Summary
Cancer cells have remarkable plasticity allowing them to acquire many biological states. Melanoma cells have the ability to switch from a proliferative melanocytic state to an invasive mesenchymal state and back again resulting in intratumoral heterogeneity. While microphthalmia-associated transcription factor (MITF) promotes the melanocytic phenotype, it is unclear what transcription factors drive the mesenchymal phenotype, and what mechanisms regulate the switch from the proliferative state to the mesenchymal state. We show that nuclear localization of the MITF paralog TFE3 correlates positively with metastatic potential in melanoma cell lines and tumors, and that deletion of TFE3 in MITF-low melanoma cell lines eliminates migration and metastatic ability. Further, we find that MITF suppresses the mesenchymal phenotype by activating expression of FNIP2, which encodes a component of an mTORC1-stimulated pathway promoting cytoplasmic retention and lysosomal degradation of TFE3. These findings point to the mTOR pathway and TFE3 as key regulators of melanoma plasticity.
Introduction
Cellular plasticity is a critical facet of tumor biology, impacting cancer progression and aggressiveness. Plasticity of cancer cells additionally drives therapy resistance via differentiation to treatment-resistant states (Torborg et al., 2022). Targeting cellular plasticity is a potentially effective therapeutic approach but requires further understanding into the mechanisms critical for these cell state transitions. Melanoma, the most lethal cutaneous malignancy, can convert from a proliferative, pigmented, and relatively immotile status to a mesenchymal, invasive status with metastatic potential, a transition known as phenotype-switching (Hoek and Goding, 2010, Hoek et al., 2008, Carreira et al., 2006). The proliferative-to-mesenchymal phenotype switching is characterized by the re-emergence of a transcriptional state resembling that of the neural crest, the embryonic precursor population from which melanocytes are derived (Kaufman et al., 2016). Phenotype switching is critical for initiation of melanoma metastasis (Campbell et al., 2021, Diener and Sommer, 2021, Heppt et al., 2018, Kaufman et al., 2016, Hoek et al., 2008, Hoek and Goding, 2010, Wouters et al., 2020, Carreira et al., 2006).
Microphthalmia-associated transcription factor (MITF) plays a major role in differentiation of melanocytes and regulates phenotype switching in melanoma (Levy et al., 2006, Carreira et al., 2006). MITF promotes survival and differentiation of melanoblasts by directly regulating a cohort of pigmentation genes (Kenny et al., 2022, Goding and Arnheiter, 2019, Seberg et al., 2017). Mitf-null mice lack melanocytes, suffer from associated deafness, and have unpigmented retinal pigment epithelia (reviewed in (Goding and Arnheiter, 2019)). In melanomas, high levels of MITF activity correlate with a proliferative and pigmented phenotype, while low levels of MITF activity are associated with metastasis and drug resistance (Rambow et al., 2018, Carreira et al., 2006, Hoek et al., 2008). The canonical BRAF / MAPK pathway is activated in MITF-low melanoma and many studies have documented a characteristic transcriptional profile that includes expression of genes that promote epithelial mesenchymal transition (EMT), invasion, and stem-cell-like or neural-crest-cell-like properties (Carreira et al., 2006, Hoek et al., 2008, Wouters et al., 2020, Karras et al., 2022). However, it is unclear how activation of these genes is achieved. Based on the proximity of MITF peaks to genes expressed at higher levels in MITF-low vs. MITF-high melanoma, we predicted that MITF functions as a transcriptional repressor at some enhancers (Dilshat et al., 2021), a possibility that we explore here.
To investigate the mechanism of upregulated gene expression in MITF-low melanoma, we performed CUT&RUN with antibodies to MITF and to chromatin marks indicative of active and repressed chromatin in SKMEL28 melanoma cells that were wild-type or harbored loss of function mutations in the MITF gene (i.e., MITF knock-out, KO). In contrast to our prediction, we detected very few enhancers with evidence of being directly repressed by MITF, but thousands of novel anti-MITF peaks in MITF mutant cells that were absent from MITF wild-type cells. However, the presence of MITF peaks in MITF KO cells suggested the antibody was cross-reacting with a related transcription factor. Moreover, these peaks were enriched near genes whose expression was upregulated in MITF mutant cells relative to MITF wild-type cells and whose functions were associated with cell migration and epithelial-to-mesenchymal transition (EMT). We found that the novel anti-MITF peaks correspond to chromatin binding by the MITF-paralog Transcription Factor E3 (TFE3), and that TFE3 activates many of the genes characteristic of the MITF-low expression profile. Expression of TFE3 and the presence of the peaks correlated with invasive behavior in a panel of melanoma cell lines and tumor samples, and deletion of TFE3 blocked invasive behavior of the MITF-low cell lines A375 and RPMI. We identify FNIP2 as an MITF-dependent member of an E3 ligase complex that excludes TFE3 from the nucleus and targets it to the proteosome. Expression of MITF and FNIP2 are strongly correlated in melanoma and inhibition of FNIP2 expression in an MITF-high cell line promoted nuclear transport of TFE3; in contrast elevating FNIP2 expression in MITF KO cells lead to a reduction of TFE3-dependent gene expression. These findings help to explain why MITF-low melanoma tends towards an invasive phenotype.
Results
Chromatin profiling suggests MITF directly activates but does not directly repress gene expression.
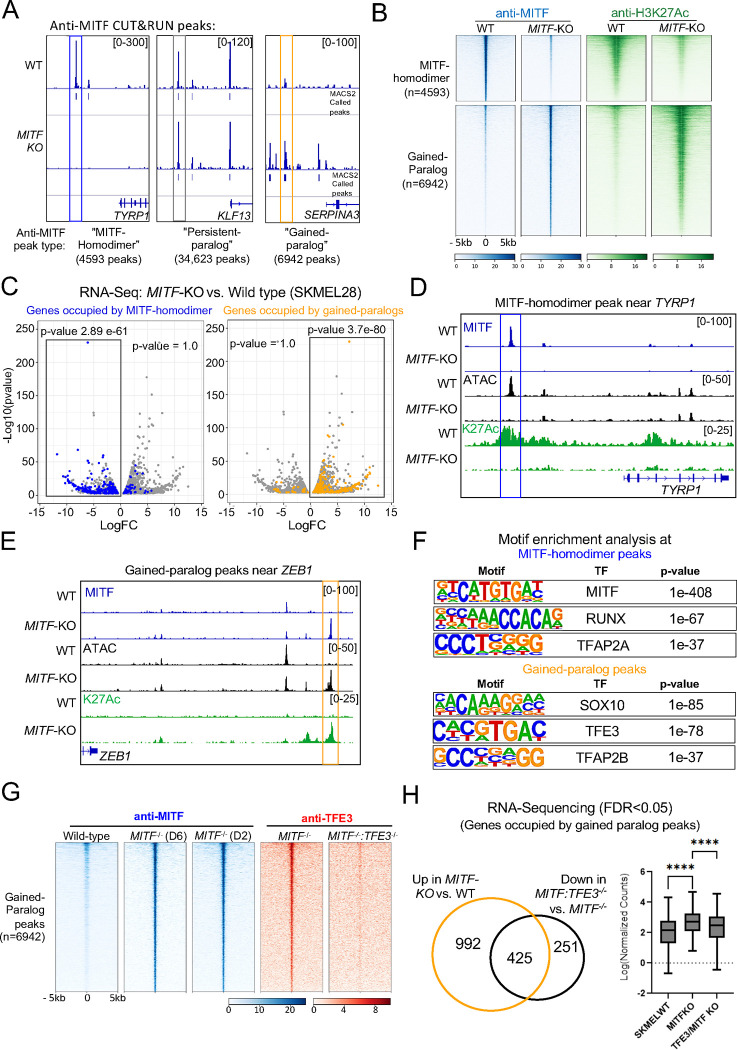
To identify enhancers directly activated or directly inhibited by MITF, we assessed the changes in several chromatin marks underlying anti-MITF CUT&RUN peaks in SKMEL28 melanoma cells that were wild-type or MITF loss-of-function mutants. In this study, we used ΔMITF-X6 SKMEL28 cells, which lack functional MITF alleles, and ΔMITF-X2 SKMEL28 cells, which also lack functional MITF alleles, but retain a small molecular weight protein that reacts with anti-MITF antibody by Western blot analysis (Dilshat et al., 2021). RNA-Seq analyses of ΔMITF-X6 and wild-type SKMEL28 cells revealed 1203 genes downregulated at least 2-fold, and 1790 genes up-regulated at least 2-fold in ΔMITF-X6 vs. wild-type SKMEL28 cells (i.e., MITF-activated genes and MITF-repressed genes, respectively); the sets of differentially expressed genes in ΔMITF-X2 were similar. For CUT&RUN, we used a commercial anti-MITF antibody that was generated against a peptide sequence present in MITF; slight variants of this peptide are also present in its paralogs TFEB, TFE3 and TFEC (Figure S1A). We performed CUT&RUN with this antibody in wild type SKMEL28, ΔMITF-X6 and ΔMITF-X2 melanoma cells (Dilshat et al., 2021). Of 46,519 anti-MITF peaks in wild-type cells, about 10% (4,593) were unique to wild-type cells, which we termed MITF-homodimer peaks, about 75% (34,623) were present in both wild-type and MITF-KO cells, which we termed persistent-paralog peaks, and about 15% (6,943) were unique to MITF-KO cells, which we termed gained-paralog peaks (Figure 1A) (84% of anti-MITF peaks in ΔMITF-X6 were shared in ΔMITF-X2 (Figure S2A)). Across duplicate CUT&RUN experiments, MITF-homodimer peaks were highly correlated (r = 0.97, Pearson correlation), and persistent-paralog peaks and gained-paralog peaks also, although slightly less so (r = 0.84 and 0.88, respectively).
Figure 1: MITF inhibits TFE3 occupancy at thousands of genomic loci.
(A) Screenshot of IGV genome browser (GRCH37/hg19) visualization of bigwig files generated from anti-MITF CUT&RUN sequencing in SKMEL28 cells that are wild-type or MITF-KO (delta 6). Three groups of anti-MITF peaks were identified based on the presence or absence of an anti-MITF peak in MITF-KO cells. The frequency of each peak type; “MITF-homodimer peaks”, “persistent-paralog peaks” and “gained paralog peaks”, are labeled. Peaks were called using MACS2 software using FDR < 0.05 with IgG serving as background control (B) Density plot of MITF homodimer peaks (upper panel) and Gained-paralog peaks (lower panel) showing anti-MITF (blue) and anti-H3K27Ac (Green) CUT&RUN signal in SKMEL28 WT and MITF-KO cells. Peaks are sorted on anti-MITF-signal in wild-type SKMEL28 cells (C) Volcano plot showing 2136 DEGs with qval <0.5 among which 1516 genes with log2FC≥|1| fold change in expression ΔMITF-X6 vs. EV-SkMel28 (Dilshat et al., 2021). Hypergeometric analysis was used to determine the association of MITF-homodimer peaks and gained-paralog peaks with DEG, p-values are as labeled, significant associations are indicated with black rectangles. (D) IGV screenshot illustrating a MITF-homodimer peak near the MITF-activated gene TYRP1 and (E) a gained-paralog peak near the MITF-inhibited gene ZEB1. Additional tracks include ATAC-Seq (Black), and anti-H3K27Ac CUT&RUN (Green) in WT and MITF-KO cells. Bigwig files were generated using deeptools. (F) Enrichment of transcription factor motifs using HOMER at MITF-homodimer peaks (upper panel) and gained-paralog peaks (lower panel). (G) Density plot of anti-MITF CUT&RUN-seq in WT and two clones of MITF-KO cells and anti-TFE3 CUT&RUN-seq in MITF-KO and double MITF:TFE3-KO SKMEL28 cells at gained-paralog peaks. (H) Venn diagram illustrating the overlap of genes occupied by gained TFE3 peaks and upregulated in MITF-KO cells compared to WT SKMEL28 cells (depicted by the orange circle), with the genes downregulated in double TFE3:MITF-KO compared to MITF-KO cells (depicted by the black circle). Additionally, box plot visualization displays the expression profiles (Log normalized counts) of the overlapping genes (n=425).
