Abstract
Tagmentation combines DNA fragmentation and sequencing adapter addition by leveraging the transposition activity of the bacterial cut-and-paste Tn5 transposase, to enable efficient sequencing library preparation. Here we present an open-source protocol for the generation of multi-purpose hyperactive Tn5 transposase, including its benchmarking in CUT&Tag, bulk and single-cell ATAC-seq. The OpenTn5 protocol yields multi-milligram quantities of pG-Tn5E54K, L372P protein per liter of E. coli culture, sufficient for thousands of tagmentation reactions and the enzyme retains activity in storage for more than a year.
Introduction
Tn5 is a bacterial cut-and-paste transposase, which can simultaneously fragment target DNA and append its ends with custom DNA adapter sequences in a process referred to as tagmentation[1] (Fig. 1A). In the reaction, Tn5 homodimer first binds two double-stranded DNA molecules encoding the mosaic end sequence[2], referred to as MEDS adapters[3], to form a transposome competent for target DNA binding. In the presence of Mg2+, the transposome-target synapse complex then performs a strand transfer reaction, in which the 3’OH groups of the MEDS adapters perform a nucleophilic attack on the target DNA backbone offset by 9 base pairs. The resulting target DNA ends are now appended with adapter sequences and contain 9 bprecessed 3’OH ends. This simultaneous target DNA cleavage and adapter attachment activity of Tn5 is referred to as tagmentation. Through decades of research pioneered by William S. Reznikoff and colleagues, many gain-of-function Tn5 mutations have been isolated that greatly increase the rate of Tn5 transposition, and are commonly referred to as hyperactive mutants[1]. Two of these, E54K[4] and L372P[5], are sufficient for a dramatic increase in activity and are the basal hyperactive mutations historically used in tagmentation applications. Subsequent studies have also investigated the effects of additional Tn5 mutations on further augmentation of tagmentation activity[6], as well as altering target DNA sequence insertion bias[7–9].
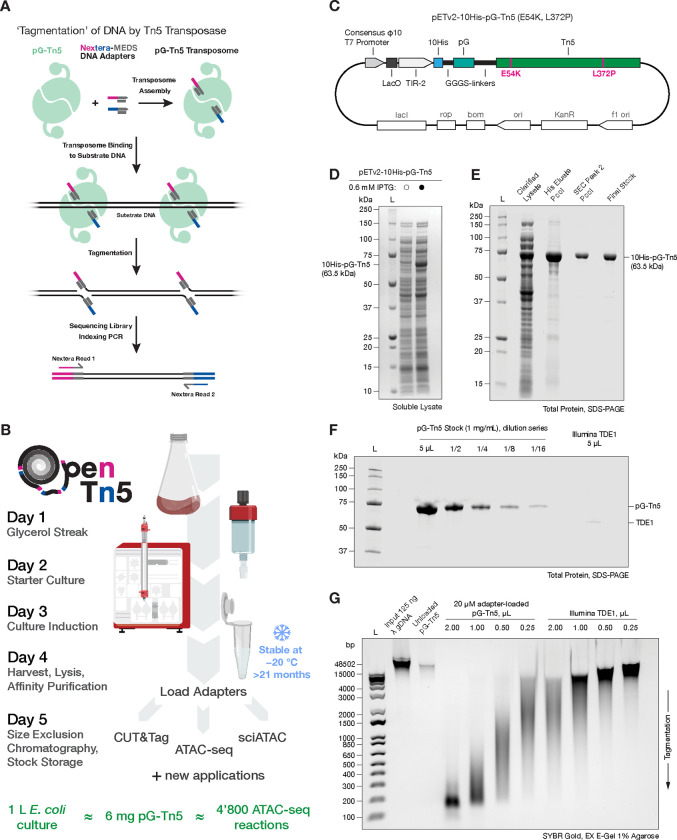
Figure 1: OpenTn5 enables robust, scalable, open-source Tn5 transposase purification for multiple applications.
A. Diagram of simultaneous DNA fragmentation and addition of DNA sequencing adapters in vitro by Tn5 transposition reaction, referred to as tagmentation. B. Expression and purification of pG-Tn5 can be carried out within 5 days and yields multi-milligram amounts of enzyme, sufficient for thousands of tagmentation reactions. Less than 1.25 μg of pG-Tn5 is required per 50’000 cell input ATAC-seq reaction. C. Schematic of 10His-pG-Tn5(E54K, L372P) expression plasmid, VR124 (Addgene #198468). D. Robust over-expression of soluble 10His-pG-Tn5(E54K, L372P) protein. E. His-tag affinity and size exclusion chromatography (SEC) enable efficient purification of pG-Tn5 homodimer. F. Comparison of pG-Tn5 adapter unloaded stock at 1 mg/mL with commercial Illumina TDE1 enzyme (Cat #20034198). G. Comparison of tagmentation activity of pG-Tn5 (1 mg/mL) and Illumina TDE1 using equal transposome volumes on Lambda genomic DNA (λ gDNA).
Tagmentation has been leveraged for streamlined preparation of high-throughput DNA sequencing libraries from sample DNA using custom sequencing adapter sequences. The efficiency and versatility of Tn5 tagmentation has given rise to a variety of genomics technologies for investigating chromatin organization, transcription and cell identity, with innovations continuing to be made in novel Tn5 protein fusions as well as custom MEDS adapter designs[10–19]. Despite the accomplishments of tagmentation-based methods to date, in-house production of high-quality Tn5 enzyme remains a persistent challenge[10] which hinders both ongoing efforts in the development of novel tagmentation applications and the equitable, broad adoption of existing methods.
