Abstract
To test the hypothesis that changing neutralizing antibody responses against human immunodeficiency virus type 1 (HIV-1) during chronic infection were a response to emergence of neutralization escape mutants, we cloned expressed and characterized envelope clones from patients in the Multicenter AIDS Cohort Study (MACS). Pseudotyped HIV-1 envelope clones obtained from differing time points were assessed for sensitivity to neutralization by using sera from different times from the same and different patients. Clones from early and late time points during chronic infection had similar neutralization sensitivity, and neutralizing antibody responses cross-reacted with early, late, and heterologous envelopes. The potential for broadly effective HIV-1 immunization is supported.
The phenotypic evolution of lentiviruses is thought to be significant in disease pathogenesis. Mutations in the human immunodeficiency virus type 1 (HIV-1) envelope gene alter cellular host range and neutralization epitopes of the virus (3, 4, 6). A general, progressive broadening of the neutralizing antibody response after HIV-1 seroconversion is well documented (1, 16, 21, 24). Whether this broadening is a response to envelope mutations causing antigenic variation or a progressive response to antigenically stable, infecting virus is pertinent to strategies for broadly effective HIV-1 immunization. HIV-1 envelope mutants emerging through escape from neutralization in vivo or in vitro in the presence of sera from infected people have been described previously (15). Mutations in variable regions of the envelope which change specificity of interaction with antibodies have been observed during the early postseroconversion time period or under the selective pressure of monoclonal antibodies (8, 10, 11, 14, 28). Later during infection or under the selective pressure of polyclonal human serum, mutations have been observed at sites which are distant from neutralization epitopes but which, nevertheless, alter general sensitivity to neutralization (2, 17–20, 22, 23). Resistance to neutralization mediated by nonepitope mutations can result from mutations that alter gp120 conformation or insertional mutations which add glycosylation sites in the V2 and V4 regions of the envelope (2, 18–20, 22, 23, 29).
Previously, we reported a study demonstrating the evolution of the specificity of neutralizing antibodies in 10 homosexual men monitored over a 5-year period (21). Sera from each patient from multiple time points were tested for neutralization of nine different strains of HIV-1. Increasing neutralizing antibody titers against one or more of the virus strains developed in each patient, while in the same patients titers against other strains remained unchanged or declined. The participants included in the study were males who had enrolled in the Multicenter AIDS Cohort Study (MACS) in 1984, who were infected with HIV-1 at the time of their enrollment, and who had been continuously monitored approximately every 6 months since then (9, 21). The participants were also selected from the MACS cohort because their CD4+ cell counts were >400/mm3 at entry and they remained clinically well, with counts above 200/mm3, for 5 years of study. These characteristics indicated that these patients were likely to be in the postacute, early phase of chronic HIV-1 infection at the time they entered the study. Patients in the early stages of chronic HIV-1 infection are competent to develop antibody responses to viral vaccines and should be competent to develop similar responses to antigenically variant escape mutants during this period of infection (30). Neutralizing antibodies generally develop within 6 months of initial HIV infection, and responses to new antigenic variants in these patients may have developed in a similar time period (23). If the neutralizing antibody responses we had observed in this previous study were induced by emergence of antigenically variant escape mutants, we anticipated that these variants would have developed approximately during the 6-month interval before the responses occurred.