To understand the function of these different categories of anti-MITF peaks in wild-type and MITF-KO cells, we performed ATAC-seq and CUT&RUN using antibodies targeting post-translational marks on histone H3, including the activation marks chromatin lysine 27 acetylation (H3K27Ac) and lysine 4 tri-methylation (H3K4Me3) and the repressive mark lysine 27 tri-methylation (H3K27Me3). All MITF-homodimer peaks were located within active chromatin (i.e., positive for ATAC-seq and H3K27Ac signals and lacking H3K27Me3 signal) in wild type cells, and at 92% of these sites, H3K27Ac signal was reduced in MITF-KO cells relative to what was seen in wild-type cells (Figure 1B and Figure S2B). The set of genes associated (i.e., transcription start site within 100 kb) with this large subset of MITF-homodimer peaks was enriched for MITF-activated genes (Figure 1C) and for gene ontology (GO) terms related to melanocyte development (e.g., DCT, OCA2, GPR143, TRPM1 and TYRP1) (Figure S2D). An example of an MITF-homodimer occupied enhancer near TYRP1, a gene involved in melanin synthesis, is shown in Figure 1D. At the remaining 8% of MITF-homodimer peaks, the levels of ATAC-seq and H3K27Ac signal were equivalent in WT and MITF-KO cells; importantly, in no case were these signals elevated in the knockout cells, which would have indicated MITF homodimers serving as a repressor at these sites (Figure S2B–C). The set of genes associated with such peaks was weakly enriched for MITF-activated genes. We infer that when engaging DNA as a homodimer, MITF activates but does not inhibit chromatin.
By contrast, despite the anti-MITF signal at persistent paralog peaks being equivalent in wild type and MITF-KO cells, 27% of those peaks occupied enhancers where the ATAC and H3K27Ac signal was stronger in MITF-KO cells (Figure S2B). However, such peaks were weakly associated with MITF-activated or MITF-repressed genes. We infer that an MITF-paralog binds at most of these sites in both WT and MITF KO cells, perhaps as a heterodimer with MITF in some instances.
Finally, all gained-paralog peaks were located within chromatin that is inactive in WT cells but active in MITF-KO cells (Figure 1B, 1E). Moreover, the genes associated with gained-paralog peaks were enriched for MITF-repressed genes (Figure 1C) and for GO terms such as cell migration, cell motility, extracellular remodeling, and focal adhesion (e.g., TGFA, ITGA2, MMP15, FGF1, SERPINA3, ZEB1) (Figure S2E). An example of a gained-paralog peak near ZEB1, which is associated with EMT, is shown in Figure 1E. These findings indicate that in the absence of MITF, an MITF-paralog binds and activates enhancers near genes associated with migratory behavior.
TFE3 binds at gained-paralog peaks
To determine the identity of the MITF paralog that binds at gained-paralog peaks we performed a motif enrichment analysis of chromatin underlying such peaks using Hypergeometric Optimization of Motif EnRichment (Heinz et al., 2010). Homodimer peaks were strongly enriched for the binding site of MITF and MITF co-factor, TFAP2A (Figure 1F) (Kenny et al., 2022). By contrast, chromatin elements underlying gained-paralog peaks and associated with MITF-repressed genes were most strongly enriched for the binding site of TFE3, and for that of SOX10 and TFAP2B, among other transcription factor binding sites (Figure 1F). Next, we used the anti-MITF antibody to immuno-precipitate protein from lysates of wild-type and MITF-KO SKMEL28 cells. In Western blots of protein immunoprecipitated from SKMEL28 cells of all genotypes, anti-TFE3 antibody recognized bands at the appropriate size (~72–82 kDa) (Figure S3A–C). We cut out these bands, extracted protein, subjected it to mass spectrometry, and detected peptides derived unambiguously from TFE3 (Figure S3D). Finally, we deleted the TFE3 gene from ΔMITF-X2 cells and then conducted anti-TFE3 CUT&RUN in ΔMITF-X2 and MITF/TFE3 double KO cells. In ΔMITF-X2 cells there was almost complete overlap between anti-TFE3 peaks and anti-MITF (i.e., gained paralog) peaks, while in MITF/TFE3 double KO cells, no anti-TFE3 peaks were detected (Figure 1G). These results suggest that TFE3 binds to novel enhancers and drives a unique transcriptional program in MITF-KO cell lines. We will refer to gained paralog peaks as gained-TFE3 peaks.
TFE3 activates expression of genes in the MITF-low transcriptional profile
We predicted that a subset of MITF-repressed genes would be directly activated by TFE3. We performed RNA-sequencing in WT, ΔMITF-X2, and MITF/TFE3 double KO cells and, consistent with this prediction, about 30% (425/1417) of MITF-repressed genes were down-regulated in MITF/TFE3 double KO cells relative to in ΔMITF-X2 cells (i.e., meaning they were TFE3-activated genes) (Figure 1H). Moreover, all such genes were flanked (within 100 kb) by gained-TFE3 peaks, indicating that most or all were directly activated by TFE3. This set of MITF-repressed/TFE3-activated genes was enriched for GO terms associated with cell migration (e.g., ZEB1) (Figure S4A), cell differentiation, and angiogenesis (Figure S4B). The remaining 70% (992/1417) of MITF-repressed genes that were not TFE3-activated were enriched for GO term associated with multicellular organismal processing and basement membrane assembly but not for GO terms related EMT (Figure S4C). Interestingly, several MITF-repressed/TFE3-activated genes are known TFE3 targets in Naive Human Embryonic Stem Cells (hESCs) (Mathieu et al., 2019), including SOX2, TBX3, TBX6, PRDM12, FOXDF2, FOXD1, FOXD3, as well as various members of the KLF transcription factor family and the WNT signaling pathway (Table S2). In summary, in an MITF-high cell line deleted of MITF, TFE3 drives expression of genes that control cell migration and stem-cell features.
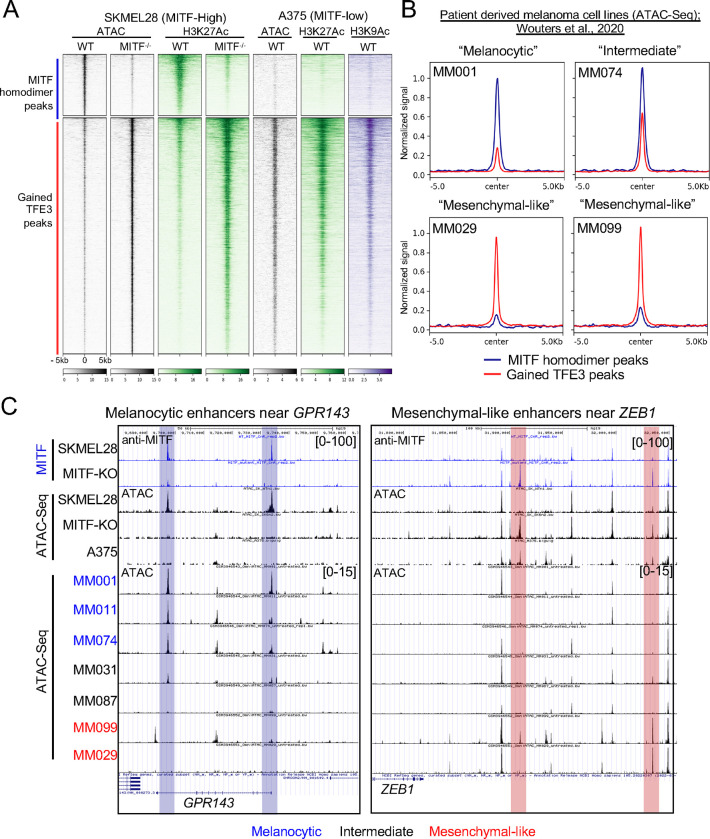
As expression of cell migration and stem-cell genes is characteristic of MITF-low cell lines, we next evaluated what other features of MITF-KO cells are shared with MITF-low cell lines. The invasive A375 cell line expresses far lower levels of MITF than the proliferative SKMEL28 cells (Figure S4E–F). In published ATAC-seq and in our H3K27Ac and H3K9Ac CUT&RUN-seq data from A375 cells, we found that, as expected, virtually all (98%) enhancers bound and activated by MITF-homodimers in SKMEL28 cells are inactive in A375 cells (Figure 2A). Conversely and remarkably, the majority (65%) of gained-TFE3 peaks in MITF-KO cells overlapped active enhancers in A375 cells (Figure 2A). We next examined ATAC-Seq data from melanoma cell lines classified as melanocytic (three cell lines, MM001, MM011, MM074), intermediate (two cell lines, MM031, MM087), or mesenchymal (two cell lines, MM099, MM029) based on expression of pigmentation genes (e.g., MITF, GPR143, and TYR) and genes characteristic of mesenchyme (ZEB1, TGFBI, and SERPINE1) (Wouters et al., 2020). In melanocytic cells, the ATAC-seq signal was high at enhancers activated by MITF-homodimers in wild-type SKMEL28 cells, and in mesenchymal cells it was high at enhancers activated by gained-TFE3 peaks in MITF-KO SKMEL28 cells (examples shown in Figure 2B). In intermediate cell lines, ATAC-seq signal was moderately high at both (Figure 2B). Examples of ATAC-seq peaks in each category of melanoma cell line near the MITF-activated gene GPR143 and the TFE3-activated gene ZEB1 are shown in Figure 2C. In summary, many enhancers that are activated upon deletion of MITF from an MITF-high cell line are also active in MITF-low melanoma cell lines.
Figure 2: Gained TFE3 peaks in MITF-KO cells overlap with active enhancers in A375 and patient-derived “melanocytic-like” cell lines, distinguishing them from MITF-homodimer bound “melanocytic” enhancers.
(A) Density plot depicting ATAC-Seq (black) and anti-H3K27Ac (green) CUT&RUN-seq in WT and MITF-KO SKMEL28 cells, alongside ATAC-Seq (black), anti-H3K27Ac (green), and H3K9Ac (purple) CUT&RUN-seq in A375 cells. The density heatmap is ordered by MITF-homodimer peaks (upper panel) and gained-TFE3 peaks (lower panel) as identified in WT and MITF-KO SKMEL28 cells, respectively. (B) Summary plots illustrating normalized ATAC-Seq read counts at MITF-homodimer (blue line) and Gained-TFE3 peaks (Red line) for “Melanocytic” (MM001), “Intermediate” (MM074), and two “mesenchymal-like” (MM029, MM099) human patient derived melanoma cell lines (Wouters et al., 2020). (C) Genome browser screenshot of enhancers activated by MITF-homodimers (Blue highlight) near the melanocytic gene GPR143 and enhancers activated by gained TFE3-peaks (red highlight) near the mesenchymal-like gene ZEB1. Bigwigs of anti-MITF CUT&RUN-seq and ATAC-Seq from SKMEL28 and MITF-KO cells as well as ATAC-Seq traces from Melanocytic (blue), Intermediate (black) and mesenchymal-like (red) are shown.