Here we present our current integration of optimizations into a “start-to-finish”, open-source protocol for the generation of a robust, all-purpose hyperactive Tn5 transposase, including validation across multiple sequencing-based methods that rely on transposition[9, 20, 21]. We demonstrate the functional equivalence of the produced pG-Tn5E54K, L372P to commercially-sourced Tn5 used for CUT&Tag, but importantly we show the same enzyme can also be readily used for assays not requiring protein G for antibody-directed recruitment of Tn5, such as bulk and single-cell ATAC-seq, underscoring its versatility. Notably, pG-Tn5E54K, L372P can be readily loaded with custom DNA adapters. We hope that this OpenTn5 protocol will further disseminate existing applications of tagmentation, as well as facilitate future method development efforts.
Results
We sought to develop a robust protocol for production of high quality, hyperactive Tn5 which could be reproducibly carried out using generally available molecular biology laboratory equipment. To this end, the OpenTn5 protocol can be carried out within one work week (Fig. 1B). At 1x scale, the protocol uses 500 mL E. coli culture to yield roughly 3 milligrams of final pure pG-Tn5E54K, L372P homodimer stock (Fig. S1A–F), which is stable for > 21 months at −20 °C (Fig. S1G–H). This amount is sufficient for several thousand reactions, e.g. one 50’000 cell input ATAC-seq reaction consumes 1.25 μg of pG-Tn5E54K, L372P.The transposase stock is stored unloaded, and adapter loading is carried out at time of use, ensuring stock stability and allowing versatility for different tagmentation applications.
We provide a detailed step-by-step protocol, including an assay for measuring Tn5 tagmentation activity as supplementary materials. The expression plasmid is available from Addgene in two E. coli strains DH5α (#198467) and NEB T7 Express lysY/Iq (#198468), for plasmid propagation and protein expression, respectively.
E. coli expression, purification and storage of 10His-pG-Tn5E54K, L372P
We began by focusing on optimizing the expression and purification of the 10His-pG-Tn5E54K, L372P construct previously described by Xu and colleagues[21], which showed good over-expression in E. coli. Xu and colleagues demonstrated that Tn5 does not tolerate C-terminal tagging[21], likely due to the sensitivity of the nearby dimerization domain that is critical for transposition[22]. We note that the pTXB1-derived Tn5 expression plasmids[3] employ a C-terminal intein tag, which may contribute to the difficulty in obtaining active Tn5 protein using these constructs. The N-terminal 10His-protein G tag we use serves to promote solubility, facilitates straightforward affinity purification, and can be used for antibody-mediated Tn5 recruitment in CUT&Tag. We cloned an E. coli codon-optimized 10His-pG-Tn5E54K, L372P protein coding sequence into an optimized derivative of IPTG-inducible pET28a protein expression vector described by Shilling and colleagues[20] (Fig. 1C). We refer to this expression plasmid as pETv2–10His-pG-Tn5 E54K, L372P.
The resulting construct showed robust IPTG-dependent over-expression of soluble 10His-pG-Tn5 at 18 °C when introduced into the NEB T7 Express lysY/Iq E. coli strain. Notably, we did not encounter problems with plasmid toxicity in E. coli neither during cloning in the NEB 5-alpha strain nor during protein expression. Crucially, the majority of the over-expressed protein reliably remained in the soluble lysate fraction (Fig. 1D). We optimized E. coli culture conditions; using richer Terrific Broth (TB) instead of Lysogeny Broth (LB) media[23], supplemented with an anti-foam agent in double-baffled 2 L flasks to maximize the culture aeration. These changes further improve the soluble expression yield independently of construct design, and maximize the E. coli biomass harvested, reducing culture volumes required to be handled.
We use a standard sonication lysis without adding exogenous nucleases to minimize the risk of their carryover throughout and adapted a standard immobilized metal affinity chromatography (IMAC), specifically Ni-NTA resin affinity purification without subsequent cleavage of the 10His-pG tag, which streamlines the protocol. We extensively wash the resin-bound Tn5 using a high salt buffer to minimize non-specific binding of E. coli nucleic acid contaminants to Tn5 (Fig. S1A). We do not routinely perform any additional nucleic acid removal during the purification, such as polyethylenimine (PEI) precipitation, ion or heparin exchange, as in our experience the trace amounts of E. coli genomic DNA impurities in the final Tn5 stock do not significantly interfere in typical sequencing assay applications (Fig. S2B).
We polish the Ni-NTA column eluate using size exclusion chromatography (SEC), following the protocol developed by Hennig and colleagues[9] (Fig. 1E). We confirmed that SEC successfully separates full-length 10His-pG-Tn5 homodimers away from Tn5 aggregates, heterodimers containing truncated Tn5 and spuriously cleaved 10His-pG tag (Fig. S1B). We pool the SEC fractions corresponding to the 10His-pG-Tn5 homodimer peak and concentrate them to a concentration of 2 mg/mL in the SEC buffer. We prepare the final Tn5 stock by adding an equal volume of pure glycerol to the concentrated protein (1 mg/mL protein in 55% glycerol, final) and store the liquid stock at −20 °C in enzyme cooler blocks. We see a negligible loss of 10His-pG-Tn5 activity after almost 2 years of storage under these conditions (21 months, (Fig. S1G)). We do not load Tn5 with MEDS DNA adapters prior to storage, as we have found the assembled transposomes to be unstable. This is likely due to the change in the effective final buffer composition resulting from diluting the protein with adapters resuspended in a low-salt buffer.
We believe the SEC step, often omitted from many existing Tn5 purification protocols, is likely critical to the long-term stability of Tn5 as the aggregates present in the final protein glycerol stock may seed further aggregation and thereby lead to enzyme deterioration[24]. Additionally, SEC purification is likely important for reproducible performance in antibody-based tagmentation assays in which an excess of 10His-pG proteolytic degradation fragments may significantly compete with 10His-pG-Tn5 for antibody binding, thereby reducing assay sensitivity.