We hypothesized that the changes in neutralizing antibody specificity we had observed were induced by escape mutants with antigenically altered neutralization epitopes. To test this hypothesis in the present study, envelope genes from peripheral blood mononuclear cells (PBMC) from four of the same patients (patients 3, 4, 6 and 8 in the earlier study) were cloned, expressed on pseudoviruses, and characterized. These four patients were selected from among the 10 on the basis of increases in their neutralizing antibody titers that began more than 1 year after enrollment in the study. Plasma samples and PBMC collected from these patients during their first 5 years of participation in the MACS were used. The plasma samples that were used in this study were obtained at entry into the MACS and approximately at annual intervals thereafter (MACS visits 1, 3, 5, 7, 9, and 11). The cryopreserved PBMC were selected to correspond to the earliest PBMC samples available (early samples corresponded to either visit 1 or 2) or to the samples collected at the visit immediately preceding a visit at which increases in neutralizing antibody titers had been observed (late samples corresponded to either visit 3 or 4). The two PBMC samples from each individual were selected from samples collected at visits at least 1 year apart. Patient PBMC were cocultivated with normal human PBMC to obtain virus replication (13, 21). RNA was extracted from reverse transcriptase (RT)-positive cell culture fluids. The earliest culture fluid extracts which yielded positive results on RT-PCR were used as sources of genes for cloning. The env genes were cloned from DNA synthesized by RT-PCR as previously described (20, 21). The plasmids pNL4-3.Luc.E-R- (N. Landau, Aaron Diamond AIDS Research Center, the Rockefeller University) and pSV7d (P. Luciw, University of California, Davis, and R. L. Burke, Chiron Corp.) were used for envelope gene expression and pseudovirus construction (5, 25). The 293T cell line was used for transfections (Rockefeller Institute) (12). The HOS cell lines expressing CD4 and various coreceptors for HIV-1 (National Institutes of Health AIDS Research and Reference Reagent Program [ARRRP], provided by N. Landau) were used for infections and neutralization assays (25). Cloned genes were expressed on pseudoviruses by transfection of 293T cells in 24-well plates. Plasmids carrying inserts of appropriate size were screened for function by infection of HOS-CD4-CCR5 cells. Genes encoding envelopes which mediated virus entry sufficient to produce luminescence of ≥100 times background were selected for use in this study. The number of functional genes obtained from each PBMC culture varied from 4 to 11; approximately one-half of these yielded luminescence results in the screening assay of ≥100 times background. Of 676 plasmid clones which carried inserts of appropriate size that were screened for function, 52 (7.7%) were functional. Functional envelopes were tested in pseudovirus neutralization assays, as described previously (20, 21). The reference HIV-1 neutralizing sera 1 and 2 and the normal control serum were used as positive and negative controls in these assays (provided by L. Vujcic and G. Quinnan, ARRRP, catalog no. 1983, 1984, and 2411) (26, 27). Each neutralization assay was performed in triplicate, and each assay was conducted three times. The 90% inhibitory endpoints were calculated by a modified Reed-Munch method, as described previously (20, 21). This endpoint determination method yields good test-to-test consistency and titers similar to those obtained in conventional virus neutralization assays with the same virus strains (21). The cloning and expression of envelope genes from pNL4-3 and pNL(SF162) and patients 9 and 10, prepared in our earlier study, and methods for DNA sequencing have been previously described (21). The primers used for nucleotide sequencing encompassed a region beginning in C2 and extending into V4. Sequencing was conducted on both strands.
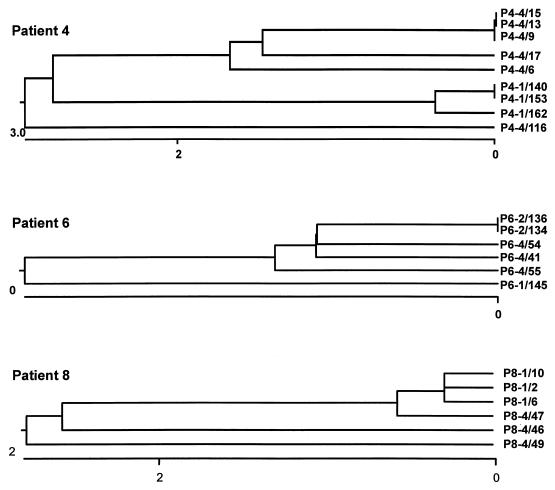
RT was detected in and functional envelope clones were obtained from the cultures of cells from the early samples from patients 4, 6, and 8 and from the late samples from all four patients. In each case the genes which produced the highest luminescence in the screening assay were selected for further study. Six envelope gene clones were selected from the late PBMC culture from patient 4, and three envelope clones were selected from each of the other samples. There was a high level of similarity among the late clones from patient 3 and from the early clones from each of patients 4, 6, and 8; the percentage of divergence in nucleotide sequence was 0.0 to 0.2, 0.0 to 0.2, 0.0 to 1.1, and 0.0 to 0.6 for the four sets of clones, respectively, calculated by the methods of Higgins and Sharp (7). All the late clones from patients 4, 6, and 8 differed from their respective early clones, varied among themselves somewhat more than the respective early clones, and in each case appeared to have evolved from a progenitor common to the early clones, as illustrated in Fig. 1. The percentages of divergence in comparisons of these sets of late clones to the early clones from the same patients were 1.7 to 2.5, 0.2 to 1.4, and 0.9 to 6.6, respectively. Thus, even though the early clones appeared to represent a highly homogeneous population of viruses in each case, the changes observed in the late clones appeared to represent the diversity that existed in the quasispecies mixture that preceded the early clones and did not demonstrate the emergence of dominant mutants from the populations represented by the early clones.