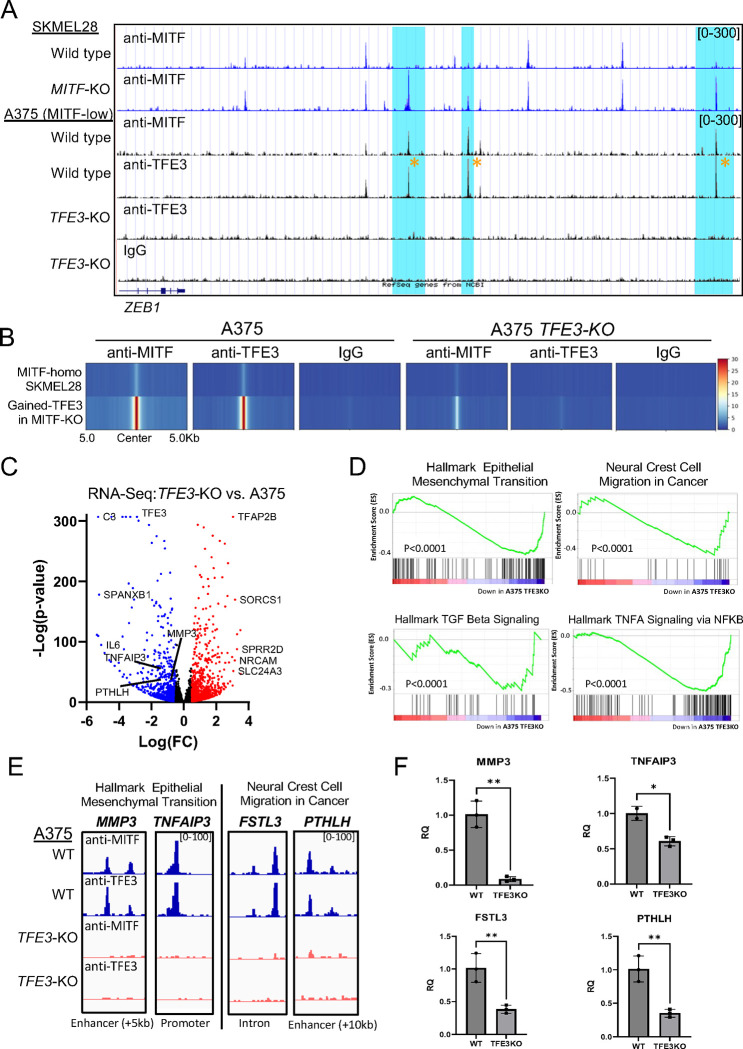
To test the importance of TFE3 in a model MITF-low melanoma cell line, we created three independent A375 TFE3-KO clones (Figure S5A–C). Using anti-TFE3 CUT&RUN in wild-type A375 cells, we identified 30,342 anti-TFE3 peaks, 66% of which were located within active chromatin (i.e., marked with H3K27Ac). In TFE3-KO cells, there were no anti-TFE3 peaks, confirming the specificity of the TFE3 antibody (Figure 3A–B and Figure S5D). The large majority of active anti-TFE3 peaks in A375 cells coincided with active gained-TFE3 peaks in MITF-KO cells (Figure 3A–B). Examples of such enhancers are found near ZEB1 (Figure 3A). RNA-Seq analysis showed that 3,344 genes were downregulated and 3,206 were upregulated in A375 TFE3-KO cells when compared to A375 wild-type cells (TFE3-activated and TFE3-repressed genes, respectively) (Figure 3C). Of the TFE3-activated genes, 32% (1023) were directly bound by TFE3 (i.e. within 100kb of an Anti-TFE3 peak). Direct TFE3-activated genes were associated with “epithelial mesenchymal transition” and “neural crest cell migration in cancer” gene sets by GSEA analysis; “TGF-beta signaling,” and “TNF-alpha signaling via NFKB” were also enriched among such genes (Figure 3D–F). Examples of TFE3 occupancy and subsequent gene expression changes at EMT (MMP3 and TNFAI3) and neural crest migration genes in cancer (FSTL3 and PTHLH) are illustrated in Figures 3E and 3F, respectively. In summary, TFE3 activates an invasive gene program in several MITF-low melanoma cell lines.
Figure 3: TFE3 establishes a mesenchymal-like transcriptional profile in A375 cell lines:
(A) Screenshot of IGV genome browser (GRCH37/hg19) visualization of bigwig files generated from anti-MITF CUT&RUN sequencing in SKMEL28 cells that are wild-type or MITF-KO (delta 6), as well as anti-MITF, anti-TFE3, and IgG CUT&RUN sequencing in A375 cells that are wild-type or TFE3-KO. Gained paralog peaks in MITF-KO cells near the ZEB1 gene are highlighted in blue, and yellow asterisks indicate TFE3 occupancy at gained paralog peaks in A375 cells. (B) Density heatmap illustrating anti-MITF, anti-TFE3, and IgG CUT&RUN bigwig signals in A375 cells that are wild-type or TFE3-KO at genomic regions bound by MITF-homodimer and gained-TFE3 peaks in SKMEL28 wild-type and MITF-KO cell lines, respectively. (C) Volcano plot showing differentially expressed genes (DEGs) with qval < 0.5, among which 3344 genes were downregulated (blue) and 3206 genes upregulated (red) with log2 fold change (log2FC) ≥ |1| in TFE3-KO vs. A375 cells. (D) Gene Set Enrichment Analysis (GSEA) was used to determine differentially regulated gene ontology and signaling pathways using DEGs from TFE3-KO and wild-type A375 cell lines. Enriched ontology terms and pathways are labeled. The enrichment score is indicated by green lines, the number of genes by vertical black lines, and gene expression changes (positive or negative) by the red to blue heatmap (high to low). (E) Screenshot of IGV genome browser (GRCH37/hg19) visualization of bigwig files generated from anti-MITF and anti-TFE3 CUT&RUN sequencing in A375 cells that are wild-type (blue) or TFE3-KO (orange) near TFE3-activated genes associated with epithelial to mesenchymal transition and neural crest cell migration in cancer. (F) Validation of RNA-Seq by qPCR Analysis. Bar chart displaying the expression levels of genes identified as directly TFE3-dependent in A375 cells. The comparison is made between wild-type (WT) and TFE3 knockout (TFE3-KO) cells. The y-axis represents the relative expression levels, while the x-axis lists the validated genes. Error bars indicate the standard deviation from triplicate experiments.
TFE3 promotes invasive behavior of melanoma cell lines and patient-derived xenografts.
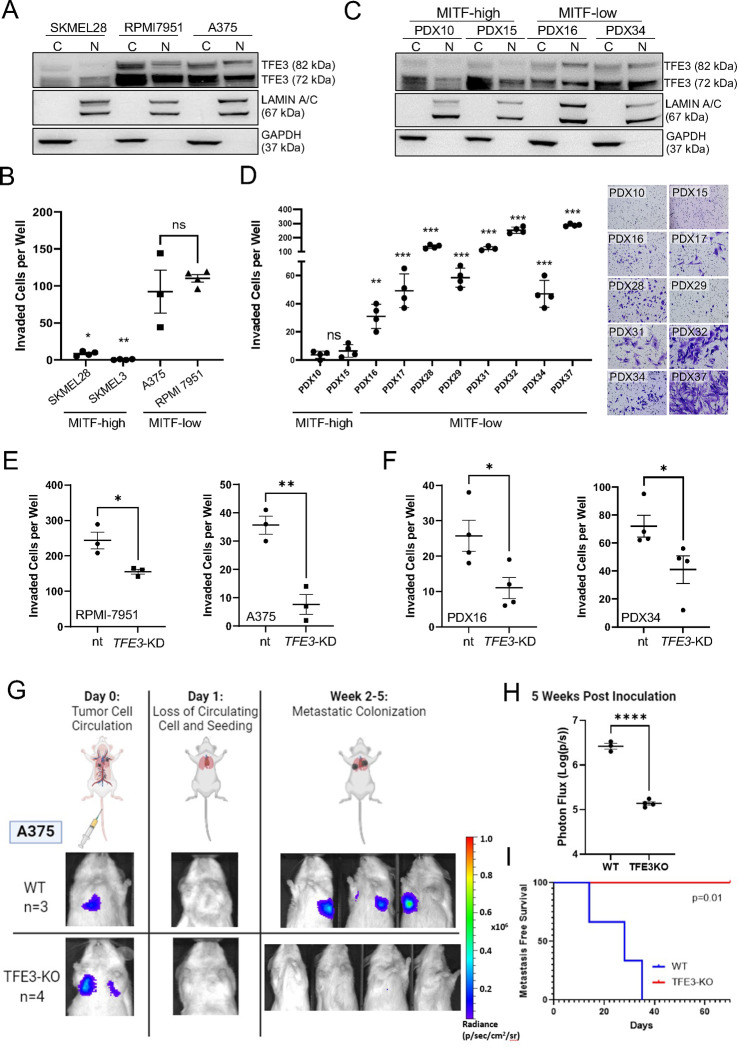
We predict that MITF-low melanoma cellular phenotype is related to TFE3 activity. We performed a transwell invasion assay to assess effects on migratory phenotype. Consistent with predictions based on gene expression profiles (Ghandi et al., 2019) and Western blot analysis of cytoplasmic and nuclear cell fractions (Figure 4A), the two TFE3-high cell lines (RPMI-7951 and A375) were more invasive than the three TFE3-low cell lines (SKMEL28, SKMEL24 and SKMEL3) (Figure 4B and Figure S4E–F). This was also true of MITF-low patient-derived melanoma cell lines, where the presence of nuclear TFE3 correlated with increased metastatic potential by transwell assays (Figure 4C–D). Moreover, knockdown of TFE3 in A375 (and in TFE3-KO clones), RPMI, and two PDX cell lines inhibited cell migration (Figure 4E–F, Figure S6A–C) but had no effect on cell proliferation (in MITF-low A375 cells) (Figure S6D), suggesting loss of TFE3 in MITF-low cells alters migration independent of cell viability. Similarly, knockout of TFE3 in MITF-KO SKMEL28 cell lines had no effect on proliferation (Figure S6E). Unexpectedly, however, MITF-KO SKMEL28 cells, which are TFE3 high, behaved similar to MITF/TFE3 double KO cells in transwell invasion assays and in colony formation assays (Figure S6F–G). Moreover, as we recently reported (Dilshat et al., 2021), unlike MITF-low melanoma cells, MITF-KO cells did not exhibit increased migration compared to SKMEL28 cells. We expect that this result is due to the non-physiologic nature of a complete knockout of MITF. In summary, TFE3 expression and nuclear localization largely correlated with invasive behavior in an in vitro assay.
Figure 4: TFE3 enhances invasion and metastasis in MITF-Low melanoma cell lines:
(A) Nuclear and cytoplasmic protein lysates were extracted from MITF-high SKMEL28 cells and MITF-low RPMI-7951 and A375 cell lines. The localization and expression of TFE3 was analyzed using Western blot. LaminA/B and GAPDH served as loading controls for the nuclear and cytoplasmic fractions, respectively. (B) Scatterplot representing the comparative cell invasion of SKMEL28, A375, and RPMI-7951 through Matrigel-coated Boyden chambers over 24 hours. Individual dots represent biological experiments (n=3) with three technical replicates. Statistical analysis was performed using the Student’s t-test. **P-value < 0.01. (C) Nuclear and cytoplasmic protein lysates were extracted from patient-derived xenograft (PDX) cell lines characterized as MITF-high (PDX10, PDX15) or MITF-low (PDX16, PDX34). The localization and expression of TFE3 was analyzed using Western blot. LaminA/B and GAPDH served as loading controls for the nuclear and cytoplasmic fractions, respectively. (D) Scatterplot representing the comparative cell invasion of MITF-high and MITF-low PDX cell lines through Matrigel-coated Boyden chambers over 24 hours. Individual dots represent biological experiments (n=4) with four technical replicates. Statistical analysis was performed using the Student’s t-test. **P-value < 0.01. (E) Knockdown of TFE3 using siRNA in A375 and RPMI cells, as well as in (F) PDX16 and PDX32, was performed and cell invasion was assessed over 24 hours using Matrigel-coated Boyden chambers. Scatterplots of the results indicate a profound effect of TFE3 on cell invasion for all cell lines tested. Individual dots represent biological experiments (n=3) with three technical replicates. Statistical analysis was performed using the Student’s t-test. **P-value < 0.01. (G) Tail vein metastatic colonization assay design. Tumor size measured via photon flux (radiance). (H) Comparison of metastasis size at 5 weeks post inoculation. Statistical analysis using Student’s t-test. **** P-value <0.0001. (I) Kaplan Meier curve of metastasis free survival. Statistical analysis using Log-Rank Test.
To investigate the role of TFE3 on melanoma behavior in vivo, we transfected WT and TFE3-KO A375 cells with firefly luciferase, generating lines with stable expression, and injected them into immunocompromised mice. To test primary tumor formation, we injected the cells into the flank, and to test metastatic colonization potential we injected them into the lateral tail vein; cancer cells injected in this way preferentially colonize the lung (Thies et al., 2020). Animals injected in the flank with either WT or TFE3-KO A375 cells exhibited similar latency to primary tumor formation (Figure S7A–B). Animals injected in the tail vein with WT A375 cells started to exhibit tumors in the lungs after about 14 days and had a median time of tumor-free survival of just 5 weeks. By contrast, those injected in the tail vein with TFE3-KO A375 cells remained tumor free for the duration of the assay (10 weeks) (Figure 4G–I). These findings demonstrate that TFE3 expression is critical for melanoma cell migration and metastatic colonization of distant organs.
