SUMMARY
Opioid receptors are therapeutically important G protein-coupled receptors (GPCRs) with diverse neuromodulatory effects. The functional consequences of opioid receptor activation are known to depend on receptor location in the plasma membrane, but mechanisms mediating selective localization of receptors to any particular membrane domain remain elusive. Here, we demonstrate the targeting of the mu opioid receptor (MOR) to the primary cilium, a discrete microdomain of the somatic plasma membrane, both in vivo and in cultured cells. We further show that ciliary targeting is specific to MORs, requires a 17-residue sequence unique to the MOR cytoplasmic tail, and additionally requires the Tubby-like protein 3 (TULP3) ciliary adaptor protein. Our results reveal the potential for opioid receptors to undergo selective localization to the primary cilium. We propose that ciliary targeting is mediated through an elaboration of the recycling pathway, directed by a specific C-terminal recycling sequence in cis and requiring TULP3 in trans.
In brief
The primary cilium defines a unique microdomain of the plasma membrane with restricted cargo access. Fagan et al. find that the mu-type opioid receptor (MOR) localizes to neuronal primary cilia. They demonstrate that cilia targeting requires a C-terminal recycling sequence unique to MORs and the TULP3 ciliary adaptor protein.
Graphical Abstract:
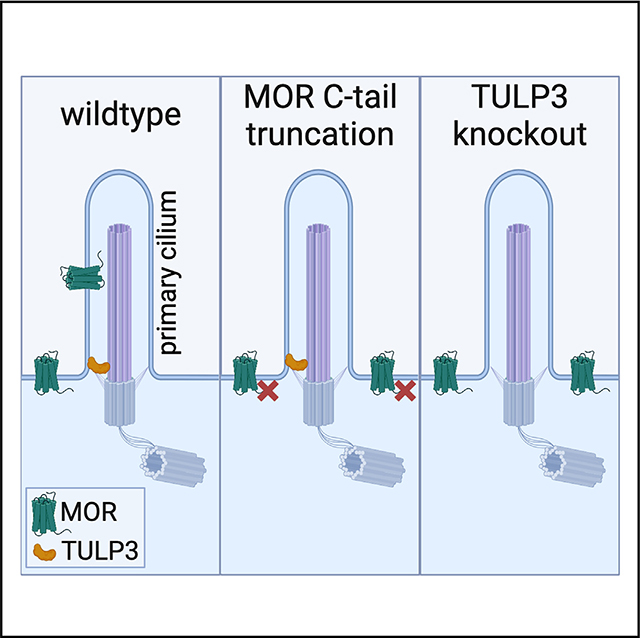
INTRODUCTION
Opioid receptors make up an important subfamily of G protein-coupled receptors (GPCRs) that determine the neuromodulatory effects of endogenously produced opioid peptides and are the targets of a large class of non-peptide drugs used clinically as analgesics.1 It is increasingly clear that opioid receptors can specifically localize to, and be activated from, multiple membrane locations in neurons.2,3 Support for this concept has recently been extended to intracellular membrane compartments relative to the plasma membrane,4,5 but evidence that opioid receptors can selectively partition between distinct domains of the plasma membrane long precedes this.6–9 However, the mechanistic basis by which opioid receptors are targeted to any particular domain of the plasma membrane remains unclear.
The primary cilium is one such cellular microdomain that extends from the plasma membrane of nearly all animal cells, including neurons in the adult brain.10 The ciliary membrane is restricted from the plasma membrane by a diffusion barrier, or transition zone, through which only select cargo can pass, including a select subset of GPCRs.11 The separated nature of the ciliary compartment necessitates the existence of tightly regulated mechanisms for cargo entry. However, we are not aware of any evidence to date that opioid receptors can localize to primary cilia.
Here, we report the serendipitous observation that the mu opioid receptor (MOR) localizes to neuronal primary cilia in vivo and in vitro. We show that the capacity for this ciliary targeting is specific to MORs among opioid receptor types, dependent on a sequence uniquely present in the MOR cytoplasmic tail (C-tail), and highly sensitive to modifications made to the receptor tail adjacent to this sequence. Additionally, we show that ciliary targeting of the MOR requires the conserved ciliary adaptor protein Tubby-like protein 3 (TULP3), and we propose a model for ciliary targeting to the primary cilium by an elaboration of the recycling pathway.
RESULTS
MOR-Venus localizes to neuronal primary cilia
Opioid receptors are known to localize to and function in distinct plasma membrane domains; however, the mechanisms by which receptors are targeted to any specific surface membrane domain remain unclear. We examined MOR membrane localiza tion in native brain tissue by leveraging a fluorescent receptor knockin mouse used previously to map receptor expression at the cellular level.12 When examined at a subcellular level of spatial resolution, we detected MORs at distinct membrane domains in adult neurons. In brain sections containing the septum, MOR-Venus expression was relatively high, and receptor-expressing cells were sufficiently well separated to allow subcellular detail to be readily visualized (Figure 1A). We observed MOR-Venus expression on the neuronal plasma membrane and internal punctate structures. We also noticed MOR-Venus localization on a single hair-like protrusion of the somatic plasma membrane, suggesting localization to the primary cilium (Figure 1B).
Figure 1. MOR-Venus and native, untagged MORs localize to neuronal primary cilia.
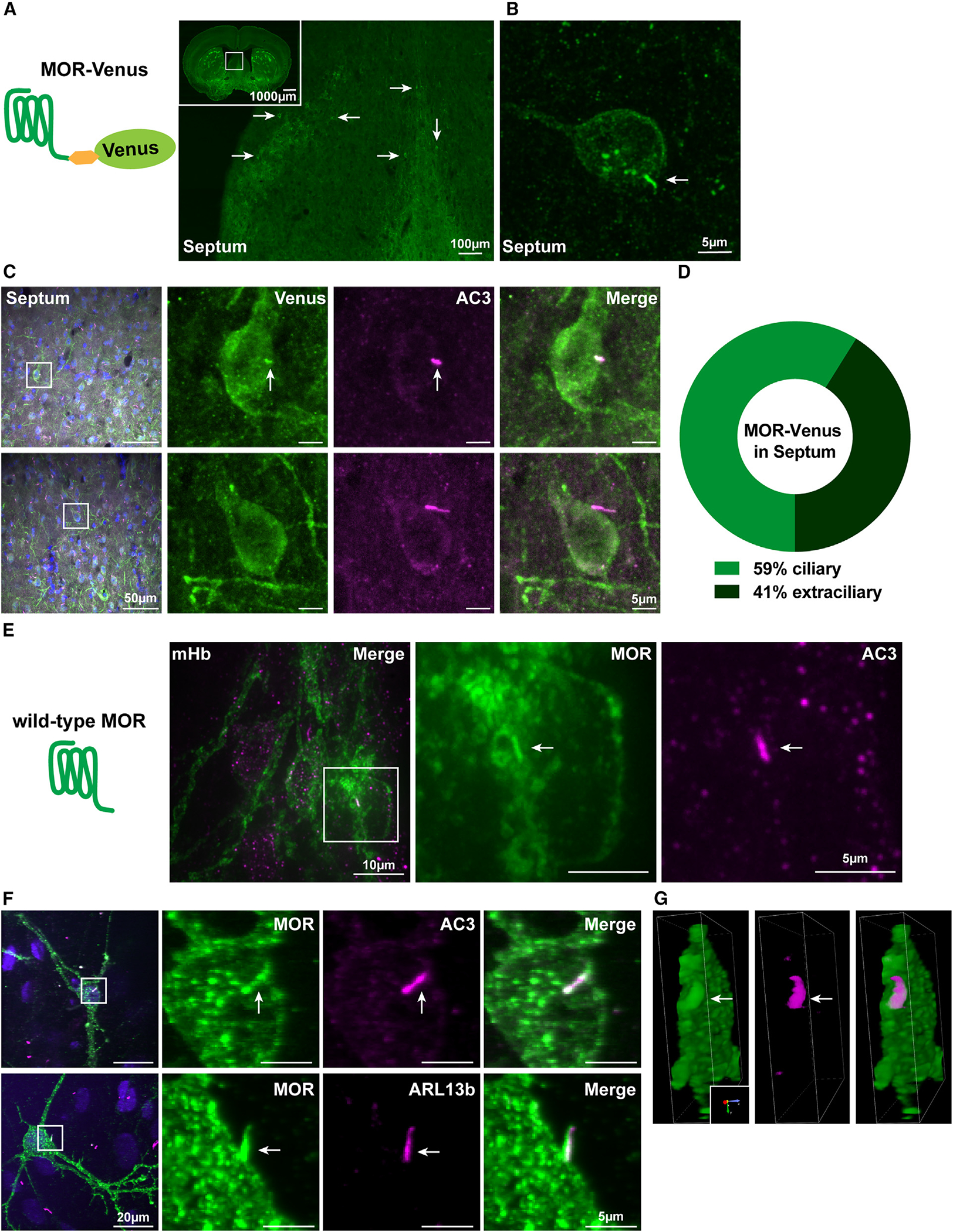
(A) Schematic representation of MOR-Venus construct expressed in the knockin mouse model and presented in (A)–(D). Representative confocal image of adult MOR-Venus coronal sections containing septum and the dorsal third ventricle. Inset: image of a whole 25 μm slice. Arrows indicate MOR-Venus(+) cell bodies. Scale bar: 100 μm (inset: 1,000 μm).
(B) Higher-magnification image of septal neuron positive for MOR-Venus. Arrow indicates membrane protrusion reminiscent of a primary cilium. Scale bar: 5 μm.
(C) Representative images of adult MOR-Venus coronal sections containing septal neurons. MOR-Venus signal was amplified using GFP antibody, neuronal cilia were labeled with anti-adenylyl cyclase III (AC3), and cell bodies were stained with DAPI. Arrows indicate MOR-Venus(+) primary cilium. Scale bars: 50 μm (left, merge) and 5 μm (right, individual channels). For additional brain regions and the MOR-mCherry mouse model, see Figure S1.
(D) Quantification of the percentage of MOR-Venus receptors localized to AC3 cilia in the septum. n = 3 mice, with approximately 9–42 neurons per animal.
(E) Schematic representation of wild-type MOR analyzed in (E)–(G).
(E) Representative confocal image of native (wild-type) adult mouse medial habenula. Slices were stained with anti-MOR (UMB3) antibody to label endogenous MOR and AC3. Arrow indicates MOR(+) primary cilium. Scale bars: 10 μm (left, merge) and 5 μm (right, individual channels).
(F) Representative confocal images of embryonic day 18 cultured rat habenula neurons stained with antibodies against MOR and AC3 (top) or MOR and ARL13B (bottom). Arrows indicate MOR(+) primary cilia. Scale bars: 20 μm (left, merge) and 5 μm (right, individual channels). For staining controls, see Figure S1.
