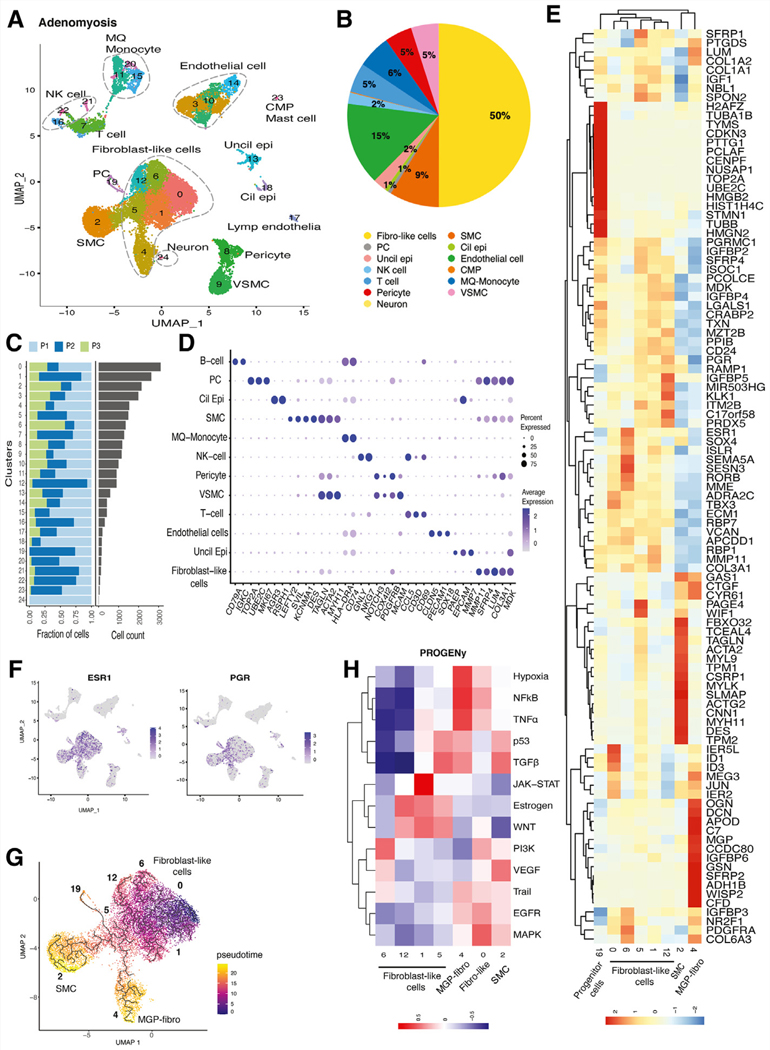
FIGURE 2.
Single-cell transcriptomic analysis of adenomyosis tissue. (A) UMAP presentation of adenomyotic cell clusters. There is a larger and more heterogeneous group of fibroblast-like cells than endometrial fibroblasts (see Fig. 1). In contrast to endometrial tissue, there is no readily recognizable connection between the pericyte progenitors and fibroblast-like cells. (B) Fibroblasts comprised a much larger portion (50%) of adenomyosis, compared with endometrial fibroblast population (36%, see Fig. 1). In contrast, epithelial cells made up only 3% of adenomyosis versus 14% of endometrial tissue (see Fig. 1). Macrophage (MQ)-monocyte clusters seem to compose the most prominent immune cell population in adenomyosis. (C) Bar graph showing the number of cells from each patient (n = 3) in clusters of integrated data. (D) Dot plot of identified markers of each cell type. (E) Heatmap of highly expressed genes in progenitor cells, fibroblast-like cells, SMCs, and MGP-expressing fibroblasts. MGP-fibroblast-like cluster (A4) prominently expresses ECM-related genes and SFRPs. SMC cluster (A2) expresses SMC markers such as ACTA2. (F) Feature plots showing the distribution of expression of ESR1 and PGR, which concentrated in fibroblast-like cells. (G) Pseudotime trajectory of fibroblast-like cells suggesting that fibroblast-like cells might originate from cluster A0, differentiate through A1/A6/A12/A5, and terminate in 2 endpoints, A2 SMCs and A4 MGP-fibroblast-like cells. (H) Heatmap of the PROGENy-predicted pathway activity scores of selected cell clusters (fibroblast-like cells, MGP-expressing fibroblasts, and SMCs). Estrogen and WNT pathways are coactivated in A1/A5/A12, consistent with high ESR1 expression in these cluster (see Fig 2F). MGP, matrix gla protein; Cil epi, ciliated epithelial cells; CMP, common myeloid progenitor; fibro-like, fibroblast-like; Lymph endothelia, lymphatic endothelial cells; NK, natural killer; PC, Progenitor-like cells; VSMC, vascular SMCs; Uncil epi, unciliated epithelial cells.
RESULTS
Endometrium: A Complex Landscape Rich in Fibroblast and Immune Cell Populations
We identified 21 cell clusters comprised of 12 distinct cell types shown in the UMAP: unciliated (3 clusters) and ciliated epithelial cells, endothelial cells (3 clusters), fibroblast-like cells (4 clusters), activated fibroblasts, pericytes, vascular smooth muscle cells, NK cells, T cells (3 clusters), B cells, macrophages, monocytes, and potential progenitor cells (Fig. 1B) The top markers for each cell type are shown in a dot blot (Fig. 1C), and the top markers for each cluster are shown in a detailed heatmap (Supplemental Fig. 1a).
Unciliated epithelial cells organized in 3 clusters (E2, E17, E19) comprised 11% of the endometrium. Epithelial cells were identified with canonical markers including EPCAM, PAEP, MMP7, and KRT18. We also identified one ciliated epithelial cell cluster, which expresses cilia-associated genes (C14), such as RSPH1, AGR3, and PIFO; this cell type was described recently in a study performing scRNA-seq of normal endometrium (27; Figs. 1B–D).
Endothelial cells expressed PECAM1, SOX18 and CLDN5 and were present in 3 clusters (E3, E9, E16), which constituted 14% of endometrium (Figs. 1B–D).
Immune cells constituted of 24% of endometrium. The dominant immune cells were NK cells (7%) expressing NKG7 and GNLY) and T cells (12%) expressing CD3D and CD69. Macrophages and monocytes were identified based on CD74 and HLA-DRA expression. A very small B cell cluster also was present (1%), which expressed CD79A and IGKC (Figs. 1B–D).