FIG. 1.
Phylogenetic relationships among env clones from patients 4, 6, and 8. The clones are designated P#-N/nn, where # indicates the patient number, N indicates the semiannual visit number, and nn indicates the clone number. The clones sequenced and their GenBank accession numbers are listed in the text. The results of analyses of the clones from patients 4, 6, and 8 are shown. The regions sequenced are described in the text. The graphs below each dendrogram indicate the percentage of divergence. Sequences were assembled and analyzed by CLUSTAL analysis with the program DNAstar (7).
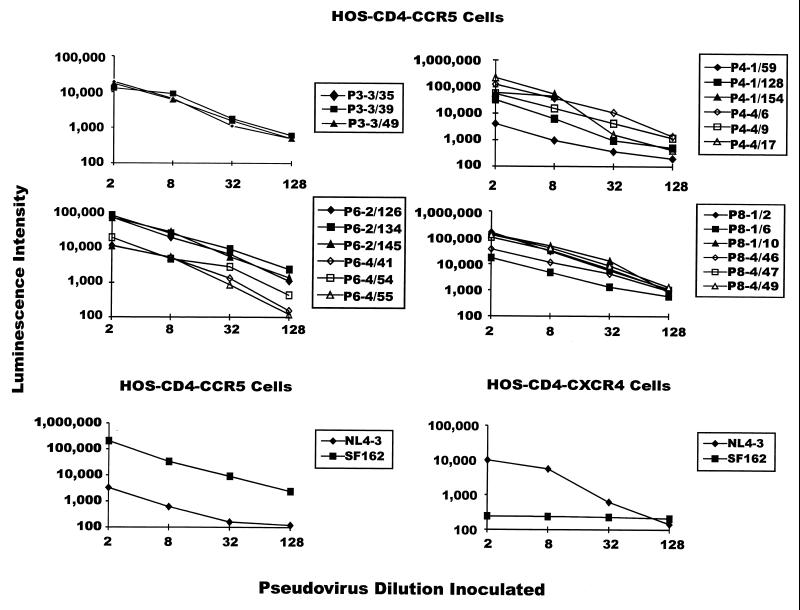
Pseudoviruses expressing each of the clones were evaluated for capacity to infect HOS-CD4 cells expressing either CCR5 or CXCR4, as exemplified by the results shown in Fig. 2. The three clones from visit 4, patient 4, which are not shown in the figure, behaved similarly to those which are shown. All of the pseudoviruses expressing envelope clones from the patients were infectious for HOS-CD4-CCR5 cells, as shown in the figure, but not for HOS-CD4-CXCR4 cells (data not shown), while the NL4-3 and SF162 pseudovirus controls were infectious for cells expressing CXCR4 and CCR5, respectively. NL4-3 pseudovirus was about 10-fold more infectious for cells expressing CXCR4 than CCR5, as expected.
FIG. 2.
Infectivity of pseudoviruses expressing primary envelopes from study patients for HOS-CD4 cells expressing CCR5 or CXCR4, as reflected in luciferase activity in infected cells. Designations take the form of P#-N/nn, indicating the patient number (#), visit number (N), and number (nn) of each clone. The NL4-3 and SF162 pseudoviruses have been previously described and were used as experimental infectivity controls (21).
Pseudoviruses expressing each of these envelopes were tested for neutralization by the HIV-1 neutralizing sera 1 and 2. More clones were neutralized by serum 2 than serum 1, as would be expected based on the known greater neutralizing cross-reactivity of the former. The clones from given samples from individual patients did not differ significantly among themselves in sensitivity to neutralization by these sera, and neither did early and late clones from patient 6 or 8 (data not shown). The late clones from patient 4 were similar to the early clones but were in three separate comparisons up to fourfold more resistant and averaged about twofold more resistant to neutralization than the early clones. These results may indicate a minor difference in neutralization sensitivity between the early and late clones from patient 4.
