Abstract
Background and purpose:
Anakinra must be injected daily due to its short half-life and this leads to lower patient compliance. Therefore, the aim of this study was to produce an interleukin-1 receptor antagonist (IL-1Ra) with albumin binding domain (ABD) as a novel fusion protein and evaluate its binding ability to albumin and its biological effects.
Experimental approach:
The three-dimensional structure of IL-1Ra-ABD was predicted by MODELLER software and its interaction with IL-1R was evaluated by the HADDOCK server. The expression of IL-1Ra-ABD was performed in E. coli in fusion with intein 1 of pTWIN1 in soluble form and then purified. The affinity of IL-1Ra-ABD to human serum albumin (HSA) was determined on native-PAGE, and its release percent toward time was evaluated. Moreover, an MTT assay was used to determine the antagonizing properties of recombinant IL-1Ra-ABD against IL-1β in A375 and HEK293 cell lines.
Findings/Results:
The stable complex of IL-1Ra-ABD with IL-1R established the absence of steric hindrance due to the addition of ABD to IL-1Ra. The expression induction of intein 1-IL-1Ra-ABD using 0.1 mM IPTG at 15 °C, and its cleavage represented bands approximately in 50 and 23 kDa. Furthermore, about 78% of IL-1Ra-ABD was attached to the HSA after 2 h of incubation, and the MTT assay showed no significant differences between the effects of IL-1Ra-ABD and native IL-1Ra in cell survival.
Conclusions and implications:
The production of soluble IL-1Ra-ABD with no significant differences in IL-1Ra antagonizing effects was successfully performed. IL-1Ra-ABD showed suitable interaction with HSA and was released over time. However, the half-life of IL-1Ra-ABD in vivo must be determined in the subsequent investigations.
Keywords: Albumin binding domain, Human serum albumin, IL-1Ra, Intein
INTRODUCTION
IL-1Ra (interleukin 1 receptor antagonist), known as anakinra (Kineret®), was produced by Amgen Company for the first time, and the FDA approved it for the treatment of rheumatoid arthritis, as well as the deficiency of IL-1Ra, a rare auto-inflammatory disease with involving skin and bones (1).
This protein has 153 amino acid residues with a molecular weight of 17.3 kDa and is produced in the Escherichia coli expression system by recombinant DNA technology. The drug is recommended as the SC (subcutaneous) injection of 100 mg daily with a half-life between 4 to 6 h (2).
The rapid clearance of biomolecules from the bloodstream can limit their clinical effects and lead to frequent administration. As mentioned, anakinra must be administered once a day by the SC injection. Although it is generally safe and well tolerated, the reactions of the injection site are the most concern in anakinra administration, and result in the low acceptance of patients (3). Therefore, trying to produce derivatives with an extended half-life is essential.
Pegylation is one of the most prevalent ways to extend the half-life of drugs and reduce the frequency of administration. However, alternative methods have been considered in recent years due to the complexity of the pegylated formulations and the possibility of polyethylene glycol side effects (4). In this regard, one study showed that anakinra binding to the biodegradable hydroxyethyl starch in comparison to its pegylated form resulted in increasing its thermal stability during the shelf-life (5).
Furthermore, many proteins have been engineered by consensus sequence design methods to enhance drug stability. In evidence, Podust et al. used the XTEN sequence to increase the half-life of various recombinant drugs (6). On the other hand, serum albumin is an attractive option to increase the half-life of bio-drugs due to the biological features including the long half-life (19 days in humans), the highest blood concentration compared to the other plasma proteins, and the biological distribution in inflammation or tumor site (7). One study produced a recombinant form of IL-1Ra protein in fusion to human serum albumin (HSA) and showed that this fusion protein is able to bind to the related receptor with no difference to the native protein (8). However, the attachment of a large molecule such as albumin to the target protein may interfere with its receptor binding, in addition to increasing the gene synthesis cost and disrupting the vital processes of the host cell. Thus, the recombinant production of drugs in fusion with the albumin binding domains (ABD) is a suitable approach to overcome these drawbacks (9). Literature has reported several biological drugs such as exenatide (10), hirodin (11), and glucagon-like peptide-1 GLP1 receptor agonist (12) whose half-lives increased via the addition of ABD. One study investigated several peptides with G148 ABD of streptococci strains as a template. Finally, a 44- amino acid peptide with eukaryotic origin was introduced with the most similarity to the ABD sequence (13).
Accordingly, the aim of the present study was to design an IL-1Ra-ABD fusion protein and produce it by recombinant DNA technology with an intein tag (Ssp intein) from the pTWIN1 plasmid. Furthermore, the current study evaluated the IL-1Ra-ABD antagonizing efficacy of IL-1β compared to the native IL-1Ra and determined its affinity to HSA and release efficacy.