MITF promotes cytoplasmic retention and lysosome-mediated degradation of TFE3
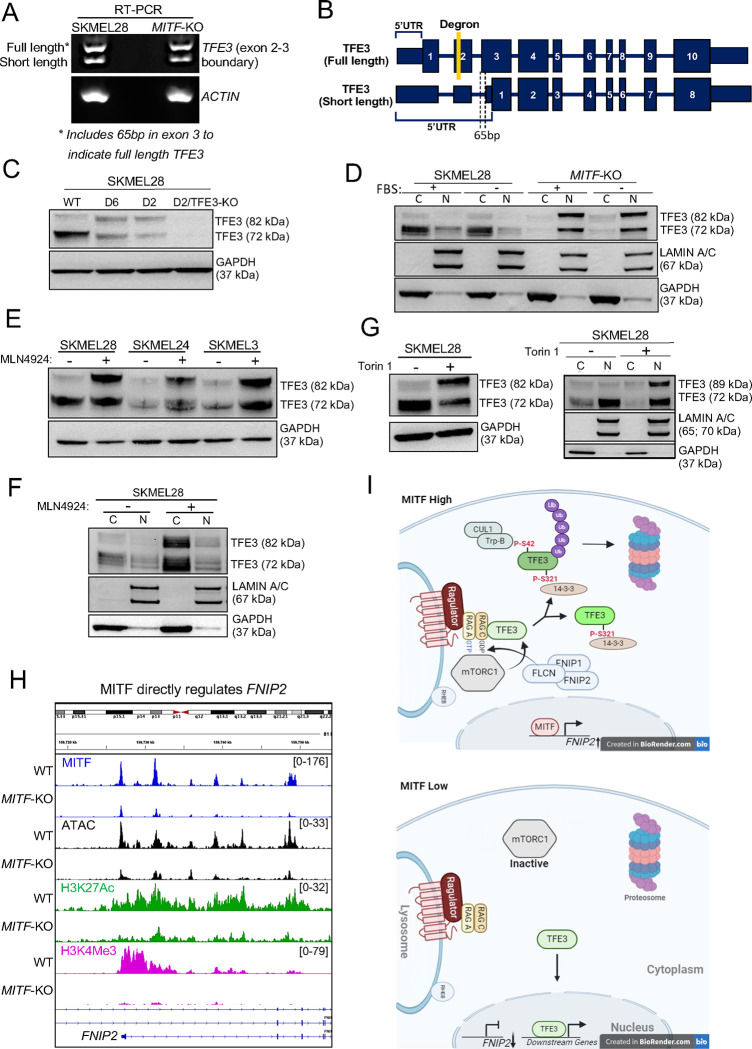
How does the loss of MITF lead to increased TFE3 activity? TFE3 mRNA expression by RNA-Seq was comparable in wild-type and MITF-KO SKMEL28 cells, arguing against direct effects of MITF on TFE3 transcription. During profiling of multiple melanoma cell lines, we noted two isoforms of TFE3 (72 kDa and 82 kDa), with the 82 kDa isoform correlating with MITF-low melanoma (Figure 4A and 4C, Figure S8A). RT-PCR and sequencing revealed two splice variants, differing by 65 base pairs at the exon2-exon3 boundary with both isoforms expressed in equal ratios in wild-type and MITF-KO SKMEL28 cells (Figure 5A–B). Western blot analysis of lysates from cells of both genotypes revealed full-length TFE3 at 82 kDa and a 72 kDa isoform presumably derived from the shorter splice variant (Figure 5C). Although overall levels of TFE3 protein were comparable in wild-type and MITF-KO cells, the fraction of full-length TFE3, and the proportion of TFE3 localized to the nucleus, was higher in MITF-KO cells (Figure 5D). Following the same trend, in a MITF-high cell line (SKMEL28), TFE3 expression was less nuclear than in two MITF-low cell lines (RPMI-7952 and A375) (Figure 4A). Among the patient-derived melanoma xenograft (PDX) cell lines, PDX-10 and PDX-15 were characterized as MITF-high/TFE3-low, while PDX-16, PDX-28, PDX-32, PDX-34, and PDX-37 were MITF-low/TFE3-high (TFE3-high referring to the 82 kDa isoform). PDX-31 expressed high levels of MITF and TFE3 whereas PDX-17 and PDX-29 showed relatively low expression of both proteins (Figure 4C, Figure S8A). Morphologically, MITF-high PDX cell lines resembled melanocytic melanoma cells, whereas MITF-low cell lines had an elongated and spindly appearance (Figure S8B), as described in mesenchymal-like melanoma cell lines (Wouters et al., 2020). While there was variability of bulk TFE3 protein levels across the PDX cell lines there was a strong trend of higher nuclear-to-cytoplasm ratio of TFE3 in MITF-low cell lines relative to MITF-high cells, and full-length (82 kDa) TFE3 protein was observed exclusively in MITF-low cell lines (Figure 4C), except for the PDX31 cell line which likely representing the intermediate melanoma subtype (Figure S8A). In summary, loss of MITF leads to a shift towards full-length, nuclear-localized TFE3, and this trend was observed in MITF-low melanoma cells.
Figure 5: MITF regulates TFE3 protein stability and nuclear localization through activation of FNIP2 and mTORC1 signaling.
(A) Gel image of RT-PCR showing two distinct bands separated by 65 base pairs (bps), corresponding to different transcript variants of TFE3. Both bands are present in equal ratios within and between the SKMEL28 and MITF-KO cell lines, indicating consistent expression levels of the TFE3 transcript variants across these cell lines. Primers targeting actin mRNA were used as a loading control to ensure equal RNA input and efficient cDNA synthesis across samples. (B) Schematic of two TFE3 variants, labeled as full-length and short-length TFE3. The full-length TFE3 contains a degron sequence in exon 2, represented by a yellow rectangle. Both TFE3 variants share the same transcriptional start site, but have different ribosome entry sites, leading to two distinct protein products with molecular weights of 72 kDa and 82 kDa. (C) Immunoblotting of anti-TFE3 on bulk protein lysates of SKMEL28 cells, two MITF-KO cell lines (delta 2 and delta 6; (Dilshat et al., 2021)), and one double MITF/TFE3-KO cell line. Immunoblotting of anti-GAPDH was used as a loading control. The upper ~82 kDa band represents the full-length TFE3, while the lower ~72 kDa band represents the short-length TFE3. (D) Immunoblotting of anti-TFE3 from SKMEL28 and MITF-KO (delta 6) nuclear and cytoplasmic cell fractions +/− fetal calf serum (FBS) for 24 hours. Immunoblotting of LAMIN A/B and GAPDH were used as nuclear and cytoplasmic loading controls, respectively. (E) Immunoblotting of anti-TFE3 from bulk protein lysates of SKMEL28, SKMEL24, and SKMEL3 cell lines, −/+ 1 μM MLN4924. (F) Immunoblotting of anti-TFE3 from SKMEL28 nuclear and cytoplasmic cell fractions, treated with −/+ 1 μM MLN4924 for 24 hours. Immunoblotting of LAMIN A/B and GAPDH were used as nuclear and cytoplasmic loading controls, respectively. (G) Immunoblotting of TFE3 in bulk (left) or nuclear and cytoplasmic cell fractions (right) for SKMEL28 cells −/+ 500 nM Torin1 for 24 hours. (H) Screenshot of IGV genome browser (GRCH37/hg19) visualization of bigwig files generated from anti-MITF, anti-H3K27Ac, and anti-H3K4Me3 CUT&RUN-seq and ATAC-seq at the FNIP2 locus in SKMEL28 cells that are wild-type or MITF-KO (delta 6). (I) Schematic representation of MITF-mediated regulation of TFE3 protein stability and nuclear localization in MITF-high and MITF-low conditions.
How does MITF affect TFE3 cellular localization? In HEK-293T cells, it was recently shown that mTOR-mediated phosphorylation of full-length TFE3 activates a degron for Cullin-RING E3 Ubiquitin Ligase 1 (CUL1) which subsequently targets TFE3 for lysosomal degradation (Nardone et al., 2023). This degron is absent from the smaller isoform of TFE3 and is lost in TFE3 translocations present in renal cell carcinoma, thus accounting for its stable expression and oncogenicity. Of note, it is also absent from the variant of MITF present in melanocytes and melanoma (Nardone et al., 2023, Goding and Arnheiter, 2019). To test if TFE3 levels are sensitive to the CUL1 pathway in melanoma, we treated three TFE3-low cell lines (SKMEL28, SKMEL24 and SKMEL3) with MLN4924, a selective NEDD8-Activating Enzyme inhibitor, which inactivates CUL activity (Harper and Schulman, 2021). MLN4924-treatment led to an increase in both full length and short form TFE3 and an increased fraction of the full-length form (Figure 5E), but TFE3 cellular localization was not altered by MLN4924-treatment of SKMEL28 cells (Figure 5F). The mTOR pathway responds to cellular nutrition, and it has been shown that in an immortalized human retinal pigmented epithelial (RPE) cell line, TFE3 nuclear localization was induced upon 24-hour serum deprivation (Martina et al., 2014). To test if mTOR pathway regulates TFE3 in melanoma, we subjected MITF-KO cells to serum starvation, but did not detect a change in nuclear localization of TFE3 (Figure 5D). However, when we inhibited the mTOR pathway with Torin, nuclear localization and ratio of full length to small form of TFE3 were both increased (Figure 5G).
One remaining question is how MITF affects mTOR activity. In naïve pluripotent stem cells, a complex containing Folliculin (FLCN) and Folliculin interacting proteins 1 and 2 (FNIP1 and FNIP2) promotes phosphorylation and cytoplasmic retention of TFE3 (Mathieu et al., 2019). Interestingly, expression of several members of the FLCN and mTOR pathway is significantly lower in MITF-KO cells compared to wild-type (Table S2). In particular, expression of a short length FNIP2 variant is strongly MITF-dependent and appears to be directly MITF-activated (Figure 5H). It is therefore plausible that when MITF is low, the activity of the FLCN/FNIP1/FNIP2 and CUL1-dependent degron pathway is low, resulting in stabilization of the TFE3 protein. Together these findings support the model that mTOR pathway acts at the top of a cascade that stimulates both cytoplasmic retention (through interaction with 14-3-3) and FLCN/CUL1-mediated lysosomal degradation (Figure 5I, schematic). Of note, both YWHAH (14-3-3 Eta) and YWHAQ (14-3-3 Tau) are directly regulated by MITF in SKMEL28 cells and significantly downregulated by loss of MITF, providing further evidence that MITF negatively regulates TFE3 activity.
Discussion
A knowledge of cancer plasticity and intra-tumor heterogeneity in melanoma and other cancers is central to understanding the cellular mechanisms governing tumor invasiveness and therapy resistance (Fanelli et al., 2020). The cancer stem cell (CSC) model, whereby a minority of cancer cells exist with self-renewing capacities, provides an explanation for maintenance of tumor heterogeneity and hierarchy. CSCs exhibit plasticity, switching between tumor-initiating stem-cell like, rapidly-proliferating and differentiated states through environmental, epigenetic, and transcriptional stimuli (“Reviewed in”(Fanelli et al., 2020, Torborg et al., 2022)). The ability to selectively target these cell populations, or plasticity in general, therapeutically remains in its infancy. Whether core transcriptional programs exist that define plasticity across many cancer types, and the relationship between plasticity and invasive phenotypes, remain unknown (Torborg et al., 2022). Cutaneous melanoma exemplifies these characteristics. Melanoma tumors are heterogeneous, comprising phenotypically, transcriptionally, and epigenetically distinct cell populations that also differ in their sensitivity to therapy (Wouters et al., 2020, Rambow et al., 2018, Hoek et al., 2008, Hoek and Goding, 2010). For instance, a common finding among these studies is that proliferative cell states have high levels of MITF and SOX10 expression and/or activity while invasive, dedifferentiated cells have low levels of MITF expression and/or activity. Phenotype switching refers to the phenomenon of cells transitioning among these states through transcriptional and epigenetic reprogramming. Elucidating the mechanisms governing phenotype switching may help in the design of novel therapies to target invasive and therapy-resistant melanoma.