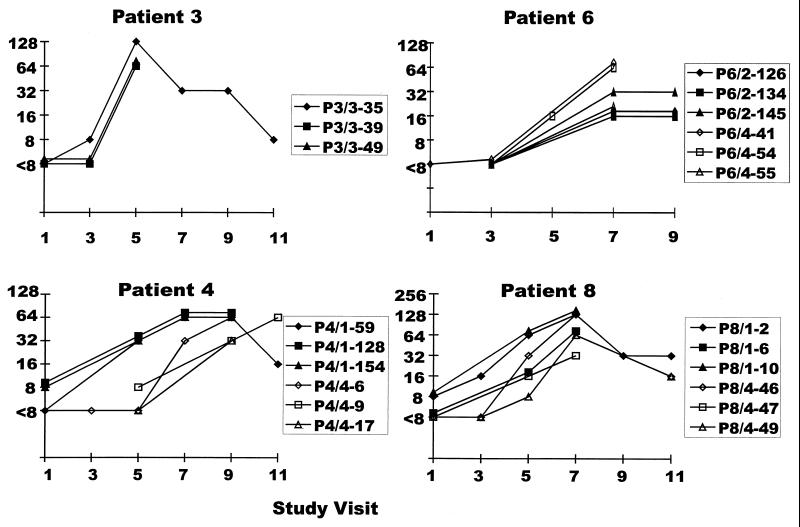
Neutralization of pseudoviruses expressing the different envelope genes by homologous sera from different time points is shown in Fig. 3. Each patient developed increases in neutralizing antibody titers against all homologous pseudoviruses tested, including the three early and three late clones from patient 4 which are not shown in the figure. The three clones from patient 3 were each sensitive to increases in neutralizing antibodies during the same time period, as were the early and late clones from patients 6 and 8. The three early clones from patient 4 detected increases in neutralizing activity at an earlier test date than the late clones. However, the neutralizing activity detected by these early clones continued to increase during the time in which increases were detected with the clones from visit 4. Thus, in each case the early and late clones were generally similar regarding the detection of neutralizing antibody responses occurring after the late clones were obtained.
FIG. 3.
Neutralization of pseudoviruses expressing HIV-1 envelopes from each study patient by sera from the same patients from which the envelopes were derived. These sera had been collected at approximately annual intervals since the patients’ enrollment in the MACS.
The extent of cross-reactivity of the neutralizing antibody responses detected was evaluated by testing sera from the different patients against pseudoviruses from the different patients, as shown in Table 1. For each patient, preresponse (year 1 or 2) and postresponse (year 3 or 4) sera were selected and tested for neutralization of pseudoviruses obtained from the other three patients. The neutralization by these sera of pseudoviruses from patients 9 and 10, which was described previously, is also shown for comparison (21). In the sera from patient 3 there was a 16-fold increase in titer against homologous pseudovirus and 4-, 4-, and 8-fold increases in titer against pseudoviruses from patients 4, 6, and 9, respectively. The changes against the other pseudoviruses tested were ≤twofold. In the sera from patient 4 there was a 16-fold increase in titer against homologous pseudovirus and 8-, 4-, 32-, and 4-fold increases against pseudoviruses from patients 3, 6, 8, and 10, respectively. In the sera from patient 6 there was a 32-fold increase in homologous neutralization, and increases of 8-, 16-, 8-, and 4-fold against pseudoviruses from patients 3, 4, 8, and 9, respectively. In the sera from patient 8 there was an 8-fold increase in homologous neutralization, and 4-, 32-, and 16-fold increases in neutralization titers against pseudoviruses from patients 4, 9, and 10, respectively. Thus, each of the patient’s responses cross-reacted with three or four of the heterologous pseudoviruses tested, and each of the pseudoviruses was recognized by the increase in antibody titers in two, three, or four of the patients.
TABLE 1.
