Abstract
Since their initial discovery in maize, transposable elements (TEs) have emerged as being integral to the evolution of maize, accounting for 80% of its genome. However, the repetitive nature of TEs has hindered our understanding of their regulatory potential. Here, we demonstrate that long-read chromatin fiber sequencing (Fiber-seq) permits the comprehensive annotation of the regulatory potential of maize TEs. We uncover that only 94 LTR retrotransposons contain the functional epigenetic architecture required for mobilization within maize leaves. This epigenetic architecture degenerates with evolutionary age, resulting in solo TE enhancers being preferentially marked by simultaneous hyper-CpG methylation and chromatin accessibility, an architecture markedly divergent from canonical enhancers. We find that TEs shape maize gene regulation by creating novel promoters within the TE itself as well as through TE-mediated gene amplification. Lastly, we uncover a pervasive epigenetic code directing TEs to specific loci, including that locus that sparked McClintock’s discovery of TEs.
Transposable elements (TEs), first described as ‘controlling elements’ by Barbara McClintock (1–7), have the potential to shape the regulation of the host genome (8–11). For example, the insertion of a TE in a regulatory region of the maize domestication gene teosinte branched1 (tb1) enhances its expression, contributing to the increased apical dominance of maize compared to its ancestor teosinte (12). Although over 80% of the maize genome is annotated as intact TEs or TE fragments (13), a comprehensive analysis of their regulatory potential is lacking. Commonly used methods to map regulatory elements (i.e. accessible chromatin regions, ACRs) have relied on short sequence reads which rarely map uniquely within TEs. Here, we use the long-read method Fiber-seq to overcome this limitation and map ACRs across the maize B73 genome. Fiber-seq uses a non-specific DNA N6-adenine methyltransferase to methylate accessible adenines (14) – a modification that is extremely sparse in plants (15) – followed by single-molecule PacBio sequencing of ~18 kb maize chromatin fibers, enabling the synchronous detection of accessible adenines (m6A) and endogenous cytosine methylation (5mCpG).
Assessing the single-molecule regulatory landscape of maize with Fiber-seq
We compared Fiber-seq and ATAC-seq using paired samples of leaf protoplasts isolated from 14-day-old dark-grown maize seedlings (Fig. 1A, fig. S1A–C). The use of leaf protoplasts minimized cell-type heterogeneity as leaf tissue is enriched in mesophyll cells. We observed that Fiber-seq-derived m6A and 5mCpG calls showed the expected signals at ATAC-seq-derived ACRs and CAGE-defined transcription start sites (TSSs), in addition to the expected correlation of signal intensity with gene expression at TSSs (fig. S1D–G). However, unlike ATAC-seq, Fiber-seq also revealed periodic m6A signals downstream of the TSS that were most pronounced for highly expressed genes, reflecting promoter-proximal well-positioned nucleosomes typically measured by MNase-seq (fig. S1E, F) (16).
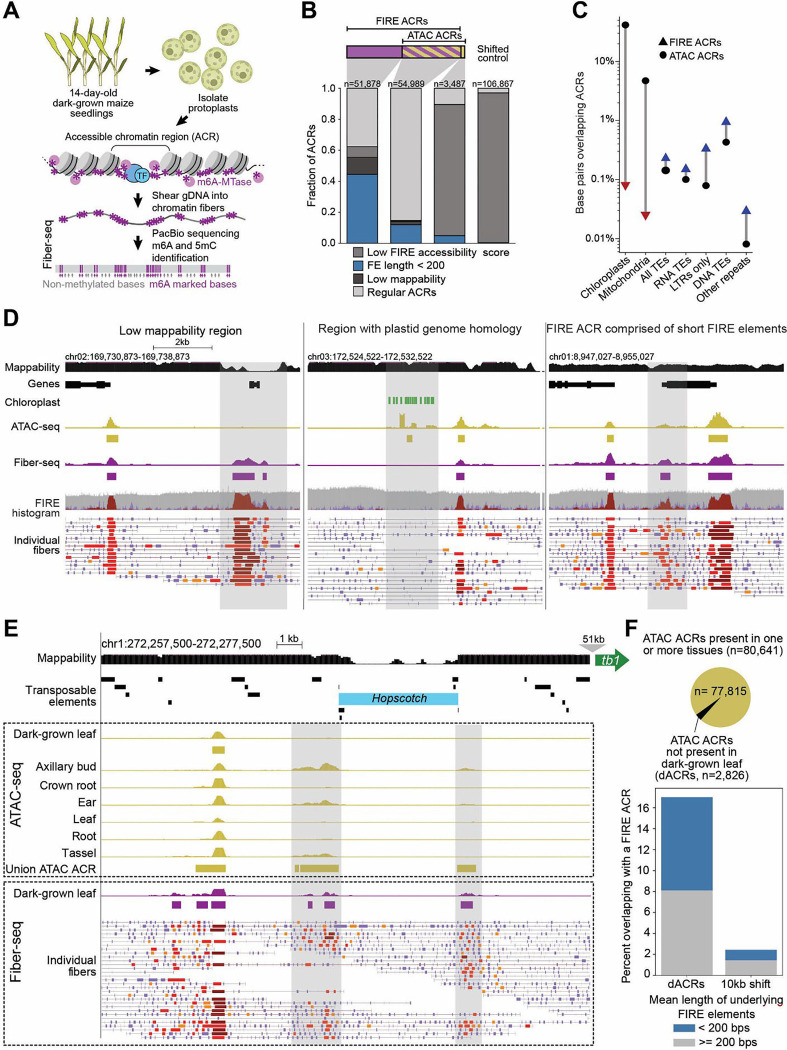
Fig. 1. Fiber-seq captures the regulatory landscape of maize comprehensively.