Benchmarking 10His-pG-Tn5 E54K, L372P in vitro activity
We compared the protein concentrations of our 1 mg/mL pG-Tn5 stock against commercially available Illumina TDE1 enzyme (CAT: #20034198, LOT: 20541598), provided at an unspecified concentration. We loaded equal volumes (5 μL) of both enzymes, alongside a dilution series of the adapter-unloaded 1 mg/mL pG-Tn5 stock and measured total protein content by SDS-PAGE followed by Coomassie staining (Fig. 1F). Illumina TDE1 protein concentration was roughly 16-fold lower than that of the 1 mg/mL pG-Tn5 stock, giving an estimate of 62.5 μg/mL protein concentration of the adapter pre-loaded commercial TDE1 enzyme.
To measure the tagmentation activity of Tn5, we use commercially available Lambda phage genomic DNA (λ gDNA) as substrate. λ gDNA serves as a reproducible assay substrate; it is supplied as double-stranded, high-molecular weight (48.5 kb) DNA. The large size of λ gDNA allows for a sensitive, yet simple readout of tagmentation activity by gel electrophoresis (Fig. 1G, Fig. S1G). We load the pG-Tn5 stock with an equal volume of 20 μM Nextera MEDS adapters, corresponding to a modest stoichiometric excess relative to Tn5, to ensure saturated adapter loading (see Supplementary Protocol for details on adapter loading and tagmentation assay). The resulting adapter- loaded pG-Tn5 transposome protein concentration is half that of unloaded stock (0.5 mg/mL). As noted above, we load pG-Tn5 with adapters on a single-use basis to maintain long-term activity in storage.
We compared the tagmentation activity of matched-volume serial dilutions of pG-Tn5 and TDE1 transposomes using the Lambda assay, which showed that 0.5 mg/mL pG-Tn5 transposome has an approximately 8-fold higher tagmentation activity than Illumina TDE1, on volume-to-volume basis (Fig. 1G). This 8-fold difference is therefore within the magnitude expected based on the 16-fold difference in Tn5 protein stock concentration, as determined by SDS-PAGE (Fig. 1F), followed by a two-fold pG-Tn5 dilution upon adapter loading.
CUT&Tag
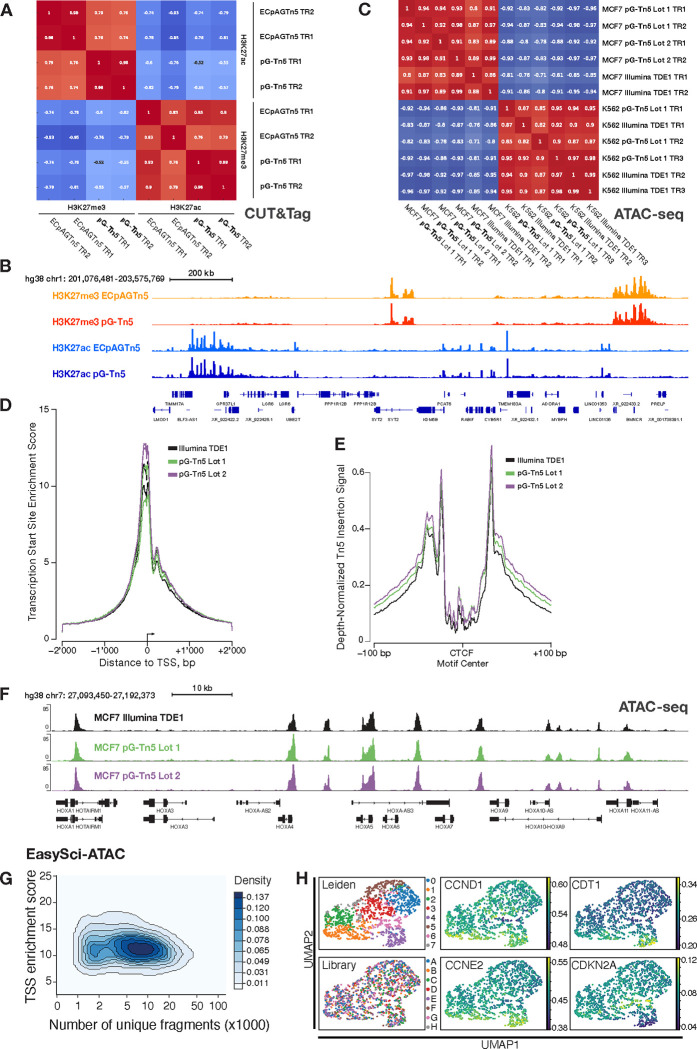
To determine if the performance of in-house pG-Tn5 is comparable to commercial pAG-Tn5 from Epicypher (SKU: 15–1017), we performed CUT&Tag using both enzymes to profile the distribution of H3K27me3 and H3K27ac histone modifications in MCF7 cells. Two replicates per condition were generated to assess reproducibility. After confirming that the peaks called were highly reproducible across replicate and enzyme, Spearman correlation coefficients were calculated on the rlog normalized counts matrix. The sample correlation matrix shows that samples cluster by histone modifications and different enzymes for the same histone modification clustered with each other (Fig. 2A). The correlation coefficient within each histone modification was between 0.75–1.0. Genome wide coverage tracks also show a similar pattern of distribution across different Tn5 enzymes for each histone modification (Fig. 2B).
Figure 2: Application and validation of pG-Tn5 in CUT&Tag, ATAC-seq and EasySci-ATAC assays.