Here we showed that TFE3 promotes the expression profile and behaviors characteristic of MITF-low melanoma samples, and identified a mechanism by which MITF normally suppresses these qualities. Through investigating the transcription factor binding profiles and chromatin accessibility in wild type and MITF-KO melanoma cells, we found that in an MITF-high melanoma cell line, MITF activates enhancers at genes associated with pigmentation and differentiation, but that upon deletion of MITF, its paralog TFE3 activates expression of genes promoting migration and stem-cell like features. Further, we found that knockdown or knockout of TFE3 in aggressive MITF-low cell lines suppressed their migration in vitro and colonization in vivo. It may also have increased their stem-cell like qualities, as TFE3 directly activated genes associated with pluripotency (SOX2, KLF and WNT family members), as it does in naïve embryonic stem cells (Mathieu et al., 2019). MITF and TFE3 levels were reported to be negatively correlated (Möller et al., 2019), and we found that TFE3 protein stability and nuclear localization were both altered by the knockout of MITF. We found that MITF directly activates expression of members of the 14-3-3 protein family, which potentially bind and sequester TFE3 to the nucleus, and expression of FNIP2 which together with FNIP1 and FLCN recruit mTORC1 and TFE3 to the lysosome membrane, resulting in cleavage and lysosome degradation of full length TFE3 (Nardone et al., 2023). FLCN-mediated degradation of TFE3 also occurs in hESC (Mathieu et al., 2019). Together, the data show that TFE3 promotes an invasive phenotype, while MITF promotes a differentiated phenotype and, by activating FNIP2 expression, represses the invasive, stem-cell like phenotype.
This work can be extended in at least two ways. First, not all enhancers that gain activity in MITF-KO cells have gained TFE3 peaks, which points to other mechanisms or factors that activate genes in the absence of MITF. Many, but not all, persistent paralog peaks were absent in TFE3-KO cells, implying that that there may be homologs other than TFE3 bound by the anti-MITF antibody and regulating gene expression; roles of other MITF paralogs were not considered here. Importantly, however, knocking out TFE3 completely inhibited melanoma metastasis suggesting that TFE3 plays a major role in this process. Second, we have not discovered how the invasive phenotype suppresses the differentiated one. Transcriptional regulation of MITF expression (Kubic et al., 2008, Lang et al., 2005) and post-transcriptional modification of MITF protein (Bertolotto et al., 2011), as well as cooperating transcription factors (Kenny et al., 2022) have been described and are potential loci of control, and likely under the influence of the tumor microenvironment.
While many studies report success of inhibiting the mTOR pathway in preclinical models of cancer, our findings suggest a more nuanced approach may be more successful still. mTORC1 is a signaling hub that integrates environmental cues, including nutrients, metabolic intermediates, and growth factors to regulate growth and metabolism (Ling et al., 2022). Recently, canonical and non-canonical activities have been described, which differ by their mechanisms of substrate recruitment (Napolitano et al., 2022). Substrates of canonical mTORC1 activity are p70 ribosomal S6 kinase (S6K) and eukaryotic initiation factor 4E-binding protein 1 (4EBP1), while TFE3 is a substrate of non-canonical activity (Napolitano et al., 2022). Clearance of FLCN suppresses mTORC1-mediated phosphorylation, and thereby inactivation, of TFE3, but not of its canonical substrates (Gosis et al., 2022). Multiple studies have shown mTOR activity in malignant melanoma (Karbowniczek et al., 2008), and that rapamycin, which inhibits mTOR kinase activity, inhibits melanoma tumor growth (Gong and Xia, 2020, Wang et al., 2024). In one recent case, this was shown to occur by stimulating autophagy (Wang et al., 2024). Our results suggest that mTORC1 activity in melanoma cells destabilizes TFE3 and thereby suppresses metastatic behavior. Therefore, an optimal melanoma therapy may include an inhibitor of canonical mTORC1 activity that permits non-canonical mTORC1 activity, thus shutting down nuclear transport of TFE3 in MITF-low tumors.
Materials and Methods
Cell Culture
SKMEL28 (HTB-72), A375, RPMI-7951 (HTB-66), SKMEL3 (HTB-69), and SKMEL24 (HTB-71) cell lines was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and maintained in the appropriate medium as recommended by the manufacturer. MITF-KO Δ2 and MITF-KO Δ6 cell lines were a gift from the laboratory of Eirikur Steingrimsson (University of Iceland). Patient derived melanoma cell lines (PDX10, PDX15, PDX16, PDX34) were a gift from the laboratory of Michael Henry (University of Iowa). SKMEL28, and PDX cell lines were maintained at 37°C and 5% CO2 in RPMI 1640 medium (#11875093, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. A375 cells were maintained at 37°C and 5% CO2 in DMEM medium (#11965092, Thermo Fisher Scientific) supplemented with 10% FBS and 1% penicillin-streptomycin. SKMEL3 and SKMEL24 cell lines were maintained in EMEM medium (30–2003, ATCC) supplemented with 15% FBS and 1% penicillin-streptomycin. RPMI 7951 cells were maintained in EMEM medium (30–2003, ATCC) supplemented with 10% FBS and 1% penicillin-streptomycin. PDX-derived melanoma cell lines were developed directly from the patient biopsy (PDX#28, PDX#29, PDX#32, PDX#34, and PDX#37) or from a first generation PDX (PDX#10, PDX#15, PDX#16, PDX#17, and PDX#31). Cells were cultured in RPMI1640 supplemented with 10% FBS, 1%NEAA and grown at 37 °C in a 5% CO2 atmosphere.
Generation and culture of PDX cell lines
Patient tumor samples to be used in PDX models were collected and processed by the University of Iowa College of Medicine Tissue Procurement Core under IRB# 200804792. Tissue collection and distribution was performed in accordance with the guidelines of the University of Iowa Institutional Review Board. Briefly, following collection tumor samples were placed in DMEM containing 5% FBS, 1X Pen/Strep, and 1% fungizone. For initiation of PDX tumors, patient tumor samples were processed by mincing until they passed through an 18 G needle easily. The cells were then suspended in a 1:1 mixture of DMEM (GIBCO) and Matrigel (Corning, 354230), and subcutaneously injected into 2 or 3 NOD.Cg-PrkdcscidIl2rgtmWjl/SzJ (NSG) (Jackson Laboratory) mice. For the propagation of samples, tumors were dissociated into single-cell suspensions and each mouse was injected subcutaneously with 1×106 cells. Single-cell suspensions were generated by placement of the tumor into a sterile Petri dish containing 10mL collagenase IV solution (9 mL HBSS (Ca2+- and Mg2+-free), 1 mL collagenase IV (Worthington Biochemical Corp.), and 5 mM CaCl2). The tumor samples were first minced and incubated for 1 hr at 37°C. Following this, samples were further digested in 10 mL 0.05% trypsin (at 37°C for 10 min), and passed through an 18 G needle 30 times, after which the suspension was filtered using a 70 μm cell strainer.
Generation of TFE3-KO cells and validation of mutations using Sanger Sequencing
CRISPR/Cas9 strategies were utilized to generate knock out mutations in the TFE3 gene in A375, RPMI 7951 and PDX cell lines. Guide RNA was designed targeting exon 4 of TFE3, which encodes the helix-loop-helix DNA binding domain (Beckmann and Kadesch, 1991) (Figure S5A–C). The gRNA used was: CTTCTCTGAGCTGGACCCGA (Integrated DNA Technologies INC, IDT). gRNA was conjugated 1:1 with tracerRNA (crRNA) (IDT) by incubating at 95°C for 5 minutes and letting rest till room temperature. Cas9 nuclease (#10007807, IDT) was used to form ribonucleoprotein complex (RNPs). 1 × 10^6 cells were electroporated with RNPs in Gene Pulser electroporation buffer (#1652676, Bio-Rad, Hercules, CA) with electroporation enhancer (#1075915, IDT) using 140V, 2mm pulse gap. Nontargeting negative control gRNA (#1072544, IDT) was used as control for all CRISPR/Cas9 knockouts.
For sanger sequencing, genomic DNA was isolated from the TFE3 knockout cell lines using DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). Regions of interest was amplified using region specific primers: Fwd GCTAGCTCCATGGCTTAGCG, Rev GAGACGCCAACCACAGAGAT. The amplified DNA was run on a 2% agarose gel at 100V for 20 minutes. The bands were cut out of the gel and DNA was extracted using QIAquick Gel Extraction Kit (#28704, Qiagen) and QIAquick PCR purification kit (#28104, Qiagen). Purified DNA fragments were cloned into pcDNA3.1 vector and transformed into 5-alpha competent cells (#C2987H, New England Biolabs, Ipswich, MA) and 10 colonies were picked for each cell line, DNA isolated, and sequenced using Sanger Sequencing.
RNA isolation, cDNA synthesis and RT-qPCR
Cells were grown in 6-well culture dishes to 70–80% confluency and RNA was isolated with RNeasy Plus kit (#74034, Qiagen). CDNA was generated using High Capacity cDNA Reverse Transcription Kit (#4368814, Applied Biosystems) using 1 μg of RNA. RT-qPCR was performed with Taqman RT PCR reagents (ThermoFisher Scientific, 4304437) and the following probes: MMP3 (ThermoFisher Scientific, Hs00968305_m1), FSTL3 (ThermoFisher Scientific, Hs00610505_m1), TNFAIP3 (ThermoFisher Scientific, Hs00234713_m1), PTHLH (ThermoFisher Scientific, Hs00174969_m1) and ZEB1 (ThermoFisher Scientific, Hs01566408_m1). qPCR reactions were performed in triplicate and relative gene expression was calculated using D-ΔΔCt method (Livak and Schmittgen, 2001). The geometric mean of 18S Ribosomal subunit was used to normalize gene expression of target genes.
Wound scratch Assay
8 × 10^5 cells were seeded per well of a 6-well plate to reach confluent monolayer. Scratches were made with a P200 pipet tip in a cross configuration. 4 areas per well were chosen in the cardinal directions for image analysis. Images were recorded at each chosen area using 4x magnification. Images were analyzed using ImageJ (National Institutes of Health) with Wound healing plug-in. Area of wound was quantified at time points 0h, 24h, 36h, 48h, 60h, 72h and 84h to assess gap closure.
Transwell Invasion Assay
Invasion assay 24 well plate transwell chambers with 8um pore size (#353097, Corning, Corning, NY) were coated with growth factor reduced matrigel matrix (354230, Corning) at 1:10 dilution with serum free medium. Cell suspension of 15,000 cells in appropriate seum free medium was added to the matrigel coated upper chamber. Medium containing 10% FBS was added to the lower chamber to serve as a chemoattractant. Cells were allowed to invade for 24 hours after which cells that migrated to other side of the membrane were fixed with Crystal violet (with 4% PFA) and let to dry. Images were acquired using EVOS microscope (ThermoFisher Scientific) with 10x magnification and cells were counted. Student’s t test was used to compare conditions.
RNA sequencing and data analysis
RNA-sequencing experiments were performed on SKMEL28 WT, MITF-KO Δ2, A375 and A375 TFE3-KO cell lines in triplicate. RNA was isolated using RNeasy Plus Kit (#74034, Qiagen) and submitted to the Novogene Co (China) for library construction and RNA sequencing with 75 bp paired-end reads on Novaseq 6000 platform. Data analysis was performed on UseGalaxy. All raw sequencing data was assess by FastQC for quality control. Reads were mapped with RNAStar to human reference genome hg38 with default parameters. FeatureCounts was used to calculate read counts for each individual gene from STAR output bam files. Differential gene expression analysis was performed using DEseq2. Adjusted p-value <0.05 was used for statistically significant difference. Gene Set Enrichment Analysis (GSEA) was performed following the instructions using all available signatures in the MSigDB (v2023.1).