Cross-reactivity of increasing HIV-1 neutralizing antibodies against heterologous primary pseudoviruses
| Pseudovirus clone(s) | Reciprocal neutralization titers of early/late sera from indicated patient
|
|||
|---|---|---|---|---|
| 3 | 4 | 6 | 8 | |
| P3-4/49 | 8/128 | <8/32 | <8/32 | 256/256 |
| P4-4/116 | 8/32 | <8/64 | <8/64 | 32/128 |
| P6-4/41 | 32/128 | <8/16 | <8/128 | 256/512 |
| P8-4/49 | 8/8 | <8/128 | <8/32 | 32/256 |
| P9-1 | <8/32 | 16/8 | <8/16 | 8/256 |
| P10-1 | 16/32 | 8/64 | <8/<8 | 16/256 |
The broadening of the neutralizing antibody response that characterizes chronic HIV-1 infection could result from the accumulation of responses to antigenically variant forms of virus and/or slow development of responses against stable epitopes (15, 16, 23). If the changes in neutralizing antibodies found in our patients were induced by antigenically variant epitopes resulting from escape mutation, early clones which lacked the variant epitopes would not have been recognized by the responses induced by the later variant clones. Levels of neutralizing activity against the early clones would have remained unchanged or decreased, while those against the late clones, expressing the epitope against which the response was directed, would have increased. Our finding that the early envelope clones were recognized by the neutralizing antibody responses just as well and, in some cases, earlier than the late clones obtained just before the responses, is inconsistent with the responses having been induced by new antigenically variant epitopes. Rather, the data are more consistent with the responses being directed against epitopes which are conserved throughout the duration of antecedent infection studied. The conserved nature of these epitopes is further evidenced by the cross-reactivity of each of the four patients’ responses against the majority of heterologous, primary virus envelope pseudotyped viruses tested.
While we did not find evidence that escape mutants with antigenically variant epitopes had emerged antecedent to the neutralizing antibody responses, the development of neutralization resistance as a result of mutations at sites distant from neutralization epitopes is well documented in work by our laboratory and others (20, 22, 23, 29). There is substantial concern that the use of envelope proteins of primary, neutralization-resistant viruses in vaccines may be necessary to induce neutralizing antibody responses that are broadly cross-reactive among primary viruses. The neutralizing antibody responses we describe here, using CCR5 tropic primary envelopes, were temporally coincident with those we have described previously in the same patients with laboratory-adapted T and M tropic strains of HIV-1 (21). Thus, these responses were directed at epitopes conserved among strains of varying tropism, laboratory passage history, and neutralization sensitivity. Definition of the cognate epitopes for these responses may reveal epitopes that could be presented in vaccines to achieve broadly effective immunity. If HIV can present its envelope to infected patients in such a way as to achieve these responses, it should be possible to present antigen to uninfected individuals and achieve similar responses.
Nucleotide sequence accession numbers.
The clones sequenced and their GenBank accession numbers (in parentheses) are as follows: for patient 3, P3-3/35 (AF130388) and P3-3/39 and P3-3/49 (AF130389); for patient 4, P4-1/140 and P4-1/153 (AF130390), P4-1/162 (AF130391), P4-4/6 (AF130395), P4-4/9, P4-4/13, and P4-4/15 (AF130393), P4-4/17 (AF130394), and P4-4/116 (AF130392); for patient 6, P6-2/126 (AF130396), P6-2/134 and P6-2/145 (AF130397), P6-4/41 (AF130398), P6-4/54 (AF130399), and P6-4/55 (AF130400); for patient 8, P8-1/2 (AF130402), P8-1/6 and P8-1/10 (AF130401), P8-4/46 (AF130403), P8-4/47 (AF130404), and P8-4/49 (AF130405).
Acknowledgments
This work was supported by USUHS grant R087EZ and NIH grant RO1-AI37438.
