Abstract
Twelve G protein-coupled receptors, including chemokine receptors, act as coreceptors and determinants for the cell tropisms of human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus (SIV). We isolated HIV-1 variants from T-cell-line (T)- and macrophage (M)-tropic (i.e., dualtropic) (R5-R3-X4) HIV-1 strains and also produced six HIV-1 mutants carrying single-point amino acid substitutions at the tip of the V3 region of the Env protein of HIV-1. These variants and three mutants infected brain-derived CD4-positive cells that are resistant to M-, T-, or dualtropic (R5, X4, or R5-X4) HIV-1 strains. However, a factor that determines this cell tropism has not been identified. This study shows that primary brain-derived fibroblast-like cell strains, BT-3 and BT-20/N, as well as a CD4-transduced glioma cell line, U87/CD4, which were susceptible to these HIV-1 variants and mutants and the HIV-2ROD strain, expressed mRNA of an orphan G protein-coupled receptor (GPCR), GPR1. When a CD4-positive cell line which was strictly resistant to infection with diverse HIV-1 and HIV-2 strains was transduced with GPR1, the cell line became susceptible to these HIV-1 variants and mutants and to an HIV-2 strain but not to T- or dualtropic HIV-1 strains, and numerous syncytia formed after infection. These results indicate that GPR1 functions as a coreceptor for the HIV-1 variants and mutants and for the HIV-2ROD strain in vitro.
Twelve G protein-coupled receptors (GPCRs), including six chemokine receptors (CKRs), have been shown to act as coreceptors for human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus (SIV) (15, 32). Coreceptors mediate the infection processes initiated by the binding of HIV-SIV virions to the principal receptor, the CD4 molecule (10, 15, 32). GPCRs are constituted with diverse cell membrane receptors, such as CKRs, peptide hormone receptors, and light receptors, are evolutionarily related to one another, and all take the seven-transmembrane architecture (2, 18, 26). Chemokines and CKRs play important roles in the chemotaxis of leukocytes in vivo (2, 18, 26). A CXC-CKR, CXCR4, acts as a major coreceptor for T-cell-line (T)-tropic HIV-1 and HIV-2 strains (13, 15). The CC-CKRs CCR5, CCR3, and CCR2b specifically facilitate the infection with primary or macrophage (M)-tropic HIV-1, HIV-2, or SIV strains (4, 5, 7, 8, 39). Cell tropism of HIV-1, HIV-2, and SIV is mostly explained by their uses of specific CKRs as coreceptors for infection. In addition to the GPCRs described above, a CC-CKR (CCR8), a cytomegalovirus-encoded CKR, (US28), and five orphan GPCRs (APJ, GPR1, GPR15, STRL33/Bonzo, and v28) were shown to serve as coreceptors for HIV-1, HIV-2, or SIV strains (7, 11, 14, 19, 22, 30, 33).
We isolated three HIV-1 strains, GUN-1WT, GUN-4WT, and GUN-7WT, which show both T- and M-tropisms (i.e., dualtropism) (41, 42). From these wild-type (WT) strains, HIV-1 variants GUN-1V, GUN-4V, and GUN-7V, which infect brain-derived cells such as the CD4-positive, established glioma cell line U87/CD4 and the primary brain-derived fibroblast-like cell strains BT-3 and BT-20/N were further isolated (38, 41, 42). Isolation of HIV-2, SIV, and primary HIV-1 strains which infect U87/CD4 cells has also been reported (9, 44), suggesting the presence of these types of viruses in vivo. BT-20/N cells were freshly isolated from macroscopically normal tissues next to glioblastoma tissue, and BT-3 cells were isolated from meningioma by the explant culture method (41, 42). These two primary cell strains stop growing when they are passaged more than 10 to 15 times. They were morphologically fibroblast-like and negative for the expression of specifically differentiated cell markers present in the brain, such as factor VIII-related antigens (endothelial cell marker) or glial fibrillary acid protein (astrocyte marker). Because monoclonal antibodies against BT-3 cells stain the brain blood vessels in brain tissue, BT-3 or BT-20/N cells may have been derived from cells associated with them (unpublished data).