CUT&RUN
To identify direct MITF and TFE3 target genes and histone 3 modifications, we performed Cleavage Under Targets and Release Using Nuclease (CUT&RUN) sequencing in SKMEL28, MITFΔ6, MITFΔ2, MITFΔ2/TFE3-KO, A375 and A375 TFE3-KO cell lines as previously described (Kenny et al., 2022, Skene and Henikoff, 2017). Cells in log-phase culture (60–80% confluent) were harvested by cell scraping, centrifuged at 300 g and washed twice in calcium-free wash-buffer (20 mM HEPES, pH7.5, 150 mM NaCl, 0.5 mM spermidine and protease inhibitor cocktail, cOmplete Mini, EDTA-free Roche). Cells were counted and 500,000 cells per antibody per condition were used for CUT and RUN. CUT and RUN experiments were performed in biological duplicates. Preactivated Concanavalin A-coated magnetic beads (Bangs Laboratories Inc) were added to cell suspension and incubated at 4°C for 10 minutes. Antibody buffer (wash buffer with 2mM EDTA and 0.025% digitonin) containing anti-MITF (Sigma, HPA003259), anti-TFE3 (Cell Signaling Technologies (CST), 14779), Anti-H3K27Ac (CST, #8173), Anti-H3K27Me3 (CST, #9733), H3K4Me3 (CST, #9751), H3K9Ac (CST, #9649), or Rabbit IgG (Millipore, 12–370) was added and cells were incubated overnight at 4°C on nutator. The following day, cells were washed in dig-wash buffer (wash buffer containing 0.03% digitonin) and pAG-MNase was added at a concentration of 500 μg/mL. The pAG-MNase enzyme was purified following a previously described protocol (Meers et al., 2019). The pAG-MNase reactions were quenched with 2X Stop buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 0.05% Digitonin, 100 μg/mL RNAse A, 50 μg/mL Glycogen, and 2 pg/mL sonicated yeast spike-in control). Released DNA fragments were Phosphatase K (1 μL/mL, Thermo Fisher Scientific) treated for 1 hr at 50°C and purified by phenol/chloroform-extracted and ethanol-precipitated. CUT&RUN experiments were performed in parallel as positive control and fragment sizes analyzed using a 2100 Bioanalyzer (Agilent).
Library preparation and data analysis
CUT and RUN libraries were prepared using the KAPA Hyper Prep Kit (Roche). Quality control post-library amplification was conducted using the 2100 Bioanalyzer for fragment analysis. Libraries were pooled to equimolar concentrations and sequenced with paired-end 150 bp reads on a NovoSeq HiSeq X instrument. Paired-end FastQ files were processed through FastQC (Andrews, 2010) for quality control. Reads were trimmed using Trim Galore Version 0.6.3 (Developed by Felix Krueger at the Babraham Institute) and Bowtie2 version 2.1.0 (Langmead and Salzberg, 2012) was used to map the reads against the hg19 genome assembly. The mapping parameters were performed as previously described (Meers et al., 2019).
Assay for Transposase-Accessible Chromatin with sequencing (ATAC-Seq)
ATAC-seq was performed according to (Kenny et al., 2022) with minor alterations. Briefly, 70,000 SKMEL28 wild type and ΔMITF-X6 (four replicates) were lysed in ice-cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40: Sigma). Transposition was performed directly on nuclei using 25 μl tagmentation reaction mix (Tagment DNA Buffer #15027866, Tagment DNA Enzyme #15027865 from Illumina Tagment DNA kit #20034210). Tagged DNA was subjected to PCR amplification and library indexing, using the NEBNext High-Fidelity 2x PCR Master Mix (New England Biolabs #M0451S) with Nextera DNA CD Indexes (Illumina #20015882), according to the following program: 72°C for 5 minutes; 98°C for 30 seconds, 12 cycles of 98°C for 10 seconds, 63°C for 30 seconds, and 72°C for 1 minute. The PCR product was purified with 1.8 times the volume of Ampure XP beads (Beckman Coulter #A63881). Library quality was assessed using a BioAnalyzer 2100 High Sensitivity DNA Chip (Agilent Technologies). All DNA libraries that exhibited a nucleosome pattern were pooled and processed for 150 bp paired-end sequencing.
Cytoplasmic and Nuclear fractionation
Cytoplasmic and nuclear protein fractionation and extraction was performed in cell lines using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Cat# 78835. ThermoFisher Scientific). Cytoplasmic and nuclear protein components were subsequently run in 4–12% Bis-Tris Gel with appropriate primary and secondary antibody incubation for analysis as described below. Anti-GAPDH (Santa Cruz, sc-47724) was utilized as cytoplasmic protein loading control while anti-Lamin A/C (Cell Signaling Technologies, #4777) was used as nuclear protein loading control.
Immunoblotting
Cells were resuspended in RIPA cell lysis buffer (R0278, Sigma-Aldrich) with Halt Protease and Phosphatase Inhibitor Cocktail (#78440, Thermo Fisher Scientific). Protein concentration was assessed by the Pierce BCA Protein Assay Kit (#23225, Invitrogen). NuPAGE LDS Sample Buffer and Reducing Buffer (NP0007 & NP0009, Invitrogen) was added to equal amounts of protein (15–25 mg) and samples were boiled at 95 °C for 10 minutes. Protein samples were loaded to 4–12% Bis-Tris Gel (NP0335BOX, Invitrogen), then transferred to polyvinylidene fluoride (PVDF) membranes (IB24001, Invitrogen), which were incubated with primary antibodies overnight at 4°C. After TBS-T washing, membrane was incubated with horseradish peroxidase-conjugated anti-rabbit or anti-mouse (#7074S & #7076S, Cell Signaling Technology), and developed using SuperSignal West (#34095, Thermo Fisher Scientific). The following antibodies were used for western blots:
| Antibody (Concentration) | Company | Catalog Number |
|---|---|---|
| HRP-linked Anti-Rabbit IgG (1:1000) | Cell Signaling Technologies | #7074 |
| HRP-linked Anti-Mouse IgG (1:1000) | Cell Signaling Technologies | #7076 |
| Rabbit Anti-TFE3 (1:1000) | Cell Signaling Technologies | #14779 |
| Mouse Anti-TFE3 (1:1000) | Sigma-Aldrich | SAB4200824 |
| Rabbit Anti-MITF (1:1000) | Sigma Aldrich | SAB003259 |
| Rabbit Anti-ZEB1 (1:1000) | Cell Signaling Technologies | #70512 |
| Mouse Anti-GAPDH (1:1000) | Santa Cruz | sc-47724 |
| Mouse Anti-Lamin A/C (1:1000) | Cell Signaling Technologies | #4777 |
Cell Proliferation Assay (MTT)
Cells were seeded at 1,000 cells per well of a 96-well plate in a set of 6 technical replicates. Every 24 hours, cells were grown in fresh medium with 0.5mg/ml MTT reagent (# M6494, Thermo Fisher) for 2 hours at 37°C and 95% CO2, followed by 10 minutes of solubilization in 200 ul DMSO. Measurements were obtained on an Infinity 200 Pro (Tecan: Switzerland) plate reader at an absorbance of 540 nm. Measurements were taken daily for one week and normalized to Day 0 absorbance measurements.
Soft Agar Assay
Bottom layer of 0.5% agarose in 12% FBS of appropriate media was plated. Top layer of 0.35% agarose in 12% FBS of appropriate media was created and 10,000 cells per well were seeded and incubated for 2–4 weeks at 37°C and 95% CO2 to allow for colony formation. Soft agar assay was performed in triplicate with 3 representative images per replicate quantified and averaged and conditions compared using Student’s t tests.
Tail Vein and flank injection
Cell lines with stable luciferase expression were grown in subconfluent conditions. To assess metastatic colonization, 1 × 10^6 cells were harvested and injected into the lateral tail vein of immunocompromised NOD/SCID mice. Mice were imaged with in vivo small animal imaging (IVIS) at day 0 with visualization of circulating tumor cells confirming injection accuracy. Mice were imaged on day 1 to confirm loss of signal. Imaging of mice was performed weekly to assess for metastatic colonization and metastasis growth. To assess primary tumor formation, 1 × 10^6 cells were harvested and injected into the flanks of NOD/SCID mice to create xenografts. Mice were imaged weekly to assess tumorigenesis and growth. Mice were euthanized for body condition score approaching 2 or less or flank tumor exceeding 2 cm in diameter. All mouse experiments were approved by and performed in accordance with the University of Iowa IACUC.
Supplementary Material
Figure S1: Multiple sequence alignment of the MIT/TFE transcription factor family and the anti-MITF antibody immunogen sequence by CLUSTALW. Amino acid sequences of MITF, TFE3, TFEB, and TFEC were recovered from UniProt and aligned to the immunogen sequence of the anti-MITF (HPA003259, Sigma) using by CLUSTALW (Thompson et al., 1994).
Figure S2: (A) Venn Diagram of overlapping anti-MITF CUT&RUN-seq peaks identified by MACS2 in delta 2 and delta 6 MITF-KO cell lines. (B) Density heatmap representing anti-MITF and anti-H3K27Ac CUT&RUN-Seq as well as ATAC-Seq in SKMEL28 WT and MITF-KO (delta 6) cell lines. Peak regions represent MITF-homodimer peaks, gained-paralog peaks, and persistent paralog. (C) Pie-chart of MITF-homodimer peaks, gained-paralog peaks, and persistent paralog peaks that overlap MITF-regulated enhancers, enhancer subtypes as labeled. Gene Ontology (GO) analysis of (D) MITF-homodimer occupied genes and (E) Gained-paralog occupied genes (peaks are within 100 kb of a genes TSS).
Figure S3: (A) Protein lysates from SKMEL28 WT and MITF-KO (Delta6) cell lines were subjected to immunoprecipitation using an anti-MITF antibody. The immunoprecipitates were analyzed by Western blot to detect TFE3 protein. A distinct band corresponding to the expected molecular weight of TFE3 (~82 kDa) was observed in the immunoprecipitated samples, including those from the MITF-KO cell line. This suggests potential cross-reactivity of the anti-MITF antibody with nondenatured TFE3 protein. (B) The same immunoprecipitated lysates were analyzed by Western blot to detect the MITF protein. A band at the expected molecular weight of MITF (~60 kDa) in the SKMEL28 WT cell line using the anti-MITF antibody was observed. However, no/weak MITF positivity was detected in the MITF-KO cell line. Suggesting, the anti-MITF antibody does not have strong immunoreactivity with denatured TFE3. (C) Protein lysates from SKMEL28 WT and MITF-KO cell lines were also subjected to immunoprecipitation using an anti-TFE3 antibody, followed by Western blot analysis to detect TFE3. Two distinct bands at the expected molecular weight for TFE3 (~72, 82 kDa) was observed in both cell lines with the larger form more prominent in MITF-KO cells. (D) Mass spectrometry analysis of protein lysates from SKMEL28 WT and MITF-KO (Delta6) cells following immunoprecipitation using an anti-MITF antibody.
Figure S4: (A) Investigation of ZEB1 expression differences. qPCR comparing expression of ZEB1 in SKMELWT, ΔMITF-X2 control CRISPR clones and ΔMITF-X2/ΔTFE3KO Clones. Relative expression is graphed on y-axis. Immunoblotting of ZEB1 and TFE3 protein level in ΔMITF-X2 vs ΔMITF-X2/ ΔTFE3KO clones and A375 WT vs TFE3-KO clones. GAPDH is utilized as loading control. (B) Gene ontology analysis of MITF-Repressed genes. (C) Gene Ontology analysis of MITF-activated genes. (D) Immunoblotting of TFE3 and MITF in A375 and SKMEL28 WT cells. GAPDH is utilized as loading control. (E) Transwell invasion assay of A375 vs SKMEL28 cells. Number of invaded cells is plotted. **P-value <0.01.
Figure S5: (A) Schematic of CRISPR gRNA target region for TFE3 knockout system. (B) Immunoblotting of TFE3 in A375 WT vs individually isolated TFE3-KO clones. GAPDH is utilized as loading control. (C) Sanger sequencing of CRISPR target region of TFE3 gene with associated translated amino acid sequence. (D) Screenshot of UCSC (hg19) genome browser visualization of bigwig files generated from anti-MITF and anti-TFE3 CUT&RUN demonstrating representative TFE3 gained-paralog peak at F2RL1 enhancer.