A. Spearman rank correlation of anti-H3K27me3 and anti-H3K27ac CUT&Tag signal under common peaks generated using pG-Tn5 and Epicypher pAG-Tn5 (ECpAGTn5) enzyme (Cat #15–1017) in human MCF7 cells. B. Genome track visualization of combined two technical replicates of above conditions, as in panel A. H3K27me3 and H3K27ac signal from pG-Tn5 and pAG-Tn5 experiments plotted on same sequencing-depth scaled y-axes, respectively. C. Spearman rank correlation of signal under common peaks, generated using ATAC-seq protocol generated libraries from human K562 and MCF7 cells using pG-Tn5 and Illumina TDE1, using matched transposome volumes. D. Transcription start site enrichment score of ATAC-seq signal in human MCF7 cells, comparison of two purification lots of pG-Tn5 and Illumina TDE1 enzyme, two technical replicates each. E. Sequencing-depth normalized Tn5 insertion signal around occupied CTCF sites in human MCF7 cells for ATAC-seq signal from pG-Tn5 and Illumina TDE1, as above in panel D. F. Genome track visualization of ATAC-seq signal from pG-Tn5 and Illumina TDE1 generated libraries, combined two technical replicates of above conditions, as in panel D and E. G. Single-cell accessible chromatin mapping using pG-Tn5 loaded with six unique barcoded adapter combinations, using the EasySci-ATAC protocol in human MCF7 cells. TSS enrichment scores for 2’672 single cells, majority of the cells showing score >10. H. Leiden clustering annotations (0–7) of EasySci-ATAC data from cycling human MCF7 cells from eight library replicates (A-H), showing no library-based bias in clustering. Signal values from several cell cycle-associated genes is shown.
ATAC-seq
To compare the performance of pG-Tn5 with commercially available Tn5 (Illumina TDE1, CAT: #20034198), we conducted ATAC-seq experiments in MCF7 and K562 human cell lines with matched enzyme volumes. Both enzymes generated high-quality ATAC-seq libraries with similar performance (Supplementary Table 1, Fig. S2A–D). Libraries from the same cell type were highly correlated by peaks and show highly similar genomic coverage (Fig. 2C, F). TSS Enrichment Scores, commonly used as a signal-to-noise QC metric, were comparable across replicates (Fig. 2D). In MCF7 cells, the average TSS scores were 10.9 with Illumina TDE1 and 11.5 with pG-Tn5 (Fig. 2D). In K562 cells, the scores were 12.7 with Illumina TDE1 and 11.7 with pG-Tn5 (Fig. S2C). CTCF site insertion and coverage profiles were highly similar, indicating that protein G does not appreciably sterically interfere with Tn5 transposition, making pG-Tn5 suitable for investigating regulatory landscapes(Fig. 2E, Fig. S2D).
We performed a pG-Tn5 transposome titration in K562 cells to understand how varying pG-Tn5 enzyme concentration influences ATAC-seq signal globally, over 1 kb-sized genomic bins (see Methods). We found samples tagmented with 2.5, 5.0 and 7.5 μL of pG-Tn5 were most correlated, with samples made with 1 μL pG-Tn5 to be most correlated with Illumina TDE1 (Fig. S2A). TSS scores remained high even when using as little as 1 μL of pG-Tn5 S2C).
EasySci-ATAC
Cycling MCF7 cells were barcoded with six separately loaded Tn5 N5/N7 combinations as part of the EasySci-ATAC protocol[25], which requires custom loading of adapters. We profiled the chromatin accessibility of 2’672 single cells passing quality control filters for read depth (>1’000) and overall alignment (>80% reads mapped). The majority of cells had TSS enrichment scores >10 with coverage between 5’000–10’000 fragments (Fig. 2G).
The SnapATAC2[26] framework was used for analysis, applying the Leiden community detection algorithm to identify subclusters (see Methods). As the underlying data is derived from individual cells taken from a single flask of cycling MCF7 breast cancer cells, we expected to find minimal separation of clusters, consistent with what we observed (Fig. 2H). Using Leiden annotations as a means to partition cells, we generated pseudobulk libraries to compare standard quality control metrics with bulk libraries; we observed each Leidenbased pseudobulk dataset had the same characteristic fragment length distributions and TSS enrichment profiles as seen for bulk ATAC libraries (Fig. 2E,F). Cells profiled across eight different PCR libraries distributed evenly across the landscape, revealing no library-based bias. Although generally cell-specific chromatin accessibility was not variable enough to drive distinct clustering, imputed gene expression values based on gene body accessibility exhibited distinct patterning among select cell cycle progression genes (Cyclin D1, CCND1), (Cyclin E2, CCNE2), (Chromatin Licensing and DNA Replication Factor 1, CDT1, which was anti-correlated with CDKN2A, the gene encoding the endogenous CDK inhibitor p16 (Fig. 2H).
Discussion
OpenTn5 offers a robust and scalable solution for the production of Tn5 transposase, addressing key challenges in enzyme expression, purification, and storage. Our optimized, open-source protocol synthesizes the existing knowledge base into a robust protocol that yields multi-milligram quantities of soluble, long-term stable 10His-pG-Tn5E54K, L372P per liter of E. coli culture. The produced Tn5 demonstrates functional equivalence to commonly used commercial enzymes, applied both in techniques which use antibody recruitment e.g. CUT&Tag, and those which do not, such as ATAC-seq. We demonstrated pG-Tn5 can be readily applied to sensitive emerging applications that require custom adapter loading, such as EasySci-ATAC[25], underlining its versatility. We hope that the OpenTn5 protocol provides a robust foundation for facilitating further developments of tagmentation, such as leveraging the development of novel Tn5 mutants, functionalization with custom protein tags, as well as DNA adapter design for emerging DNA sequencing platforms.