(A) Experimental scheme. (B) ACRs called in paired Fiber-seq and ATAC-seq experiments are shown in three bar graphs representing FIRE ACRs (purple) that did not overlap with ATAC ACRs (n=51,878), FIRE ACRs that overlapped with ATAC ACRs (purple/gold, n=54,989), ATAC ACRs that did not overlap with FIRE ACRs (gold, n=3,487), in addition to a bar graph representing shifted control regions (10kb downstream of FIRE ACRs, n=106,867). ACRs in each category were hierarchically classified as having (i) a FIRE accessibility score of less than 0.25, (ii) a mean FIRE element (FE) length less than 200 bp, (iii) low short-read mappability, or (iv) none of the above (Regular ACR). Stacked bar charts indicate the distribution of these classifiers for each ACR category. (C) Percentage of base pairs (Y-axis) overlapping with ACRs called by either Fiber-seq (FIRE ACRs, triangles) or ATAC-seq (ATAC ACRs, circles) for distinct genomic regions (X-axis). As expected, fewer base pairs in regions with homology to plastid and mitochondrial genomic sequence overlap with FIRE ACRs (red triangles) than with ATAC ACRs, while more base pairs in regions annotated as transposable elements or repeats overlap with FIRE ACRs (blue triangles) than with ATAC ACRs. (D) Screenshots of three genomic regions illustrating marked differences between FIRE and ATAC ACR calls. Top to bottom, each panel shows the following tracks: genomic location, mappability calculated as in fig. S2A, annotated genes, chloroplast sequence, ATAC-seq signal with ATAC ACRs indicated below as rectangles in gold, Fiber-seq signal with FIRE ACRs indicated below as rectangles in purple, FIRE histogram, and individual chromatin fibers with FIRE elements in shades of red, with darker shades indicating greater significance. Left, the region highlighted in grey contains two FIRE ACRs but no ATAC ACRs because of low mappability. Middle, the chloroplast sequence track indicates high sequence homology at this nuclear locus with the plastid genome. The ATAC ACR in the highlighted region is a false positive due to incorrect mapping of short sequence reads. Right, the highlighted region shows a FIRE ACR with underlying short FIRE elements. No ATAC ACR was called. In all panels, ATAC-seq signal is a sliding window histogram displaying the number of Tn5 insertion sites, with the height of each 20 bp bar representing the number of Tn5 insertions within a 150 bp window centered on these 20 bp (minimum=0, maximum=210 mapped reads/16,662,983 total mapped reads=1.26e-5). ATAC ACRs are MACS2 derived peaks (q<0.01) (45). Fiber-seq signal is a per-nucleotide average of the scaled, log-transformed ML-model-derived probability of each underlying fiber containing a FIRE element at that nucleotide. Features used by the ML model include m6A density, length of methyltransferase sensitive patch, and A/T content (minimum=0, maximum=100). FIRE ACRs are called by fiberseq-FIRE (FDR<0.01) (46). FIRE histogram is three overlaid histograms: inferred nucleosome x-coverage (gray), inferred MSP x-coverage (light purple), inferred FIRE element x-coverage (red). Individual fibers are annotated with MSPs (light purple) and FIRE elements (reds, FDR<=5%, and oranges, 5%<FDR<=10%). (E) Loci lacking ATAC ACRs in dark-grown maize leaves that show ATAC ACRs in other tissues often overlap with FIRE ACRs comprised of short FIRE elements. A screenshot is shown for the region upstream of the tb1 gene (green arrow, Zm00001eb054440) in which a hopscotch TE insertion (light blue) generated an enhancer. Top to bottom, tracks are genomic locus, mappability as in fig.S2A, in first dotted box, ATAC-seq data: ATAC-seq signal (gold) for dark-grown leaves (this study), subsequent tracks pseudo-bulked single-cell ATAC-seq signal for indicated tissues (19), last track, union ATAC ACRs (present in at least one tissue) as golden rectangles, in second dotted box, Fiber-seq data: Fiber-seq signal (purple) in dark-grown leaves and indicated below FIRE ACRs as purple rectangles, individual fibers with short FIRE elements in shades of red. In dark-grown leaves, ACRs were detected by Fiber-seq but not ATAC-seq in the loci flanking the hopscotch TE. However, ATAC ACRs were detected in these loci in axillary bud, ear, and tassel tissue. (F) Of the 80,641 union ATAC ACRs across these seven tissues, 2,826 were not detected in dark-grown leaves (differentially accessible ACRs, dACRs, see Methods for details). About 17% of the loci overlapping with these differentially accessible ACRs overlap with FIRE ACRs, and about half of these are FIRE ACRs comprised of short FIRE elements.
To rigorously distinguish regions with elevated exogenous m6A signal (methyltransferase-sensitive patches, MSP) due to nucleosome linkers from regions representing ACRs (fig. S1E, F), we called FIRE elements (Fiber-seq Inferred Regulatory Elements) with the semi-supervised machine learning classifier fiberseq-FIRE (17). After recalibrating fiberseq-FIRE for maize, 4.6 million methyltransferase-sensitive patches were classified as actuated FIRE elements (precision >0.9), with the remaining 150 million classified as nucleosome linkers. By aggregating single-molecule FIRE elements across the genome, we called 106,867 FIRE ACRs (FDR <0.01, Fig. 1B, table S1). In contrast, we called only 51,817 ACRs with ATAC-seq (q-value <0.01, table S2), consistent with Fiber-seq revealing a more comprehensive regulatory landscape of maize. Fiber-seq identified the vast majority of ACRs called with ATAC-seq in paired samples (Fig. 1B), added ACRs in repeat regions with low mappability, and corrected for false-positive ATAC ACRs, such as those in nuclear genomic regions with homology to plastid or mitochondrial genomes (18) (Fig. 1C, D, tables S3, 4). Signal intensity at ACRs in the paired bulk ATAC-seq strongly correlated with the Fiber-seq signal (fig. S1H). However, for a set of ~40,000 shared ACRs, most were detected with Fiber-seq on half or more of the sequenced chromatin fibers, whereas fewer than 5% of cells showed Tn5 insertions in these ACRs in single-cell ATAC-seq (19) (fig. S1I, table S5). This comparison illustrates the limitations of single-cell ATAC-seq as a quantitative measure of per-molecule chromatin accessibility. Taken together, our results show that Fiber-seq accurately captures chromatin accessibility and 5mCpG in maize with single-molecule and single-nucleotide precision.
FIRE ACRs comprised of short FIRE elements can mark ATAC ACRs detected in other tissues
Although Fiber-seq identified nearly twice as many ACRs as paired ATAC-seq experiments, this increase was only in part explained by low mappability of ATAC-seq reads (fig.S2A, B). Rather, over half of the FIRE ACRs missed by ATAC-seq were comprised of short FIRE elements (<200 bp) (fig.1B, fig. S2C–F). These FIRE ACRs shared the features typical of ACRs comprised of long FIRE elements identified by both methods such as enrichment of the 6mA signal, depletion of the 5mCpG signal (fig. S2E) and genomic distribution (fig. S2F).
We detected ACRs comprised of short FIRE elements flanking the TE insertion introduced a tb1 enhancer (Studer et al, 2011) but failed to detect these by ATAC-seq (Fig 1E). However, these flanking regions were detected as ATAC ACRs in embryonic and reproductive tissues (axillary bud, tassel, and ear) (19), suggesting that ACRs comprised of short FIRE elements may mark genomic loci with tissue-specific chromatin accessibility in maize (Fig. 1E). To systematically evaluate this possibility, we identified ATAC ACRs present in one or more tissues (19), and then filtered for the subset of these ACRs for which the corresponding genomic loci showed only background ATAC signal in dark-grown leaves (2,826 dACRs/80,641 union ACRs, tables S6, S7). Of the 2,826 dACRs, 480 overlapped with a FIRE ACR (17%), and over half of the 480 overlapped with FIRE ACRs comprised of short FIRE elements (251/480, Fig. 1F), consistent with the short ACRs representing functional regulatory elements in maize that may display tissue-selective activity.
Distinctive patterns of ACRs mark functional LTR retrotransposons
We next sought to interrogate ACRs in TEs, focusing on long terminal repeat (LTR) retrotransposons because of their prevalence in the maize genome (74.4%) (13). Intact LTR retrotransposons are class I TEs with bilateral LTRs that flank an internal region (fig. 2A, B). Each of the bilateral LTRs are thought to contain the regulatory elements, promoters and adjacent enhancers, that drive expression of the TE genes encoded in the internal region (20). LTR retrotransposons mobilize through reverse transcription of their mRNA and integration of the cDNA into another genomic location. They are divided into autonomous (which encode the proteins needed for transposition) and non-autonomous (which require proteins encoded by other elements for transposition) LTR retrotransposons. It has been challenging to determine the functional activity of individual LTR retrotransposons because of their high sequence identity (9, 11, 20).