Figure S6: (A) Transwell invasion assay of A375 WT vs nontargeting control CRISPR clones vs CRISPR TFE3-KO clones performed with n=4 replicates. Invaded cells per well graphed on y-axis. **P-value <0.01, ***P-value <0.001. (B) Representative 10x brightfield images taken from transwell invasion assay with A375 vs RPMI vs PDX 16 vs PDX 34. (C) Scratch wound healing assay using A375 WT and TFE3-KO cells. 4x brightfield images taken at 0, 24, 48 hours post scratch. Area of scratch and % wound open quantified via ImageJ. *P-value<0.05, ***P-value<0.001. (D) Cell proliferation assay with A375 WT vs TFE3-KO clones. 1000 cells are plated at day 0 in 96 well plates with n=6. Normalized absorbance @ 540nm is plotted on the Y-axis. (E) Cell proliferation assay of SKMEL28 WT vs ΔMITF-X2 vs ΔMITF-X6 vs ΔMITF-X2/ΔTFE3KO. 1000 cells are plated at day 0 in 96 well plates with n=6. Normalized absorbance at 540nm is plotted on the Y-axis. (G) Transwell invasion assay with SKMEL28 WT vs delta 2 vs delta 6 vs delta 2/TFE3-KO. Invaded cells per well plotted. (H) Soft agar colony formation assay with SKMEL28 WT vs delta 2 vs delta 6 vs delta 2/TFE3-KO. Average colonies per high powered field plotted.
Figure S7: (A) In Vivo small animal imaging of luciferase expressing A375 WT and TFE3-KO tumor xenografts in immunocompromised NOD/SCID mice. Xenografts formed by flank injection of 1 × 10^6 tumor cells. Imaging performed at 60 seconds of exposure 10 minutes post injection of luciferin. (B) Quantification of bioluminescence from IVIS imaging of luciferase expressing A375 WT and TFE3-KO tumor xenografts (N=4).
Figure S8: (A) Western blot analysis for MITF and TFE3 protein using bulk lysates from PDX cell lines. GAPDH staining was used as loading control. Red dashed boxes highlight the 82 kDa TFE3 isoform. (B) Brightfield images of PDX cell lines taken at 10x magnification.
Highlights.
In melanoma, high MITF activity correlate with lower levels of the full-length form of TFE3 and with TFE3 being in the cytoplasm.
In an MITF-high melanoma cell line, deletion of MITF results in increased levels of the full-length form of TFE3 and with its nuclear localization.
In an MITF-low melanoma cell line, deletion of TFE3 reduces expression levels of genes promoting epithelial-mesenchymal transition and prevents distant cell colonization and metastasis in a xenograft model.
MITF promotes expression of FNIP2 which encodes a component of the folliculin complex. This complex activates RagC/D guanosine triphosphatase which recruits mTOR and TFE3 to the lysosomal surface, resulting in cytoplasmic localization and degradation of full-length TFE3.
Inhibition of mTORC1 signaling in MITF-high cells reverses the effect of MITF on TFE3 stability and nucular localization.
Funding
This work was supported by grants from the Holden Comprehensive Cancer Center (HCCC) at The University of Iowa through the Center for Advancement and Cancer Strategic Investment to CK. Additional grants from the Institutional Research Grant (IRG-18-164-43) from the American Cancer Society, administered through the HCCC to CK, and by the National Institutes of Health (NIH) to RAC (R01-AR062457). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Inclusion and diversity statement:
One or more of the authors of this manuscript self-identifies as an underrepresented ethnic minority, gender minority and member of the LGBTQIA+ community. We support inclusive, diverse and equitable conduct of research.
Footnotes
Declaration of Interests: All authors declare no competing interests.
Data and materials availability:
All processed and raw high throughput sequencing from ATAC-seq, RNA-seq and CUT&RUN-seq experiments were uploaded to NCBI GEO. GEO accession number pending.
References
- BECKMANN H. & KADESCH T. 1991. The leucine zipper of TFE3 dictates helix-loop-helix dimerization specificity. Genes Dev, 5, 1057–66. [DOI] [PubMed] [Google Scholar]
- BERTOLOTTO C., LESUEUR F., GIULIANO S., STRUB T., DE LICHY M., BILLE K., DESSEN P., D’HAYER B., MOHAMDI H., REMENIERAS A., MAUBEC E., DE LA FOUCHARDIÈRE A., MOLINIÉ V., VABRES P., DALLE S., POULALHON N., MARTIN-DENAVIT T., THOMAS L., ANDRY-BENZAQUEN P., DUPIN N., BOITIER F., ROSSI A., PERROT J. L., LABEILLE B., ROBERT C., ESCUDIER B., CARON O., BRUGIÈRES L., SAULE S., GARDIE B., GAD S., RICHARD S., COUTURIER J., TEH B. T., GHIORZO P., PASTORINO L., PUIG S., BADENAS C., OLSSON H., INGVAR C., ROULEAU E., LIDEREAU R., BAHADORAN P., VIELH P., CORDA E., BLANCHÉ H., ZELENIKA D., GALAN P., AUBIN F., BACHOLLET B., BECUWE C., BERTHET P., BIGNON Y. J., BONADONA V., BONAFE J. L., BONNET-DUPEYRON M. N., CAMBAZARD F., CHEVRANT-BRETON J., COUPIER I., DALAC S., DEMANGE L., D’INCAN M., DUGAST C., FAIVRE L., VINCENT-FÉTITA L., GAUTHIER-VILLARS M., GILBERT B., GRANGE F., GROB J. J., HUMBERT P., JANIN N., JOLY P., KEROB D., LASSET C., LEROUX D., LEVANG J., LIMACHER J. M., LIVIDEANU C., LONGY M., LORTHOLARY A., STOPPA-LYONNET D., MANSARD S., MANSUY L., MARROU K., MATÉUS C., MAUGARD C., MEYER N., NOGUES C., SOUTEYRAND P., VENAT-BOUVET L., ZATTARA H., CHAUDRU V., LENOIR G. M., LATHROP M., DAVIDSON I., AVRIL M. F., DEMENAIS F., BALLOTTI R. & BRESSAC-DE PAILLERETS B. 2011. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature, 480, 94–8. [DOI] [PubMed] [Google Scholar]
- CAMPBELL N. R., RAO A., HUNTER M. V., SZNURKOWSKA M. K., BRIKER L., ZHANG M., BARON M., HEILMANN S., DEFORET M., KENNY C., FERRETTI L. P., HUANG T. H., PERLEE S., GARG M., NSENGIMANA J., SAINI M., MONTAL E., TAGORE M., NEWTON-BISHOP J., MIDDLETON M. R., CORRIE P., ADAMS D. J., RABBIE R., ACETO N., LEVESQUE M. P., CORNELL R. A., YANAI I., XAVIER J. B. & WHITE R. M. 2021. Cooperation between melanoma cell states promotes metastasis through heterotypic cluster formation. Dev Cell, 56, 2808–2825.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARREIRA S., GOODALL J., DENAT L., RODRIGUEZ M., NUCIFORO P., HOEK K. S., TESTORI A., LARUE L. & GODING C. R. 2006. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev, 20, 3426–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIENER J. & SOMMER L. 2021. Reemergence of neural crest stem cell-like states in melanoma during disease progression and treatment. Stem Cells Transl Med, 10, 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DILSHAT R., FOCK V., KENNY C., GERRITSEN I., LASSEUR R. M. J., TRAVNICKOVA J., EICHHOFF O. M., CERNY P., MÖLLER K., SIGURBJÖRNSDÓTTIR S., KIRTY K., EINARSDOTTIR B., CHENG P. F., LEVESQUE M., CORNELL R. A., PATTON E. E., LARUE L., DE TAYRAC M., MAGNÚSDÓTTIR E., ÖGMUNDSDÓTTIR M. H. & STEINGRIMSSON E. 2021. MITF reprograms the extracellular matrix and focal adhesion in melanoma. Elife, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FANELLI G. N., NACCARATO A. G. & SCATENA C. 2020. Recent Advances in Cancer Plasticity: Cellular Mechanisms, Surveillance Strategies, and Therapeutic Optimization. Front Oncol, 10, 569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GHANDI M., HUANG F. W., JANÉ-VALBUENA J., KRYUKOV G. V., LO C. C., MCDONALD E. R. 3RD, BARRETINA J., GELFAND E. T., BIELSKI C. M., LI H., HU K., ANDREEV-DRAKHLIN A. Y., KIM J., HESS J. M., HAAS B. J., AGUET F., WEIR B. A., ROTHBERG M. V., PAOLELLA B. R., LAWRENCE M. S., AKBANI R., LU Y., TIV H. L., GOKHALE P. C., DE WECK A., MANSOUR A. A., OH C., SHIH J., HADI K., ROSEN Y., BISTLINE J., VENKATESAN K., REDDY A., SONKIN D., LIU M., LEHAR J., KORN J. M., PORTER D. A., JONES M. D., GOLJI J., CAPONIGRO G., TAYLOR J. E., DUNNING C. M., CREECH A. L., WARREN A. C., MCFARLAND J. M., ZAMANIGHOMI M., KAUFFMANN A., STRANSKY N., IMIELINSKI M., MARUVKA Y. E., CHERNIACK A. D., TSHERNIAK A., VAZQUEZ F., JAFFE J. D., LANE A. A., WEINSTOCK D. M., JOHANNESSEN C. M., MORRISSEY M. P., STEGMEIER F., SCHLEGEL R., HAHN W. C., GETZ G., MILLS G. B., BOEHM J. S., GOLUB T. R., GARRAWAY L. A. & SELLERS W. R. 2019. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature, 569, 503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GODING C. R. & ARNHEITER H. 2019. MITF-the first 25 years. Genes Dev, 33, 983–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GONG C. & XIA H. 2020. Resveratrol suppresses melanoma growth by promoting autophagy through inhibiting the PI3K/AKT/mTOR signaling pathway. Exp Ther Med, 19, 1878–1886. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- GOSIS B. S., WADA S., THORSHEIM C., LI K., JUNG S., RHOADES J. H., YANG Y., BRANDIMARTO J., LI L., UEHARA K., JANG C., LANZA M., SANFORD N. B., BORNSTEIN M. R., JEONG S., TITCHENELL P. M., BIDDINGER S. B. & ARANY Z. 2022. Inhibition of nonalcoholic fatty liver disease in mice by selective inhibition of mTORC1. Science, 376, eabf8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARPER J. W. & SCHULMAN B. A. 2021. Cullin-RING Ubiquitin Ligase Regulatory Circuits: A Quarter Century Beyond the F-Box Hypothesis. Annu Rev Biochem, 90, 403–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEINZ S., BENNER C., SPANN N., BERTOLINO E., LIN Y. C., LASLO P., CHENG J. X., MURRE C., SINGH H. & GLASS C. K. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell, 38, 576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEPPT M. V., WANG J. X., HRISTOVA D. M., WEI Z., LI L., EVANS B., BEQIRI M., ZAMAN S., ZHANG J., IRMLER M., BERKING C., BESCH R., BECKERS J., RAUSCHER F. J. 3RD, STURM R. A., FISHER D. E., HERLYN M. & FUKUNAGA-KALABIS M. 2018. MSX1-Induced Neural Crest-Like Reprogramming Promotes Melanoma Progression. J Invest Dermatol, 138, 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOEK K. S., EICHHOFF O. M., SCHLEGEL N. C., DÖBBELING U., KOBERT N., SCHAERER L., HEMMI S. & DUMMER R. 2008. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res, 68, 650–6. [DOI] [PubMed] [Google Scholar]
- HOEK K. S. & GODING C. R. 2010. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res, 23, 746–59. [DOI] [PubMed] [Google Scholar]
- KARBOWNICZEK M., SPITTLE C. S., MORRISON T., WU H. & HENSKE E. P. 2008. mTOR is activated in the majority of malignant melanomas. J Invest Dermatol, 128, 980–7. [DOI] [PubMed] [Google Scholar]