Methods
Expression, Purification, and Storage of 10His-pG-Tn5E54K L372P
VR124 pETv2–10His-pG-Tn5 (E54K, L372P) E. coli strain (Addgene #198468) glycerol stock was streaked onto LB agar plates containing 50 μg/mL kanamycin and incubated overnight at 37 °C. A single colony was picked and inoculated into 25 mL TB media supplemented with kanamycin (50 μg/mL), and the culture was grown overnight at 37 °C with shaking at 300–350 rpm. The next morning, the culture was scaled up to 500 mL TB media in a 2 L double-baffled flask, supplemented with kanamycin (50 μg/mL) and antifoam-204. The culture was grown at 37 °C until OD600 reached approximately 1.0. IPTG was then added to a final concentration of 0.6 mM to induce protein expression, and the culture was incubated overnight at 18 °C with shaking. Cells were harvested by centrifugation at 8,000 ×g for 30 minutes at 4 °C, subsequently the pellets were washed by resuspension with ice-cold DPBS, pelleted and frozen at −80 °C. For lysis, the frozen cell pellets were resuspended on ice in ice-cold lysis buffer LysEQ (20 mM HEPES pH 7.8, 800 mM NaCl, 20 mM Imidazole, 10% Glycerol, 1 mM EDTA, 2 mM TCEP-HCl, 1x EDTA-free cOmplete protease inhibitor) and lysed using sonication in a salt ice bath. The lysate was clarified by centrifugation at 20,000 ×g for 35 minutes at 3 °C and filtered through a 0.45 μm PES syringe filter. The cleared lysate was applied to a HisTrap HP 5 mL column (Cytiva) pre-equilibrated with buffer LysEQ , and the column was washed with buffer WashB1 (20 mM HEPES pH 7.8, 800 mM NaCl, 30 mM Imidazole, 10% Glycerol, 1 mM EDTA, 2 mM TCEP-HCl) and buffer WashB2 (20 mM HEPES pH 7.8, 800 mM NaCl, 45 mM Imidazole, 10% Glycerol, 1 mM EDTA, 2 mM TCEP-HCl) before elution with elution buffer EluB (20 mM HEPES pH 7.8, 800 mM NaCl, 400 mM Imidazole, 10% Glycerol, 1 mM EDTA, 2 mM TCEP-HCl). The eluted protein fractions were analyzed by SDS-PAGE, and the fractions containing 10His-pG-Tn5 were pooled and dialyzed overnight against SECB buffer (50 mM Tris-HCl pH 7.5, 800 mM NaCl, 10% glycerol, 0.2 mM EDTA, 2 mM DTT) in preparation for size exclusion chromatography (SEC). The following morning, the dialyzed protein was filtered through a 0.45 μm PES syringe filter. SEC was carried out on a ÄKTA pure™ FPLC system using a HiLoad 16/600 Superdex 200 pg column (Cytiva). 5 mL of filtered dialyzed protein was injected manually onto the pre-equilibrated column, followed by chromatography with 1.5 column volumes of buffer SECB at a flow rate of 1 mL/min, collecting 0.8 mL fractions from 40 mL to 110 mL elution volumes in a 96 deep-well plate. Peak fractions were analyzed by SDS-PAGE, fractions corresponding to the pure homodimer peak at 71.6 mL were pooled, and concentrated using a using a 30 MWCO Amicon Ultra concentrator to 2 mg/mL The concentrated pG-Tn5 homodimer was gently mixed with an equal volume of 100% UltraPure glycerol to a final concentration of 55% glycerol, aliquoted (1 mL) into 1.5 mL protein LoBind® tubes (Eppendorf), passively frozen and stored at −20 °C in an enzyme cooler block.
10His-pG-Tn5E54K L372P in vitro tagmentation activity characterization
Nextera Mosaic End Double Stranded (MEDS) adapters were used, ordered as desalted annealed duplexes. The activity was dependent on the proper preparation of the MEDS adapters. Lambda phage genomic DNA (λ gDNA) from NEB was used as the substrate for tagmentation, and the activity was performed in the final 1xTD tagmentation buffer. To prepare the transposomes, 10 μL of pG-Tn5 stock was mixed with 10 μL of MEDS adapters and incubated at 25 °C for 10 minutes. Unless otherwise specified, the adapters were diluted to 20 μM in terms of MEDS sequence common to both N5 and N7 Nextera adapters. The tagmentation reaction was performed by mixing the transposomes with λ gDNA and incubating at 55 °C for 10 minutes. The reaction was quenched with 4% SDS, and the samples were analyzed using a genomic DNA Screentape on an Agilent Tapestation instrument or a 1% agarose EX E-Gel. One Tagment Unit (TU) of Tn5 was defined as the amount needed to uniformly tagment 125 ng of λ gDNA to <250 bp size.
Cell culture
K562 cells were cultured in Iscove’s Modified Dulbecco’s Medium with 10% Fetal Bovine Serum and 1% PenicillinStreptomycin.
MCF-7 cells were cultured in in 75 cm2 flasks DMEM/F-12 (ThermoFisher 10565018) supplemented with Gluta-MAX (Gibco 10565018), 10% Fetal Bovine Serum and 1% Penicillin-Streptomycin at 37 °C and 5% CO2. Cells were subcultured at a 1:3 ratio every 3 days. Prior to their use, MCF7 cell identity was verified by STR profiling through LabCorp Cell Line Authentication service.
Library Preparation
pG-Tn5 adapter loading:
20 μM Nextera sequencing adapters were in a 1:1 volume with pG-Tn5, mixed gently by pipetting, incubated at 23 °C for 10 minutes, and used same-day.