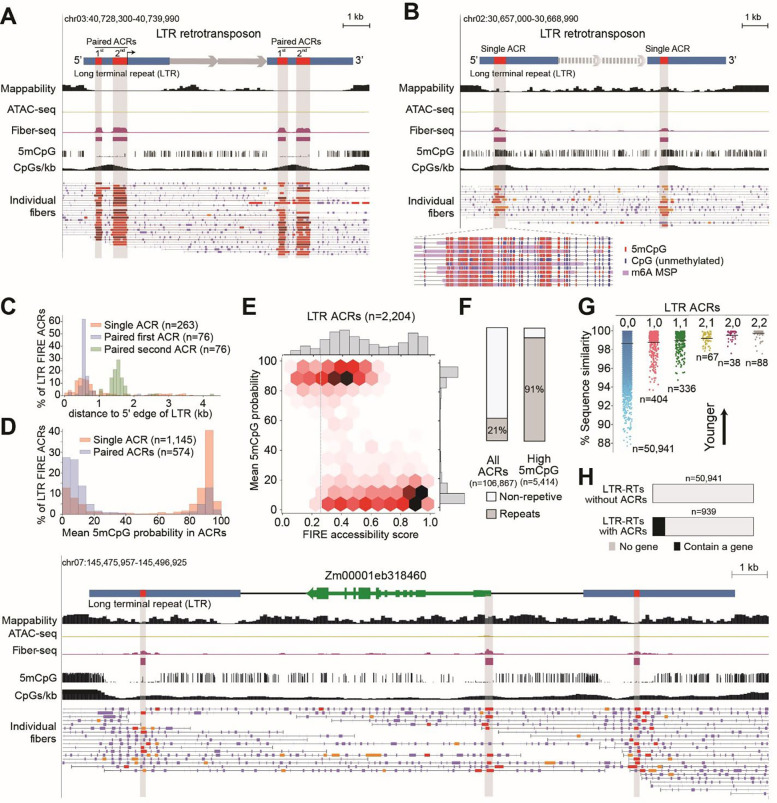
Fig.2. FIRE ACRs in intact LTR retrotransposons identify functional retrotransposons, TE-derived regulatory elements and TE-enabled host gene amplification.
(A) Representative example of an intact LTR retrotransposon with paired FIRE ACRs both in the left and the right long terminal repeats (paired bilateral ACRs in LTRs, ID=LTRRT_14411). Top indicates left and right LTR in blue and paired bilateral FIRE ACRs in red. First and second paired ACRs are labeled, representing the putative enhancer and the putative promoter, respectively. Putative transcription start site is indicated with black arrow, and genes in internal region are indicated. Top to bottom, screenshot shows tracks for genomic location, mappability calculated as in fig. S2A, ATAC-seq signal in gold, Fiber-seq signal in purple with FIRE ACRs indicated as purple rectangles below, 5mCpG methylation as the per-CpG methylation probability calculated by pb-CpG-tools (47).The vertical axis of the 5mCpG track represents the methylation probability at individual CpG sites, expressed as a percentage, minimum=0, maximum=100. The CpGs/kb track represents a sliding window histogram displaying the number of CpG dinucleotides, with the height of each 100 bp bar representing the number of CpG dinucleotides within a 1 kb window centered on these 100 bp. Paired bilateral ACRs in LTR retrotransposons tended to be hypo-methylated as expected for accessible regions. (B) Representative example of an intact LTR retrotransposon with one FIRE ACR both in the left and the right LTR (single bilateral ACRs in LTRs, ID=LTRRT_8308). Tracks as in (A). The single FIRE ACRs in this LTR retrotransposon showed high levels of 5mCpG methylation coinciding with the m6A signal, magnified detail below shows methylated 5mCpGs (red) and unmethylated CpGs (blue) and m6A methyltransferse-sensitive patches (purple) on individual fibers. (C) LTR retrotransposons with single bilateral FIRE ACRs tended to maintain the ACRs marking putative enhancers. Histogram of FIRE ACR location relative to the 5’ edge of a given retrotransposon, stratified by type of ACR. (D) Single FIRE ACRs in LTR retrotransposons were more likely to be 5mCpG-methylated than paired bilateral FIRE ACRs, regardless of their position. (E) Hexbin plot shows FIRE accessibility scores (X-axis) and mean 5mCpG probabilities (Y-axis) for 2,204 ACRs in LTRs. Shades of red denote frequency, also shown in plotted histograms (top, right). 57% (1261/2204) of FIRE ACRs in LTRs were highly 5mCpG-methylated and 37% (733/1978) of high-confidence FIRE ACRs within LTRs (FIRE accessibility score greater than 0.25, grey dotted line) were highly 5mCpG-methylated. (F) Percentage of all FIRE ACRs and FIRE ACRs with high 5mCpG methylation (mean 5mCpG methylation over 50%) that overlap with an annotated repeat by more than 50 bp. (G) The presence of FIRE ACRs correlates with the sequence similarity of left and right LTRs, a measure of evolutionary age. LTRs with paired bilateral FIRE ACRs showed the greatest sequence similarity while those without FIRE ACRs showed the least. 0,0, no ACRs; 1,0, single unilateral ACR; 1,1, single bilateral ACRs; 2,1, paired ACR in one LTR, single in the other; 2,0, paired ACR in one LTR, none in the other; 2,2, paired bilateral ACRs. Rare instances of other configurations are omitted. (H) Fraction of intact LTR retrotransposons with or without at least one FIRE ACR that contain an annotated gene. (I) The highly duplicated, well-annotated gene (Zm00001eb318460, green) within an LTR retrotransposon is a candidate for TE-enabled gene amplification. Tracks as in (A). There are single bilateral ACRs present in the LTRs, in addition to a FIRE ACR marking the transcription start site of this gene (highlighted in grey).
Using Fiber-seq, we mapped ACRs residing within each of the 51,882 intact LTR retrotransposons in the maize genome (table S8) as well as for ACRs in solo LTRs (table S9). Only about 2% (941/51,882) of intact LTR retrotransposons contained at least one FIRE ACR entirely within one of their bilateral LTRs (table S8), consistent with widespread epigenetic silencing by RNA-mediated DNA methylation, a plant-specific pathway that targets TEs (21). Of the 941 ACR-containing LTR retrotransposons, 21% (201/941) contained two adjacent ACRs (paired ACRs, fig. 2A, table S8) in one or both of their LTRs, likely corresponding to the putative LTR promoter and the enhancer elements (20). Nearly half (94/201) contained the two adjacent ACRs in both LTRs (paired bilateral ACRs, table S8). The paired bilateral ACRS almost always exhibited single-molecule co-accessibility and hypo-5mCpG methylation (fig. 2A, fig. S3A). In short, maize leaves contain only 94 LTR retrotransposons that contain the functional regulatory elements required for transposon mobilization, with only 76 of these being autonomous LTR retrotransposons.
LTR with single ACRs are putative enhancers that display a novel epigenetic signature
Most ACR-containing intact LTR retrotransposons contained only a single ACR in one or both LTRs (fig. 2B, table S8). Intact LTR retrotransposons with single ACRs were enriched for containing a single ACR in both of their bilateral LTRs (i.e., single bilateral ACRs, 499/941, 53%). The single ACRs exhibited far greater single-molecule heterogeneity than paired ACRs (fig. S3A). Specifically, while paired ACRs showed a bimodal actuation distribution with over half being supported by FIRE elements called in 75% of underlying fibers, only 7% of single ACRs crossed this actuation threshold (fig. S3A).
Next, we examined whether single ACRs preferentially localized to the putative LTR enhancer or to the promoter by analyzing ACR distance to the 5’ LTR edge. Thus, this analysis was limited to the 268 ACR-containing autonomous LTR retrotransposons, in which strandedness was inferred. Nearly all LTR retrotransposons with a single ACR selectively retained the ACR that corresponds to the putative LTR enhancer element (fig. 2C), suggesting that chromatin accessibility is lost at putative LTR promoters. Consistent with this interpretation, single ACRs had a transcription factor motif profile more similar to the putative LTR enhancers than to the putative LTR promoters in paired ACRs (fig. S3B, tables S10–13).