- KARRAS P., BORDEU I., POZNIAK J., NOWOSAD A., PAZZI C., VAN RAEMDONCK N., LANDELOOS E., VAN HERCK Y., PEDRI D., BERVOETS G., MAKHZAMI S., KHOO J. H., PAVIE B., LAMOTE J., MARIN-BEJAR O., DEWAELE M., LIANG H., ZHANG X., HUA Y., WOUTERS J., BROWAEYS R., BERGERS G., SAEYS Y., BOSISIO F., VAN DEN OORD J., LAMBRECHTS D., RUSTGI A. K., BECHTER O., BLANPAIN C., SIMONS B. D., RAMBOW F. & MARINE J. C. 2022. A cellular hierarchy in melanoma uncouples growth and metastasis. Nature, 610, 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAUFMAN C. K., MOSIMANN C., FAN Z. P., YANG S., THOMAS A. J., ABLAIN J., TAN J. L., FOGLEY R. D., VAN ROOIJEN E., HAGEDORN E. J., CIARLO C., WHITE R. M., MATOS D. A., PULLER A. C., SANTORIELLO C., LIAO E. C., YOUNG R. A. & ZON L. I. 2016. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science, 351, aad2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KENNY C., DILSHAT R., SEBERG H. E., VAN OTTERLOO E., BONDE G., HELVERSON A., FRANKE C. M., STEINGRÍMSSON E. & CORNELL R. A. 2022. TFAP2 paralogs facilitate chromatin access for MITF at pigmentation and cell proliferation genes. PLoS Genet, 18, e1010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUBIC J. D., YOUNG K. P., PLUMMER R. S., LUDVIK A. E. & LANG D. 2008. Pigmentation PAX-ways: the role of Pax3 in melanogenesis, melanocyte stem cell maintenance, and disease. Pigment Cell Melanoma Res, 21, 627–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANG D., LU M. M., HUANG L., ENGLEKA K. A., ZHANG M., CHU E. Y., LIPNER S., SKOULTCHI A., MILLAR S. E. & EPSTEIN J. A. 2005. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature, 433, 884–7. [DOI] [PubMed] [Google Scholar]
- LEVY C., KHALED M. & FISHER D. E. 2006. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med, 12, 406–14. [DOI] [PubMed] [Google Scholar]
- LING Y., LI Y. & LI L. 2022. Targeting folliculin to selectively inhibit mTORC1: a promising strategy for treating nonalcoholic fatty liver disease. Signal Transduct Target Ther, 7, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIVAK K. J. & SCHMITTGEN T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods, 25, 402–8. [DOI] [PubMed] [Google Scholar]
- MARTINA J. A., DIAB H. I., LISHU L., JEONG A. L., PATANGE S., RABEN N. & PUERTOLLANO R. 2014. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal, 7, ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATHIEU J., DETRAUX D., KUPPERS D., WANG Y., CAVANAUGH C., SIDHU S., LEVY S., ROBITAILLE A. M., FERRECCIO A., BOTTORFF T., MCALISTER A., SOMASUNDARAM L., ARTONI F., BATTLE S., HAWKINS R. D., MOON R. T., WARE C. B., PADDISON P. J. & RUOHOLA-BAKER H. 2019. Folliculin regulates mTORC1/2 and WNT pathways in early human pluripotency. Nat Commun, 10, 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MÖLLER K., SIGURBJORNSDOTTIR S., ARNTHORSSON A. O., POGENBERG V., DILSHAT R., FOCK V., BRYNJOLFSDOTTIR S. H., BINDESBOLL C., BESSADOTTIR M., OGMUNDSDOTTIR H. M., SIMONSEN A., LARUE L., WILMANNS M., THORSSON V., STEINGRIMSSON E. & OGMUNDSDOTTIR M. H. 2019. MITF has a central role in regulating starvation-induced autophagy in melanoma. Sci Rep, 9, 1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAPOLITANO G., DI MALTA C. & BALLABIO A. 2022. Non-canonical mTORC1 signaling at the lysosome. Trends Cell Biol, 32, 920–931. [DOI] [PubMed] [Google Scholar]
- NARDONE C., PALANSKI B. A., SCOTT D. C., TIMMS R. T., BARBER K. W., GU X., MAO A., LENG Y., WATSON E. V., SCHULMAN B. A., COLE P. A. & ELLEDGE S. J. 2023. A central role for regulated protein stability in the control of TFE3 and MITF by nutrients. Mol Cell, 83, 57–73.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAMBOW F., ROGIERS A., MARIN-BEJAR O., AIBAR S., FEMEL J., DEWAELE M., KARRAS P., BROWN D., CHANG Y. H., DEBIEC-RYCHTER M., ADRIAENS C., RADAELLI E., WOLTER P., BECHTER O., DUMMER R., LEVESQUE M., PIRIS A., FREDERICK D. T., BOLAND G., FLAHERTY K. T., VAN DEN OORD J., VOET T., AERTS S., LUND A. W. & MARINE J. C. 2018. Toward Minimal Residual Disease-Directed Therapy in Melanoma. Cell, 174, 843–855.e19. [DOI] [PubMed] [Google Scholar]
- SEBERG H. E., VAN OTTERLOO E. & CORNELL R. A. 2017. Beyond MITF: Multiple transcription factors directly regulate the cellular phenotype in melanocytes and melanoma. Pigment Cell Melanoma Res, 30, 454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SKENE P. J. & HENIKOFF S. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THIES K. A., STECK S., KNOBLAUGH S. E. & SIZEMORE S. T. 2020. Pathological Analysis of Lung Metastasis Following Lateral Tail-Vein Injection of Tumor Cells. J Vis Exp. [DOI] [PubMed] [Google Scholar]
- THOMPSON J. D., HIGGINS D. G. & GIBSON T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res, 22, 4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TORBORG S. R., LI Z., CHAN J. E. & TAMMELA T. 2022. Cellular and molecular mechanisms of plasticity in cancer. Trends Cancer, 8, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG P., ZHANG H., GUO K., LIU C., CHEN S., PU B., CHEN S., FENG T., JIAO H. & GAO C. 2024. Rapamycin inhibits B16 melanoma cell viability invitro and invivo by inducing autophagy and inhibiting the mTOR/p70-S6k pathway. Oncol Lett, 27, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOUTERS J., KALENDER-ATAK Z., MINNOYE L., SPANIER K. I., DE WAEGENEER M., BRAVO GONZÁLEZ-BLAS C., MAUDUIT D., DAVIE K., HULSELMANS G., NAJEM A., DEWAELE M., PEDRI D., RAMBOW F., MAKHZAMI S., CHRISTIAENS V., CEYSSENS F., GHANEM G., MARINE J. C., POOVATHINGAL S. & AERTS S. 2020. Robust gene expression programs underlie recurrent cell states and phenotype switching in melanoma. Nat Cell Biol, 22, 986–998. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Multiple sequence alignment of the MIT/TFE transcription factor family and the anti-MITF antibody immunogen sequence by CLUSTALW. Amino acid sequences of MITF, TFE3, TFEB, and TFEC were recovered from UniProt and aligned to the immunogen sequence of the anti-MITF (HPA003259, Sigma) using by CLUSTALW (Thompson et al., 1994).
Figure S2: (A) Venn Diagram of overlapping anti-MITF CUT&RUN-seq peaks identified by MACS2 in delta 2 and delta 6 MITF-KO cell lines. (B) Density heatmap representing anti-MITF and anti-H3K27Ac CUT&RUN-Seq as well as ATAC-Seq in SKMEL28 WT and MITF-KO (delta 6) cell lines. Peak regions represent MITF-homodimer peaks, gained-paralog peaks, and persistent paralog. (C) Pie-chart of MITF-homodimer peaks, gained-paralog peaks, and persistent paralog peaks that overlap MITF-regulated enhancers, enhancer subtypes as labeled. Gene Ontology (GO) analysis of (D) MITF-homodimer occupied genes and (E) Gained-paralog occupied genes (peaks are within 100 kb of a genes TSS).
Figure S3: (A) Protein lysates from SKMEL28 WT and MITF-KO (Delta6) cell lines were subjected to immunoprecipitation using an anti-MITF antibody. The immunoprecipitates were analyzed by Western blot to detect TFE3 protein. A distinct band corresponding to the expected molecular weight of TFE3 (~82 kDa) was observed in the immunoprecipitated samples, including those from the MITF-KO cell line. This suggests potential cross-reactivity of the anti-MITF antibody with nondenatured TFE3 protein. (B) The same immunoprecipitated lysates were analyzed by Western blot to detect the MITF protein. A band at the expected molecular weight of MITF (~60 kDa) in the SKMEL28 WT cell line using the anti-MITF antibody was observed. However, no/weak MITF positivity was detected in the MITF-KO cell line. Suggesting, the anti-MITF antibody does not have strong immunoreactivity with denatured TFE3. (C) Protein lysates from SKMEL28 WT and MITF-KO cell lines were also subjected to immunoprecipitation using an anti-TFE3 antibody, followed by Western blot analysis to detect TFE3. Two distinct bands at the expected molecular weight for TFE3 (~72, 82 kDa) was observed in both cell lines with the larger form more prominent in MITF-KO cells. (D) Mass spectrometry analysis of protein lysates from SKMEL28 WT and MITF-KO (Delta6) cells following immunoprecipitation using an anti-MITF antibody.
Figure S4: (A) Investigation of ZEB1 expression differences. qPCR comparing expression of ZEB1 in SKMELWT, ΔMITF-X2 control CRISPR clones and ΔMITF-X2/ΔTFE3KO Clones. Relative expression is graphed on y-axis. Immunoblotting of ZEB1 and TFE3 protein level in ΔMITF-X2 vs ΔMITF-X2/ ΔTFE3KO clones and A375 WT vs TFE3-KO clones. GAPDH is utilized as loading control. (B) Gene ontology analysis of MITF-Repressed genes. (C) Gene Ontology analysis of MITF-activated genes. (D) Immunoblotting of TFE3 and MITF in A375 and SKMEL28 WT cells. GAPDH is utilized as loading control. (E) Transwell invasion assay of A375 vs SKMEL28 cells. Number of invaded cells is plotted. **P-value <0.01.
Figure S5: (A) Schematic of CRISPR gRNA target region for TFE3 knockout system. (B) Immunoblotting of TFE3 in A375 WT vs individually isolated TFE3-KO clones. GAPDH is utilized as loading control. (C) Sanger sequencing of CRISPR target region of TFE3 gene with associated translated amino acid sequence. (D) Screenshot of UCSC (hg19) genome browser visualization of bigwig files generated from anti-MITF and anti-TFE3 CUT&RUN demonstrating representative TFE3 gained-paralog peak at F2RL1 enhancer.
Figure S6: (A) Transwell invasion assay of A375 WT vs nontargeting control CRISPR clones vs CRISPR TFE3-KO clones performed with n=4 replicates. Invaded cells per well graphed on y-axis. **P-value <0.01, ***P-value <0.001. (B) Representative 10x brightfield images taken from transwell invasion assay with A375 vs RPMI vs PDX 16 vs PDX 34. (C) Scratch wound healing assay using A375 WT and TFE3-KO cells. 4x brightfield images taken at 0, 24, 48 hours post scratch. Area of scratch and % wound open quantified via ImageJ. *P-value<0.05, ***P-value<0.001. (D) Cell proliferation assay with A375 WT vs TFE3-KO clones. 1000 cells are plated at day 0 in 96 well plates with n=6. Normalized absorbance @ 540nm is plotted on the Y-axis. (E) Cell proliferation assay of SKMEL28 WT vs ΔMITF-X2 vs ΔMITF-X6 vs ΔMITF-X2/ΔTFE3KO. 1000 cells are plated at day 0 in 96 well plates with n=6. Normalized absorbance at 540nm is plotted on the Y-axis. (G) Transwell invasion assay with SKMEL28 WT vs delta 2 vs delta 6 vs delta 2/TFE3-KO. Invaded cells per well plotted. (H) Soft agar colony formation assay with SKMEL28 WT vs delta 2 vs delta 6 vs delta 2/TFE3-KO. Average colonies per high powered field plotted.
Figure S7: (A) In Vivo small animal imaging of luciferase expressing A375 WT and TFE3-KO tumor xenografts in immunocompromised NOD/SCID mice. Xenografts formed by flank injection of 1 × 10^6 tumor cells. Imaging performed at 60 seconds of exposure 10 minutes post injection of luciferin. (B) Quantification of bioluminescence from IVIS imaging of luciferase expressing A375 WT and TFE3-KO tumor xenografts (N=4).
Figure S8: (A) Western blot analysis for MITF and TFE3 protein using bulk lysates from PDX cell lines. GAPDH staining was used as loading control. Red dashed boxes highlight the 82 kDa TFE3 isoform. (B) Brightfield images of PDX cell lines taken at 10x magnification.
Data Availability Statement
All processed and raw high throughput sequencing from ATAC-seq, RNA-seq and CUT&RUN-seq experiments were uploaded to NCBI GEO. GEO accession number pending.