The third variable domains (V3) of the Env proteins of HIV-1, HIV-2, and SIV are thought to interact with coreceptors (28, 43). The GPGR (glycine-proline-glycine-arginine) tip sequence of the V3 region of the Env protein present in the GUN-1WT, GUN-4WT, and GUN-7WT strains was replaced with GS(serine)GR, GA(alanine)GR, and GT(threonine)GR in GUN-1V, the GUN-4V, and GUN-7V variants, respectively (mutated amino acid residues are underlined) (38, 41). Therefore, the cell tropism characteristic of these HIV-1 variants is probably determined by the specific amino acid sequences of the V3 domain which will interact with a coreceptor. To confirm the effects of amino acid changes of the V3 tip sequences on the cell tropisms of HIV-1, we constructed six mutants of the GUN-1WT strain, GUN-1/A, GUN-1/L, GUN-1/P, GUN-1/R, GUN-1/S, and GUN-1/T, as described below. These mutants were also used in this study to examine whether a given GPCR functions as a coreceptor for them.
We tried to identify CKR-related genes expressed in brain-derived cells by reverse transcription (RT)-PCR using degenerated primers and found that CCR8/TER1 acts as a coreceptor for several HIV-1 strains, such as GUN-1WT, GUN-1V, GUN-4WT, GUN-4V, GUN-7WT, and GUN-7V (19). However, CCR8/TER1 was not considered to be a specific coreceptor for these variants, because not only GUN-1V, GUN-4V, and GUN-7V but also GUN-1WT, GUN-4WT, and GUN-7WT use it as a coreceptor (19). Other than CCR8/TER1, CCR1 was found to be expressed in the brain-derived cells by RT-PCR using the degenerated primers (19). NP-2/CD4 cells expressing CCR1 are resistant to all HIV-1 strains tested including these HIV-1 variants (19), which indicates that CCR1 is not a coreceptor specifically used by the HIV-1 variants and mutants which infect the brain-derived cells. An orphan G protein-coupled receptor (GPCR), APJ, has recently been shown to be widely expressed in the brain (7). However, APJ could be excluded as a possible coreceptor used by these variants because the expression of APJ mRNA was not detected in the brain-derived cells we used, as described below (Table 1).
TABLE 1.
Expression of mRNA for HIV-SIV coreceptors in various cells as detected by RT-PCR
| Core-ceptor | Level of expression bya:
|
||||||
|---|---|---|---|---|---|---|---|
| BT-20/N | BT-3 | U87/CD4 | C8166 | NP-2/CD4 | Macrophages | PBLs | |
| APJ | − | − | − | − | − | − | + |
| CCR2b | − | − | − | − | − | − | + |
| CCR3 | − | − | − | − | − | + | ± |
| CCR5 | − | − | − | − | − | ++ | − |
| CCR8 | − | − | − | + | − | − | ± |
| CCR9 | − | − | − | − | − | − | − |
| CXCR4 | − | − | − | ++ | − | ± | ++ |
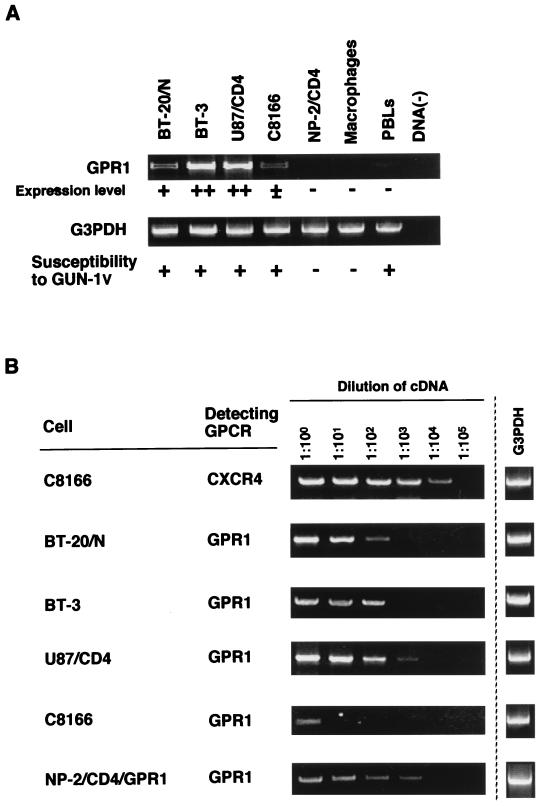
| GPR1 | + | ++ | ++ | + | − | − | ± |
| GPR15 | − | − | − | + | − | − | ++ |
| STRL33/ Bonzo | ± | + | + | − | ± | + | ++ |