MATERIALS AND METHODS
Protein design and molecular docking
To compare the binding of anakinra and IL1Ra-ABD to IL-1R, the prediction of the three-dimensional structure of IL-1R, IL-1Ra, and ABD was performed by type-1 interleukin-1 receptor complexed with interleukin-1 beta (PDB code: 1TTB), the X-ray structure of interleukin-1 receptor antagonist at 2.0 Å resolution (PDB code: 1ILT), and the crystal structure of the GA module complexed with HSA (PDB code: 1TF0), respectively, using the software of Modeller 9.24 and by multiple homology modeling strategies. In each case, among 1000 predicted models, one with the lowest value of discrete optimized protein energy (DOPE) was considered the best model for protein docking. The analysis of the best model was performed by ProSA, PROCHECK, and Verify 3D web servers and finally, the formation of the salt bridge was predicted using the software of visual molecular dynamics. The web server of HADDOCK 2.2 was then used to compare the affinity of two types of IL-1Ra for the interaction with its receptor. The active residues for each molecule were prepared according to the study of Barkestani et al. (14).
Expression of IL-lRa-ABD fusion protein
The coding sequence of IL-1Ra was obtained from DrugBank (https://go.drugbank.com) with accession number DBCAT002727 and fused to the amino acid sequence of ABD obtained from Jacob’s study (13). The pTWIN1-IL-1Ra-ABD plasmid was synthesized by BioMatik Company (Canada) in fusion to intein 1 and a chitin-binding domain (CBD) at the amino end which was used as the affinity tag which can bind to chitin resin to facilitate the purification process (Fig. 1A). This construct was transformed to the E. coli BL21 (DE3) (Pasteur Institute, Tehran, Iran) as the host cells, followed by selecting the recombinant colonies on Luria Bertani (LB) agar plates (HiMedia, India) containing 100 μg/mL of ampicillin (Sigma, Germany). The selected colony was then cultivated overnight and used to inoculate fresh culture to reach an optical density at 600 nm (OD600) of 0.4 to 0.6. Next, the expression of intein1-IL-1Ra-ABD was induced by isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma, Germany) in various concentrations (0.1, 0.5, and 1 mM) for 4 h at different temperatures (15, 25, and 37 °C). The cells were harvested via centrifuging at 7000 g at 4 °C for 10 min. For each expression condition, the separation of soluble protein and inclusion bodies was performed by cell lysis via sonicating. The soluble part of the protein in the supernatant and the inclusion body in the cell plate was separated as a precipitate by centrifuging. Finally, the supernatant of various conditions was evaluated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Fig. 1.
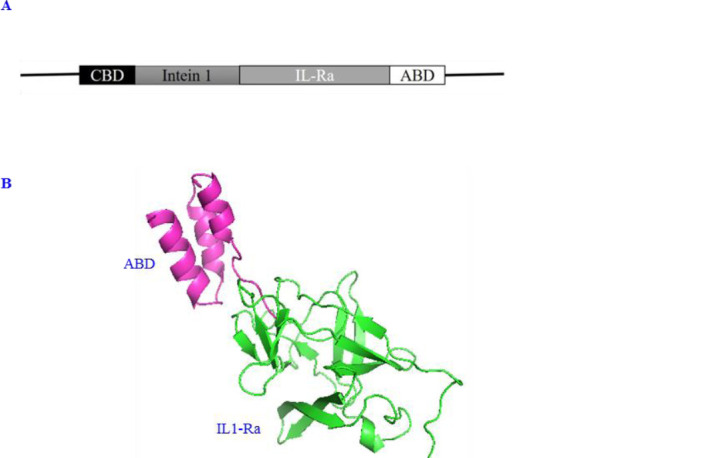
(A) Schematic structure of the gene encoding the IL-lRa-ABD in fusion to the intein 1; (B) three-dimensional structure of IL-1Ra-ABD visualized by PyMol. CBD, Chitin-binding domain; IL-1Ra, interleukin-1 receptor antagonist; ABD, albumin binding domain.
Self-cleavage inducing of Ssp intein
On-column cleavage and the purification of IL-1Ra-ABD were mediated by the intein-mediated purification with an affinity chitin-binding tag (IMPACT™, New England Biolabs, USA) using the CBD at the N-terminal of the intein 1 with affinity to chitin resin. Briefly, the cell pellets related to the most soluble expression and the least auto-cleavage (determined in the previous stage) were re-suspended in B1 buffer (Tris-HCl 20 mM, NaCl 500 mM, and EDTA 1 mM, pH 8.5), and followed by sonicating for the cell lysis. The sample was then centrifuged at 7000 g at 10 °C for 15 min and finally, the supernatant was loaded on the chitin column. The flow-through was discarded, and the column washing procedure was repeated five times with B1 buffer and then replaced with B2 buffer (Tris-HCl 20 mM, NaCl 500 mM, and EDTA 1 mM, pH 6.5). The column was incubated for 24 h at 25 °C. Eventually, various elutions were collected and analyzed by 15% SDS-PAGE. All elutions were mixed and subjected to dialysis against phosphate-buffered saline (PBS), pH 7.4 for 24 h at 4 °C. Finally, the protein concentration was determined using the Bradford method against various concentrations of HSA (Octapharma, Sweden).