(G) 3D-rendered visualization of ARL13B (F, bottom) rotated 90° depicts side view of primary cilium co-localized with MOR.
For full visualization of 3D rendering of samples from (F), see Videos S1 and S2.
To test if MOR-Venus localizes to cilia, we co-stained brain sections with an antibody against adenylyl cyclase type 3 (AC3), an established marker of primary cilia in neurons.13 MOR-Venus clearly co-localized with AC3 in some receptor-expressing neurons (Figure 1C, top) but not all of them (Figure 1C, bottom). Quantification revealed that, in the septum, MOR-Venus localized to the primary cilium in 59% of the receptor-positive neurons and was extraciliary in 41% of MOR-Venus-expressing neurons (Figure 1D).
MOR-Venus ciliary localization was not restricted to the septum. When surveyed across eight brain regions known to express MORs, we observed MOR-Venus in AC3-positive cilia in some neurons in each region (Figure S1A). In brain regions known to have low MOR-Venus cellular expression—including the dorsal striatum (Figure S1B, top), nucleus accumbens, and various cortical regions (agranular insular area and prefrontal cortices)—we observed qualitatively low levels of MOR-Venus in primary cilia and overall weak fluorescence intensity. In regions expressing MOR-Venus at higher levels—including the medial habenula (Figure S1B, bottom), septum, lateral hypothalamus, and paraventricular nucleus of the thalamus—MOR-Venus localization to cilia was clearly visible in a subset of neurons. As an independent approach to examine MOR ciliary localization, we used a distinct reporter mouse in which the MOR is labeled with mCherry rather than Venus (MOR-mCherry; Figure S1C).14 We focused on dorsal root ganglia (DRGs), in which we and others showed that most primary afferent nociceptive neurons express MORs.15–17, We found that MOR-mCherry also localized in AC3-positive primary cilia in a subpopulation of DRG neurons (Figure S1C′). In other DRG neurons, we could not detect a signal in the cilia, despite clear MOR-mCherry expression elsewhere on the plasma membrane (Figure S1C′′). Together, these results suggest that endogenously tagged MOR ciliary localization is widespread in native neurons of both the central and peripheral nervous systems and that the degree of ciliary receptor localization may vary between individual neurons, including between neurons in the same region.
Localization of native MORs in neuronal primary cilia
We then asked if the native MOR also localizes to primary cilia. To this aim, we immunostained tissue sections from adult wildt-ype mice using an anti-MOR antibody with validated specificity in knockout (KO) mice18 and an anti-AC3 antibody. We observed co-localization of endogenous MOR with AC3 in medial habenula brain sections (Figure 1E). Due to the high density of MOR-positive cell bodies in this brain region, it was difficult to determine the fraction of neurons in which the native MOR is targeted to primary cilia. We therefore examined MOR localization in primary cultures prepared from embryonic day 18 rat habenula. The native MOR endogenously expressed in these cells clearly localized in the primary cilium, as indicated by co-localization with AC3 (Figures 1F, top, and S1H, top; Video S1) as well as ARL13B (Figures 1F, bottom, 1G, S1F, and S1H, bottom; Video S2), another marker of primary cilia. Control labeling experiments demonstrate that this co-localization is only seen in the presence of native MOR labeling (Figures S1D, S1E, and S1G). This co-localization was observed in >90% of MOR(+) neurons using either cilia marker (Figure S1I). These data indicate that native, untagged MOR localizes to neuronal primary cilia both in brain tissue and in dissociated cell cultures.
Ciliary targeting of MORs in IMCD3 cells is sensitive to modification of the receptor’s C-tail
We next investigated if ciliary localization of MORs can also be observed in inner medullary collecting duct (IMCD3) cells, a well-established ciliated cell model system used widely for mechanistic studies.19–21 The localization of MOR-Venus in ARL13B-marked primary cilia was evident in these cells (Figure 2A, left), and receptors were enriched in cilia relative to the extraciliary plasma membrane as measured using a fold enrichment metric (Figure 2B, left bar). This indicates that ciliary localization of MOR-Venus can indeed be replicated in this cell model. However, we noticed that removing the C-terminal fusion from MORs diminished ciliary targeting (Figure 2A, middle), such that visible receptor localization to cilia was only observed occasionally (Figure S2A), and receptors were not enriched (Figure 2B, middle bar). Ciliary localization of MOR-Venus was not simply a consequence of adding a bulky fluorescent protein tag because fusing only the 5-residue linker sequence (GSIAT) used in the MOR-Venus construct was sufficient to increase receptor localization to cilia (Figure 2A, right, MOR+linker). Moreover, the estimated degree of ciliary enrichment was significantly higher for this construct than for the receptor construct containing both the linker and Venus in tandem (Figure 2B). These results indicate that MOR targeting to the primary cilium can be observed in IMCD3 cells and that the degree of ciliary targeting is sensitive to modification of the receptor C-tail.
Figure 2. MOR cilia targeting in IMCD3 cells is sensitive to modification of the receptor’s C-tail and is specific to MORs.
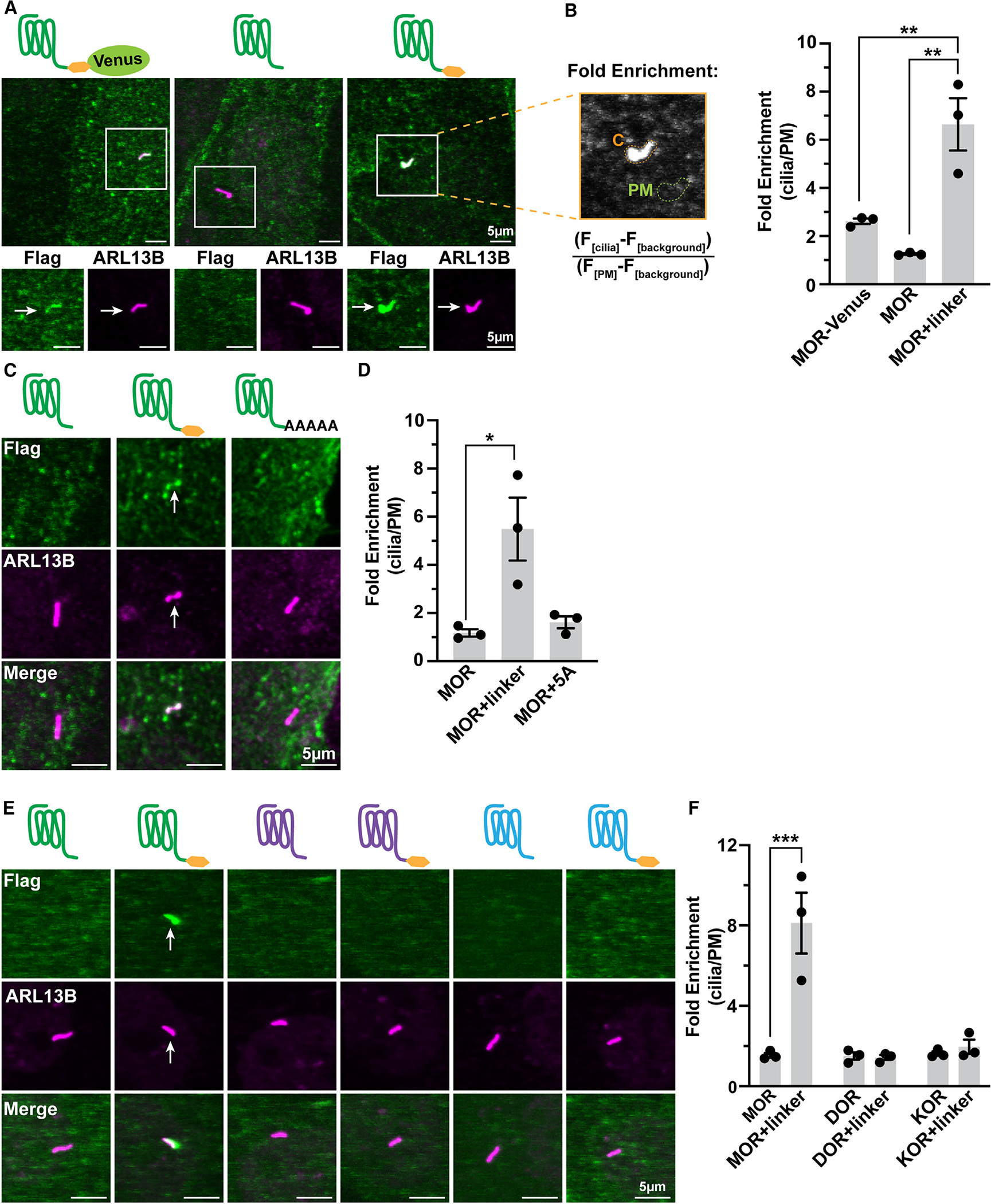
(A) Schematics and representative confocal images of IMCD3 cells transiently expressing FLAG-tagged MOR-Venus, MOR, or MOR+linker (GSIAT, yellow hexagon). Cilia were stained with anti-ARL13B antibody. Arrows indicate cilia-localized receptor. Cells were incubated with anti-M1 (FLAG) antibody conjugated to Alexa Fluor 647 to label surface receptor prior to fixation and permeabilization.
(B) Fold enrichment was calculated by dividing the background-subtracted receptor ciliary fluorescence by the background-subtracted plasma membrane (PM) fluorescence. MOR+linker was significantly increased by the linker extension as compared to MOR and MOR-Venus. One-way ANOVA (**p = 0.002) with Tukey’s multiple comparisons test; data are represented as mean ± SEM from 3 independent experiments, 30–48 cells; **p < 0.01. For lower-magnification and expression controls, see Figure S2.
(C) Representative confocal images of transiently transfected IMCD3 cells expressing MOR, MOR+linker, or MOR+5 alanines assessed and measured as done for (A) and (B).
(D) MOR cilia enrichment is completely lost upon replacement of the linker with alanines. One-way ANOVA (*p = 0.014) with Dunnett’s multiple comparisons test; data are represented as mean ± SEM from 3 independent experiments, 32–39 cells; *p < 0.05.
(E) Representative confocal images of IMCD3 cells transiently expressing FLAG-tagged MOR, DOR, and KOR ± linker assessed and measured as done for (A) and (B).
(F) MOR cilia enrichment is significantly enhanced by the C-tail linker, but DOR and KOR are not. Two-way ANOVA interaction ***p = 0.0003; main effect of receptor ***p = 0.0004; main effect of linker ***p = 0.0010. Tukey’s multiple comparisons test; data are represented as mean ± SEM from 3 independent experiments, 31–53 cells; ***p = 0.0001. Scale bars: 5 μm. Arrows indicate cilia-localized receptor.