| US28 | − | − | − | − | − | − | − |
| v28 | − | − | − | ± | − | − | − |
DNA amplified by RT-PCR was examined by agarose gel electrophoresis. Intensities of specific bands detected as shown in Fig. 1A were expressed as follows: ±, faint; +, weak; ++, strong; −, not detected.
In this paper, to identify a GPCR which functions as a coreceptor for HIV-1 and HIV-2 strain infection of brain-derived cells, the expression of mRNA for 12 reported HIV-SIV coreceptors (APJ, CCR2b, CCR3, CCR5, CCR8, CCR9, CXCR4, GPR1, GPR15, STRL33/Bonzo, US28, and v28) was examined in various cells, including the two primary brain-derived cell strains.
The HIV-1 strains GUN-1WT, GUN-1V (42), GUN-1/P, GUN-1/S, GUN-1/T, GUN-1/A, GUN-1/L, GUN-1/R, IIIB (46), GUN-4WT, GUN-4V, GUN-7WT, and GUN-7V (38) and an HIV-2 strain, ROD (17), were propagated in C8166 cells (35). GUN-1/A, GUN-1/L, GUN-1/P, GUN-1/R, GUN-1/S, and GUN-1/T strains were constructed by changing the GPGR sequence of GUN-1WT to GAGR, GLGR, GPGR (mutated but synonymous with that in GUN-1WT), GRGR, GSGR, and GTGR, respectively (unpublished data). We noticed that GUN-1/L, GUN-1/P, and GUN-1/R mutants infect both T-cell lines and macrophages but not the BT-3, BT-20/N, or U87/CD4 brain-derived cells, while GUN-1/A, GUN-1/S, and GUN-1/T mutants infect the brain-derived cells but not macrophages (unpublished data). These mutants were propagated in C8166 cells. The M-tropic (R5) HIV-1 strains SF162 (6) and BaL (47) were propagated in human peripheral blood lymphocytes (PBLs) isolated from healthy donors by a method described elsewhere (25). The culture supernatants of the cells infected with HIV-1 and HIV-2 were harvested when numerous syncytia were observed microscopically, and the supernatants were stored as viral stocks.
NP-2/CD4, NP-2/GPR1, and NP-2/CD4/GPR1 cells were established by introducing CD4 and/or GPR1 genes into a human glioma cell line, NP-2 (37), by using a retrovirus vector as described below. These NP-2-derived cells and a CD4-introduced, established human glioma cell line, U87/CD4 (41, 45), were maintained in E minimal essential medium (Nissui Co., Ltd., Tokyo, Japan) supplemented with 10% (vol/vol) fetal calf serum (FCS). C8166 cells were cultured in RPMI 1640 medium (Nissui) containing 10% (vol/vol) FCS. Primary brain-derived fibroblast-like BT-20/N and BT-3 cells were cultured in RPMI 1640 medium containing 10% (vol/vol) FCS, 10 μg of endothelial cell growth supplement per ml, and 10 ng of epidermal growth factor per ml. BT-20/N cells had been derived from macroscopically normal tissues next to glioblastoma tissue and BT-3 cells were from a meningioma (37, 38, 41, 42). They were isolated by the explant culture method and showed senescent phenotypes after 10 to 15 passages in vitro.