Evaluation of the albumin binding ability of IL-lRa-ABD
To determine the ability of IL-lRa-ABD fusion protein to attach to the HSA, the equal molar ratios (0.2 μM of the fusion protein and HSA) were shaken in PBS (pH 7.4) in the 10 mL of final volume for 2 h on ice. This content was then transferred to an Amicon filter (Millipore, USA) with a 30 kDa cut-off followed by centrifuging at 7000 g at room temperature for 15 min. The supernatant and the flow-through were analyzed by 12% native-PAGE and quantified by the Bradford method. By measuring the amount of IL-lRa-ABD passed through the Amicon filter after the centrifugation, the relative amount of albumin-bound protein was calculated compared to the total used protein.
Evaluation of the release of IL-lRa-ABD from the HSA
As mentioned above, the equal molar ratios of the recombinant IL-lRa-ABD and HSA (0.2 μM of each protein) were added to the 10 mL of PBS as the protein diluent and shaken for 2 h on ice. At various times, the tube content was seeded into the Amicon filter (30 kDa cut-off) and subjected to the centrifuge (7000 g for 15 min at room temperature). The flow-through was analyzed by the Bradford method to determine detached protein concentration. Finally, the cumulative release graph of IL-1Ra-ABD was drawn based on the percent of released IL-1Ra-ABD toward the time.
Biological assay
To determine the antagonizing properties of recombinant IL-1Ra-ABD on A375 (as IL-1 receptor-positive cells) and human embryonic kidney 293 cells (HEK293) (as IL-1 receptor-negative cells) 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay was performed. Briefly, 160 μL of the medium containing 3 × 104 cells/mL of each cell line was poured into each well of 96-well microplates and incubated for 24 h at 37 °C. Then, 20 μL of the IL-1β (Pepro Tech, Canada) with the final concentration of 2 ng/mL was added to each well except the blank well. After a 1-h incubation, the purified IL-1Ra-ABD or native IL-1Ra at the concentrations of 0.95, 1.9, 3.75, 7.5, and 15 μg/mL produced according to the previous study (15) was added into plates. Next, the pl ates were incubated in 5% CO2 and at the temperature of 37 °C for 48 h. After that, 20 μL of MTT (5 mg/mL) was added to each well followed by a 3-h incubation period. Then, the content of wells were discarded, and 150 μL of dimethyl sulfoxide (DMSO) was added to each well for dissolving formazan crystals. Finally, the plates were subjected to read the absorbance at 570 nm by a microplate reader (Bio-Rad, USA).
Statistical analysis
To ensure the accuracy and reproducibility of data, the MTT test was performed for each cell line as a triple-independent exp eriment. Cell culture media, PBS-treated cells, and cells treated with 2 ng/mL of IL-1β were assigned as blank, negative control, and positive control, respectively. The software of SPSS, version 25, was used for statistical analysis. The analysis of variance (ANOVA) followed by Tukey’s post hoc test was used to determine the differences among groups. P < 0.05 was considered the statistical significance.
RESULTS
Molecular docking
Among 1000 predicted models for IL-1Ra-ABD by multiple homology modeling, one with the lowest molpdf value as 2821.44116, the lowest DOPE score as -16103.19336, and the highest GA341 score as 1 was selected as the best model which was visualized by PyMol software, version 2.3.2 (Fig. 1B). Table 1 shows the interaction between two types of antagonists (native form versus the form with ABD) and the IL-1R. As shown, the attachment of ABD to anakinra molecule had no effects on its ability to interact with its receptor according to the HADDOCK and Z score with no meaningful difference (-1.6 versus -1.4).
Table 1.
HADDOCK results of molecular docking.
| Parameters | IL-1Ra/IL-1R complex | IL-1Ra-ABD/IL-1R complex |
|---|---|---|
| HADDOCK score | -163.6 ± 8.2 | -149.6 ± 7.9 |
| Cluster size | 138 | 58 |
| RMSD | 0.8 ± 0.4 | 0.5 ± 0.2 |
| Einter | -332 | -466.1 |
| Evdw | -51.1 ± 4.4 | -43 ± 2.2 |
| Eelec | -300.9 ± 60.5 | -403.1 ± 21.7 |
| Edesolv | -68.2 ± 9.4 | 52.5 ± 7.3 |
| Erv | 258.7 ± 31.03 | 235.4 ± 36.42 |
| Total BSA | 1975.1 ± 64.6 | 13284.7 ± 52.6 |
| Z-Score | -1.6 | -1.4 |
Einter, Intermolecular energy (sum of the van der Waals and electrostatic energies); Evdw, van der Waals energy; Eelec, electrostatic energy; Edesolv, desolvation energy; Erv, restrain violation energy; BSA, buried surface energy; BSA. IL-1Ra, interleukin 1 receptor antagonist; IL-1R, interleukin 1 receptor; ABD, albumin binding domain.