CUT&Tag:
CUT&Tag on MCF7 cells was performed by following the protocol from Kaya-Okur et al, Nature Protoc 2020[27]. MCF7 cells were briefly centrifuged followed by incubation with Nuclear Extraction buffer on ice for 10 minutes. After resuspension in Wash Buffer, nuclei were slow frozen in cryogenic vials with 10% DMSO for storage at −80 °C before resuming the rest of the protocol. Approximately 100,000 cells were used per sample for overnight incubation with primary antibody (Tri-Methyl-Histone H3 (Lys27) (C36B11) Rabbit mAb, 9733T, Cell Signaling Technology 1:50, Anti-Histone H3 (acetyl K27) antibody - ChIP Grade (ab4729), Abcam 1:100). Secondary antibody (Guinea Pig anti-Rabbit IgG (Heavy & Light Chain) antibody, Guinea Pig, ABIN101961 1:100) was incubated for 30 minutes at room temperature followed by addition of commercial pAG-Tn5 (EpiCypher SKU: 15–1017) or in-house pG-Tn5 preloaded with adapters. Phenol chloroform DNA extraction was performed using Qiagen’s Maxtract High Density phase-lock tubes. Libraries were PCR amplified for 12 cycles with Illumina i5 or i7 barcoded primers and Ampure XP beads were used for post-PCR cleanup.
ATAC-seq:
ATAC assay was performed in duplicate or triplicate on 50,000 cells per technical replicate following Omni-ATAC protocol with the following modifications[11]. Prior to tagmentation, cells were treated in culture medium with DNase (Worthington cat# LS002007) at a final concentration of 200 U/mL at 37 °C for 30 mins, then washed twice with PBS at 500g, RT. For Illumina enzyme tagmented material, 2.5 μL of Illumina TDE1 enzyme was used. K562 transposome titration experiment: pG-Tn5 tagmentation was titrated with the following amounts of loaded transposome: 1.0, 2.5, 5.0, 7.0 μL. 7 cycles of PCR were performed during library preparation for K562 experiment. MCF7: 8 cycles of PCR were performed during library preparation of MCF7 experiment. Prior to sequencing, fragment length distribution and DNA concentration were determined via Tapestation and Qubit respectively.
EasySci-ATAC
For harvesting, flasks were washed with ice cold PBS then cells were dissociated with TrypLE Express enzyme. Nuclei were extracted and single-cell indexing was performed following the EasySci-ATAC protocol, as described in Supplemental protocol 2 of Sziraki, et al., 2023[25]. Briefly, cells were resuspended in 1 mL Nuclei Isolation Buffer (NIB) + 0.1% IGEPAL CA-630 and filtered through a 40 μm filter. Nuclei were stained with Trypan Blue and counted with the EVE Automated Cell Counter; the final concentration was adjusted to 1’000 nuclei/μL using NIB. For each barcode combination, 5’000 nuclei were mixed with 4 μL of barcoded pG-Tn5 and tagmentation was carried out in a thermomixer at 55 °C for 10 minutes with gentle shaking at 350 RPM. Following tagmentation, nuclei were put directly on ice to prevent over tagmentation, followed by addition of a stop buffer consisting of spermidine-EDTA. Nuclei were then pooled and redistributed to 96-wells to allow multiplexing via barcoded ligation, followed by a third round of pooling and redistribution to a final 96-well reaction for barcoding via PCR amplification and library preparation. Libraries underwent size selection via E-gel prior to sequencing. All libraries were sequenced to 0.5–10M reads on the NextSeq 1000 with 58,8,8,60 sequencing parameters.
Custom loading of Tn5 with combinatorial barcodes for EasySci-ATAC:
For uniquely barcoded tagmentation reactions, two N5 barcoded oligo duplexes were mixed in combination with three N7 barcoded oligo duplexes. Per barcode combination: 1 μL of 10 μM N5 duplex, 1 μL of 10 μM N7 duplex, and 2 μL of pG-Tn5 (Lot 2) were mixed and incubated for 15 minutes at 25 °C. Loaded pG-Tn5 was kept on ice until nuclei were ready for tagmentation.
Data Analysis for bulk CUT&Tag and ATAC-seq
Adapter trimming:
Adapters were trimmed from pair-end FASTQ files using a custom script adapted from Corces et al. 2017[11] (https://github.com/riscalab/pipeSeq/blob/master/scripts/pyadapter_trimP3V2.py) (https://github.com/riscalab/pipeSeq/blob/master/scripts/pyadapter_trimP3V2.py) , then FASTQC and FASTQC Screen performed per-base sequence content QC and screened for mycoplasma genome representation.
Alignment to hg38
CUT&Tag:
Reads were aligned to hg38 using bowtie2[28] using the following parameters--end-to-end –very-sensitive --no-mixed --no-discordant --phred33 -I 10 -X 700 for mapping of inserts 10–700 bp in length.
ATAC-seq:
Reads were aligned using -X 2000 parameters on bowtie2. Reads were shifted to account for Tn5 insertion bias using deepTools[29] alignmentSieve command with —ATACshift parameter set.
For both CUT&Tag and ATAC-seq data, duplicate reads, reads mapping to the mitochondrial genome and black-listed regions were removed using picardtools[30] to generate the final bam file. Bam files were then sorted using picardtools. Libraries sequenced across multiple flow cells were merged as bams, then sorted using picardtools.
Peak Calling
CUT&Tag:
Using bedtools, bam files were converted to bedpe files which were then converted to bedgraph format. Bedgraph files were used as input into SEACR peak caller[31], where peaks were called with parameters specified to non-normalized and stringent threshold set to 0.01. Afterwards, a master peak set between technical replicates was created keeping only peaks that overlapped with each other between technical replicates and merging overlapping peaks into a single peak. A master non-redundant peak set was created by merging overlapping peaks of peaksets from technical replicates of different Tn5 conditions (Epicypher’s pAG-Tn5 vs in house pG-Tn5) using GenomicRanges package[32] in R.
ATAC-seq:
Peaks were called from bam files with MACS2[33] using the following parameters -B --SPMR --nomodel --shift −37 --extsize 73 --nolambda --keep-dup all --call-summits --slocal 10000. Next, a non-redundent master peakset was made by intersecting technical replicate narrowPeaks in bedtools, then merging intersected narrowPeak files in bedtools for each cell type. Cell-type masterpeak sets were merged in bedtools[34] to create a non-redundant peakset across all ATAC samples.