We wondered if increased ciliary enrichment of MOR+linker might result as an artifact of excessive receptor expression in the plasma membrane. If this were the case, then we predicted that MOR+linker would only localize to primary cilia in cells with high, but not low, surface expression of the receptor. However, receptor expression level did not appear to correlate with ciliary localization when examined across large fields of surface-labeled cells expressing the MOR, MOR+linker, or dopamine DRD1 (an established ciliary receptor20,22–24) at different relative levels (Figure S2B). Rather, ciliary localized receptors (MOR+linker and DRD1) were observed in primary cilia in cells expressing surface receptors over a wide range, whereas the MOR was rarely observed in primary cilia regardless of the level of surface expression. Moreover, when we measured surface receptor expression quantitatively using flow cytometry, all of these receptor constructs were expressed at a comparable level (Figure S2C). Immunoblot analysis indicated that all of the receptor constructs resolved in the expected molecular mass range. MORs and MOR+linker receptors resolved as a heterogeneous species of similar apparent molecular mass (~90 kDa), consistent with heterogeneous N-linked glycosylation. MOR-Venus resolved similarly but in a higher apparent molecular mass range (~125 kDa), as expected due to the addition of mVenus (Figure S2D). MOR+linker was consistently detected by immunoblotting in a higher amount than MORs or MOR-Venus, despite its similar level of surface expression. This suggests that linker fusion results in an increase in the fraction of receptors retained intracellularly at steady state, a difference that might be relevant to the mechanism of ciliary targeting (see below). However, as the surface expression of all of the receptor constructs was comparable, we are confident that the observed differences in ciliary localization are not an artifact of excessive plasma membrane expression.
Although the primary focus of the present study is to investigate the mechanism of opioid receptor localization to the primary cilium, we wondered if the observed differences in ciliary localization among the engineered receptor constructs might provide some insight into signaling function. As a first step to investigate this, we compared the ability of MORs and MOR+linker to inhibit adenylyl cyclase activity in IMCD3 cells. We evaluated this by measuring whole-cell, MOR-dependent cAMP inhibition using a genetically encoded fluorescent cAMP biosensor, cADDis, in living IMCD3 cells expressing either the MOR or MOR+linker (Figures S2E and S2F). Forskolin (FSK; 1 μM), an activator of adenylyl cyclase, produced a rapid elevation of cytoplasmic cAMP concentration, indicated by increased cADDis fluorescence, as expected. A similar effect was observed irrespective of which receptor construct was expressed. Co-application of the MOR agonist DAMGO (1 μM) together with FSK inhibited this response, also irrespective of which receptor construct was expressed, indicating that both the MOR and MOR+linker functionally couple to adenylyl cyclase regulation (Figure S2E). Interestingly, the effects of the two receptor constructs were not identical. MORs produced a nearly complete inhibition of the cAMP signal that persisted over a 20 min time course of continuous DAMGO exposure, whereas the inhibition produced by MOR+linker was initially strong but decayed over time (Figure S2F). These results indicate that MOR+linker is functional to couple to adenylyl cyclase inhibition in intact cells but that it produces a signaling effect differing from MORs in its duration. We are intrigued by this effect but note that significant additional study will be required to determine if it results specifically as a consequence of ciliary localization.
Remarkably, the ability of the fused 5-residue linker sequence to increase ciliary targeting of the MOR was specific because fusing 5 alanine residues to the MOR C terminus (GSIAT→AAAAA) did not mimic this effect (Figures 2C and 2D). We considered two possible explanations. First, we considered the hypothesis that the linker sequence itself acts as an autonomous ciliary targeting determinant when fused to the receptor C-tail, irrespective of the MOR having any intrinsic potential for localization to the primary cilium. Second, we considered the alternate hypothesis that the MOR has an inherent potential for targeting to the cilium but that functional expression of this potential is sensitive to modification(s) of the receptor C terminus that are mimicked by fusion of the short linker sequence. As a first step toward distinguishing these possibilities, we tested the effect of fusing the same linker sequence to homologous GPCRs. If the linker sequence acts as an autonomous ciliary targeting determinant, then we would predict it to drive ciliary targeting of other receptors. We started with the delta opioid receptor (DOR) because it is a GPCR that has a very high level of homology to the MOR and was shown previously not to localize to cilia when expressed with its native C terminus.20 We verified that wild-type DOR does not accumulate in cilia and found that fusing the same linker sequence to the DOR C-tail did not result in ciliary targeting, in marked contrast to the MOR. We then tested the kappa opioid receptor (KOR), the next closest MOR homolog. The KOR also did not detectably accumulate in the primary cilium, with or without the fused linker (Figures 2E and 2F). Together, these results support the hypothesis that the MOR has an inherent potential for targeting to the primary cilium that is specific to this receptor and that the functional expression of this ciliary targeting potential is conditional, as defined by its sensitivity to modification of the receptor C-tail.
Defining a conditional ciliary targeting determinant in the MOR C-tail
As a next step to test the hypothesis that the MOR has the capacity to be specifically targeted to the primary cilium, we asked if a discrete structural determinant in MOR specifies ciliary targeting. To identify such a determinant, we constructed chimeric mutant receptors and assessed ciliary targeting in the presence of the fused C-terminal linker. We swapped corresponding receptor domains between MORs and DORs, focusing on the intracellular loop 3 (ICL3) and the C-tail because they display the highest sequence divergence and contain the cilia targeting sequences for other neuromodulatory ciliary receptors.20,25–27 Replacing the MOR ICL3 with the DOR-derived sequence had little effect on the ciliary targeting potential of mutant receptors; in contrast, replacing the MOR C-tail with the DOR-derived sequence significantly blocked cilia targeting (Figures 3A and 3B), despite the receptors retaining plasma membrane localization (Figure S3A). Conversely, we next asked whether replacing the ICL3 or C-tail of the DOR with the corresponding MOR-derived sequence is sufficient to confer receptor targeting to the cilium. Replacing the DOR ICL3 had no detectable effect, whereas replacing the DOR C-tail with the MOR-derived sequence was sufficient to confer ciliary targeting (Figures S3B‒S3D). These results further suggest that the MOR has an inherent potential for selective targeting to the primary cilium. We therefore focused our attention on the MOR C-tail amino acid residues as determinants underlying this conditional targeting.
Figure 3. The MOR conditional cilia targeting determinant is in its C-tail.
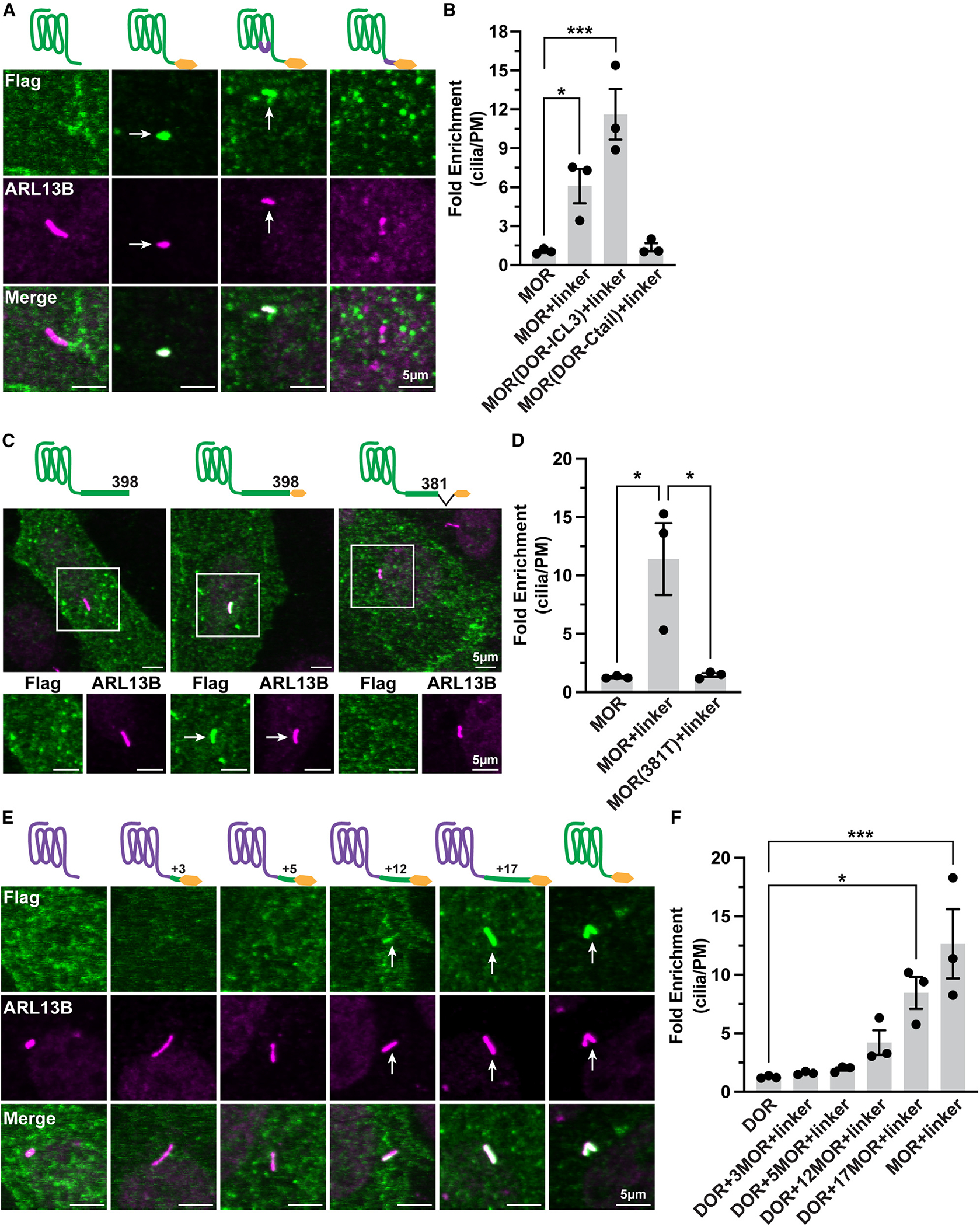
(A) Representative confocal images of IMCD3 cells transiently transfected with the indicated FLAG-tagged MORs. Cells were labeled with M1–647 antibody and fixed, and cilia were labeled with ARL13B.
(B) Quantification of receptor cilia enrichment indicates that MOR+linker and MOR(DOR-ICL3)+linker chimera have significantly higher enrichment in cilia compared to MOR, whereas MOR(DOR-C-tail)+linker does not. One-way ANOVA (***p = 0.0007) with Dunnett’s multiple comparisons; data are represented as mean ± SEM from 3 independent experiments, 29–36 cells; *p < 0.05 and ***p < 0.001.