Expression of rIL-lRa-ABD
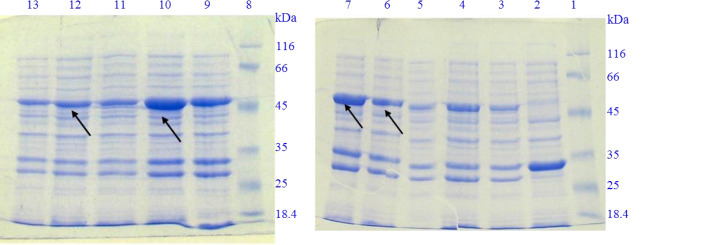
The expression of intein 1-IL-1Ra-ABD fusion protein was evaluated by 12% SDS-PAGE. As shown in Fig. 2, the induction of expression using IPTG at the concentration of 1 mM and after 4 h of incubation revealed a band of approximately 54 kDa for cells transformed with non-recombinant pTWIN1 plasmid which corresponds to the molecular weight of inteins 1 and 2 fusion protein. For E. coli BL21 (DE3) cells containing recombinant pTWIN1-IL-1Ra-ABD, on the other hand, induction with IPTG at the concentration of 1 mM represented a band approximately 50 kDa which confirmed the expression of IL-1Ra-ABD in fusion to intein 1. Furthermore, the best condition for soluble protein expression and with less auto-cleavage during the bacterial growth and protein expression was determined as 0.1 mM of IPTG at 15 °C used for about 16 h in order to earn the most soluble protein.
Fig. 2.
SDS-PAGE analysis of soluble protein expression induction of IL-1Ra-ABD in various conditions regarding IPTG concentration and temperature of incubation for 4 h of incubation. Lanes 1 and 8, protein marker; lane 2, induced E. coli BL21(DE3) cells containing non-recombinant pTWIN1 by 1 mM IPTG at 37 °C; lane 3, induced E. coli BL21(DE3) cells containing recombinant pTWIN1-IL1-R.a-ABD; lanes 4-6, induced E. coli BL21(DE3) cells containing recombinant pTWIN1-IL1-Ra-ABD by 0.1, 0.5, and 1 mM IPTG at 37 °C, respectively; lanes 7-9, induced E. coli BL21(DE3) cells containing recombinant pTWIN1-IL1-Ra-ABD by 0.1, 0.5, and 1 mM IPTG at 25 °C, respectively; lanes 10-12, induced E. coli BL21(DE3) cells containing recombinant pTWIN1-IL1-Ra-ABD by 0.1, 0.5, and 1 mM IPTG at 15 °C, respectively. SDS-PAGE, Sodium dodecyl sulfate-polyacrylamide gel electrophoresis; IL-1Ra, interleukin 1 receptor antagonist; ABD, albumin binding domain; IPTG, isopropyl β-D-1-thiogalactopyranoside.
Purification of the rIL-lRa-ABD
In Fig. 3, a band of approximately 23 kDa represented the recombinant IL-1Ra-ABD separated from intein 1 and CBD. The final yield of recombinant protein production was calculated to be 4.3 mg/L of bacterial culture medium.
Fig. 3.
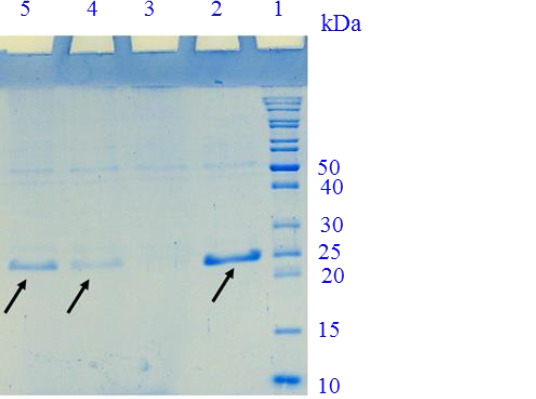
15% SDS-PAGE for the evaluation of the IL-1Ra-ABD cleavage. Lane 1, protein marker; lane 2, purified recombinant IL-1Ra after the first elution of the column with B2 buffer; lane 3, the fourth elution of the column; lane 4, the third elution of the column; lane 5, the second elution of the column. SDS-PAGE, Sodium dodecyl sulfate-polyacrylamide gel electrophoresis; IL-1Ra, interleukin 1 receptor antagonist; ABD, albumin binding domain.
Evaluation of the albumin binding ability of IL-lRa-ABD
With the appearance of a band approximately 90 kDa (equal to the IL-1Ra-ABD and HSA molecular weights) after a 2-h incubation period in the native-PAGE, it was concluded that the binding between two molecules occurred. Actually, from 4.5 mg of IL-1Ra-ABD shaken with HSA, approximately 0.98 mg passed through the Amicon filter, and consequently it was calculated that about 78% of IL-1Ra-ABD in the test solution was attached to the HSA (Fig. 4).