We examined the expression of mRNA of 12 reported HIV-SIV coreceptor genes (APJ, CCR2b, CCR3, CCR5, CCR8, CCR9, CXCR4, GPR1, GPR15, STRL33/Bonzo, US28, and v28) in various cells, including the brain-derived cells (Table 1). For this, specific PCR primer pairs for each gene were synthesized (Nihon Idenshi Kenkyujo Co., Ltd., Miyagi, Japan). GPCRs examined in this study and the nucleotide sequences, orientations, and positions of the primers in their coding regions are as follows: APJ, 5′-ATGGAGGAAGGTGGTGATTTTGACAACTAC-3′ (sense; positions 1 to 30) and 5′-CTA GTCAACCACAAGGGTCTCCTGCTGTAG-3′ (antisense; positions 1113 to 1143) (DDBJ/GenBank/EMBL accession no. U03642); CCR2b, 5′-ATGCTGTCCACATCTCGTTCTCGGTTTATCAG-3′ (sense; positions 1 to 32) and 5′-TTATAAACCAGCCGAGACTTCCTGCTCCCCAG-3′ (antisense; positions 1052 to 1083) (accession no. U03905); CCR3, 5′-GCCC GGACTGTCACTTTTGGTGTCATCACCAG-3′ (sense; positions 433 to 464) and 5′-CTTCTCACTAGGAAGGAATGGGATGTATCT-3′ (antisense; positions 982 to 1011) (accession no. U49727); CCR5, 5′-GCCAGGACGGTCACCTTTGGGGTGGTGACAA-3′ (sense; positions 415 to 445) and 5′-AGCCTCTTGCTGGAAAATAAAACAGCATTT-3′ (anti-sense; positions 964 to 993) (accession no. U54994); CCR8, 5′-GTGAGGACGATCAGGATGGGCACAACGCTGTG-3′ (sense; positions 430 to 461) and 5′-GCTCTCCCTAGGCATTTGTCTTCCTAGGTA-3′ (antisense; positions 973 to 1002) (accession no. U62556); CXCR4, 5′-CCAAGGAAGCTGTTGGCTGAAAAGGTGGTCTA-3′ (sense; positions 439 to 470) and 5′-TCCACCTCGCTTTCCTTTGGAGAGGATCTT-3′ (antisense; positions 979 to 1008) (accession no. X71635); GPR1, 5′-ATGGAAGATTTGGAGGAAACATTATTTGAA-3′ (sense; positions 1 to 30) and 5′-TTATTGAGCTGTTTCCAGGAGACACAGATTC-3′ (antisense; positions 1038 to 1050) (accession no. U13666); GPR15, 5′-ATGGACCCAGAAGAAACTTCAGTTTATTTG-3′ (sense; positions 1 to 30) and 5′-TTAGAGTGACACAGACCTCTTCCTCCTCCTGG-3′ (anti-sense; positions 1052 to 1083) (accession no. U34806); STRL33/Bonzo, 5′-AATCTCGACAAGCTCATATGTGGTTACCATG-3′ (sense; positions 520 to 550) and 5′-AGATTTCCATTGATGTGAGACCCCAAGGTAAG-3′ (antisense; positions 929 to 960) (accession no. AF007545); US28, 5′-CCGCATTTCCAGAATCGTTGCGGTGTCTCAG-3′ (sense; positions 624 to 654) and 5′-TGTGAGACGCGACACGCCTCGTCGGACAGCG-3′ (antisense; positions 1019 to 1048); v28, 5′-ATGG ATCAGTTCCCTGAATCAGTGACAGAAAAC-3′ (sense; positions 1 to 33) and 5′-TCAGAGAAGGAGCAATGCATCTCCATCACTCG-3′ (antisense; positions 1037 to 1068) (accession no. U20350).