Sample Correlation
Fragment counts for each replicate under the masterpeak set were counted using chromVAR’s getCounts function[35]. For K562 transposome dilution analysis, fragment counts under 1 kb hg38 bins were counted using chrom-VAR’s getCounts function for each replicate[35]. The resultant fragment count matrix was then used as input into DE-Seq2[36]v1.26.0 and sequencing depth was normalized across samples using the estimateSizeFactors function.For visualization purposes, the counts matrix underwent a “regularized log” transformation using the rlog function from DESeq2. Next, the distance measure between pairwise samples was calculated. 1 – distance measure was used as input to calculate the Spearman correlation coefficient between each sample pair. Custom python matplotlib code was used to plot the sample correlation matrix.
E. coli transposome contamination analysis
Reads were aligned using bowtie2 to the E. coli genome from ATAC-seq bam files. Number of E. coli mapped reads/Total mapped reads were calculated and plotted.
Genome coverage visualization
Technical replicates were combined into a single bam file using samtools merge command. Bigwig files were generated using deepTools’ bamCoverage command with the following parameters to normalize:
Coverage to the hg38 genome size CUT&Tag: --binSize 5 --normalizeUsing RPGC --effectiveGenomeSize 2913022398 --extendReads.
Coverage to the hg38 genome size ATAC-seq: --binSize 1 --normalizeUsing RPGC --effectiveGenomeSize 2913022398 --extendReads
ATAC-seq Coverage Plots
Plotting CTCF Insertions:
Merged ATAC-seq bam files from technical replicates were randomly down-sampled to 3M reads using samtools for read-depth normalized comparisons. Homo sapiens CTCF PWM from the Hoco-mocoV10 database[37] were used to plot insertions +/− 100bp around the motif using the regionPlot function from soGGI R package[38].
TSS Enrichment:
TSS enrichment profile of insertions were produced as previously described[11] using hg38 TSS annotation from gencode. Insertions were counted in 1bp bins +/− 2’000 bp surrounding annotated TSSs. Bins were then normalized by dividing by the average of the first 200 bp bins from each flank: https://github.com/riscalab/pipeSeq/blob/master/scripts/pyMakeVplot_css_v01.pyhttps://github.com/riscalab/pipeSeq/blob/master/scripts/pyMakeVplot_css_v01.py
Coverage CTCF:
IDR-thresholded CTCF narrow peak ChIP-Seq data in MCF7 cell line was downloaded from ENCODE accession number ENCFF278FNP https://www.encodeproject.org/files/ENCFF278FNP/, https://www.encodeproject.org/files/ENCFF278FNP/. Total coverage around ChIP CTCF peaks was calculated using deepTools computeMatrix function on RPGC normalized bigwigs (described above) in reference-point mode with 1 kb flanking regions. The matrix was plotted using deepTools plotHeatmap function with bilinear interpolation.
Computational procedures for processing EasySci-ATAC libraries:
Libraries were demultiplexed using custom scripts to assign base calls to individual cells based on indices from PCR, ligation, and Tn5 combinatorial barcoding. The SnapATAC2 analysis framework was followed to generate the TSS enrichment score vs. unique fragments plot, perform doublet removal, dimensional reduction, and Leiden algorithm derived clustering analysis[26]. Gene body accessibility was counted using Gencode v38 basic gene annotation, and subsequently used for visualizing gene accessibility as a proxy for expression after applying the MAGIC algorithm for imputation and data smoothing.
Supplementary Material
Acknowledgments
J.S. acknowledges support from Boehringer Ingelheim Fonds PhD Fellowship. V.I.R. acknowledges support from NIH Common Fund (1DP2GM150021-01), The Rita Allen Foundation (Scholar Award), and Irma T. His-chl/Monique Weill-Caulier Trust (Career Science Award). J. L. Y. acknowledges support from a NSERC PGS D scholarship. L.J.A acknowledges support from the NSF Graduate Research Fellowship Program (1946429). J. M. R. was supported by the American Cancer Society – Fairfield Research Council– Postdoctoral Fellowship, PF-23-1034949-01-CCB, https://doi.org/10.53354/ACS.PF-23-1034949-01-CCB.pc.gr.168157 H.A.K. was supported by the Uehara Memorial Foundation Research Fellowship and the Japan Society for the Promotion of Science Overseas Research Fellowships.
We acknowledge support from the Rockefeller University Structural Biology Resource Center RRID: SCR_017732. We acknowledge technical support from Rockefeller University high performance computing. We thank Wei Zhu for technical assistance. We thank Ziyu Lu and Junyue Cao for providing EasySci- ATAC adapters and protocol advice. We thank Oscar Alejandro Perez for graphic illustration and data visualization advice. We thank all members of the Risca laboratory, Yasuhiro Arimura and Pierre-Jacques Hamard for insightful discussions and advice. We thank Ariana Brenner Clerkin for manuscript formatting advice.