In stark contrast to paired ACRs in LTRs or ACRs elsewhere in the maize genome, single ACRs in LTRs could exhibit hyper-5mCpG methylation directly coinciding with chromatin accessibility (fig. 2B), two epigenetic marks that are thought to be mutually exclusive. Leveraging the single-molecule nature of our chromatin accessibility and 5mCpG methylation calls, we demonstrate that chromatin accessibility and hyper-5mCpG methylation co-occurred and directly overlap along the same chromatin fiber at single ACRs of LTR retrotransposons (fig. 2B, D, E, table S14). As expected, hyper-5mCpG methylation was rarely seen overlapping FIRE ACRs in the maize genome. However, the rare ACRs with simultaneous chromatin accessibility and hyper-5mCpG methylation were almost exclusively present within repeat elements, with 15% of these corresponding to single ACRs in intact LTR retrotransposons (fig. 2F). This unexpected co-occurrence of chromatin accessibility and hyper-5mCpG methylation was rare and not substantially enriched in LTR retrotransposons in humans (fig. S3C). These results point to the plant-specific RNA-mediated DNA methylation pathway as contributing to this unusual co-occurrence of these two epigenetic marks. However, further analysis of the methylation signatures typical of RNA-mediated DNA methylation or other chromatin states at these low-mappability loci was not feasible because the publicly available data sets resulted from short-read sequencing (11, 22).
Given the features of paired and single ACRs, we reasoned that LTR retrotransposons containing the former might be evolutionarily younger TEs, while TEs containing the latter might be older but still younger than the many fully silent transposons. To address evolutionary age, we examined the sequence similarity between the left and the right LTR of each intact LTR retrotransposon as a metric reflecting time since transposition. LTR retrotransposons with exactly one FIRE ACR per LTR (single ACR) showed greater mean sequence similarity than those without FIRE ACRs (99.0% vs 98.7%, p-value=0.004, Mann-Whitney U test) (fig. 2G). LTR retrotransposons with exactly two FIRE ACRs per LTR (paired ACRs) showed a mean sequence similarity of 99.8%, significantly greater than those with one FIRE ACR per LTR (p-value=6.8e-26, Mann-Whitney U test) (fig. 2G). Thus, recently transposed LTR retrotransposons have a characteristic chromatin and CpG methylation pattern that degenerates with evolutionary age.
ACRs in LTRs are co-opted as gene promoters and facilitate amplification of host genes
TEs have been long thought to shape host gene regulation by adding or disrupting promoters, enhancers, insulators and coding regions (23, 24). In humans, TE-derived promoters have been inferred via mapping of transcription start sites (25) and transcription factor binding sites overlapping TE sequences (26, 27). However, in maize attempts to infer the regulatory effects of LTR retrotransposons have largely been limited to studying gene expression patterns associated with the presence of absence of neighboring polymorphic TEs that overlap ATAC ACRs (10, 11) - an analysis that is severely limited by short-read mappability issues inherent to TEs. We sought to leverage our comprehensive maps of FIRE ACRs across intact maize LTR retrotransposons to identify LTRs that may be shaping host gene regulation (fig. S4A, B). Using this approach, we discovered that the putative target gene impacted by LTR FIRE ACRs often resided within the LTR retrotransposon itself. In fact, of the 941 LTR retrotransposons with ACRs in B73, 114 (12%) contained an annotated gene within the intact retrotransposon (fig. S4C, D), 24-fold greater than LTR retrotransposons without ACRs (fig. 2H). Of these 114, the promoters of 49 annotated genes were marked by a FIRE ACR with 48 co-opting one of the LTR ACRs as their promoter (fig. S4D). Overall, these findings indicate that one of the major ways LTRs shape maize host gene regulation is by providing novel gene promoter elements within the LTR itself.
In one case, the internal gene (Zm00001eb318460) maintained its promoter, marked by a FIRE ACR, in addition to single FIRE ACRs in the flanking bilateral LTRs (fig. 2I). This histone deacetylase complex gene is highly expressed (79% percentile in dark-grown maize leaves) (28) and has orthologs in the close maize relative Sorghum bicolor, its ancestor teosinte (Zea mays ssp. mexicana) (29) and other grasses. In human, Alu TEs have been proposed to enable segmental duplication (30), so we sought to evaluate whether the gene’s residence within the LTR retrotransposon might be associated with its duplication within the B73 genome. Consistent with this hypothesis, we found numerous Zm00001eb318460 paralogs in the B73 genome (21 amino acid blast hits with e-value<1-e50). This is a highly unusual level of gene duplication for maize genes with a similar expression level and length, as 93% of similar genes showed <5 amino acid blast hits (table S15). Taken together, these findings implicate TEs in enabling gene amplification in maize.
Diffuse chromatin accessibility marks putative insertion sites of DNA TEs
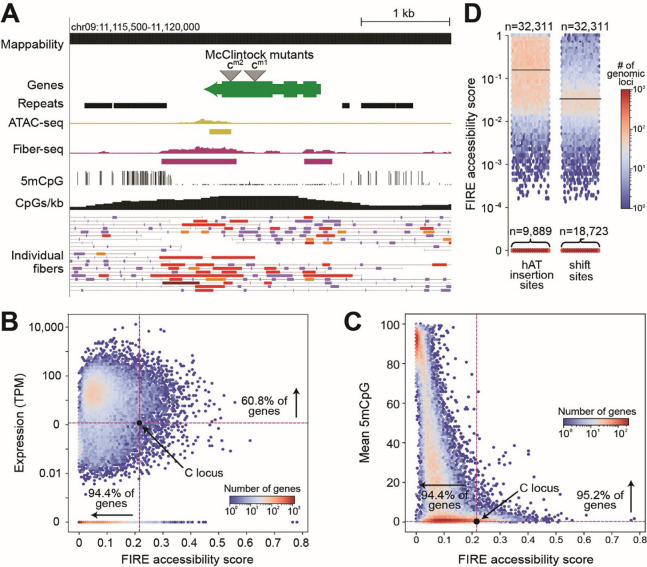
In general, transposons have been shown to preferentially insert into accessible chromatin both in vitro and in vivo (31, 32). However, the epigenetic features that predispose genomic loci to insertion of class II (DNA) TEs, in particular hAT TEs, remain unresolved. The first gene reported by McClintock to be susceptible to insertion of hAT TEs is the C locus (1, 2), now called C1 or colored aleurone 1 gene. The C1 gene body showed unusual diffuse Fiber-seq chromatin accessibility (Fig. 3A), ranking among the top 5% of all genes (Fig. 3B). We also observed hypo-5mCpG methylation across the C1 gene body, ranking among the bottom 5% of all genes (Fig. 3C). Other genes identified by McClintock as having hAT TE insertions also showed diffuse Fiber-seq chromatin accessibility and hypo-5mCpG methylation within their respective gene bodies (fig. S5) (33–37). Neither diffuse gene body chromatin accessibility (Fig. 3B) nor hypo-5mCpG methylation strongly correlated with gene expression (38–42), indicating that these features may uniquely mark preferred hAT TE insertion sites. To test this hypothesis, we identified over 32,000 loci in the B73 maize reference genome that contain hAT TE insertions in exactly one of the 25 non-B73 NAM founder lines (43). Indeed, the hAT TE insertion sites were substantially more accessible in B73 than control regions and were preferentially marked by hypo-5mCpG methylation (Fig. 3D, table S16), indicating that these are pervasive epigenetic marks guiding the insertional landscape of hAT TEs in maize, including those initially described by Dr. McClintock.