REFERENCES
- 1.Arendrup M, Sonnerborg A, Svennerholm B, Akerblom L, Nielsen C, Clausen H, Olofsson S, Nielsen J O, Hansen J E. Neutralizing antibody response during human immunodeficiency virus type 1 infection: type and group specificity and viral escape. J Gen Virol. 1993;74:855–863. doi: 10.1099/0022-1317-74-5-855. [DOI] [PubMed] [Google Scholar]
- 2.Back N K T, Smit L, Schutten M, Nara P L, Tersmette M, Goudsmit J. Mutations in human immunodeficiency virus type 1 gp41 affect sensitivity to neutralization by gp120 antibodies. J Virol. 1993;67:6897–6902. doi: 10.1128/jvi.67.11.6897-6902.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergeron L, Sullivan N, Sodroski J. Target cell-specific determinants of membrane fusion within the human immunodeficiency virus type 1 gp120 third variable region and gp41 amino terminus. J Virol. 1992;66:2389–2397. doi: 10.1128/jvi.66.4.2389-2397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N L. Identification of a major co-receptor for primary isolates of HIV. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 6.Haigwood N L, Shuster J R, Moore G K, Lee H, Skiles P V, Higgins K W, Barr P J, George-Nascimento C, Steimer K S. Importance of hypervariable regions of HIV-1 gp120 in the generation of virus neutralizing antibodies. AIDS Res Hum Retroviruses. 1990;6:855–869. doi: 10.1089/aid.1990.6.855. [DOI] [PubMed] [Google Scholar]
- 7.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. CABIOS. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 8.Ho D D, Fung M S C, Cao Y, Li X L, Sun C, Chang T W. Another discontinuous epitope on glycoprotein gp120 that is important in human immunodeficiency virus type 1 neutralization is identified by a monoclonal antibody. Proc Natl Acad Sci USA. 1991;88:8949–8952. doi: 10.1073/pnas.88.20.8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaslow R A, Ostrow D G, Detels R, Phair J P, Polk B F, Rinaldo C R., Jr The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. Am J Epidemiol. 1987;126:310–318. doi: 10.1093/aje/126.2.310. [DOI] [PubMed] [Google Scholar]
- 10.Korber B, Wolinsky S, Haynes B, Kunstman K, Levy R, Furtado M, Otto P, Myers G. HIV-1 intrapatient sequence diversity in the immunogenic V3 region. AIDS Res Hum Retroviruses. 1992;8:1461–1465. doi: 10.1089/aid.1992.8.1461. [DOI] [PubMed] [Google Scholar]
- 11.Langedijk J P, Zwart G, Goudsmit J, Meloen R H. Fine specificity of antibody recognition may predict amino acid substitution in the third variable region of gp120 during HIV type 1 infection. AIDS Res Hum Retroviruses. 1995;11:1153–1162. doi: 10.1089/aid.1995.11.1153. [DOI] [PubMed] [Google Scholar]
- 12.Liou H C, Sha W C, Scott M L, Baltimore D. Sequential induction of NF-kappa B/Rel family proteins during B-cell terminal differentiation. Mol Cell Biol. 1994;14:5349–5359. doi: 10.1128/mcb.14.8.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mascola J, Louwagie J, McCutchan F E, Fischer C L, Hegerich P A, Wagner K F, Fowler A K, McNeil J G, Burke D S. Two antigenically distinct subtypes of human immunodeficiency virus type 1: viral genotype predicts neutralization serotype. J Infect Dis. 1994;169:48–54. doi: 10.1093/infdis/169.1.48. [DOI] [PubMed] [Google Scholar]
- 14.McKeating J A, Giw J, Goudsmit J, Pearl L H, Mulder C, Weiss R A. Characterization of HIV-1 neutralization escape mutants. AIDS. 1989;3:777–784. doi: 10.1097/00002030-198912000-00001. [DOI] [PubMed] [Google Scholar]
- 15.McKnight A, Clapham P R. Immune escape and tropism of HIV. Trends Microbiol. 1995;3:356–361. doi: 10.1016/s0966-842x(00)88975-1. [DOI] [PubMed] [Google Scholar]
- 16.McKnight A, Clapham P R, Goudsmit J, Cheingsong-Popov R, Weber J N, Weiss R A. Development of HIV-1 group-specific neutralizing antibodies after seroconversion. AIDS. 1992;6:799–802. doi: 10.1097/00002030-199208000-00005. [DOI] [PubMed] [Google Scholar]