Fibroblast-like cells constituted the majority of the endometrium (36%). We identified 4 closely grouped clusters of fibroblast-like cells (E0, E1, E11, E13), which express collagen and matrix-related genes (Figs. 1B–D). When comparing endometrial tissues across the 3 patients, all 3 tissues contained the majority of cellular clusters, although at varying levels (Fig. 1E).
Fibroblast clusters E0 and E1 had similar gene expression profiles, with both expressing SFRP4, COL1A1, LUM, MMP11, SFRP1, ECM1, and IGF1 (Fig. 1F). Feature plots also revealed that both clusters also expressed MME, ESR1, and PGR, which are expressed by endometrial stromal fibroblasts (Fig. 1G). Cluster E11 showed high expression of adhesion-related genes, such as WISP2, CCDC80, GSN, OGN, and SERPINF1. Cluster E13 was slightly segregated from the group and uniquely expressed differentiation and mitogenesis-related genes, such as FOSB, EGR1, NR4A1, FOS, RO RB, THBS1, ESR1, JUN, and PDGFRA. Among all fibroblast-like cells, ESR1 expression was the highest in cluster E13, which we identified as differentiated fibroblasts. Some of the key genes expressed in the fibroblast-like populations are shown as feature plots in Figures 1F and 1G.
A distinct cluster (E7) that closely mapped to fibroblasts showed unique expression of the pericyte markers COX4I2, MCAM, NOTCH3, and PDGFRB. Attached to E7 was a vascular smooth muscle cell cluster (E6) expressing markers, such as MYL9, TINAGL, BCAM, MYH11, MYLK, and ACTA2 (Fig. 1F). Because stem/progenitor type cells were detected consistently among pericytes, we performed a pseudotime analysis starting from cluster E7 (33). Intriguingly, the differentiation trajectory initiating from pericytes progressed through fibroblast-like cells in E0 and E1, then through actively differentiating fibroblast-like cells (E13) and ended in ciliated epithelium (E14; Fig. 1H). Endometrial mesenchymal stem cells were reported previously to be clonogenic and multipotent pericytes, which may respond to conditions during endometrial regeneration (34). Therefore, our findings suggest that fibroblast-like cells might originate from pericytes with mesenchymal stem cell characteristics. The scRNA-seq data also suggested that ciliated epithelial cells may be derived from pericytes through mesenchymal–epithelial transition (MET; Fig. 1H). The PROGENy analysis of this MET trajectory showed that 2 distinct signaling pathways, namely PI3K and P53, were upregulated in differentiated fibroblast-like cells (E13) and the ciliated epithelial cluster (E14), respectively (Fig. 1I). We also used DoRothEA to examine differential expression of TFs in MET differentiation = trajectories = (E7→E0→E1→E11→E14), = which = is detailed in Supplemental Fig. 1b (available online).
Adenomyosis is Dominated by Fibrosis and a Highly Heterogeneous Fibroblast Population
We identified 24 clusters including 13 cell types in adenomyotic tissue, as shown in UMAP (Fig. 2A). Compared with endometrium, adenomyotic tissue had a higher proportion of fibroblasts and lower proportion of epithelial cells. Fibroblast-like cells constituted 50% of the tissue and showed a high degree of heterogeneity, gathering in 6 distinct clusters (A0, A1, A4, A5, A6, A12). A smooth muscle cell cluster (A2), which closely grouped with the fibroblast populations, constituted 9% of adenomyotic tissue. Epithelial cells constituted 3% of the tissue including ciliated and unciliated epithelium (A13, A18; Figs. 2A, 2B). Unlike endometrium, we identified a small neuron-like cluster (A24) grouped with fibroblast-like cells and a cluster with myeloid progenitor cells grouped with mast cells (A23). The relative contributions of each cluster per patient and the top gene markers for each cell type are shown in Figures 2C and 2D and Supplemental Figure 2a (available online). Immune cells constituted 13% of adenomyosis, compared with 24% in endometrium. Macrophages and monocytes constituted 6% of cells in adenomyotic tissue followed by T cells (5%) and NK cells (2%). Endothelial cells constituted approximately 15% of the tissue (A3, A10, A14), with a small cluster including lymphatic endothelial cells (A17). In contrast with endometrium, pericytes (A8) and vascular smooth muscle cells (A9) clustered separately from the fibroblast-like cells. Similar to the endometrium, we identified a potential progenitor cell cluster (A19), which associated closely with fibroblast-like cells and constituted 1% of adenomyotic tissue (Figs. 2A, 2B).
The fibroblast-like clusters A0, A6, A5, A1 and A12 showed transcriptomic similarities and represented a group distinct from A2, which was uniquely enriched in smooth muscle cell markers, and A4 that had a strong signature for extracellular matrix-related genes (Fig. 2E). In A0, a TF analysis demonstrated increased activity of ESR2, ETS2, FLI1, and FOXL2 (Supplemental Fig. 2b), whereas PROGENy showed activation of the MAPK and EGFR pathways, both of which are associated with cell proliferation (Fig. 2H). The neighboring A6 cluster had high expression of PDGFRA, COL6A3, MME, ESR1, RORB and HAND2, classical markers for mesenchymal progenitors as well as endometrial stromal cells (Fig. 2E). The PDGFRA together with ESR1 genes were expressed in A6, suggestive of actively proliferating and differentiating mesenchymal cells driven by estrogen. Interestingly, A6 showed a pattern of maximum ESR1 expression indicative of high estrogen responsiveness, whereas the highest PGR expression was seen in A12 and A5 (Fig. 2F). This pattern of progression of ESR1/PGR expression is consistent with a pseudotime analysis showing differentiation in the direction of A0→A6→A12→A5 (Figs. 2F, 2G). The TF analysis suggested that A6 and A12 have increased ATF3 activity, which is related to cellular response to injury, and A6 showed MXI1 and THAP11 upregulation, which are related with pluripotency and mesenchymal cell differentiation (Supplemental Fig. 2b).