Fig. 3. hAT TEs tend to insert in regions with diffuse chromatin accessibility detected by Fiber-seq.
(A) Screenshot of the locus containing the C1 or colored aleurone 1 gene (green, Zm00001eb373660; called the C locus by McClintock). Tracks as in fig. 2. hAT TE insertions identified by McClintock as mutant alleles cm-1 and cm-2. The C1 gene in B73, which does not contain a hAT TE, showed diffuse gene body accessibility detected in Fiber-seq (purple) coinciding with unusual gene body hypo-5mCpG methylation. (B) The C1 gene body showed higher FIRE accessibility scores than 94.4% of other genes while showing only slightly above average gene expression. (C) The C1 gene body showed lower 5mCpG gene body methylation than 95.2% of genes. (D) Loci identified as hAT TE insertions sites in exactly one of the 25 non-B73 NAM strains (43) were more likely to show diffuse gene body accessibility (shown as FIRE accessibility scores) than control loci (same-size regions 10 kb shifted to the right in the B73 genome). Mean and median FIRE accessibility scores for hAT TE insertion sites were 0.159 and 0.046. Mean and median FIRE accessibility scores for shifted control regions were 0.035 and 0. Horizontal lines indicate mean FIRE accessibility scores. Thousands of hAT TE insertion sites showed FIRE accessibility scores of 0 (n= 9,889).
Discussion
The contemporary maize genome landscape is comprised of nearly 85% transposons, with its ~40,000 genes clustered in tiny islands of non-repetitive sequence (44). Here, we use the long-read method Fiber-seq to delineate ACRs across the entire maize genome. We discover that only 94 LTR retrotransposons contain the functional regulatory elements required for transposon mobilization within dark-grown maize leaves, with only 76 of these being autonomous LTR retrotransposons. These 94 represent only a fraction of the recently transposed LTRs in maize, highlighting the efficiency of the plant-specific RNA-mediated DNA methylation pathway in checking the activity of LTRs in maize.
Furthermore, we demonstrate that LTR retrotransposons that lose chromatin accessibility at both regulatory elements required for transposon mobilization preferentially maintain an ACR at the putative enhancer elements. However, the epigenetic pattern at these LTR enhancer elements markedly diverges from that of canonical maize enhancer elements genome-wide. Specifically, these LTRs with a single ACR exhibit heterogeneous per-molecule chromatin accessibility, and contain the unexpected single-molecule co-occurrence of chromatin accessibility and hyper-5mCpG methylation, two epigenetic marks that are widely thought to be mutually exclusive. Consequently, as the epigenetic pattern of recently transposed LTR retrotransposons degenerates with evolutionary age, these LTRs are preferentially adopting regulatory architectures that diverge from that of canonical maize enhancer elements, suggesting that plant-specific DNA methylation pathways may be actively altering the epigenome at these LTRs to avoid the widespread occurrence of TE exaptation as gene enhancers. In contrast, we present evidence that one of the primary mechanisms by which LTRs shape maize gene regulation is by creating novel gene promoter elements within the LTR itself, or via TE-mediated host gene amplification.
Finally, we find that the loci in which Barbara McClintock discovered TE insertions show unusually low gene body methylation and unusually high gene body accessibility, consistent with the common assumption that gene body methylation protects against TE insertion. While the mechanistic underpinnings of these correlated epigenetic features are unclear, this finding adds to our understanding of the complexity of epigenetics and genome evolution in plants.
Further Fiber-seq studies across divergent maize lines, hybrid lines and ancestral species as well as across different tissues and conditions will help further illuminate the regulatory potential of TEs in plants, as well as their role in shaping maize genome expression and evolution (44). Furthermore, we anticipate that the future addition of CHH and CHG methylation data to Fiber-seq will enable a more mechanistic understanding of how the plant-specific RNA-mediated DNA methylation pathway is modulating TEs. Taken together, our results demonstrate the promise that Fiber-seq holds for understanding how TEs have shaped the structure, function, and evolution of angiosperm genomes.
Materials and Methods
Maize mesophyll protoplast generation
We used the PEG transformation method of maize mesophyll protoplasts as described in (48). Maize (Zea mays L. cultivar B73) seeds were soaked in water overnight at 25°C. The seeds were germinated in soil for 3 days under long day conditions (16 hours light, 8 hours dark) at 25°C, then moved to complete darkness at 25°C for 10–11 days. From each seedling, 10 cm sections from the second and third leaf were cut into thin 0.5 mm strips perpendicular to veins and immediately submerged in 10 ml of protoplasting enzyme solution (0.6 M mannitol, 10 mM MES ph 5.7, 15 mg/ml cellulase R10, 3 mg/ml macerozyme, 1 mM CaCl2, 0.1% [w/v] BSA, and 5 mM beta-mercaptoethanol). The mixture was covered in foil to keep out light, vacuum infiltrated for 3 min at room temperature (RT), and incubated on a shaker at 40 rpm for 2.5 hours at RT. Protoplasts were released by incubating an extra 10 min at 80 rpm. To quench the reaction, 10 mL ice-cold MMG (0.6 M Mannitol, 4 mM MES ph 5.7, 15 mM MgCl2) was added to the enzyme solution and the whole solution was filtered through a 40 μM cell strainer. To pellet protoplasts, the filtrate was split into equal volumes of no more than 10 mL in chilled round-bottom glass centrifuge vials and centrifuged at 100 × g for 4 min at RT. Pellets were resuspended in 1 mL cold MMG each and combined into a single round-bottom vial. To wash, MMG was added to make a total volume of 5 mL and the solution was centrifuged at 100 × g for 3 min at RT. This wash step was repeated two more times. The final pellet was resuspended in 1–2 mL of MMG. A sample of the resuspended protoplasts was diluted 1:20 in MMG and used to count the number of viable cells using Fluorescein Diacetate as a dye.
ATAC-seq data collection
An aliquot of 50,000 isolated protoplasts was added to new tubes and spun down at 4°C 2000g for 10 min. Supernatant was discarded and the pellet of protoplasts was washed with 750μl of lysis buffer (0.4M Sucrose, 10mM MgCl2, 25mM Tris-HCL pH 8.0, 0.1x Protease inhibitor, 0.5% TritonX). Samples were then spun down at 4°C 1500g for 5min and the supernatant discarded. Samples were then washed once more with buffer (0.4M Sucrose, 10mM MgCl2, 25mM Tris-HCL pH 8.0, 0.1x Protease inhibitor) at 4°C 1500g for 3 min to remove the lysis buffer. The nuclear pellet was then resuspended in 22.5 ddH20 followed by adding 25μl of 2x TD buffer (20mM Tris-HCl pH7.6, 10mM MgCl2, 20% vol/vol DMF) and 2.5μl of Tn5. Samples were then incubated at 37°C for 5 minutes. Reaction was stopped by adding 250μl of Zymo Research DNA Binding Buffer and DNA was purified using Zymo research Clean and concentrator kit. Samples were size selected using 1.8X ampure beads and barcoded with Illumina Nextera Index primers. Final library concentrations were determined using Qubit DNA HS assay and average fragment length was determined using TapeStation D1000 ScreenTape Assay.