(C) Representative confocal images of transiently transfected IMCD3 cells expressing MOR, MOR+linker, or MOR(381T)+linker. Cells were incubated with M1–647 antibody, fixed, and stained with anti-ARL13B to identify primary cilia.
(D) Quantification of receptor cilia enrichment revealed that deleting the MOR distal 17 amino acids completely abolishes cilia enrichment, One-way ANOVA (*p = 0.011) with Tukey’s multiple comparisons test; data are represented as mean ± SEM from 3 independent experiments, 45–58 cells; *p < 0.05.
(E) Representative confocal images of IMCD3 cells transiently expressing the indicated constructs and labeled as described above.
(F) Fold enrichment quantification of receptor cilia expression determined that adding the last 17 amino acids of MOR to DOR significantly enhanced DOR cilia enrichment. See Figure S3 for plasma membrane localization validation of constructs. One-way ANOVA (***p = 0.0004) with Dunnett’s multiple comparisons test; data are represented as mean ± SEM from 3 independent experiments, 41–51 cells; *p < 0.05 and ***p < 0.001. Scale bars: 5 μm. Arrows indicate cilia-localized receptor.
The MOR and DOR C-tails are identical or highly similar until aspartic acid D381 in the MOR (D364 in the DOR), after which the sequences become highly divergent. Accordingly, we hypothesized that the information required to specify the ciliary targeting potential of MORs is contained within this distal portion of the C-tail. Indeed, truncating the MOR C-tail after D381 abrogated MOR localization in primary cilia (Figures 3C and 3D). This limited distal portion of the MOR C-tail, comprising only 17 amino acid residues, was also sufficient to confer ciliary targeting potential on the DOR when fused on the C-tail (Figures 3E and 3F; see also Figure S3E). Ciliary enrichment required the entire 17-residue MOR-derived sequence, as ciliary targeting diminished sharply upon sequence shortening (Figure 3F), despite the constructs displaying normal surface localization (Figure S3F).
Ciliary targeting directed in cis by the MOR C-tail requires TULP3 in trans
Having defined a minimal sequence in the MOR C-tail that is necessary and sufficient to confer conditional ciliary targeting of the MOR in cis, we next sought to gain some insight into the trans-acting mechanism mediating receptor targeting to the cilium. In particular, we asked if the conditional targeting property of MORs is mediated through a similar downstream targeting machinery as other ciliary receptors. We focused on TULP3, a key adaptor protein that is widely required for ciliary targeting of many GPCRs.19,28 We first tested a mutant version of TULP3 previously shown to inhibit ciliary receptor targeting in a dominant-negative manner.19 The targeting of the linker-fused MOR in ARL13B-marked cilia was significantly reduced in cells expressing the mutant TULP3 when compared to cells overexpressing wild-type TULP3 at a similar level, suggesting that the conditional targeting of MORs to the primary cilium is TULP3 dependent (Figures 4A and 4B).
Figure 4. Cilia targeting directed in cis by the MOR C-tail requires TULP3 in trans.
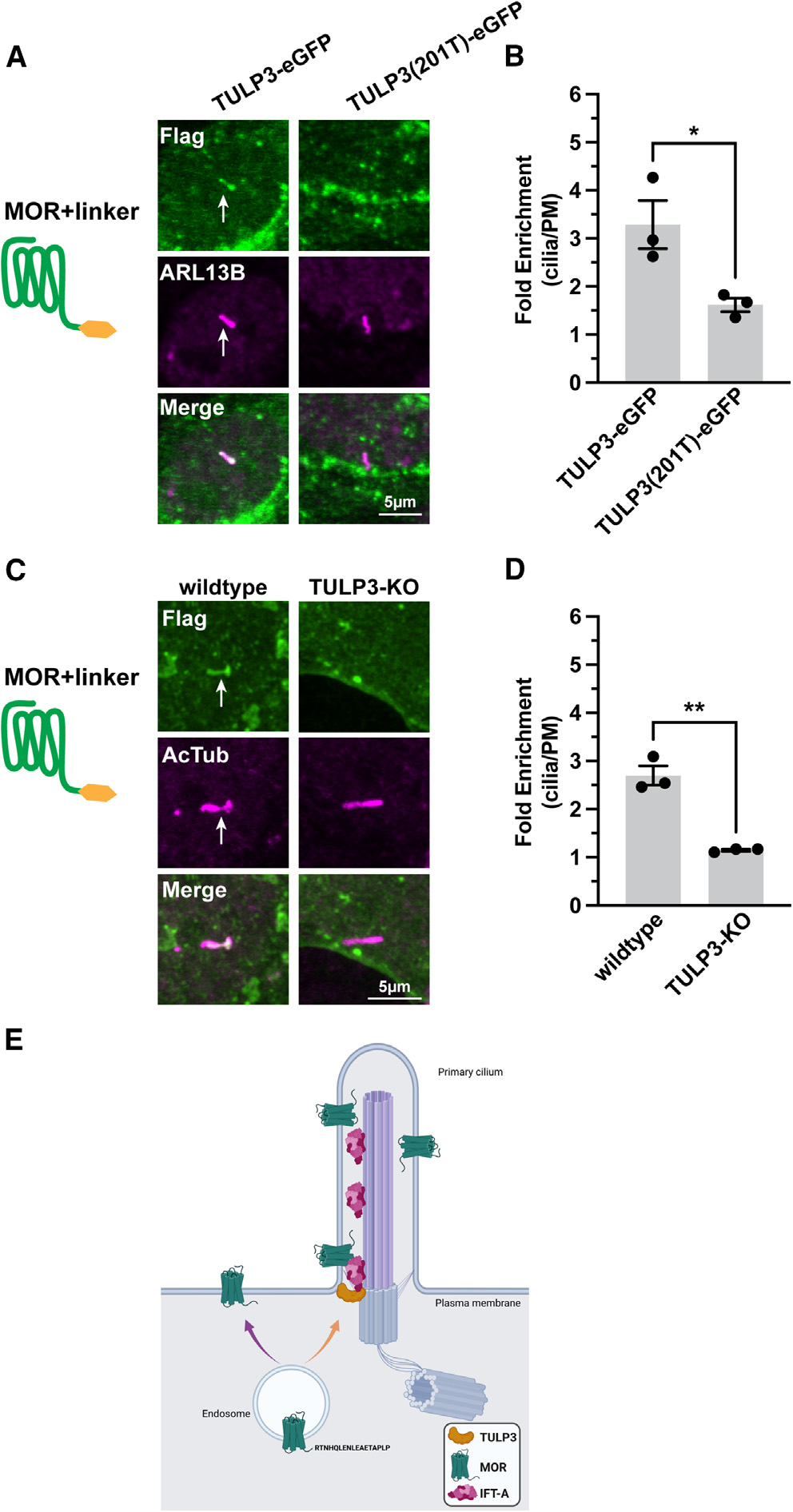
(A) Representative confocal images of IMCD3 co-transfected with MOR+linker and either TULP3-EGFP or truncation mutant, TULP3(201T)-EGFP. Cells were fixed, permeabilized, and stained with unconjugated M1 FLAG antibody to label receptor and ARL13B antibody to mark cilia. EGFP expression was confirmed.
(B) Quantification of fold receptor enrichment in cilia revealed that co-expression of the dominant-negative TULP3 mutant significantly reduced MOR+linker cilia enrichment. Student’s t test; data are represented as mean ± SEM from 3 independent experiments, 43–44 cells; *p < 0.05.
(C) Representative images of wild-type (left) and TULP3-knockout (KO) #1 (right) IMCD3 cells transiently transfected with MOR+linker. Cells were fixed and stained with anti-FLAG and anti-AcTub (acetylated tubulin) antibodies.
(D) MOR+linker cilia enrichment was more significantly reduced in TULP3-KO #1 IMCD3 cells than in wild-type cells (see also Figure S4). Student’s t test; **p < 0.0015; data are represented as mean ± SEM; n = 3 experiments, 33–40 cells. Scale bars: 5 μm. Arrows indicate cilia-localized receptor.
(E) Proposed model depicting MOR delivery to the primary cilium. The cytoplasmic sequence “LENLEAE” that facilitates MOR recycling29 overlaps with the sequence “RTNHQLENLEAETAPLP” required for cilia delivery (purple arrow). TULP3 promotes entry into the cilium presumably via its interaction with IFT-A (intraflagellar transport complex A)19 (orange arrow).
We next tested the effect of depleting the endogenous adaptor protein by generating TULP3-KO IMCD3 cells using CRISPR. For these experiments, we utilized acetylated tubulin (AcTub) as a ciliary marker because deletion of endogenous TULP3 protein results in primary cilia lacking ARL13B.30,31 Due to these antibody constraints, we used a different labeling technique to stain for MOR+linker that effectively measures ciliary enrichment, albeit with a worse signal-to-noise ratio (Figures S4A‒S4C). Consistent with the dominant-negative results, TULP3-KO blocked conditional ciliary targeting of the MOR, as indicated by a full suppression of the ciliary accumulation of linker-fused receptors above the level in the extraciliary plasma membrane (Figures 4C and 4D). We observed an identical effect in TULP3-KO cells generated and validated in our laboratory, as well as in an independent TULP3-KO clone generated and validated previously by others21 (Figures S4D and S4E). TULP3 gene deletion in both TULP3-KO cell clones was validated by western blotting for endogenous TULP3 expression (Figure S4F). These data suggest that MOR conditional ciliary targeting, which is defined by sensitivity to modification of the C-tail, likely engages a similar machinery to that engaged by GPCRs constitutively targeted to cilia.
DISCUSSION
The present results demonstrate localization of the MOR to neuronal primary cilia. We first observed this localization by examining the subcellular distribution of endogenously expressed, tagged MORs in neural tissue and verified ciliary localization of native, untagged MORs in tissue sections and primary neuronal culture. Despite clear evidence that the MOR has the potential to localize to primary cilia, a key observation from our in vivo data is that the degree to which the MOR accumulates in primary cilia is variable, both across brain regions and between individual neurons in the same region. This variability suggests that MOR localization to the primary cilium is not constitutive; rather, it appears to be conditional. Our studies of ciliary localization in IMCD3 cells support this concept and establish that ciliary targeting of opioid receptors is specific to MORs, relative to homologous DORs and KORs.