Cluster A2 showed clear expression of smooth muscle cell markers, such as ACTA2, DES, ACTG2, and MYLK; therefore, we identified it as the smooth muscle cells (SMCs) cluster (Fig. 2E). Pseudotime analysis suggested that this SMC cluster seems to originate from an adenomyotic fibroblast cluster after a differentiation pattern of A0→A6→A12→A5→A2 (Fig. 2G). The TF analysis of A2 showed high activity of MEF2A, TEAD1, and SMAD4, which are associated with EMT regulation and muscle hypertrophy. Matrix gla protein (MGP) was the top marker in A4; these cells showed high expression of genes associated with matrix remodeling including decorin (DCN), which is a collagen binding proteoglycan of extracellular matrix, asporin (ASPN), which has a key role in osteoblast driven collagen mineralization activity, and other genes related to macrophage response, such as AEBP1. The TFs associated with immune response (STAT3, SPI) and muscle hypertrophy (SMAD4), and the related TNF and NFKB pathways, were predicted to be activated in this cluster (Supplemental Fig. 2b). The MGP-expressing A4 cluster may represent terminally differentiated fibroblasts that are resident in adenomyosis.
In the fibroblast-like cell population, the pseudotime analysis initiated from A0 showed a pro-proliferative transcriptomic profile similar to that of the small progenitor cluster (A19, Fig. 2E); the differentiation trajectory diffusely transitioned via A1, A6, A12, and A5, and segregated to reach 2 clear end points, namely A2 (SMCs) and A4 (fibroblast-like cells expressing matrix-associated genes including MGP, thus designated MGP fibroblast-like; Fig. 2G). Intriguingly, this analysis also demonstrated a trajectory between the small progenitor cluster (A19) and the entire congregated fibroblast-like population (Fig. 2G). A19 expressed mitosis and cell division-associated genes and was closely related transcriptomically to the progenitor cluster E18 in endometrium. Fibroblast-like cells were clustered tightly in adenomyosis, and unlike endometrial tissue, pericytes (A8) and vascular SMCs (A9) were not linked directly to this fibroblast-like population (Figs. 1B, 2A). A pseudotime analysis starting from pericytes did not show a continuation to fibroblast-like cells in adenomyotic tissue.
Myometrium: Fibroblast-Like Clusters Form a Population Distinct From Smooth Muscle Cell Clusters
Overall, there were 20 clusters including 10 cell types in myometrium (Supplemental Fig. 3a, available online). As in endometrium and adenomyosis, the dominant cell type in myometrium comprised 5 closely bundled fibroblast-like cell clusters (40%), followed by SMCs (18%; Supplemental Fig. 3b). Intriguingly, there were 2 small clusters of epithelial cells (<1%) in myometrium (M17, M20) and 4 clusters of endothelial cells (M2, M12, M14) including a lymphatic endothelial cell cluster (M13). Similar to endometrium and adenomyosis, pericytes (M7) and vascular SMCs (M8, M9) clustered together. T cells made up the dominant immune cell cluster, similar to endometrium, followed by macrophages and monocytes. Relative contributions of clusters by each patient are shown in (Supplemental Fig. 3c).
The fibroblast-like cells in myometrial tissue were comprised of a close group of 5 clusters (M0, M1, M4, M11, M16). We performed subcluster analysis on these groups, as demonstrated in a heatmap (Supplemental Fig. 3d). The OGN, CTGF, CYR61, and LUM genes were the top markers in M0; these genes are associated with collagen formation and remodeling (35–37). Cluster M0 showed similarity to M11, which uniquely expressed MGP and was designated a “MGP fibroblast-like” cluster, similar to A4 (Fig. 2E). Cluster M11 also showed high expression of matrix-associated proteins, such as gelsolin (GSN), decorin (DCN), and fibulin-1 (FBLN1) (38).
Fibroblast clusters M1 and M4 had high expression of MMP11, SFRP4, CRABP2, and VCAN; however, unlike M1, M4 also showed upregulation of WNT2, ECM1, and COL3A1 as well as retinoic acid metabolism-associated genes, such as ALDH1A2, RORB, and RBP1 (39). The MDK gene, another highly expressed gene in M4, encodes a protein that responds to retinoic acid and promotes cell survival, migration, and tumorigenesis (40). Cluster M16 was distinct from all other clusters and showed a unique gene expression profile of differentiation and early response genes, such as FOS, JUN, and EGR1 (41). The PROGENy analysis showed upregulation of hypoxia, TGFB, P53, NFκB, TNF, and JAK-STAT pathways in M11 (MGP fibroblast-like cells), similar to A4 in adenomyosis (Supplemental Fig. 3e and Fig. 2H). A pseudotime analysis originating from the myometrial fibroblast cluster M4 showed a differentiation map through M1 and M0 that ended in M11 (MGP fibroblast-like cells; Supplemental Fig. 3f), similar to adenomyosis fibroblasts (Fig. 2G). Cluster-specific markers and a list of regulatory TFs in fibroblast-like cell clusters in myometrium are shown in a detailed heatmap (Supplemental Figs. 4a, 4b, available online).
We also interrogated the similarities and the differences between the 2 SMC clusters M3 and M5 and the SMC cluster (A2) in adenomyosis (Supplemental Fig. 4c). The transcriptomic profiles of M3 and A2 are quite similar with high expression of SFRP4, a modulator of the WNT pathway (Supplemental Fig. 4c). These data collectively suggest that in myometrial and adenomyotic tissues, SMCs likely originate from a fibroblast-like cluster.
Comparative Analysis of Fibroblast Populations in Adenomyosis, Endometrium, and Myometrium
We next evaluated differential gene expression patterns in fibroblast clusters that distinguish adenomyosis from endometrium and myometrium (Figs. 3A, 3B). Among the fibroblast-like clusters in adenomyosis and endometrium, the most striking observation was that A0 is very similar to E0 and E1 endometrial fibroblasts (Fig. 3A). This suggested that A0 fibroblasts represent a population that migrated from endometrium to invade myometrial tissue and contributed to adenomyosis (see Figs. 1B, 2A). Pseudotime analyses suggested that A0, E0, and E1 all have progenitor-like features, from which more differentiated fibroblasts originate (see Figs. 1H, 2G).