Fiber-seq data collection
1–5 million Isolated protoplasts were spun down at 2000g and resuspended in a 100uL working buffer (400mM sucrose, 15mM Tris-Cl, 15mM NaCl, 60 mM KCl, 1mM EDTA, 0.5mM EGTA, 0.5mM Spermidine), with 1.5uL of 32mM SAM added to a final concentration of 0.8 mM along with 0.5 uL of Hia5 MTase (100U), then carefully mixed by pipetting the 10 times with wide bore tips. Reactions were incubated for 10 minutes at 25°C then stopped with 3 ul of 20% SDS (1% final concentration) and transferred to a new 1.7 mL microfuge tubes. High molecular weight DNA was then extracted using the Promega Wizard HMW DNA extraction kit A2920. PacBio SMRTbell libraries were then constructed using the manufacturer’s SMRTbell prep kit 3.0 procedure.
Fiber-seq data processing
Fibertools (49) was used to call m6A methylation and label regions as MSPs and nucleosomes on individual reads. Fiber-seq FIRE (46) was used to assign FDR values to MSPs and call Fiber-seq ACRs. For ACR calling we used the set of peaks identified by the FIRE pipeline with an FDR threshold of 1%.
ATAC-seq data processing
ATAC-seq read pairs were aligned to the MaizeV5 reference genome (13) using bwa v0.7.17-r118 (50). The resulting bams were filtered using samtools view (51) to discard reads that were unmapped (-F 4), had map quality of zero (-q 1). ACRs were called using MACS2 v2.2.7.1 (45), and the narrowPeaks output was merged to generate a non-overlapping set of ACRs. The ATAC-seq signal track is a sliding window histogram displaying the number of ATAC read ends, with the height of each 20-bp bar representing the number of Tn5 insertions within a 100-bp window centered on that 20-bp.
RNA-seq data used to define expression quantiles and transcription start sites (TSS)
66,143,401 publicly available RNA-seq reads were obtained from NCBI SRA: ERR3322830. These reads were derived from the second leaves of 9-day old, etiolated seedlings (Stelpflug SC et al 2016). Reads were aligned to the maize V5 annotation using hisat2 and counts were tallied using htseq-count. Transcripts Per Million (TPM) were calculated for each gene. 13,542 genes had a TPM of zero. The remainder were split into deciles by expression level, with each decile containing 2991 or 2992 genes. TSS positions were obtained using CAGE data (52)
Methylation rate (m6A and m5CpG)
For each genomic locus being aggregated, at each 20 bp bin, the number of possible methylation sites was calculated from the individual fiber sequences. The observed methylation events were tallied and divided by the number of possible sites to get a fraction of sites methylated.
MSP score and FIRE accessibility score
MSP score is the fraction of fiber-bases within a given region that are annotated as MSPs. FIRE accessibility score is the fraction of fiber-bases within a given region that are annotated as FIRE element (see fig. S1A).
Percent actuation
For a given genomic region, the number of unique reads (fibers) with at least one FIRE element overlapping the region, divided by the total number of unique reads overlapping the region.
Comparing single-cell ATAC-seq to Fiber-seq
The sparse matrix containing binary (cut or no cut) information for all cells from all tissues and all peaks reported in Marand et al 2021 were downloaded
(https://ftp.ncbi.nlm.nih.gov/geo/series/GSE155nnn/GSE155178/suppl/GSE155178%5FACR%5Fx%5Fcell.binary.sparse.txt.gz). We then generated a bed file consisting of only peaks with at least one Tn5 insertion in a leaf-designated cell, and reported, for each peak, the fraction of total leaf cells having one or more Tn5 insertion at that site. We used liftOver (53) to convert the genomic positions of the peaks in this file from V4 to V5 coordinates, then filtered peaks to retain those that (1) overlap by MACS2 peaks by 100 bp or more, and (2) overlap our FIRE-peaks by 100 bp or more. 39,132 peaks remain. Percent of cells with one or more Tn5 insertion is plotted against percent of fibers containing a fire element (%actuation) in Figure 1g. For this analysis, 58,712 MACS2 narrowPeaks were called on an alignment file (bam) containing 94,945,002 mapped 50-bp paired-end reads. These peaks were merged (bedops -m), resulting in 50,349 MACS2 peaks used above.
Short read mappability analysis
2.1 billion fragments were generated evenly distributed across the B73 reference genome chromosomes 1–10. Fragment lengths were sampled from a log-normal distribution fit to one of our ATAC-seq data sets. For each simulated fragment a paired-end read was generated with 50 base reads on either end of the fragment. The true start and end of the fragment was encoded in the read name. We did not simulate per-base errors in these reads; each read matches exactly the reference sequence from which it was generated. These reads were then mapped back to the genome using BWA. The ‘fraction mapped’ for a given region or window was calculated as the number of correctly mapped reads with mapq score > 0 divided by the total number of simulated reads with the outer end (Tn5 insertion) falling in the region. See fig. S2A.
Annotation of repetitive regions, including all transposable elements
Annotation file Zm-B73-REFERENCE-NAM-5.0.TE.gff3.gz was downloaded from maizegdb.org (54)
Annotation of regions of the nuclear genome with homology to organellar genomes
Regions of homology within the nuclear genome to organellar genomes were identified as follows for each of the chloroplast and mitochondrial genomes, separately. Paired-end reads were simulated to achieve 100x coverage (142,724 and 579,124 read-pairs, respectively), then mapped to the MaizeV5 reference genome (13) using bwa v0.7.17-r118 (50). The resulting bams were filtered using samtools view (51) to discard reads that were unmapped (-F 4), had map quality of zero (-q 1), or mapped to the centromere (55). Alignment files (bams) were then converted to bed files, and overlapping regions were merged.
Classification of ACRs
ACRs were sorted hierarchically as follows: (1) all ACRs with low FIRE accessibility score were included in the ‘low FIRE score’ set (medium gray), (2) ACRs with FE length < 200 bp and high FIRE accessibility score were included in the ‘FE length < 200’ set (blue), (3) ACRs with mappability < 80% and both high FIRE accessibility score and FE length >= 200 bp were included in the ‘Low mappability’ set (dark gray), (4) ACRs with high FIRE accessibility score, high FE length, and high mappability are in the ‘Regular ACRs’ set (light gray).
Enrichment of GWAS SNPs within different classes of ACRs
SNPs associated with 41 distinct phenotypes (56) were used to assess if newly called FIRE ACRs have a similar enrichment of GWAS SNPs to ATAC called ACRs. GWAS SNPs with RMIP<0.05 were removed as described in the paper. FIRE ACRs were split into two categories based on whether they overlap ATAC-seq ACRs as in Figure 0C. For both categories an enrichment was calculated by comparing the fraction of ACR bases covered by GWAS SNPs to the fraction covered in the shifted control category. FIRE ACRs overlapping and not overlapping ATAC-seq ACRs were found to have enrichment values of 3.37 and 3.16 respectively.
Calling differential ACRs (dACRs)
ATAC-seq reads from the following six tissues were downloaded from the NCBI Gene Expression Omnibus (Marand et al 2021): Tassel (GSM4696882), Ear (GSM4696883), GSM4696884 (Root1), Axillary_bud1 (GSM4696886), Crown_root1 (GSM4696888), Leaf2 (GSM4696890). For each sample, 100 million read pairs were downloaded, trimmed to 50 bp. Each of the six downloaded samples as well as reads from our in-house dark leaf protoplast sample were aligned to the MaizeV5 reference genome (13) using bwa v0.7.17-r118 (50). The resulting bams were filtered using samtools view (51) to discard reads that were unmapped (-F 4), had map quality of zero (-q 1), or mapped to the centromere (55). Because the number of MACS2 peaks is correlated with the number of mapped reads, for each of the seven samples, the number of aligned reads was subsampled to 16M. Peaks were called using MACS v2.2.7.1 (45), and the narrowPeaks output was merged to generate a non-overlapping set of peaks for each of the seven samples. A union set of 80,641 peaks was generated by merging the seven sets of peaks (bedops -m) (57). TN5 insertions were tallied in each unionpeak for each of the seven samples and per-bp accessibility was calculated by dividing by the peak length. Because our aim was to find differential ACRs that were inaccessible in dark leaf protoplast, we defined differential ACRs as those that (1) had fewer per-bp TN5 insertions than twice the minimum DLP cutcounts in a union peak overlapping a called DLP peak, and (2) the difference between the accessibility of most-accessible sample and the dark leaf protoplast sample was in the 75th percentile or greater. These 2,826 dACRs are in table S7.