Our results support a molecular logic for the determination of opioid receptor targeting to the primary cilium that differs from that of previously identified ciliary GPCRs.32,33 In particular, the specific ciliary targeting determinant that we identify in the MOR distal C-tail confers only the potential for receptors to localize to cilia but does not determine its degree. The degree to which ciliary targeting occurs is highly sensitive to modifications of the distal C-tail. To our knowledge, such a conditional logic of ciliary receptor targeting has not been described previously, as most other GPCRs known to localize in the primary cilium appear to be targeted to cilia constitutively.32,34,35 Despite this distinction, our results indicate that ciliary targeting of MORs requires the conserved TULP3 adaptor protein that is also engaged by various other GPCRs.19,25,28
We note that the sequence required for MOR ciliary targeting overlaps a determinant previously shown to sort receptors from endosomes into the recycling pathway.29 Accordingly, we speculate that the targeting of MORs to the primary cilium may occur through an elaboration of the recycling pathway in ciliated cells. A model consistent with the present data is that the MOR leverages the sequence-directed recycling pathway to deliver receptors from endosomal membranes to TULP3 at the cilia base (Figure 4E). Interestingly, GPCRs have been shown to use vesicular machinery for ciliary targeting, and TULP3 is important for cilia delivery of many receptors.28,34,36 However, it is unclear whether the MOR engages TULP3 directly when exiting the recycling pathway or if TULP3 acts as a scaffold protein for receptor trafficking to the cilia.
Finally, we do not know the functional significance of MOR localization to the primary cilium. A number of GPCRs activated by neuromodulators are now known to localize on primary cilia.10,20,22,23,37–42 Emerging functional and anatomical evidence suggests that localization of GPCRs to primary cilia can have a significant impact on neuronal transmission.24,43,44 However, to our knowledge, it remains unclear specifically how ciliary targeting impacts GPCR-mediated neuromodulation for any of these examples. Intriguingly, decreased gene expression of primary cilia components was found in addition to alterations in neuronal excitability and opioid signaling in MOR neurons derived from morphine abstinent animals.45 Opioid receptors are well known to mediate different physiological effects depending on their localization in presynaptic and postsynaptic domains in neurons.46–50 Notably, our initial examination of the cilia-targeted MOR+linker indicates that the same modification that drives cilia localization also reduces its capacity to inhibit cAMP (Figures S2E and S2F); however, whether this effect is solely a consequence of MOR subcellular localization remains undetermined. The present findings motivate investigating how primary cilia localization impacts MOR neuromodulation and, more broadly, the extent to which MOR ciliary localization influences opioid-dependent functions including analgesia, addiction, and respiratory depression.
Limitations of the study
A limitation of the present study is that it remains unclear how modification of the MOR C terminus determines the degree of ciliary receptor targeting. Our results indicate that the degree of ciliary targeting enabled by this sequence varies depending on the specific peptide or protein sequence fused. However, it does not reflect a spurious ciliary targeting activity of the fused sequence itself, as ciliary targeting remains dependent on the native recycling sequence present in the MOR C-tail. Accordingly, we propose that fusion of the C-terminal linker—while being artificial itself—likely mimics the effect of a post-translational modification in the receptor tail that occurs naturally but is presently undefined. In addition, further experiments are necessary to determine whether MOR ciliary localization impacts its downstream signaling capacity and how primary cilia on adult medial habenula MOR neurons may contribute to opioid-specific physiology.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Aliza Ehrlich (Aliza.Ehrlich@ucsf.edu).
Materials availability
Reagents generated in this study will be made available by request to the lead contact.
Data and code availability
Original and raw data are available upon request from the lead contact.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Mice
Homozygous MOR-Venus12 animals (MORVenus/Venus) were bred by crossing heterozygous animals. Animals were housed in the Neurophenotyping Center, McGill University/Douglas Hospital Research Institute, Montreal, Canada) or at the University of California, San Francisco. Animals were group housed on a 12-h light/dark schedule with constant access to food and water and handled in accordance with guidelines set by The University. All animals were genotyped twice, before and after experimental endpoint. Immunohistochemistry experiments were conducted on male and female mice aged 8–14 weeks. All experimental procedures were performed in accordance with the guidelines of the Canadian Council of Animal Care, and all animal procedures were approved by the McGill University/Douglas Hospital Animal Care Committee, by the Institutional Animal Care and Use Committee (IACUC) at the University of North Carolina at Chapel Hill or University of California, San Francisco’s IACUC. MOR-mCherry mice were housed 2–5 per cage and maintained on a 12-h light/dark cycle with ad libitum access to food and water.
Rat primary neuron cultures
Primary habenula neurons were prepared from embryonic day 18 Sprague-Dawley rats (Charles River) dissected from a timed-pregnant female following euthanasia as previously reported.2 The habenula was identified using a stereomicroscope and dissected into ice-cold Hank’s buffered saline solution calcium/magnesium/phenol red free (Thermo Fisher Scientific cat# 14–175-145). Habenula was dissociated in 0.05% trypsin/EDTA (Gibco) for 15 min at 37°C and washed 2x in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, University of California, San Francisco, Cell Culture Facility) and 30 mM HEPES (Gibco). Neurons were further dissociated by trituration through flame-polished Pasteur pipettes. Neurons were seeded onto 35 mm glass bottom dishes with 20 mm micro-well (Cellvis cat# D35–20-1.5-N) coated with poly-D-lysine (Sigma) at a density of 1.5 × 10^5 cells. Medium was exchanged on day in vitro 4 (DIV4) for phenol red free Neurobasal medium (Gibco) supplemented with Glutamax 1X (Gibco), B27 1X (Gibco), and cytosine arabinosine 1 μM (Sigma-Aldrich). On DIV8, half of the cultured medium was exchanged with fresh, equilibrated medium. Neurons were maintained in a humidified incubator with 5% CO2 at 37°C and imaged at DIV12. All experiments were performed on at least 3 independent cultures.
METHOD DETAILS
Plasmids
Mouse MOR-Venus (codon-optimized MOR) was a gift from Michel Bouvier (University of Montreal). All constructs were cloned into pGCGFP-G418 (addgene, renamed pCAGG-G418) that was modified to include an in-frame signal sequence and Flag sequence in between EcoRI and NotI sites following which constructs were cloned into NotI site using In-Fusion Cloning (Takara) using primers (see Table SI). Mouse DRD1 was synthesized commercially (IDT) and cloned into pCAGG-G418 at the NotI site. Mouse TULP3 (OriGene Technologies) was cloned into pCAGG-G418, truncated using In-Fusion Cloning, and both wildtype and truncated constructs were tagged with eGFP on the C terminus. Human ARL13B (Twist Bioscience) together with mKate (Twist Bioscience) was cloned into pCAGG-G418 between EcoRI and NotI sites without a signal or Flag sequence. All constructs were validated by DNA sequencing.
Cell culture, and transfection
Inner medullary collecting duct (IMCD3; ATCC) cells were grown in DMEM/F-12 Medium (Thermo Fisher Scientific cat# 11668019) supplemented with 10% FBS (UCSF Cell Culture Facility, San Francisco, California, USA) at 37°C, 5% CO2. Transfection of IMCD3 cells was performed using Lipofectamine 2000 (Thermo Fisher Scientific cat# 11668019) in accordance with manufacturer’s instructions. Cells were cultured for 24 h post-transfection prior to experimentation.
CRISPR/Cas9 TULP3 knock out cells
Two different generations of TULP3 knock out cells were used in this paper. One set of TULP3 CRISPR/Cas9 knock out cells were a gift from Maxence Nachury (UCSF).21 A distinct TULP3 CRISPR/Cas9 knock out line was generated by preparing Cas9 ribonucleo-proteins (RNPs) as follows.53 sgRNAs (Synthego) were resuspended in RNAse-free TE buffer at 53.3μM. 3μL of sgRNAs were mixed with 2 μL of Cas9 (UC Macrolab) and the complex was incubated at room temperature for 10 min to form RNPs. RNPs were used immediately or stored at −80°C. RNPs were nucleofected into low passaged IMCD3 cells using the Nucleofector 4D system (Lonza) and Nucleofection kit SF (Lonza cat# V4XC-2032) according to the manufacturers protocol. IMCD3 cells were lifted with PBS/EDTA (UCSF Cell Culture Facility) and counted. 300,000 cells were pelleted and resuspended in 20μL nucleofection buffer combined with 5μL of RNP and placed in a well of the nucleofector dish and immediately nucleofected using program DS-137. Following nucleofection, 80 μL of DMEM/F-12 medium supplemented with 10% FBS was added to each well and incubated at 37°C for 5 min. The nucleofected cells were then transferred to a 6 well dish and cultured in 2mL of media. Monoclonal cells were extracted by sorting individual cells into a 96-well plate using an Aria II (BD biosciences). Clones were grown in a 96-well plate until mutational efficiency was quantified using TIDE analysis. Genomic DNA was extracted from 10,000 cells using 250μL Quick Extract DNA Extraction Solution (Lucigen). Crude lysate was vortexed, heated at 65°C for 15 min, vortexed, heated at 65°C for 10 min, heated to 98°C for 2 min and stored at −20 or used immediately. Primers were designed with COSMID (https://crispr.bme.gatech.edu/) with indels and mismatch set to 0. TIDE PCR was performed using Q5 polymerase kit (NEB). PCR yielding 600 bp products around the cut site were agarose gel purified (Macherey Nagel) and sequenced. Resulting sequences were analyzed using ICE analysis (Synthego). Additional validation of monoclonal knockout cells was carried out by western blotting for TULP3 (detailed below).