Footnotes
Supplementary Files
1. OpenTn5 Protocol 10His-pG-Tn5E54K, L372P Purification and Lambda Tagmentation Assay
2. OpenTn5 Protocol Buffer Table
3. OpenTn5 Protocol Reagents Table
4. Supplementary Table 1 (Sequencing QC metrics)
5. Plasmid sequence as .genbank and .dna file
6. Raw and Processed Sequencing Data (GEO)
References
- 1.Reznikoff W. S. Transposon Tn5. en. Annu. Rev. Genet. 42, 269–286 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Zhou M., Bhasin A. & Reznikoff W. S. Molecular genetic analysis of transposase-end DNA sequence recognition: cooperativity of three adjacent base-pairs in specific interaction with a mutant Tn5 transposase. en. J. Mol. Biol. 276, 913–925 (Mar. 1998). [DOI] [PubMed] [Google Scholar]
- 3.Picelli S. et al. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. en. Genome Res. 24, 2033–2040 (Dec. 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou M. & Reznikoff W. S. Tn5 transposase mutants that alter DNA binding specificity. en. J. Mol. Biol. 271, 362–373 (Aug. 1997). [DOI] [PubMed] [Google Scholar]
- 5.Weinreich M. D., Mahnke-Braam L. & Reznikoff W. S. A functional analysis of the Tn5 transposase. Identification of domains required for DNA binding and multimerization. en. J. Mol. Biol. 241, 166–177 (Aug. 1994). [DOI] [PubMed] [Google Scholar]
- 6.Vaezeslami S., Sterling R. & Reznikoff W. S. Site-directed mutagenesis studies of tn5 transposase residues involved in synaptic complex formation. en. J. Bacteriol. 189, 7436–7441 (Oct. 2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H. et al. Comprehensive understanding of Tn5 insertion preference improves transcription regulatory element identification. en. NAR Genom Bioinform 3, lqab094 (Dec. 2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kia A. et al. Improved genome sequencing using an engineered transposase. en. BMC Biotechnol. 17, 6 (Jan. 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hennig B. P. et al. Large-Scale Low-Cost NGS Library Preparation Using a Robust Tn5 Purification and Tagmentation Protocol. en. G3 8, 79–89 (Jan. 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adey A. C. Tagmentation-based single-cell genomics. en. Genome Res. 31, 1693–1705 (Oct. 2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corces M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. en. Nat. Methods 14, 959–962 (Oct. 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaya-Okur H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. en. Nat. Commun. 10, 1–10 (Apr. 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu B. et al. Transposase-assisted tagmentation of RNA/DNA hybrid duplexes. en. Elife 9 (July 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li N. et al. Tn5 Transposase Applied in Genomics Research. en. Int. J. Mol. Sci. 21 (Nov. 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nanda A. S. et al. Direct transposition of native DNA for sensitive multimodal single-molecule sequencing. en. Nat. Genet., 1–10 (May 2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khyzha N., Henikoff S. & Ahmad K. Profiling RNA at chromatin targets in situ by antibody-targeted tagmentation. en. Nat. Methods 19, 1383–1392 (Nov. 2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mangiameli S. M. et al. Photoselective sequencing: microscopically guided genomic measurements with subcellular resolution. en. Nat. Methods 20, 686–694 (May 2023). [DOI] [PubMed] [Google Scholar]
- 18.Thornton C. A. et al. Spatially mapped single-cell chromatin accessibility. en. Nat. Commun. 12, 1274 (Feb. 2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cusanovich D. A. et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shilling P. J. et al. Improved designs for pET expression plasmids increase protein production yield in Escherichia coli. en. Commun. Biol. 3, 214 (May 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu T., Xiao M. & Yu L. Method for efficient soluble expression and purification of recombinant hyperactive Tn5 transposase. en. Protein Expr. Purif. 183, 105866 (July 2021). [DOI] [PubMed] [Google Scholar]
- 22.York D. & Reznikoff W. S. Purification and biochemical analyses of a monomeric form of Tn5 transposase. en. Nucleic Acids Res. 24, 3790–3796 (Oct. 1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikaido H. The Limitations of LB Medium https://schaechter.asmblog.org/schaechter/2009/11/the-limitations-of-lb-medium.html. Accessed: 2024-6-22. Nov. 2009.
- 24.Vagenende V., Yap M. G. S. & Trout B. L. Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. en. Biochemistry 48, 11084–11096 (Nov. 2009). [DOI] [PubMed] [Google Scholar]
- 25.Sziraki A. et al. A global view of aging and Alzheimer’s pathogenesis-associated cell population dynamics and molecular signatures in human and mouse brains. en. Nat. Genet. 55, 2104–2116 (Dec. 2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K., Zemke N. R., Armand E. J. & Ren B. A fast, scalable and versatile tool for analysis of single-cell omics data. en. Nat. Methods 21, 217–227 (Feb. 2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaya-Okur H. S., Janssens D. H., Henikoff J. G., Ahmad K. & Henikoff S. Efficient low-cost chromatin profiling with CUT&Tag. en. Nat. Protoc. 15, 3264–3283 (Sept. 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B. & Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramírez F., Dündar F., Diehl S., Grüning B. A. & Manke T. deepTools: a flexible platform for exploring deep-sequencing data. en. Nucleic Acids Res. 42, W187–91 (July 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picard https://broadinstitute.github.io/picard/. Accessed: 2024-7-1. [Google Scholar]
- 31.Meers M. P., Tenenbaum D. & Henikoff S. Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling. en. Epigenetics Chromatin 12, 42 (July 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence M. et al. Software for computing and annotating genomic ranges. en. PLoS Comput. Biol. 9, e1003118 (Aug. 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaspar J. M. Improved peak-calling with MACS2 en. Dec. 2018.
- 34.Quinlan A. R. & Hall I. M. BEDTools: a flexible suite of utilities for comparing genomic features. en. Bioinformatics 26, 841–842 (Jan. 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schep A. N., Wu B., Buenrostro J. D. & Greenleaf W. J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. en. Nat. Methods 14, 975–978 (Oct. 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. en. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulakovskiy I. V. et al. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. en. Nucleic Acids Res. 46, D252–D259 (Jan. 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dharmalingam G., Barrows D. & Carroll T. soGGi: Visualise ChIP-seq, MNase-seq and motif occurrence as aggregate plots Summarised Over Grouped Genomic Intervals en. https://rdrr.io/bioc/soGGi/. Accessed: 2024-7-1. Oct. 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.