- 17.Nara P L, Smit L, Dunlop N, Hatch W, Merges M, Waters D, Kelliher J, Gallo R C, Fischinger P J, Goudsmit J. Emergence of viruses resistant to neutralization by V3-specific antibodies in experimental human immunodeficiency virus type 1 IIIB infection of chimpanzees. J Virol. 1990;64:3779–3791. doi: 10.1128/jvi.64.8.3779-3791.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Brien W A, Chen I S Y, Ho D D, Daar E S. Mapping genetic determinants for human immunodeficiency virus type 1 resistance to soluble CD4. J Virol. 1992;66:3125–3130. doi: 10.1128/jvi.66.5.3125-3130.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Overbaugh J, Rudensey L M. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J Virol. 1992;66:5937–5948. doi: 10.1128/jvi.66.10.5937-5948.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park E J, Vujcic L K, Anand R, Theodore T S, Quinnan G V. Mutations in both gp120 and gp41 are responsible for the broad neutralization resistance of variant HIV-1 MN to antibodies directed at V3 and non-V3 epitopes. J Virol. 1998;72:7099–7107. doi: 10.1128/jvi.72.9.7099-7107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinnan G V, Zhang P F, Fu D W, Dong M, Margolick J. Episodic evolution of neutralizing antibody response in patients with HIV-1 infection. AIDS Res Hum Retroviruses. 1998;14:939–949. doi: 10.1089/aid.1998.14.939. [DOI] [PubMed] [Google Scholar]
- 22.Reitz M S, Wilson C, Naugle C, Gallo R C, Robert-Guroff M. Generation of a neutralization resistant variant of HIV-1 is due to selection for a point mutation in the envelope gene. Cell. 1988;54:57–63. doi: 10.1016/0092-8674(88)90179-1. [DOI] [PubMed] [Google Scholar]
- 23.Robert-Guroff M, Reitz M S, Robey W G, Gallo R C. In vitro generation of an HTLV-III variant by neutralizing antibody. J Immunol. 1986;137:3306–3309. [PubMed] [Google Scholar]
- 24.Sawyer L A, Katzenstein D A, Hendry R M, Boone E J, Vujcic L K, Williams C C, Zeger S L, Saah A J, Rinaldo C R, Jr, Phair J P, Giorgi J V, Quinnan G V., Jr Possible beneficial effects of neutralizing antibodies and antibody-dependent cell-mediated cytotoxicity in human immunodeficiency virus infection. AIDS Res Hum Retroviruses. 1990;6:341–356. doi: 10.1089/aid.1990.6.341. [DOI] [PubMed] [Google Scholar]
- 25.Stuve L L, Brown-Shimer S, Pachl C, Naharian R, Diaz D, Burke R L. Structure and expression of the herpes simplex virus type 2 glycoprotein gB gene. J Virol. 1987;61:326–335. doi: 10.1128/jvi.61.2.326-335.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vujcic L K, Quinnan G V., Jr Preparation and characterization of human HIV type 1 neutralizing reference sera. AIDS Res Hum Retroviruses. 1995;11:783–787. doi: 10.1089/aid.1995.11.783. [DOI] [PubMed] [Google Scholar]
- 27.Vujcic L K, Shepp D H, Klutch M, Wells M A, Hendry R M, Wittek A E, Krilov L, Quinnan G V. Use of a sensitive neutralization assay to measure the prevalence of antibodies to the human immunodeficiency virus. J Infect Dis. 1988;157:1047–1050. doi: 10.1093/infdis/157.5.1047. [DOI] [PubMed] [Google Scholar]
- 28.Wahlberg J, Albert J, Lundeberg J, Von Gegerfelt A, Broliden K, Utter G, Fenyo E M, Uhlen M. Analysis of the V3 loop in neutralization-resistant human immunodeficiency virus type 1 variants by direct solid-phase DNA sequencing. AIDS Res Hum Retroviruses. 1991;7:983–990. doi: 10.1089/aid.1991.7.983. [DOI] [PubMed] [Google Scholar]
- 29.Watkins B A, Buge S, Aldrich K, Davis A E, Robinson J, Reitz M S, Jr, Robert-Guroff M. Resistance of human immunodeficiency virus type 1 to neutralization by natural antisera occurs through single amino acid substitutions that cause changes in antibody binding at multiple sites. J Virol. 1996;70:8431–8437. doi: 10.1128/jvi.70.12.8431-8437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson J C, Hadler S C, Dykewicz C A, Reef S, Phillips L. Measles, mumps, and rubella—vaccine use and strategies for elimination of measles, rubella, and congenital rubella syndrome and control of mumps: recommendations of the Advisory Committee on Immunization Practices (ACIP) Morbid Mortal Weekly Rep. 1998;47(RR-8):1–57. [PubMed] [Google Scholar]