Immunostaining and confocal imaging
IMCD3 cells
5 × 104 cells were seeded on glass coverslips and transfected after 24 h. To label surface Flag-tagged receptors, cells were incubated with Alexa Fluor-conjugated M1 anti-Flag antibody (M1–647, see key resources table) or unconjugated M1 anti-Flag antibody (M1) diluted 1:1000 in media, 15 min, 37°C. Cells were then washed with phosphate-buffered saline with calcium and magnesium (PBS, Life Technologies cat# 14040216) and fixed for 10 min at RT in 4% paraformaldehyde/PBS (Fisher Scientific cat# AAJ19943K2). Cells were permeabilized for 10 min at RT in 0.2% Triton X-100/PBS and then blocked in 0.2% Triton X-100 and 3% bovine serum albumin (BSA, Sigma-Aldrich cat# A7030) diluted in PBS for 1 h at RT shaking gently. Primary antibodies were diluted in blocking solution and incubated overnight at 4°C rocking gently. Cells were washed three times in 0.2% Triton X-100 in PBS, 10 min, at RT shaking gently. Secondary antibodies were diluted in blocking solution 1:2000. Secondary labeling was performed at RT for 1 h, shaking gently. Cells were washed one time, 10 min with 0.2% Triton X-100 in PBS, labeled for 10 min with DAPI (Sigma)/PBS (diluted 1:20,000), and washed 1X PBS prior to mounting with ProLong Diamond antifade reagent (Thermo Fisher Scientific cat# P36970). Cells were imaged by confocal microscopy using a Nikon Ti inverted fluorescence microscope with CSU-22 spinning disk confocal with a 60 × 1.4 numerical aperture oil objective.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-MOR (UMB3) | Abcam | Cat#Ab134054; RRID: AB_2313773 |
| Anti-ACIII | EncorBio | Cat#CPCA_ACIII; RRID: AB_2744500 |
| Anti-RFP | Abcam | Cat#Ab62341; RRID: AB_945213 |
| Anti-GFP | Novus Biologicals | Cat#NB100-1614; RRID: AB_10001164 |
| Anti-ARL13B | ProteinTech | Cat#66739-1-Ig; RRID: AB_2882088 |
| Anti-AcTub | Sigma-Aldrich | Cat#T7451; RRID: AB_609894 |
| Anti-ARL13B | ProteinTech | Cat#17711-1-AP; RRID: AB_2060867 |
| Anti-ACIII | Abcam | Cat#Ab125093; RRID: AB_10975307 |
| Anti-Flag | Sigma-Aldrich | Cat#F-7425; RRID: AB_439687 |
| Anti-Flag (conjugated: M1-647) | Sigma-Aldrich | Cat#F-2040; RRID: AB_2313773 |
| Anti-ARL13B-647 | ProteinTech | Cat#CL647-17711; RRID: AB_2920234 |
| Anti-chicken Alexa Fluor 647 | Invitrogen | Cat#A-78952; RRID: AB_2921074 |
| Anti-chicken Alexa Fluor 647 | Thermo Fisher Scientific | Cat#A-21449; RRID: AB_2535866 |
| Anti-mouse Alexa Fluor 594 | Thermo Fisher Scientific | Cat#A-21203; RRID: AB_2535789 |
| Anti-mouse Alexa Fluor 633 | Thermo Fisher Scientific | Cat#A-21050; RRID: AB_2535718 |
| Anti-rabbit Alexa Fluor 488 | Thermo Fisher Scientific | Cat#A-11008; RRID: AB_143165 |
| Anti-rabbit Alexa Fluor 555 | Invitrogen | Cat#A-31572; RRID: AB_162543 |
| Anti-rabbit Alexa Fluor 647 | Thermo Fisher Scientific | Cat#A-21245; RRID: AB_2535813 |
| Anti-ACTIN | Santa Cruz Biotechnology | Cat#Sc-56459; RRID: AB_830981 |
| Anti-GAPDH | Cell Signaling Technology | Cat#5174; RRID: AB_10622025 |
| Anti-GFP | Sigma-Aldrich | Cat#11814460001; RRID: AB_390913 |
| Anti-TULP3 | Norman et al.51 | N/A; RRID: AB_2313773 |
| Anti-mouse IRDye 800CW | LI-COR Biosciences | Cat#926-32212; RRID: AB_621847 |
| Anti-rabbit IRDye 680RD | LI-COR Biosciences | Cat#926-68073; RRID: AB_10954442 |
| Anti-mouse IgG (H + L) HRP | Thermo Fisher Scientific | Cat#31430; RRID: AB_228307 |
| Anti-rabbit IgG (H + L) HRP | Thermo Fisher Scientific | Cat#31460; RRID: AB_228341 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| E. coli Stellar™ Competent Cells | Takara | Cat#636766 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| DAMGO | Sigma-Aldrich | Cat#E7384-10MG; RRID: |
| Forskolin | Sigma-Aldrich | Cat#F6886-25mg; RRID: |
| IBMX | Sigma-Aldrich | Cat#i5879-1G; RRID: |
| Cas9 | UC QB3 Macrolab | N/A |
|
| ||
| Critical commercial assays | ||
|
| ||
| cAMP Assay: Green Up cADDis cAMP Assay Kit (#U0200G) | Montana Molecular | https://montanamolecular.com/live-cell-camp-assay-caddis/ |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Mouse: IMCD3 cells | ATCC | RRID:CVCL_0429 |
| Mouse: TULP3KO-IMCD3 cells | This paper | N/A |
| Mouse: TULP3KO-IMCD3 cells | Ye et al.21 | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: MOR-Venus | Ehrlich et al.12 | RRID:IMSR_JAX:035787 |
| Mouse: MOR-mCherry | Erbs et al.14 | RRID:IMSR_JAX:029013 |
| Rat: Sprague Dawley | Charles River | RRID:RGD_734476 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primers for plasmid cloning, see Table S1 | This paper | N/A |
| sgRNA: TULP3: GGCAUCUGAGGGUCCACCAG | Synthego | https://www.synthego.com/ |
|
| ||
| Recombinant DNA | ||
|
| ||
| Plasmid: pGCGFP-G418 | Addgene | Addgene_31264 |
| Plasmid: pCAGGS-SSF-MOR | This paper | N/A |
| Plasmid: pCAGGS-SSF-MOR+linker | This paper | N/A |
| Plasmid: pCAGGS-SSF-MOR-Venus | This paper | N/A |
| Plasmid: pCAGGS-SSF-MOR+5A | This paper | N/A |
| Plasmid: pCAGGS-SSF-DOR | This paper | N/A |
| Plasmid: pCAGGS-SSF-DOR+linker | This paper | N/A |
| Plasmid: pCAGGS-SSF-KOR | This paper | N/A |
| Plasmid: pCAGGS-SSF-KOR+linker | This paper | N/A |
| Plasmid: pCAGGS-SSF-MOR(DOR-ICL3)+linker | This paper | N/A |
| Plasmid: pCAGGS-SSF-MOR(DOR-Ctail)+linker | This paper | N/A |
| Plasmid: pCAGGS-SSF-DOR(MOR-ICL3)+linker | This paper | N/A |
| Plasmid: pCAGGS-SSF-DOR(MOR-Ctail)+linker | This paper | N/A |
| Plasmid: pCAGGS-SSF-MOR(381T)+linker | This paper | N/A |
| Plasmid: pCAGGS-SSF-DOR+MOR3+linker | This paper | N/A |
| Plasmid: pCAGGS-SSF-DOR+MOR5+linker | This paper | N/A |
| Plasmid: pCAGGS-SSF-DOR+MOR12+linker | This paper | N/A |
| Plasmid: pCAGGS-SSF-DOR+MOR17+linker | This paper | N/A |
| Plasmid: pCAGGS-TULP3-eGFP | This paper | N/A |
| Plasmid: pCAGGS-TULP3(201T)-eGFP | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ | Schindelin et al.52 | https://imagej.net/software/fiji/#publication |
Rat habenula neurons
Habenula neurons were fixed on DIV12 in 4% paraformaldehyde and 4% sucrose in PBS for 10 min at RT and washed 1X with PBS prior to permeabilization in ice-cold, 100% methanol for 10′ at −20°C. Neurons were washed 1X with PBS and blocked in 3% normal goat serum, 0.3% Triton X-100 diluted in PBS at RT for 1 h. Primary antibodies (see key resources table) were diluted into blocking solution (1:1000) and incubated overnight at 4°C. Neurons were washed, incubated in secondary antibodies, washed again, and stored in PBS until imaged as described above. Two staining methods were used to evaluate wild-type MOR ciliary localization with cilia markers AC3 and ARL13B. The conventional method of co-application of primary antibodies was used for data collected in Figures S1H and S1I. A second sequential staining method to allow for staining with primary antibodies of the same species was used in Figures 1F and 1G, Videos S1 and S2, and Figure S1F (see Table S2). The second method began with labeling of MOR with primary antibodies on day 1, followed by 3 washes in PBS-T (0.2% Triton X-100 in PBS), and secondary antibody detection of MOR on the morning of day 2. Subsequently, neurons were washed 3 times in PBS-T and addition of the primary antibody for the cilia marker was added for overnight incubation. On day 3, following 3 washes in PBS-T, the secondary antibody detection of cilia markers was carried out. For staining with the conjugated ARL13b antibody (Figure S1F), the same method was employed: MOR primary and secondary antibodies were followed by the ARL13B-647 conjugated antibody (1:500, diluted in blocking solution overnight). To control for erroneous detection of MOR outside of MOR(+) neurons by the sequential staining method, controls were performed (Figures S1D, S1E, and S1G) wherein no primary antibody for MOR was included, but the remaining steps were done as in Figures 1F and 1G. We observed no ciliary staining in the channel used to detect MOR (FITC) in the absence of the MOR primary antibody. Neurons were washed in PBS before being stored in PBS until imaging on a Nikon Ti (CSU-22) spinning disk confocal microscope as stated above.
Mouse brain sections
Mice were anesthetized with i.p. injections of Ketamine/Xylazine/Acépromazine. Animals were perfused transcardially with PBS followed by 4% paraformaldehyde/PBS (ThermoFisher) as detailed previously.12 30% sucrose in PBS was used to cryoprotect brains. Cryoprotected brains embedded in O.C.T. (Sakura Finetek) were sectioned at 30 μm using a Cyrostat (Leica CM3050 S). Sections were rehydrated in PBS prior to blocking in 3% normal goat serum, 0.3% Triton X-100 diluted in PBS at RT for 1 h. Primary antibodies were incubated overnight followed by 3 washes with PBS-T. Secondary antibodies were added for 2 h at RT (See key resources table). Slices were mounted using ProLong Diamond and imaged using a Nikon Ti inverted fluorescence microscope with CSU-W1 large field view with a 100 × 1.4 numerical aperture oil objective and Nikon 6D/High Throughput widefield microscope with automated image stitching 10x.
Mouse dorsal root ganglia (DRG)
Mice were deeply anesthetized using isoflurane and perfused transcardially with 0.1M PBS followed by 4% formaldehyde solution. DRG were dissected and cryo-protected overnight in 30% sucrose in PBS at 4C. Frozen DRG tissue was then cut at 20 μm and incubated with a blocking solution (5% NDS in 0.3% Triton X-100 solution in 0.1M PBS) for at least 1 h. The primary antibodies were diluted in 1% normal donkey serum in 0.3% Triton X-100 solution in 0.1M PBS and incubated with DRG sections overnight at 4C. After washing the primary antibody 3 times for 10 min with staining solution, sections were incubated with secondary antibody solution at room temperature for 2 h. Sections were then mounted on a glass slide with Fluoromount-G (SouthernBiotech, cat no. 0100–01) after washing with 0.1M PBS 3 times for 10 min. Images were acquired with a confocal microscope (Zeiss LSM 780).