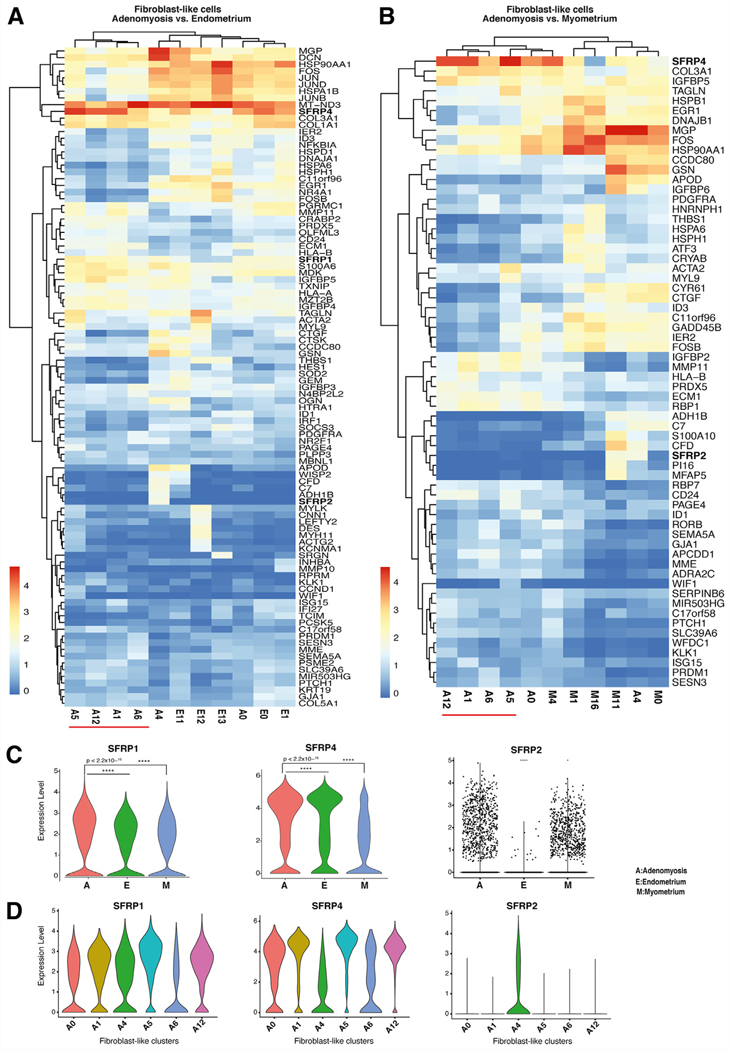
FIGURE 3.
Comparative analysis of differential gene expression in fibroblast populations across the 3 tissue types. (A) Heatmap of highly expressed genes in fibroblast-like cell populations in adenomyosis versus endometrium. The adenomyotic fibroblasts population A0 clustered together with the endometrial fibroblast types, suggesting that it may be of endometrial origin. (B) Heatmap of highly expressed genes in fibroblast-like cell populations in adenomyosis versus myometrium. The adenomyotic fibroblast-like population A4 clustered together with the myometrial fibroblast populations, suggesting that A4 might originate from myometrial tissue. (C) Violin plots displaying statistically significant differential expression of SFRPs (SFRP1, SFRP4, SFRP2) between the fibroblast-like clusters of adenomyosis (A), endometrium (E), and myometrium (M). (D) Violin plots displaying statistically significant differential gene expression of SFRPs (SFRP1, SFRP4, SFRP2) between selected fibroblast-like clusters in adenomyosis.
When adenomyotic and myometrial fibroblast clusters were compared (Fig. 3B), A4 showed striking similarities to M0 and M11, all of which expressed the highest levels of MGP and seemed to represent a terminally differentiated population among the fibroblast-like clusters in each tissue (see Fig. 2G and Supplemental Fig. 3f). The MGP is a structural extracellular matrix constituent (42), and our finding resonates with the observed increase in extracellular matrix in adenomyotic tissue.
Intriguingly, the gene family SFRP, which are modulators of the WNT pathway, were expressed prominently in fibroblast-like clusters, showing cross-tissue similarities between the endometrium, adenomyosis, and myometrium (Figs. 3A, 3B). Therefore, we first determined the differential expression of the members of this gene family across the 3 tissues (Fig. 3C). Both SFRP1 and SFRP4 levels were higher in adenomyosis than in endometrium or = myometrium (Fig. 3C). Gene SFRP2 was expressed only in adenomyosis and myometrium (A4, M11; Figs. 3A–C). It has been reported that SFRP2, which is a WNT antagonist, downregulates estrogen-dependent β-catenin activity, inhibits cell growth, and promotes myogenesis (43). The lack of SFRP2 expression in endometrium might permit epithelial cell growth and invasiveness. Thus, SFRP2 might be a key predictive marker of epithelial cell growth potential in adenomyosis. Interestingly, SFRP1 and SFRP4 are expressed in all fibroblast-like clusters in adenomyosis, whereas SFRP2 is expressed only in A4 (Fig. 3D).
The cells in clusters A1, A5, A6, and A12 seemed to be distinct within adenomyotic tissue (Figs. 3A, 3B); analysis of adenomyotic fibroblast-like cells showed uniquely upregulated genes involved in the WNT pathway, ribosome biogenesis, and protein targeting to endoplasmic reticulum (Supplemental Fig. 5, available online). Expression of genes in the ribosome biogenesis pathway was strikingly prominent in A12. Ribosome biogenesis is an established pathway that supports tumor cell growth and proliferation (44).
Analysis of Ciliated and Unciliated Epithelial Clusters in Endometrium and Adenomyosis
The epithelium represents the defining cell type in adenomyosis, and adenomyosis is thought to originate from the endometrium. Therefore, we further analyzed differentially expressed genes in epithelial cells between matched adenomyotic tissue and endometrium (5). First, we used scatter plot analysis to define the ciliated and unciliated epithelial cells in our tissues (Fig. 4A). As expected, classical markers, such as PAEP and MMP7, were conspicuously higher in unciliated epithelial cells, whereas a much larger number of genes were prominently expressed in ciliated epithelial cells (Fig. 4A) (27). Then, we identified differentially expressed genes separately in unciliated and ciliated epithelia in adenomyosis and endometrium (Figs. 4B, 4C). In general, unciliated and ciliated epithelium in adenomyotic tissue and endometrium showed similar transcriptomic profiles. In unciliated epithelium, however, expression of a number of genes was significantly higher in endometrium, including markers of differentiation, such as PAEP, PLCG2, HSPA1A, and HSP90AA1 (Fig. 4B). The PAEP marker, also known as glycodelin, is a secretory glycoprotein that is regulated by progesterone and associated with differentiation and healthy implantation (45, 46). This finding may suggest the presence of progesterone resistance in adenomyotic unciliated epithelium (39, 47). In unciliated and ciliated epithelial cells, expression of heat shock proteins (HSPA1A, HSP90AA1) was significantly lower in adenomyosis than in endometrium (Figs. 4B, 4C). Because some heat shock proteins are important for estrogen action, which seems to have a role in adenomyosis (5), this finding requires further investigation.