Identification of solo LTRs
LTR sequences from intact LTR retrotransposons containing at least one FIRE ACR within either LTR were aligned to the maize genome using blastn (58). Matches with bitscore greater than 1400 and length greater than 1000 bps were retained and merged (bedops -m). Next, we identified matches that (i) did not overlap another intact LTR retrotransposon, and (ii) contained a FIRE ACR. These are listed in table S9.
Identification of hAT insertion sites
hAT insertion sites are defined as 200-bp windows centered on the location of a hAT transposon polymorphism in which B73 lacks the hAT transposon and exactly one of the 25 NAM lines contains a hAT transposon (43).
Supplementary Material
Acknowledgments.
We thank Stanley Fields, Olivia Waltner, Michelle Stitzer, Edward Buckler, and Jeffrey Ross-Ibarra for helpful data analysis suggestions and discussion of results as well as detailed manuscript comments.
Funding.
This work was supported by the National Science Foundation (PlantSynBio grant no. 2240888 to C.Q., NSF Postdoctoral Research Fellowship in the Biology Program (Grant Number 2305660) to B.R-C.), the National Institutes of Health (NIGMS MIRA grant no. 1R35GM139532 to C.Q.), and the United States Department of Agriculture (NIFA postdoctoral fellowship no. 2023-67012-39445 to N.A.M). A.B.S. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund and is a Pew Biomedical Scholar. This study was supported by National Institutes of Health (NIH) grants 1DP5OD029630, and UM1DA058220 to A.B.S. M.R.V. was supported by a training grant (T32) from the NIH (2T32GM007454-46).
Footnotes
Competing interests. A.B.S. is a co-inventor on a patent relating to the Fiber-seq method (US17/995,058).
Data and materials availability.
Raw and processed sequencing data is available from the NCBI Short Read Archive (SRA) under Bioproject PRJNA1119563.
References
- 1.McClintock B., The origin and behavior of mutable loci in maize. Proceedings of the National Academy of Sciences 36, 344–355 (1950). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClintock B., Induction of Instability at Selected Loci in Maize. Genetics 38, 579–599 (1953). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McClintock B., The significance of responses of the genome to challenge. Science 226, 792–801 (1984). [DOI] [PubMed] [Google Scholar]
- 4.Fedoroff N. V., Transposable Genetic Elements in Maize. Sci. Am. 250, 84–99 (1984).6422549 [Google Scholar]
- 5.Jones R. N., McClintock’s controlling elements: the full story. Cytogenet. Genome Res. 109, 90–103 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Fedoroff N. V., McClintock’s challenge in the 21st century. Proceedings of the National Academy of Sciences 109, 20200–20203 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mcclintock B., Controlling elements and the gene. Cold Spring Harb. Symp. Quant. Biol. 21, 197–216 (1956). [DOI] [PubMed] [Google Scholar]
- 8.Cui X., Cao X., Epigenetic regulation and functional exaptation of transposable elements in higher plants. Curr. Opin. Plant Biol. 21, 83–88 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Makarevitch I., Waters A. J., West P. T., Stitzer M., Hirsch C. N., Ross-Ibarra J., Springer N. M., Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 11, e1004915 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao H., Zhang W., Chen L., Wang L., Marand A. P., Wu Y., Jiang J., Proliferation of Regulatory DNA Elements Derived from Transposable Elements in the Maize Genome. Plant Physiol. 176, 2789–2803 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noshay J. M., Marand A. P., Anderson S. N., Zhou P., Mejia Guerra M. K., Lu Z., O’Connor C. H., Crisp P. A., Hirsch C. N., Schmitz R. J., Springer N. M., Assessing the regulatory potential of transposable elements using chromatin accessibility profiles of maize transposons. Genetics 217, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Studer A., Zhao Q., Ross-Ibarra J., Doebley J., Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43, 1160–1163 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hufford M. B., Seetharam A. S., Woodhouse M. R., Chougule K. M., Ou S., Liu J., Ricci W. A., Guo T., Olson A., Qiu Y., Coletta R. D., Tittes S., Hudson A. I., Marand A. P., Wei S., Lu Z., Wang B., Tello-Ruiz M. K., Piri R. D., Wang N., won Kim D., Zeng Y., O’Connor C. H., Li X., Gilbert A. M., Baggs E., Krasileva K. V., Portwood J. L., Cannon E. K. S., Andorf C. M., Manchanda N., Snodgrass S. J., Hufnagel D. E., Jiang Q., Pedersen S., Syring M. L., Kudrna D. A., Llaca V., Fengler K., Schmitz R. J., Ross-Ibarra J., Yu J., Gent J. I., Hirsch C. N., Ware D., Dawe R. K., De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science 373, 655–662 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stergachis A. B., Debo B. M., Haugen E., Churchman L. S., Stamatoyannopoulos J. A., Single-molecule regulatory architectures captured by chromatin fiber sequencing. Science 368, 1449–1454 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Kong Y., Cao L., Deikus G., Fan Y., Mead E. A., Lai W., Zhang Y., Yong R., Sebra R., Wang H., Zhang X.-S., Fang G., Critical assessment of DNA adenine methylation in eukaryotes using quantitative deconvolution. Science 375, 515–522 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tullius T. W., Isaac R. S., Ranchalis J., Dubocanin D., Churchman L. S., Stergachis A. B., RNA polymerases reshape chromatin and coordinate transcription on individual fibers. bioRxivorg, doi: 10.1101/2023.12.22.573133 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vollger M. R., Swanson E. G., Neph S. J., Ranchalis J., Munson K. M., Ho C.-H., Sedeno-Cortes A. E., Fondrie W. E., Bohaczuk S. C., Mao Y., Parmalee N. L., Mallory B. J., Harvey W. T., Kwon Y., Garcia G. H., Hoekzema K., Meyer J. G., Cicek M., Eichler E. E., Noble W. S., Witten D. M., Bennett J. T., Ray J. P., Stergachis A. B., A haplotype-resolved view of human gene regulation, bioRxiv (2024). 10.1101/2024.06.14.599122. [DOI] [Google Scholar]
- 18.Bubb K. L., Deal R. B., Considerations in the analysis of plant chromatin accessibility data. Curr. Opin. Plant Biol. 54, 69–78 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marand A. P., Chen Z., Gallavotti A., Schmitz R. J., A cis-regulatory atlas in maize at single-cell resolution. Cell 184, 3041–3055.e21 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Uren A. G., Kool J., Berns A., van Lohuizen M., Retroviral insertional mutagenesis: past, present and future. Oncogene 24, 7656–7672 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Erdmann R. M., Picard C. L., RNA-directed DNA methylation. PLoS Genet. 