Western blotting
IMCD3 cells were washed three times in PBS and lysed in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 1% Na deoxycholate, with Complete Mini EDTA-free protease inhibitor cocktail (Roche cat# 04693159001) shaking at 4°C for 1 h. Lysates were spun down 10 min at 4°C and protein concentration of supernatants was determined using Pierce BCA Protein Assay kit (Thermo Fisher Scientific, cat# 23227). Samples were denatured in 4x Laemmli Sample Buffer (Bio-Rad cat# 1610747) at 95°C for 5 min. Receptor samples were lysed in Triton X-100 lysis buffer (50mM Tris, pH 7.4, 150mM NaCl, 1mM CaCl2, 0.2% Triton X-100, with Complete Mini EDTA-free protease inhibitor) and denatured in 4x Laemmli Sample Buffer at RT for 30 min. Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membrane (Bio-Rad cat# 162–0112). Membranes were blocked using LICOR Intercept (TBS) Blocking Buffer (LI-COR Biosciences cat# 927–60003) or 5% milk in TBS-T (0.1% Tween 20) and probed using primary antibodies: (see Table II) rocking at 4°C overnight. Proteins were detected using Licor secondary antibodies diluted in block (1:10,000), or by HRP secondary antibodies diluted in block (1:5000). Membranes were imaged on Licor imager or ChemiDoc XRS+ with ImageLab Software, and blots were imported into ImageJ for processing.
Flow cytometry
IMCD3 cells were transiently transfected with receptor constructs or empty vector (pCAGG-G418-ssFlag). Cells were washed in PBS and receptors were surface labeled with Alexa Fluor-conjugated M1 anti-Flag antibody (M1–647) for 15 min at 37°C. Cells were lifted in TrypLE Express Enzyme (1X), no phenol red (Thermo Fisher cat# 12–604-013) to be counted. Cells were centrifuged at 1000×g for 3 min and resuspended in 1x PBS containing 2%FBS. Cells were passed through a 5 mL round-bottom tube with cell strainer cap (Falcon cat# 352235) and plated onto a 96-well V-bottom plate (Axygen cat# P-96–450V-C). Surface staining of receptors was measured using gating settings above expression of Alexa Fluor 647 for un-transfected cells on a CytoFlex (Beckman Coulter). Four independent experiments were performed.
cADDis
To measure cAMP inhibition, IMCD3 cells were plated onto 35mm coverslip embedded imaging dishes (Cellvis) and transduced with cADDis-green up (Montana Molecular cat# U0200G) as well as transiently transfected with MOR or MOR+linker, and a cilia marker construct ARL13B-mKate. Receptors were first surface labeled with Alexa Fluor-conjugated M1 anti-Flag antibody (M1–647) for 15 min at 37°C. Excess antibody was removed, and cells were imaged at 37°C in DMEM/F-12 media not containing phenol red (Thermo Fisher cat# 11039–021) on a Nikon (CSU-22) spinning disk confocal microscope with a 60 × 1.4 numerical aperture oil objective using the perfect focus system (Nikon). Drugs including vehicle, DMSO, Dimethyl sulfoxide (Fisher cat# BP231–100), forskolin (Sigma cat# F6886–25mg), DAMGO, [D-Ala2, N-Me-Phe4, Gly5-ol]-Enkephalin acetate salt (Sigma cat# E7384–10MG) or forskolin together with DAMGO) were prepared at 5x concentrated stocks and 0.5 mL was added onto 2 mL media per dish at 60 s. Maximal cAMP stimulation was achieved by the co-application of 10mM forskolin and 200mM IBMX, 3-Isobutyl-1-methylxanthine (Sigma cat# i5879–1G) at 22 min in each dish. Cells were identified using the cilia marker channel (Cy3), and the intensity of the cADDis channel was measured over the entire image by drawing an ROI in the receptor channel (Cy5) using FIJI ImageJ. Normalized fluorescence change was calculated as previously reported54: (F-F0)/(FFskIBMX−F0) where F0 is the baseline average collected over the first minute and FFskIBMX is the maximal fluorescence averaged over the last 3 min. Each biological replicate is the average of approximately 10–44 cells per dish. Integrated cAMP is shown as the area under the curve (AUC) for positive peaks.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantifications
Quantitative image analysis was performed on unprocessed 16-bit files using FIJI software (ImageJ). Receptor-positive cilia were counted in at least 10 images per condition and receptor(+) cilia positive were determined based on whether a cell expressed the receptor and was ciliated. Unciliated cells were not counted. To measure cilia enrichment, a region of interest (ROI) was defined using thresholding on the cilia marker channel (see schematic in Figure 2B). This ROI was applied to the receptor channel to determine relative signal of the receptor within the cilium and then moved to the plasma membrane region to measure receptor expression on the cell surface. The ROI was moved to a region outside of the cell to correct for background signal and this value was subtracted from both cilia and plasma membrane values. Relative cilia enrichment was determined by dividing the corrected cilia signal by plasma membrane signal. 3D rendering of wildtype MOR habenula neurons was carried out using NIS-Elements AR Ver.5.3 (Nikon).
Statistical analysis
Results are displayed as a mean of at least 3 independent experiments ±SEM and include multiple samples per experiment. Statistically significant differences were determined using ANOVA (for more than two comparisons) with appropriate multiple comparisons test (see Figure Legends), or Student’s t-test was performed using Prism 9 software (GraphPad Software). Statistical significance is denoted as: ns (not significant, p ≤ 0.05), *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Material
Highlights.
MOR is targeted to primary cilia in brain and cultured neurons
MOR localization to cilia requires a 17-residue sequence also involved in recycling
Ciliary targeting is unique to the MOR cytoplasmic tail as compared to DOR and KOR
MOR ciliary targeting requires the TULP3 ciliary adaptor protein
ACKNOWLEDGMENTS
We thank the von Zastrow lab for helpful discussions, particularly J. Li for assistance with flow cytometry and E. Blythe for help with cAMP assay data analysis. We thank the A. Manglik lab for the use of the CytoFlex machine. We thank the following core facilities for providing assistance and services to support this research: the UCSF Helen Diller Family Comprehensive Cancer Center Laboratory for Cell Analysis (S. Elmes; supported by the National Institutes of Health under award P30CA082103) and the UCSF Center for Advanced Light Microscopy (K. Herrington, S. Kim, and D. Larsen). We thank Brigitte Kieffer (University of Strasbourg) and Michel Bouvier (University of Montreal) for creation of the MOR-Venus mouse. The graphical abstract and the scheme in Figure 4 were created with BioRender.com. This work was supported by National Institutes of Health grants R01MH120212 (M.v.Z.), R01DA044481 (G.S.), and K01MH123757 (A.T.E.). Further funding was provided by a Young Investigator Award from the Brain and Behavior Research Foundation #27303 to A.T.E., the New York Stem Cell Foundation (G.S.), and NIMH 1F32MH130096–01 to R.R.F. G.S. is a New York Stem Cell Foundation Robertson Investigator. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114164.
REFERENCES
- 1.Kieffer BL, and Evans CJ (2009). Opioid receptors: from binding sites to visible molecules in vivo. Neuropharmacology 56 (Suppl 1), 205–212. 10.1016/j.neuropharm.2008.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jullié D, Stoeber M, Sibarita JB, Zieger HL, Bartol TM, Arttamangkul S, Sejnowski TJ, Hosy E, and von Zastrow M (2020). A Discrete Presynaptic Vesicle Cycle for Neuromodulator Receptors. Neuron 105, 663–677.e8. 10.1016/j.neuron.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reeves KC, Shah N, Muñoz B, and Atwood BK (2022). Opioid Receptor-Mediated Regulation of Neurotransmission in the Brain. Front. Mol. Neurosci. 15, 919773. 10.3389/fnmol.2022.919773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoeber M, Jullié D, Lobingier BT, Laeremans T, Steyaert J, Schiller PW, Manglik A, and von Zastrow M (2018). A Genetically Encoded Biosensor Reveals Location Bias of Opioid Drug Action. Neuron 98, 963–976.e5. 10.1016/j.neuron.2018.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jimenez-Vargas NN, Gong J, Wisdom MJ, Jensen DD, Latorre R, Hegron A, Teng S, DiCello JJ, Rajasekhar P, Veldhuis NA, et al. (2020). Endosomal signaling of delta opioid receptors is an endogenous mechanism and therapeutic target for relief from inflammatory pain. Proc. Natl. Acad. Sci. USA 117, 15281–15292. 10.1073/pnas.2000500117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.André A, Gaibelet G, Le Guyader L, Welby M, Lopez A, and Lebrun C (2008). Membrane partitioning of various delta-opioid receptor forms before and after agonist activations: the effect of cholesterol. Biochim. Biophys. Acta 1778, 1483–1492. 10.1016/j.bbamem.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 7.Gondin AB, Halls ML, Canals M, and Briddon SJ (2019). GRK Mediates mu-Opioid Receptor Plasma Membrane Reorganization. Front. Mol. Neurosci. 12, 104. 10.3389/fnmol.2019.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaibelet G, Millot C, Lebrun C, Ravault S, Sauliere A, Andre A, Lagane B, and Lopez A (2008). Cholesterol content drives distinct pharmacological behaviours of μ-opioid receptor in different microdomains of the CHO plasma membrane. Mol. Membr. Biol. 25, 423–435. 10.1080/09687680802203380. [DOI] [PubMed] [Google Scholar]
- 9.Qiu Y, Wang Y, Law PY, Chen HZ, and Loh HH (2011). Cholesterol regulates mu-opioid receptor-induced beta-arrestin 2 translocation to membrane lipid rafts. Mol. Pharmacol. 80, 210–218. 10.1124/mol.110.070870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anvarian Z, Mykytyn K, Mukhopadhyay S, Pedersen LB, and Christensen ST (2019). Cellular signalling by primary cilia in development, organ function and disease. Nat. Rev. Nephrol. 15, 199–219. 10.1038/s41581-019-0116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonç alves, J., and Pelletier, L. (2017). The Ciliary Transition Zone: Finding the Pieces and Assembling the Gate. Mol. Cells 40, 243–253. 10.14348/molcells.2017.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrlich AT, Semache M, Gross F, Da Fonte DF, Runtz L, Colley C, Mezni A, Le Gouill C, Lukasheva V, Hogue M, et al. (2019). Biased Signaling of the Mu Opioid Receptor Revealed in Native Neurons. iScience 14, 47–57. 10.1016/j.isci.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishop GA, Berbari NF, Lewis J, and Mykytyn K (2007). Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J. Comp. Neurol. 505, 562–571. 10.1002/cne.21510. [DOI] [PubMed] [Google Scholar]