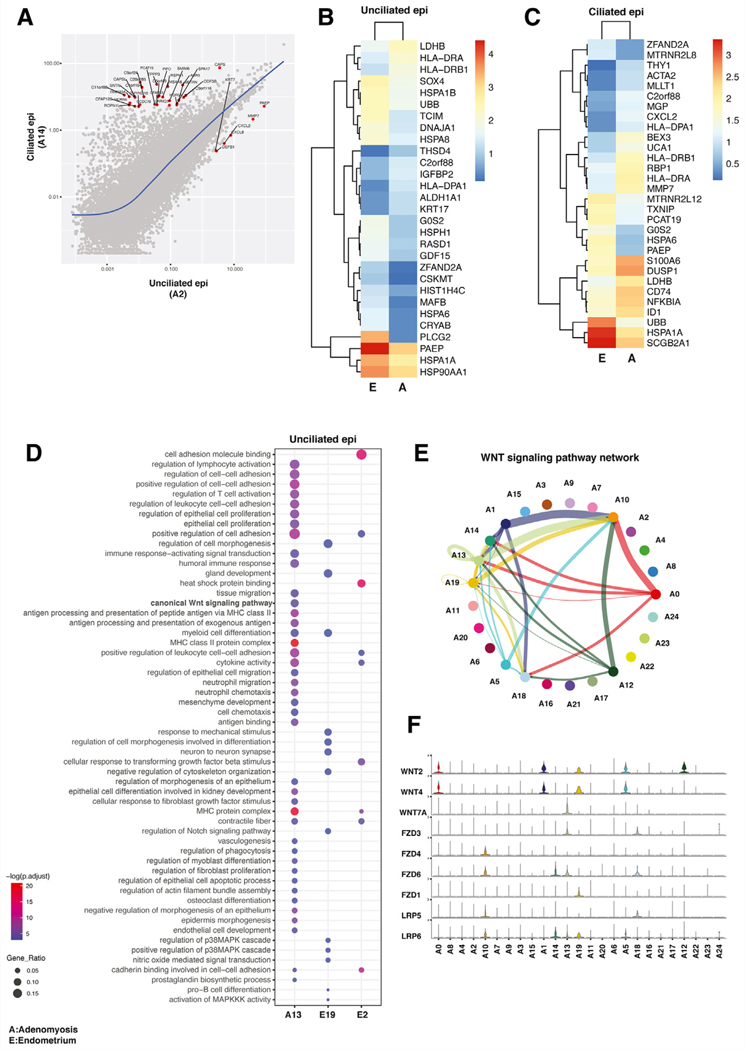
FIGURE 4.
Analysis of epithelial cell profiles in adenomyosis and endometrium. (A) Molecular characterization of epithelial cell types in adenomyosis. Scatter plot showing average log fold change in gene expression between ciliated epithelial cells and unciliated epithelial cells in adenomyotic tissue. Expression of only several select genes, such as PAEP, defined the specific transcriptional profile of unciliated epithelial cells. (B) Heatmap of highly expressed genes in unciliated epithelial cells in adenomyosis (A) versus endometrium (E). Expression of PAEP, a well-defined progesterone-responsive differentiation marker, is significantly higher in endometrial unciliated cells. (C) Heatmap of highly expressed genes in ciliated epithelial cells in adenomyosis (A) versus endometrium (E). Secretoglobin 2A1 (SCGB2A1) is prominently expressed in ciliated endometrial cells. (D) Over-representation analysis comparing the single unciliated epithelial cell cluster in adenomyosis (A13) compared with the 2 unciliated epithelial clusters in endometrium (E19 and E2). The canonical WNT pathway was uniquely activated in the adenomyotic ciliated epithelial cell cluster. (E) Inferred communication network of the entire WNT signaling pathway in adenomyosis. The general signaling direction is such that WNT ligands originate primarily from the fibroblast-like cells (A0, A1, A5 and A12), whereas the primary WNT-responsive cell types are the epithelial and endothelial cells. A13 unciliated epithelial cell cluster is unique in that it shows an autocrine signaling pattern; moreover, A13 epithelial cells also send signals to ciliated epithelial (A18) and endothelial (A10) cells. (F) Expression of WNT signaling pathway genes in each cell cluster in adenomyosis. The general pattern of expression is such that the WNT ligands are highly expressed in fibroblast-like cells, whereas their receptors or coreceptors are prominently expressed in epithelial and endothelial cells.
We also noted that immune function-related genes and pathways are upregulated in unciliated epithelial cells of adenomyosis compared with endometrium (Figs. 4B, 4D). Expression of genes associated with immune response and macrophage activation, such as HLA-DRA, HLA-DRB1, CD74, NFKBIA, and S100A6, were significantly higher in adenomyotic epithelium (Fig. 4B). These genes also are related to cell proliferation, inflammation, and cell motility. Among the upregulated pathways in adenomyosis, immune response-related processes were significantly upregulated in adenomyosis in ciliated and unciliated epithelium (Fig. 4D). Moreover, genes involved in the canonical WNT signaling pathway and regulation of epithelial cell migration pathways were upregulated only in adenomyotic unciliated epithelium.
In our analysis, WNT signaling emerged as a prominent biological process that was differentially regulated in stromal, epithelial, and SMCs; thus, we performed bioinformatics to further analyze paracrine and intracrine WNT-related interactions between all cell types in adenomyotic tissue (cell-cell communication analysis, Fig. 4E) The WNT signaling network analysis identified multiple signals originating from fibroblast clusters A0, A1, A5, and A12 toward epithelial and endothelial clusters A10, A13, A14, and A18 (Fig. 4E). The directionality of signals from stromal cells toward endothelial and epithelial cells is supported by the distribution of expression of ligands (WNT) and their receptors (FZD and LRP) in each cluster (Fig. 4F). In particular, WNT2 and WNT4 were expressed prominently in adenomyosis-specific fibroblast-like clusters, such as A0, A1, A5, and A12 (Fig. 4F; see Fig. 3). The most prominent WNT receptors expressed in endothelial or epithelial cells were FZD6 and LRP6. In summary, our scRNA-seq analysis uncovered significant roles of various fibroblast populations in endometrium and adenomyosis that seem to have central roles in adenomyosis pathophysiology via WNT signaling to endothelial and epithelial cells.