Fig. 4.
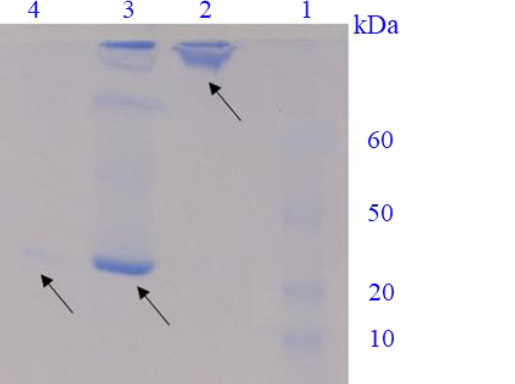
Native-PAGE for the evaluation of ABD affinity to HSA. Lane 1, protein marker; lane 2, the upper sample of Amicon filter after centrifuging after a 2-h incubation; lane 3, the mixed sample of IL-1Ra-ABD and HSA after a 30-min incubation; lane 4, the flow-through of Amicon filter after centrifuging after a 2-h incubation. PAGE, polyacrylamide gel electrophoresis; ABD, albumin binding domain; HSA, human serum albumin; IL-1Ra, interleukin 1 receptor antagonist.
Evaluation of the release of IL-lRa-ABD from the HSA
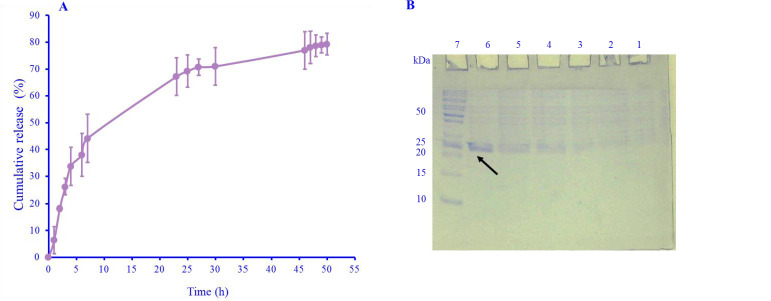
The centrifuging of the tube containing IL-1Ra-ABD and HSA via an Amicon filter at room temperature and the determining of passed IL-1Ra-ABD by Bradford method showed that about 80% of the protein was released from HSA during the first 50 h of incubation (Fig. 5).
Fig. 5.
(A) The release percent of IL-1Ra-ABD from HSA at various times; (B) native-PAGE of the released IL-IRa-ABD from the HSA from the flow-through of Amicon filter at various times. Lanes 1-6, the flow-through sample after 1-, 3-, 5-, 16-, 18-, and 25-h incubation; lane 7, protein marker. PAGE, polyacrylamide gel electrophoresis; ABD, albumin binding domain; HSA, human serum albumin; IL-1Ra, interleukin 1 receptor antagonist.
Comparison of the antagonizing efficacy of IL-lRa-ABD with IL-1Ra against IL-1β
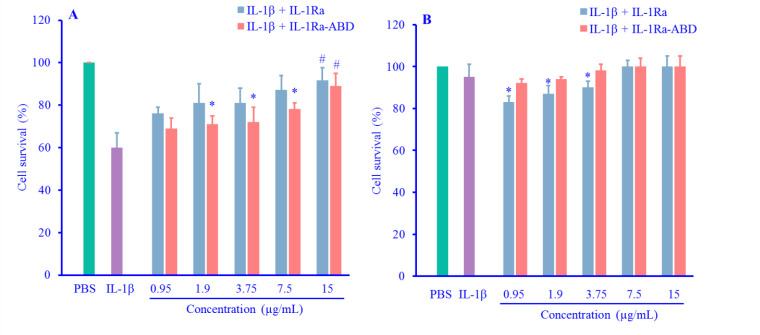
In the similar concentration of the antagonist (15 μg/mL), there were no significant differences between the effects of IL-1Ra-ABD and native IL-1Ra on A375 cells (P = 0.33). Furthermore, IL-1β showed about 60% cell survival on A375 cells which increased significantly after the addition of the antagonists at the final concentration (15 μg/mL; P = 0.015) (Fig. 6A). On the other hand, IL-1β led to less cytotoxicity (95% of cell survival) on HEK293, and both types of antagonists (native IL-1Ra and IL-1Ra-ABD) could increase the cell survival; however, this effect was less than A375 cells (Fig. 6B).
Fig. 6.
(A) Comparison of the antagonizing effect of IL-lRa-ABD and native IL-IRa on A375 survival percent after the treatment with IL-1β; (B) Comparison of the antagonizing effect of IL-lRa-ABD and native IL-1Ra on HEK293 survival percent after the treatment with IL-1β. Data were expressed as mean ± SD, n = 3. *P < 0.05 demonstrates the significant difference in comparison with cells treated with IL-1β + IL-1Ra at the same concentration; #P < 0.05 versus IL-1β (2 ng/mL) as the positive control. IL-1Ra, interleukin 1 receptor antagonist; ABD, albumin binding domain; IL-1β, interleukin 1β; HEK293, human embryonic kidney 293.