- 14.Erbs E, Faget L, Scherrer G, Matifas A, Filliol D, Vonesch JL, Koch M, Kessler P, Hentsch D, Birling MC, et al. (2015). A mu-delta opioid receptor brain atlas reveals neuronal co-occurrence in subcortical networks. Brain Struct. Funct. 220, 677–702. 10.1007/s00429-014-0717-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bardoni R, Tawfik VL, Wang D, Franç ois A., Solorzano C., Shuster SA., Choudhury P., Betelli C., Cassidy C., Smith K, et al. (2014). Delta opioid receptors presynaptically regulate cutaneous mechanosensory neuron input to the spinal cord dorsal horn. Neuron 81, 1312–1327. 10.1016/j.neuron.2014.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scherrer G, Imamachi N, Cao YQ, Contet C, Mennicken F, O’Don-nell D, Kieffer BL, and Basbaum AI (2009). Dissociation of the opioid receptor mechanisms that control mechanical and heat pain. Cell 137, 1148–1159. 10.1016/j.cell.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arvidsson U, Riedl M, Chakrabarti S, Lee JH, Nakano AH, Dado RJ, Loh HH, Law PY, Wessendorf MW, and Elde R (1995). Distribution and targeting of a mu-opioid receptor (MOR1) in brain and spinal cord. J. Neurosci. 15, 3328–3341. 10.1523/JNEUROSCI.15-05-03328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lupp A, Richter N, Doll C, Nagel F, and Schulz S (2011). UMB-3, a novel rabbit monoclonal antibody, for assessing mu-opioid receptor expression in mouse, rat and human formalin-fixed and paraffin-embedded tissues. Regul. Pept. 167, 9–13. 10.1016/j.re-gpep.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, Scales SJ, and Jackson PK (2010). TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 24, 2180–2193. 10.1101/gad.1966210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leaf A, and Von Zastrow M (2015). Dopamine receptors reveal an essential role of IFT-B, KIF17, and Rab23 in delivering specific receptors to primary cilia. Elife 4, e06996. 10.7554/eLife.06996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye F, Nager AR, and Nachury MV (2018). BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J. Cell Biol. 217, 1847–1868. 10.1083/jcb.201709041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marley A, and von Zastrow M (2010). DISC1 regulates primary cilia that display specific dopamine receptors. PLoS One 5, e10902. 10.1371/journal.pone.0010902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domire JS, Green JA, Lee KG, Johnson AD, Askwith CC, and Mykytyn K (2011). Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cell. Mol. Life Sci. 68, 2951–2960. 10.1007/s00018-010-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stubbs T, Koemeter-Cox A, Bingman JI, Zhao F, Kalyanasundaram A, Rowland LA, Periasamy M, Carter CS, Sheffield VC, Askwith CC, and Mykytyn K (2022). Disruption of dopamine receptor 1 localization to primary cilia impairs signaling in striatal neurons. J. Neurosci. 42, 6692–6705. 10.1523/JNEUROSCI.0497-22.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbeito P, Tachibana Y, Martin-Morales R, Moreno P, Mykytyn K, Kobayashi T, and Garcia-Gonzalo FR (2021). HTR6 and SSTR3 ciliary targeting relies on both IC3 loops and C-terminal tails. Life Sci. Alliance 4, e202000746. 10.26508/lsa.202000746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam BM, Moritz OL, Hurd LB, and Papermaster DS (2000). Identification of an outer segment targeting signal in the COOH terminus of rhodopsin using transgenic Xenopus laevis. J. Cell Biol. 151, 1369–1380. 10.1083/jcb.151.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berbari NF, Johnson AD, Lewis JS, Askwith CC, and Mykytyn K (2008). Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol. Biol. Cell 19, 1540–1547. 10.1091/mbc.E07-09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Badgandi HB, Hwang SH, Shimada IS, Loriot E, and Mukhopadhyay S (2017). Tubby family proteins are adapters for ciliary trafficking of integral membrane proteins. J. Cell Biol. 216, 743–760. 10.1083/jcb.201607095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanowitz M, and von Zastrow M (2003). A novel endocytic recycling signal that distinguishes the membrane trafficking of naturally occurring opioid receptors. J. Biol. Chem. 278, 45978–45986. 10.1074/jbc.M304504200. [DOI] [PubMed] [Google Scholar]
- 30.Han S, Miyoshi K, Shikada S, Amano G, Wang Y, Yoshimura T, and Katayama T (2019). TULP3 is required for localization of membrane-associated proteins ARL13B and INPP5E to primary cilia. Biochem. Bio-phys. Res. Commun. 509, 227–234. 10.1016/j.bbrc.2018.12.109. [DOI] [PubMed] [Google Scholar]
- 31.Palicharla VR, Hwang S-H, Somatilaka BN, Badgandi HB, Legué E., Tran VM., Woodruff JB., Liem KF., and Mukhopadhyay S. (2021). Interactions between TULP3 tubby domain cargo site and ARL13B amphipathic helix promote lipidated protein transport to cilia. Mol. Biol. Cell. 34, 310. https://doi.org/1101/2021.05.25.445488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McIntyre JC, Hege MM, and Berbari NF (2016). Trafficking of ciliary G protein-coupled receptors. Methods Cell Biol. 132, 35–54. 10.1016/bs.mcb.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Nemet I, Ropelewski P, and Imanishi Y (2015). Rhodopsin Trafficking and Mistrafficking: Signals, Molecular Components, and Mechanisms. Prog. Mol. Biol. Transl. Sci. 132, 39–71. 10.1016/bs.pmbts.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 34.Deretic D, and Wang J (2012). Molecular assemblies that control rhodopsin transport to the cilia. Vision Res. 75, 5–10. 10.1016/j.visres.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsiao YC, Tuz K, and Ferland RJ (2012). Trafficking in and to the primary cilium. Cilia 1, 4. 10.1186/2046-2530-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Morita Y, Mazelova J, and Deretic D (2012). The Arf GAP ASAP1 provides a platform to regulate Arf4- and Rab11-Rab8-mediated ciliary receptor targeting. EMBO J. 31, 4057–4071. 10.1038/emboj.2012.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schou KB, Pedersen LB, and Christensen ST (2015). Ins and outs of GPCR signaling in primary cilia. EMBO Rep. 16, 1099–1113. 10.15252/embr.201540530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wheway G, Nazlamova L, and Hancock JT (2018). Signaling through the Primary Cilium. Front. Cell Dev. Biol. 6, 8. 10.3389/fcell.2018.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Händel M, Schulz S, Stanarius A, Schreff M, Erdtmann-Vourliotis M, Schmidt H, Wolf G, and Höllt V (1999). Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience 89, 909–926. 10.1016/s0306-4522(98)00354-6. [DOI] [PubMed] [Google Scholar]
- 40.Hamon M, Doucet E, Lefévre K, Miquel MC, Lanfumey L, Insausti R, Frechilla D, Del Rio J, and Vergé D (1999). Antibodies and antisense oligonucleotide for probing the distribution and putative functions of central 5-HT6 receptors. Neuropsychopharmacology 21, 68S–76S. 10.1016/S0893-133X(99)00044-5. [DOI] [PubMed] [Google Scholar]
- 41.Loktev AV, and Jackson PK (2013). Neuropeptide Y family receptors traffic via the Bardet-Biedl syndrome pathway to signal in neuronal primary cilia. Cell Rep. 5, 1316–1329. 10.1016/j.celrep.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 42.Siljee JE, Wang Y, Bernard AA, Ersoy BA, Zhang S, Marley A, Von Zastrow M, Reiter JF, and Vaisse C (2018). Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat. Genet. 50, 180–185. 10.1038/s41588-017-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tereshko L, Gao Y, Cary BA, Turrigiano GG, and Sengupta P (2021). Ciliary neuropeptidergic signaling dynamically regulates excitatory synapses in postnatal neocortical pyramidal neurons. Elife 10, e65427. 10.7554/eLife.65427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheu SH, Upadhyayula S, Dupuy V, Pang S, Deng F, Wan J, Walpita D, Pasolli HA, Houser J, Sanchez-Martinez S, et al. (2022). A serotonergic axon-cilium synapse drives nuclear signaling to alter chromatin accessibility. Cell 185, 3390–3407.e18. 10.1016/j.cell.2022.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welsch L, Colantonio E, Falconnier C, Champagnol-DiLiberti C, Allain F, Ben Hamida S, Darcq E, Lutz PE, and Kieffer BL (2023). Mu Opioid Receptor-Positive Neurons in the Dorsal Raphe Nucleus Are Impaired by Morphine Abstinence. Biol. Psychiatry 94, 852–862. 10.1016/j.biopsych.2023.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bachmutsky I, Wei XP, Kish E, and Yackle K (2020). Opioids depress breathing through two small brainstem sites. Elife 9, e52694. 10.7554/eLife.52694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varga AG, Reid BT, Kieffer BL, and Levitt ES (2020). Differential impact of two critical respiratory centres in opioid-induced respiratory depression in awake mice. J. Physiol. 598, 189–205. 10.1113/JP278612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trafton JA, Abbadie C, Marek K, and Basbaum AI (2000). Postsynaptic signaling via the [mu]-opioid receptor: responses of dorsal horn neurons to exogenous opioids and noxious stimulation. J. Neurosci. 20, 8578–8584. 10.1523/JNEUROSCI.20-23-08578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams JT, Ingram SL, Henderson G, Chavkin C, von Zastrow M, Schulz S, Koch T, Evans CJ, and Christie MJ (2013). Regulation of mu-opioid receptors: desensitization, phosphorylation, internalization, and tolerance. Pharmacol. Rev. 65, 223–254. 10.1124/pr.112.005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Merrer J, Becker JAJ, Befort K, and Kieffer BL (2009). Reward processing by the opioid system in the brain. Physiol. Rev. 89, 1379–1412. 10.1152/physrev.00005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Norman RX, Ko HW, Huang V, Eun CM, Abler LL, Zhang Z, Sun X, and Eggenschwiler JT (2009). Tubby-like protein 3 (TULP3) regulates patterning in the mouse embryo through inhibition of Hedgehog signaling. Hum. Mol. Genet. 18, 1740–1754. 10.1093/hmg/ddp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hultquist JF, Schumann K, Woo JM, Manganaro L, McGregor MJ, Doudna J, Simon V, Krogan NJ, and Marson A (2016). A Cas9 Ribonucleoprotein Platform for Functional Genetic Studies of HIV-Host Interactions in Primary Human T Cells. Cell Rep. 17, 1438–1452. 10.1016/j.celrep.2016.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blythe EE, and von Zastrow M (2024). beta-Arrestin-independent endosomal cAMP signaling by a polypeptide hormone GPCR. Nat. Chem. Biol. 20, 323–332. 10.1038/s41589-023-01412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original and raw data are available upon request from the lead contact.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.