DISCUSSION
To our knowledge, this is the first detailed characterization of the transcriptomic landscape of the adenomyotic uterus using scRNA-seq to compare gene expression in distinct cell types of matched endometrium, adenomyotic tissue, and myometrium. We identified ciliated and unciliated epithelial cells, fibroblast-like cells, SMCs, endothelial cells, pericytes, vascular SMCs, potential progenitor cells, T cells, NK cells, B cells, macrophages, and monocytes, as well as small clusters including neurons and myeloid progenitors in the adenomyotic uterus. Our analyses revealed a high degree of heterogeneity in fibroblast-like cells in the adenomyotic uterus. This heterogeneity may be due to different transcriptional states driven by inflammation and fibrosis in adenomyosis. We identified fibroblast-like cell clusters that we suspect represent a disease-specific transcriptome (A1, A5, A6, and A12; Figs. 3A, 3B).
The pathophysiology of adenomyosis has been debated; however, current evidence suggests that adenomyosis occurs after the invasion of the myometrium by cellular oligoclones comprised of deep glandular epithelial cells with KRAS mutations together with attached endometrial stromal cells (5, 23). The lack of a substantial difference between epithelial cells in adenomyotic tissue and endometrium in our study supports this theory. Progesterone resistance has been demonstrated in adenomyotic tissue; Inuoe et al. (23) linked progesterone resistance to autonomously activated KRAS. Overall progesterone receptor (PGR) levels have been reported to be lower in adenomyosis compared to normal endometrium (48); however, we here identified higher PGR expression in distinct populations of fibroblast-like cells in adenomyotic tissue than in endometrium and myometrium, suggesting that progesterone resistance at the level of stromal cells is mediated by complex cellular and molecular mechanisms (Supplemental Fig. 6, available online). On the other hand, epithelial cells in adenomyotic tissue showed downregulation of PAEP compared with epithelial cells in endometrium; PAEP is an important progesterone-responsive gene (45, 46). This is suggestive that progesterone action in the context of a stromal-epithelial paracrine interaction may be deficient in adenomyosis (5, 39).
The first study comparing eutopic endometrial tissues of women with or without adenomyosis identified differentially expressed genes, which might initiate the disease pathways including eukaryotic initiation factor 2 signaling, oxidative phosphorylation, mitochondrial dysfunction, and estrogen receptor signaling (49). This study was based on microarray analysis and suggested that these abnormalities might be associated with the invasion of myometrial tissue by endometrial cells (49). A more recent study used bulk RNA sequencing to identify differentially expressed genes and molecular pathways in the eutopic endometrium from adenomyosis patients (50). These investigators found that IL-6 and ERK/MAPK-related pathways were activated in the eutopic endometrium of adenomyotic uterine tissues (50). A different study also identified the ERK/MAPK signaling pathway in association with the proliferation of uterine SMCs in women with adenomyosis (51).
The WNT pathway has been studied in the pathophysiology of adenomyosis (52); however, cell-specific details of its role have not been well understood. Here, we identified an intricate WNT signaling network between fibroblast-like cells and epithelial and endothelial cells in adenomyosis (Fig. 4E). Differentially expressed gene analysis in fibroblast-like cells from different tissues showed significant differences in expression of SFRP family members, which modulate WNT signaling (Figs. 3A, 3B). The SFRPs antagonize WNT signaling by competing with WNT proteins for receptor binding (53). We found that SFRPs are upregulated in fibroblast-like cells of adenomyosis compared with endometrium and normal-appearing myometrium, and that fibroblast-like cells in adenomyotic tissue express significantly higher SFRP1 and SFRP4. Moreover, the WNT pathway is regulated at another level with increased signaling from fibroblast-like cells to epithelial cells (Figs. 4E, 4F). This is suggestive that WNT activity may be upregulated in epithelial cells and downregulated via SFRPs in fibroblasts in adenomyotic tissue.
Another recent study compared endometrial and myometrial tissues of women with or without diffuse adenomyosis at the transcriptomic level (54). These investigators found that biological processes, including inflammation, extracellular matrix organization, collagen degradation, hyaluronan synthesis, and cell migration, were upregulated in adenomyosis. In the myometrial compartment, top biological processes in women with adenomyosis were extracellular matrix dysfunction, abnormal sensory pain perception, and γ-aminobutyric acid synaptic transmission (54). We also identified a unique cluster in adenomyosis (A4) that has a strong signature for extracellular matrix-related genes (Fig. 2E).
There are some limitations to our study. Despite being a powerful approach to establishing the transcriptomic profile of individual cells in heterogenous cell populations, single-cell sequencing has drawbacks (26). Single-cell sequencing technology allows assessment of heterogeneity in complex tissues, but low coverage prevents detection of mutations (42). Thus, a major limitation of our study is a lack of mutation information. Additionally, we did not identify cell surface markers and, thus, could not sort cells to verify the identities or functions of the cell clusters. Because adenomyosis is a disease that develops within the myometrium, we originally expected to see a higher number of SMCs but found a higher complement of fibroblasts compared with SMCs. A potential explanation is that fibroblast-like cells were more easily digested than SMCs, and thus a higher number of fibroblast-like cells than SMCs were prepared and analyzed. Our findings, on the other hand, also may reflect the in vivo nature of myometrial tissue. In fact, a recent scRNA-seq analysis of the myometrium identified fibroblasts as a major component of this tissue (55). Nevertheless, the comparative transcriptomic analysis of matched endometrium, adjacent myometrium, and adenomyotic tissue within the same uteri obtained from 3 patients via scRNA-seq is a significant strength of our study. The only other study using scRNA-seq to evaluate adenomyosis used a single patient and did not provide any comparative information regarding normal-appearing myometrium of the adenomyotic uterus (56).