DISCUSSION
In the present study, IL-1Ra in fusion to ABD was produced in the E. coli expression system as the first stage of introducing a new version of an FDA-approved drug for treating RA and purified using the intein-mediated procedure. The present results indicated that after 2 h, about 78% of this fusion protein could bind to HSA, and about 80% of the bonded protein was released during 50 h until it reached a stationary phase. On the other hand, there was no significant difference between the antagonistic efficacy of IL-1Ra-ABD and native IL-1Ra in this test when A375 and HEK293 cell lines were treated by IL-1β.
Especially during recent years, the usage of the IMPACT system has increased because of the ability of intein tags to produce the soluble form of recombinant proteins as well as the ability of their self-cleavage after protein purification. However, the optimization of conditions for soluble expression and the cleavage of intein play a critical role in earning the most active soluble protein.
Using the intein 1 (Ssp DnaB) of pTWIN1 plasmid is more convenient than intein 2, which induces the ability of its auto-cleavage by adding reducing agents. In other words, intein 1 is induced for self-splicing activity by changing pH, the same as the previous study for purifying the recombinant human granulocyte colony-stimulating factor (G-CSF) (16). However, another problem in this regard is the self-cleavage induction of intein1 during the recombinant production of target protein in the host cell cytosol. To diminish this unwanted reaction, lowering the post-induction temperature and the pH of the cleavage buffer was used in this study. In general, the auto-cleavage ability of intein 1 can improve by increasing the temperature incubation of the chitin column. In this regard, Jiang et al. reported that the best temperature was considered 37 °C among investigated temperatures ranging from 15 to 37 °C for Ssp DnaB mini-intein fused to human GLP-1/7-36 (17). It seems that lowering pH to even 4.5 can increase the time of column incubation and the temperature of incubation leading to the efficient cleavage of intein 1 (18). One study performed on the cleavage of a mutated form of insulin from the intein1 reported that pH 4.5 in comparison to pH 8, and the temperature of 25 °C was more efficient in cleavage ratios (18). The present study used the incubation temperature of 25 °C for 24 h at pH 6.5 according to the IMPACT manual.
Pharmacokinetic assays performed on IL-1Ra fused to HSA showed that native IL-1Ra was completely cleared from the blood circulation after an 8 h-injection period in healthy mice, while for the fusion protein of IL-1Ra-ALB, the data represented that about 30% of the radio-labeled fusion protein could be still observed in the circulation after a 48 h-injection period. In fact, the half-life of IL-1Ra-ALB was calculated as about 10 h in comparison to the native form with a calculated half-life of about 20.5 min (8). In the current study, ABD was used instead of full-length HSA to overcome the probable drawbacks of the expression of target protein in fusion to a large molecule such as albumin with about 66 kDa.
Several studies have used ABD to extend the half-life of biological drugs (10,11,12). To attempt to increase the half-life of exenatide using ABD, the pharmacokinetics assays showed about a 32-fold increase in the half-life of exenatide-ABD when compared to the native form (16 h vs. 30 min) (10). In vitro release tests performed in the present study demonstrated that IL-1Ra fused to ABD could be released from the HSA until 50 h, and native-PAGE revealed the stability of this fusion protein.
For the biological assay, on the other hand, this study used the potent cytotoxic and apoptotic effects of IL-1β against cells with highly expressed IL-1 receptors (19). The biological results showed that an increase in the concentration of IL-1Ra-ABD led to the inhibition of IL-1β cytotoxic effects. Liu et al. evaluated the inhibitory effects of the fusion protein of IL-1Ra with an extended half-life and showed that IL-1Ra at the concentration of 32 nM leads to 100% inhibition effects of IL-1β at the concentration of 1 ng/mL (20). In another study, Wang et al. produced several mutated forms of IL-1Ra and compared their biological activities to the native IL-1Ra. The results showed that IL-1Ra at the concentration of 25 μg/mL can inhibit the effects of IL-1 (21).
The antagonizing efficacy of IL-1Ra-ABD was not statistically significant compared to native IL-1Ra for highly expressed IL-1 receptor cells, A375. The strength of the produced recombinant protein was about 101% of the native form antagonist.
Liu et al. evaluated the antagonizing efficacy of the recombinant IL-1Ra produced in Pichia pastoris and presented that this protein could antagonize the cytolytic activity of IL-1β (1 ng/mL) when added only after the treatment of cells by IL-1β (20). However, there was a time interval of 1 h in the present study based on Adelnia et al. study (15). The present findings showed that IL-1Ra-ABD could successfully act in antagonizing the cytotoxic effects of IL-1β in a time interval of 1 h without any significant differences from native IL-1Ra.