16, e1009034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leduque B., Edera A., Vitte C., Quadrana L., Simultaneous profiling of chromatin accessibility and DNA methylation in complete plant genomes using long-read sequencing. Nucleic Acids Res. 52, 6285–6297 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colonna Romano N., Fanti L., Transposable elements: Major players in shaping genomic and evolutionary patterns. Cells 11, 1048 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen C. J., Lock W. M., Mager D. L., Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448, 105–114 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Sundaram V., Cheng Y., Ma Z., Li D., Xing X., Edge P., Snyder M. P., Wang T., Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 24, 1963–1976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medstrand P., Landry J. R., Mager D. L., Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J. Biol. Chem. 276, 1896–1903 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Dunn C. A., Medstrand P., Mager D. L., An endogenous retroviral long terminal repeat is the dominant promoter for human beta1,3-galactosyltransferase 5 in the colon. Proc. Natl. Acad. Sci. U. S. A. 100, 12841–12846 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stelpflug S. C., Sekhon R. S., Vaillancourt B., Hirsch C. N., Buell C. R., de Leon N., Kaeppler S. M., An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9, lantgenome2015.04.0025 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Yang N., Wang Y., Liu X., Jin M., Vallebueno-Estrada M., Calfee E., Chen L., Dilkes B. P., Gui S., Fan X., Harper T. K., Kennett D. J., Li W., Lu Y., Ding J., Chen Z., Luo J., Mambakkam S., Menon M., Snodgrass S., Veller C., Wu S., Wu S., Zhuo L., Xiao Y., Yang X., Stitzer M. C., Runcie D., Yan J., Ross-Ibarra J., Two teosintes made modern maize. Science 382, eadg8940 (2023). [DOI] [PubMed] [Google Scholar]
- 30.Bailey J. A., Liu G., Eichler E. E., An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 73, 823–834 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao J., Yu T., Xu B., Hu Z., Zhang X.-O., Theurkauf W. E., Weng Z., Epigenetic and chromosomal features drive transposon insertion in Drosophila melanogaster. Nucleic Acids Res. 51, 2066–2086 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fedoroff N., Wessler S., Shure M., Isolation of the transposable maize controlling elements Ac and Ds. Cell 35, 235–242 (1983). [DOI] [PubMed] [Google Scholar]
- 34.Courage-Tebbe U., Döring H. P., Fedoroff N., Starlinger P., The controlling element Ds at the Shrunken locus in Zea mays: structure of the unstable sh-m5933 allele and several revertants. Cell 34, 383–393 (1983). [DOI] [PubMed] [Google Scholar]
- 35.Shure M., Wessler S., Fedoroff N., Molecular identification and isolation of the Waxy locus in maize. Cell 35, 225–233 (1983). [DOI] [PubMed] [Google Scholar]
- 36.Fedoroff N. V., Furtek D. B., Nelson O. E., Cloning of the bronze locus in maize by a simple and generalizable procedure using the transposable controlling element Activator (Ac). Proc. Natl. Acad. Sci. U. S. A. 81, 3825–3829 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paz-Ares J., Wienand U., Peterson P. A., Saedler H., Molecular cloning of the c locus of Zea mays: a locus regulating the anthocyanin pathway. EMBO J. 5, 829–833 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elgin S. C., DNAase I-hypersensitive sites of chromatin. Cell 27, 413–415 (1981). [DOI] [PubMed] [Google Scholar]
- 39.Sullivan A. M., Arsovski A. A., Lempe J., Bubb K. L., Weirauch M. T., Sabo P. J., Sandstrom R., Thurman R. E., Neph S., Reynolds A. P., Stergachis A. B., Vernot B., Johnson A. K., Haugen E., Sullivan S. T., Thompson A., Neri F. V. 3rd, Weaver M., Diegel M., Mnaimneh S., Yang A., Hughes T. R., Nemhauser J. L., Queitsch C., Stamatoyannopoulos J. A., Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep. 8, 2015–2030 (2014). [DOI] [PubMed] [Google Scholar]
- 40.French S. L., Osheim Y. N., Cioci F., Nomura M., Beyer A. L., In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 23, 1558–1568 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takuno S., Gaut B. S., Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol. Biol. Evol. 29, 219–227 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Muyle A. M., Seymour D. K., Lv Y., Huettel B., Gaut B. S., Gene body methylation in plants: Mechanisms, functions, and important implications for understanding evolutionary processes. Genome Biol. Evol. 14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munasinghe M., Read A., Stitzer M. C., Song B., Menard C. C., Ma K. Y., Brandvain Y., Hirsch C. N., Springer N., Combined analysis of transposable elements and structural variation in maize genomes reveals genome contraction outpaces expansion. PLoS Genet. 19, e1011086 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fedoroff N. V., Transposable elements, epigenetics, and genome evolution. Science 338, 758–767 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Gaspar J. M., Improved peak-calling with MACS2, bioRxiv (2018)p. 496521. [Google Scholar]
- 46.FIRE: A Snakemake Workflow for Calling Fiber-Seq Inferred Regulatory Elements (FIREs) on Single Molecules (Github; https://github.com/fiberseq/FIRE). [Google Scholar]
- 47.Pb-CpG-Tools: Collection of Tools for the Analysis of CpG Data (Github; https://github.com/PacificBiosciences/pb-CpG-tools). [Google Scholar]
- 48.Sievers F., Higgins D. G., Clustal omega. Curr. Protoc. Bioinformatics 48, 3.13.1–3.13.16 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Tonnies J., Mueth N. A., Gorjifard S., Chu J., Queitsch C., Scalable Transfection of Maize Mesophyll Protoplasts. J. Vis. Exp., doi: 10.3791/64991 (2023). [DOI] [PubMed] [Google Scholar]
- 50.Fibertools-Rs: Tools for Fiberseq Data Written in Rust (Github; https://github.com/fiberseq/fibertools-rs). [Google Scholar]
- 51.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.SAMtools. https://samtools.sourceforge.net/.
- 53.Mejía-Guerra M. K., Li W., Galeano N. F., Vidal M., Gray J., Doseff A. I., Grotewold E., Core Promoter Plasticity Between Maize Tissues and Genotypes Contrasts with Predominance of Sharp Transcription Initiation Sites. Plant Cell 27, 3309–3320 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hinrichs A. S., Karolchik D., Baertsch R., Barber G. P., Bejerano G., Clawson H., Diekhans M., Furey T. S., Harte R. A., Hsu F., Hillman-Jackson J., Kuhn R. M., Pedersen J. S., Pohl A., Raney B. J., Rosenbloom K. R., Siepel A., Smith K. E., Sugnet C. W., Sultan-Qurraie A., Thomas D. J., Trumbower H., Weber R. J., Weirauch M., Zweig A. S., Haussler D., Kent W. J., The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 34, D590–8 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welcome to MaizeGDB. https://www.maizegdb.org/.
- 56.Wolfgruber T. K., Sharma A., Schneider K. L., Albert P. S., Koo D.-H., Shi J., Gao Z., Han F., Lee H., Xu R., Allison J., Birchler J. A., Jiang J., Dawe R. K., Presting G. G., Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic Loci shaped primarily by retrotransposons. PLoS Genet. 5, e1000743 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wallace J. G., Bradbury P. J., Zhang N., Gibon Y., Stitt M., Buckler E. S., Association mapping across numerous traits reveals patterns of functional variation in maize. PLoS Genet. 10, e1004845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neph S., Kuehn M. S., Reynolds A. P., Haugen E., Thurman R. E., Johnson A. K., Rynes E., Maurano M. T., Vierstra J., Thomas S., Sandstrom R., Humbert R., Stamatoyannopoulos J. A., BEDOPS: high-performance genomic feature operations. Bioinformatics 28, 1919–1920 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T. L., BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed sequencing data is available from the NCBI Short Read Archive (SRA) under Bioproject PRJNA1119563.