In conclusion, adenomyosis is a complex pathology associated with elaborate molecular mechanisms characterized by estrogen-dependent chronic inflammation, progesterone resistance, and epithelial KRAS mutations (23). Recent DNA-based evidence provided irrefutable support that adenomyotic lesions originate from adjacent endometrial tissue; thus, the theory of entrapped endometrium as the origin of adenomyosis pertains (23). Here, we used a cutting-edge procedure to define gene expression profiles of the individual cell types in adenomyotic tissue and adjacent endometrium and myometrium in an attempt to ascertain the cell-cell interactions and the key signaling pathways involved in the pathophysiology of adenomyosis. The WNT pathway turned out to be a candidate regulator of the critical biological processes in adenomyosis. We anticipate that our findings may facilitate the identification of new therapeutic targets for adenomyosis.
Supplementary Material
Acknowledgments
Supported in part by Grant P50-HD098580 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (S.E.B., J.J.W., and M.A.) and Grant TUBITAK-2219 International Postdoctoral Research Fellowship Program (S.Y.).
Footnotes
S.Y. and M.K. should be considered similar in author order.
S.Y. has nothing to disclose. M.K. has nothing to disclose. J.J.W. has nothing to disclose. M.M. has nothing to disclose. P.Y. has nothing to disclose. M.A. has nothing to disclose. S.E.B. has nothing to disclose.
REFERENCES
- 1.Garcia-Solares J, Donnez J, Donnez O, Dolmans MM. Pathogenesis of uterine adenomyosis: invagination or metaplasia? Fertil Steril 2018;109:371–9. [DOI] [PubMed] [Google Scholar]
- 2.Bird CC, McElin TW, Manalo-Estrella P. The elusive adenomyosis of the uterus–revisited. Am J Obstet Gynecol 1972;112:583–93. [DOI] [PubMed] [Google Scholar]
- 3.Zhai J, Vannuccini S, Petraglia F, Giudice LC. Adenomyosis: mechanisms and pathogenesis. Semin Reprod Med 2020;38:129–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Struble J, Reid S, Bedaiwy MA. Adenomyosis: a clinical review of a challenging gynecologic condition. J Minim Invasive Gynecol 2016;23:164–85. [DOI] [PubMed] [Google Scholar]
- 5.Bulun SE, Yildiz S, Adli M, Wei JJ. Adenomyosis pathogenesis: insights from next-generation sequencing. Hum Reprod Update 2021;27:1086–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benagiano G, Habiba M, Brosens I. The pathophysiology of uterine adenomyosis: an update. Fertil Steril 2012;98:572–9. [DOI] [PubMed] [Google Scholar]
- 7.Leyendecker G, Wildt L, Mall G. The pathophysiology of endometriosis and adenomyosis: tissue injury and repair. Arch Gynecol Obstet 2009;280: 529–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leyendecker G, Wildt L. A new concept of endometriosis and adenomyosis: tissue injury and repair (TIAR). Horm Mol Biol Clin Investig 2011;5:125–42. [DOI] [PubMed] [Google Scholar]
- 9.Leyendecker G, Kunz G, Noe M, Herbertz M, Mall G. Endometriosis: a dysfunction and disease of the archimetra. Hum Reprod Update 1998;4: 752–62. [DOI] [PubMed] [Google Scholar]
- 10.Fang Z, Yang S, Gurates B, Tamura M, Simpson E, Evans D, et al. Genetic or enzymatic disruption of aromatase inhibits the growth of ectopic uterine tissue. J Clin Endocrinol Meta 2002;87:3460–6. [DOI] [PubMed] [Google Scholar]
- 11.Kho KA, Chen JS, Halvorson LM. Diagnosis, evaluation, and treatment of adenomyosis. JAMA 2021;326:177–8. [DOI] [PubMed] [Google Scholar]
- 12.Yu O, Schulze-Rath R, Grafton J, Hansen K, Scholes D, Reed SD. Adenomyosis incidence, prevalence and treatment: United States population-based study 2006–2015. Am J Obstet Gynecol 2020;223:94 e1–94 e10. [DOI] [PubMed] [Google Scholar]
- 13.Maheshwari A, Gurunath S, Fatima F, Bhattacharya S. Adenomyosis and subfertility: a systematic review of prevalence, diagnosis, treatment and fertility outcomes. Hum Reprod Update 2012;18:374–92. [DOI] [PubMed] [Google Scholar]
- 14.Tellum T, Nygaard S, Lieng M. Noninvasive diagnosis of adenomyosis: a structured review and meta-analysis of diagnostic accuracy in imaging. J Minim Invasive Gynecol 2020;27:408–418 e3. [DOI] [PubMed] [Google Scholar]
- 15.Chapron C, Vannuccini S, Santulli P, Abrao MS, Carmona F, Fraser IS, et al. Diagnosing adenomyosis: an integrated clinical and imaging approach. Hum Reprod Update 2020;26:392–411. [DOI] [PubMed] [Google Scholar]
- 16.Harada T, Khine YM, Kaponis A, Nikellis T, Decavalas G, Taniguchi F. The impact of adenomyosis on women’s fertility. Obstet Gynecol Surv 2016; 71:557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sunkara SK, Khan KS. Adenomyosis and female fertility: a critical review of the evidence. J Obstet Gynaecol 2012;32:113–6. [DOI] [PubMed] [Google Scholar]
- 18.Munro MG. Uterine polyps, adenomyosis, leiomyomas, and endometrial receptivity. Fertil Steril 2019;111:629–40. [DOI] [PubMed] [Google Scholar]
- 19.Campo S, Campo V, Benagiano G. Adenomyosis and infertility. Reprod Biomed Online 2012;24:35–46. [DOI] [PubMed] [Google Scholar]
- 20.Bourdon M, Santulli P, Jeljeli M, Vannuccini S, Marcellin L, Doridot L, et al. Immunological changes associated with adenomyosis: a systematic review. Hum Reprod Update 2021;27:108–29. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Conejero JA, Morgan M, Montesinos M, Fortuno S, Meseguer M, Simon C, et al. Adenomyosis does not affect implantation, but is associated with miscarriage in patients undergoing oocyte donation. Fertil Steril 2011; 96:943–50. [DOI] [PubMed] [Google Scholar]
- 22.Vercellini P, Consonni D, Dridi D, Bracco B, Frattaruolo MP, Somigliana E. Uterine adenomyosis and in vitro fertilization outcome: a systematic review and meta-analysis. Hum Reprod 2014;29:964–77. [DOI] [PubMed] [Google Scholar]