Finally, Powers et al. used the IL-1-responsive A549 cell line to evaluate the competitive antagonizing effects of a PASylated form of IL-1Ra, produced for expanding its half-life (22). In this study, the antagonist was added to cells, and the cells received IL-1α the 1 h later. The efficacy of IL-1α (in the final concentration of 10 ng/mL) was evaluated in the release of IL-6 and found that both anakinra and its PASylated form antagonized the IL-6 secretion (22). The results of the present study also confirmed the anakinra antagonizing effects on IL-1, only before its internalizing to the cells with highly expressed receptors on their surface.
CONCLUSION
This project successfully produced and purified the recombinant IL-1Ra in fusion with the ABD in soluble form. The antagonizing effects of this protein were statistically equal to the native receptor antagonist produced in the same strategy. On the other hand, this chimeric molecule showed suitable interaction with HSA indicating its correct 3-D structure. Furthermore, in vitro release analysis confirmed the 80% cumulative release of the recombinant fusion protein from HSA. However, further in vivo pharmacokinetics analysis as well as in vitro and in vivo biological evaluations are required for this protein to act as a drug candidate for clinical trial phases.
Conflict of interest statement
All authors declared no conflict of interest in this study.
Authors' contributions
F. Shafiee designed the study, wrote the manuscript, and analyzed data; A. Yazdani performed the laboratory tests. All authors approved the finalized article.
Acknowledgments
This paper was extracted from the Pharm. D thesis submitted by Ali Yazdani financially supported by the Research Deputy of Isfahan University of Medical Sciences, with Grant No. 399095. The authors also would like to appreciate the valuable technical assistance of laboratory experts in molecular biotechnology and cell culture laboratories.
REFERENCES
- 1.Schnellbacher C, Ciocca G, Menendez R, Aksentijevich I, Goldbach-Mansky R, Duarte AM, et al. Deficiency of interleukin-1 receptor antagonist responsive to anakinra. Pediatr Dermatol. 2013;30(6):758–760. doi: 10.1111/j.1525-1470.2012.01725.x. DOI: 10.1111/j.1525-1470.2012.01725x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waugh J, Perry CM. Anakinra. BioDrugs. 2005;19(3):189–202. doi: 10.2165/00063030-200519030-00005. DOI: 10.2165/00063030-200519030-00005. [DOI] [PubMed] [Google Scholar]
- 3.Kaiser C, Knight A, Nordström D, Pettersson T, Fransson J, Florin-Robertsson E, et al. Injection-site reactions upon kineret (anakinra) administration: experiences and explanations. Rheumatol Int. 2012;32(2):295–299. doi: 10.1007/s00296-011-2096-3. DOI: 10.1007/s00296-011-2096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suk JS, Xu Q, Kim N, Hanes J, Ensign LM. PEGylation as a strategy for improving nanoparticle- based drug and gene delivery. Adv Drug Deliv Rev. 2016;99(Pt. A):28–51. doi: 10.1016/j.addr.2015.09.012. DOI: 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liebner R, Mathaes R, Meyer M, Hey T, Winter G, Besheer A. Protein HESylation for half-life extension: synthesis, characterization and pharmacokinetics of HESylated anakinra. Eur J Pharma Biopharm. 2014;87(2):378–385. doi: 10.1016/j.ejpb.2014.03.010. DOI: 10.1016/j.ejpb.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Podust VN, Balan S, Sim BC, Coyle MP, Ernst U, Perers RT, et al. Extension of in vivo half-life of biologically active molecules by XTEN protein polymers. J Control Release. 2016;240:52–66. doi: 10.1016/j.jconrel.2015.10.038. DOI: 10.1016/jjconrel.2015.10.038. [DOI] [PubMed] [Google Scholar]
- 7.Bern M, Nilsen J, Ferrarese M, Sand KMK, Gjelberg TT, Lode HE, et al. An engineered human albumin enhances half-life and transmucosal delivery when fused to protein-based biologics. Sci Transl Med. 2020;12(565):eabb0580. doi: 10.1126/scitranslmed.abb0580. 1-13. DOI: 10.1126/scitranslmed.abb0580. [DOI] [PubMed] [Google Scholar]