- 23.Inoue S, Hirota Y, Ueno T, Fukui Y, Yoshida E, Hayashi T, et al. Uterine adenomyosis is an oligoclonal disorder associated with KRAS mutations. Nat Commun 2019;10:5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L, Guo Z, Wang F, Fu L. KRAS mutation: from undruggable to druggable in cancer. Signal Transduct Target Ther 2021;6:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anglesio MS, Papadopoulos N, Ayhan A, Nazeran TM, Noe M, Horlings HM, et al. Cancer-Associated mutations in endometriosis without cancer. N Engl J Med 2017;376:1835–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell 2015;58: 610–20. [DOI] [PubMed] [Google Scholar]
- 27.Wang W, Vilella F, Alama P, Moreno I, Mignardi M, Isakova A, et al. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat Med 2020;26:1644–53. [DOI] [PubMed] [Google Scholar]
- 28.Wu B, Li Y, Liu Y, Jin K, Zhao K, An C, et al. Cell atlas of human uterus. bioRxiv 2018:267849. [Google Scholar]
- 29.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32:381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Alonso L, Holland CH, Ibrahim MM, Turei D, Saez-Rodriguez J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res 2019;29:1363–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schubert M, Klinger B, Klunemann M, Sieber A, Uhlitz F, Sauer S, et al. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat Commun 2018;9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gargett CE, Schwab KE, Deane JA. Endometrial stem/progenitor cells: the first 10 years. Hum Reprod Update 2016;22:137–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spitzer TL, Rojas A, Zelenko Z, Aghajanova L, Erikson DW, Barragan F, et al. Perivascular human endometrial mesenchymal stem cells express pathways relevant to self-renewal, lineage specification, and functional phenotype. Biol Reprod 2012;86:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall-Glenn F, Lyons KM. Roles for CCN2 in normal physiological processes. Cell Mol Life Sci 2011;68:3209–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bornstein P, Sage EH. Matricellular proteins: extracellular modulators of cell function. Curr Opin Cell Biol 2002;14:608–16. [DOI] [PubMed] [Google Scholar]
- 37.Iozzo RV, Schaefer L. Proteoglycan form and function: a comprehensive nomenclature of proteoglycans. Matrix Biol 2015;42:11–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Argraves WS, Tran H, Burgess WH, Dickerson K. Fibulin is an extracellular matrix and plasma glycoprotein with repeated domain structure. J Cell Biol 1990;111:3155–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulun SE, Yilmaz BD, Sison C, Miyazaki K, Bernardi L, Liu S, et al. Endometriosis. Endocr Rev 2019;40:1048–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang Y, Hoque MO, Wu F, Trink B, Sidransky D, Ratovitski EA. Midkine induces epithelial-mesenchymal transition through Notch2/Jak2-Stat3 signaling in human keratinocytes. Cell Cycle 2008;7:1613–22. [DOI] [PubMed] [Google Scholar]
- 41.Curran T Fos and Jun: oncogenic transcription factors. Tohoku J Exp Med 1992;168:169–74. [DOI] [PubMed] [Google Scholar]
- 42.Navin NE. Cancer genomics: one cell at a time. Genome Biol 2014;15:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hou X, Tan Y, Li M, Dey SK, Das SK. Canonical Wnt signaling is critical to estrogen-mediated uterine growth. Mol Endocrinol 2004;18: 3035–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelletier J, Thomas G, Volarevic S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer 2018;18:51–63. [DOI] [PubMed] [Google Scholar]
- 45.Uchida H, Maruyama T, Nishikawa-Uchida S, Miyazaki K, Masuda H, Yoshimura Y. Glycodelin in reproduction. Reprod Med Biol 2013;12: 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seppala M, Taylor RN, Koistinen H, Koistinen R, Milgrom E. Glycodelin: a major lipocalin protein of the reproductive axis with diverse actions in cell recognition and differentiation. Endocr Rev 2002;23:401–30. [DOI] [PubMed] [Google Scholar]
- 47.Burney RO, Talbi S, Hamilton AE, Vo KC, Nyegaard M, Nezhat CR, et al. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology 2007;148:3814–26. [DOI] [PubMed] [Google Scholar]
- 48.Vannuccini S, Tosti C, Carmona F, Huang SJ, Chapron C, Guo SW, et al. Pathogenesis of adenomyosis: an update on molecular mechanisms. Reprod Biomed Online 2017;35:592–601. [DOI] [PubMed] [Google Scholar]
- 49.Herndon CN, Aghajanova L, Balayan S, Erikson D, Barragan F, Goldfien G, et al. Global transcriptome abnormalities of the eutopic endometrium from women with adenomyosis. Reprod Sci 2016;23:1289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiang Y, Sun Y, Yang B, Yang Y, Zhang Y, Yu T, et al. Transcriptome sequencing of adenomyosis eutopic endometrium: a new insight into its pathophysiology. J Cell Mol Med 2019;23:8381–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Streuli I, Santulli P, Chouzenoux S, Chapron C, Batteux F. Activation of the MAPK/ERK cell-signaling pathway in uterine smooth muscle cells of women with adenomyosis. Reprod Sci 2015;22:1549–60. [DOI] [PubMed] [Google Scholar]
- 52.Oh SJ, Shin JH, Kim TH, Lee HS, Yoo JY, Ahn JY, et al. beta-Catenin activation contributes to the pathogenesis of adenomyosis through epithelial-mesenchymal transition. J Pathol 2013;231:210–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pawar NM, Rao P. Secreted frizzled related protein 4 (sFRP4) update: a brief review. Cell Signal 2018;45:63–70. [DOI] [PubMed] [Google Scholar]
- 54.Zhai J, Li S, Sen S, Vallve-Juanico J, Irwin JC, Vo KC, et al. Transcriptomic analysis supports collective endometrial cell migration in the pathogenesis of adenomyosis. Reprod Biomed Online 2022;45:519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goad J, Rudolph J, Zandigohar M, Tae M, Dai Y, Wei JJ, et al. Single-cell sequencing reveals novel cellular heterogeneity in uterine leiomyomas. Hum Reprod 2022;37:2334–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Z, Sun Z, Liu H, Niu W, Wang X, Liang N, et al. Single-cell transcriptomic analysis of eutopic endometrium and ectopic lesions of adenomyosis. Cell Biosci 2021;11:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.