- 8.Liu M, Huang Y, Lei Hu, Liu G, Hu X, Liu D, et al. Selective delivery of interleukine-1 receptor antagonist to inflamed joint by albumin fusion. BMC Biotechnol. 2012;12:68. doi: 10.1186/1472-6750-12-68. 1-13. DOI: 10.1186/1472-6750-12-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mester S, Evers M, Meyer S, Nilsen J, Greiff V, Sandlie I, et al. Extended plasma half-life of albumin-binding domain fused human IgA upon pH- dependent albumin engagement of human FcRn in vitro and in vivo. MAbs. 2021;13(1):e1893888. doi: 10.1080/19420862.2021.1893888. 1-13. DOI: 10.1080/19420862.2021.1893888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy OE, Jodka CM, Ren SS, Mamedova L, Sharma A, Samant M, et al. Novel exenatide analogs with peptidic albumin binding domains: potent anti-diabetic agents with extended duration of action. PLoS One. 2014;9(2):e87704. doi: 10.1371/journal.pone.0087704. 1-9. DOI: 10.1371/journal.pone.0087704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheffield WP, Smith IJ, Syed S, Bhakta V. Prolonged in vivo anticoagulant activity of a hirudin-albumin fusion protein secreted from Pichia pastoris. Blood Coagul Fibrinolysis. 2001;12(6):433–443. doi: 10.1097/00001721-200109000-00003. DOI: 10.1097/00001721-200109000-00003. [DOI] [PubMed] [Google Scholar]
- 12.Lindgren J, Refai E, Zaitsev SV, Abrahmsén L, Berggren PO, Karlström AE. A GLP-1 receptor agonist conjugated to an albumin-binding domain for extended half-life. Biopolymers. 2014;102(3):252–259. doi: 10.1002/bip.22474. DOI: 0.1002/bip.22474. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs SA, Gibbs AC, Conk M, Yi F, Maguire D, Kane C, et al. Fusion to a highly stable consensus albumin binding domain allows for tunable pharmacokinetics. Protein Eng Des Sel. 2015;28(10):385–393. doi: 10.1093/protein/gzv040. DOI: 10.1093/protein/gzv040. [DOI] [PubMed] [Google Scholar]
- 14.Barkestani MN, Naserian S, Khoddam F, Shamdanu S, Bambai B. Optimization of IL-1RA structure to achieve a smaller protein with a higher affinity to its receptor. Sci Rep. 2022;12(1):7483. doi: 10.1038/s41598-022-11100-3. 1-9. DOI: 10.1038/s41598-022-11100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adelnia R, Shafiee F. Recombinant production and one step purification of il-1ra in Escherichia coli and evaluation its il-1 antagonizing efficacy. Iran J Immunol. 2021;18(2):141–149. doi: 10.22034/iji.2021.89103.1929. DOI: 10.22034/iji.2021.89103.1929. [DOI] [PubMed] [Google Scholar]
- 16.Sima S, Shafiee F, Jahanian-Najafabadi A. Expression and one step intein-mediated purification of biologically active human G-CSF in Escherichia coli. Mol Biol Rep. 2020;47(4):2861–2869. doi: 10.1007/s11033-020-05404-8. DOI: 10.1007/s11033-020-05404-8. [DOI] [PubMed] [Google Scholar]
- 17.Jiang A, Jin W, Zhao F, Tang Y, Sun Z, Liu JN. Split Ssp DnaB mini-intein-mediated production of recombinant human glucagon-like peptide-1/7-36. Biotechnol Appl Biochem. 2015;62(3):309–315. doi: 10.1002/bab.1274. DOI: 10.1002/bab.1274. [DOI] [PubMed] [Google Scholar]
- 18.Zhang M, Zhang Y, Wu B, Peng Y, Simair AA, Siegel GW, et al. Intein-mediated recombinant expression of monomeric B22Asp desB30 insulin. BMC Biotechnol. 2020;20(1):3. doi: 10.1186/s12896-020-0598-3. 1-9. DOI: 10.1186/s12896-020-0598-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Wang MW, Tashiro SI, Onodera S, Ikejima T. IL-1β acts in synergy with endogenous IL-1β in A375-S2 human melanoma cell apoptosis through mitochondrial pathway. J Korean Med Sci. 2005;20(4):555–561. doi: 10.3346/jkms.2005.20.4.555. DOI: 10.3346/jkms.2005.20.4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu M, Huang Y, Hu L, Liu G, Hu X, Liu D, et al. Selective delivery of interleukine-1 receptor antagonist to inflamed joint by albumin fusion. BMC Biotechnol. 2012;12(1):68. doi: 10.1186/1472-6750-12-68. 1-13. DOI: 10.1186/1472-6750-12-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang YX, Yang ZX, Zhu HQ, Zhou XW, Huang PT. Construction, expression and preliminary pharmacokinetics of IL-1ra mutants. Chin J Biotechnol. 2006;22(3):472–476. doi: 10.1016/s1872-2075(06)60040-x. DOI: 1 0.1016/S1872-2075(06)60040-X. [DOI] [PubMed] [Google Scholar]
- 22.Powers NE, Swartzwelter B, Marchetti C, de Graaf DM, Lerchner A, Schlapschy M, et al. PASylation of IL-1 receptor antagonist (IL-1Ra) retains IL-1 blockade and extends its duration in mouse urate crystal-induced peritonitis. J Biol Chem. 2020;295(3):868–882. doi: 10.1074/jbc.RA119.010340. DOI: 10.1074/jbc.RA119.010340. [DOI] [PMC free article] [PubMed] [Google Scholar]