Total RNA was isolated from cells (BT-20/N, BT-3, U87/CD4, C8166, NP-2/CD4), macrophages, and PBLs by using an RNA extraction kit (SepaGene; Sanko-Junyaku Co., Ltd., Tokyo, Japan), in accordance with the manufacturer’s protocol. To remove contaminated DNA, 30 μl of the total RNA preparation was treated with 50 U of RNase-free DNase I (Boehringer Mannheim Co., Ltd., Tokyo, Japan) in a reaction mixture containing 10 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 1 mM dithiothreitol (DTT) at 37°C for 2 h. To make cDNA, 2 μg of the total RNA was reverse transcribed in 20 μl of a reaction mixture containing 0.6 μg of oligo(dT) (GIBCO BRL, Rockville, Md.), 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 2.5 mM each deoxynucleoside triphosphate (dNTP), 10 mM DTT, 10 U of RNasin (Takara Shuzo Co., Ltd., Shiga, Japan), and 100 U of Superscript II reverse transcriptase (GIBCO BRL) at 42°C for 1 h. To detect GPCR sequences in the cDNA preparation, PCR was done by using one pair of specific primers for each GPCR, in which 20 μl of a reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 2.5 mM each dNTP, 60 ng of each sense and antisense PCR primer, and 5 U of Taq DNA polymerase (Takara Shuzo) was subjected to 38 cycles of PCR as follows: 93°C for 30 s, 60°C for 1 min, and 72°C for 2 min. As a control, the expression levels of glyceraldehyde-3-phosphate dehydrogenase (G3PDH) in the cells were determined by RT-PCR. PCR-amplified cDNA was examined by 1% (wt/vol) agarose gel electrophoresis.
The expression of GPR1 and STRL33/Bonzo was detected by RT-PCR in brain-derived cells such as BT-20/N, BT-3, U87/CD4, the C8166 T-cell line, and human PBLs (Table 1; Fig. 1). The expression of the other nine HIV-SIV coreceptors was not detected in the brain-derived cells. BT-20/N, BT-3, and U87/CD4 cells were specifically susceptible to HIV-1 variants GUN-1V, GUN-4V, and GUN-7V (Fig. 2B and C; see also Fig. 5B and C) as reported previously (38, 41, 42). The expression of GPR1 mRNA was not detected in NP-2/CD4 cells and macrophages, and NP-2/CD4 cells were resistant to infection with these three HIV-1 variants (Fig. 2E; see also Fig. 5E), as were macrophages (19). These results suggest the possibility that GPR1 acts as a coreceptor for the HIV-1 variants. The expression of STRL33/Bonzo was detected not only in the brain-derived cells but also in NP-2/CD4 cells and macrophages. It is unlikely that STRL33/Bonzo serves as a coreceptor for these variants because its mRNA was detected in NP-2/CD4 cells, which are resistant to all HIV-1 strains tested, including these variants (Table 1), and in macrophages, which are resistant to these variants (25, 38).
FIG. 1.
Expression of GPR1 mRNA in various types of cells. (A) The expression of GPR1 mRNA in three cell lines (U87/CD4, C8166, and NP-2/CD4), two fibroblast-like cell lines (BT-20/N and BT-3), and macrophages was detected by RT-PCR. As a control, the expression of G3PDH mRNA was detected by RT-PCR. Total RNA without reverse transcription gave no bands (data not shown). Similar RT-PCR experiments were done as for the other 11 HIV-SIV coreceptors. Expression levels were estimated by the intensities of PCR bands and are summarized in Table 1. (B) Determination of the relative amounts of GPR1 mRNA in the brain-derived cells. The cDNA reverse transcribed from 2 μg of the total RNA extracted from the cells was serially 10-fold diluted and amplified by PCR to detect the GPR1 sequences by using the specific primers. As a control, the level of the expression of CXCR4 mRNA in C8166 cells was determined by the same method. G3PDH mRNA in each sample detected by RT-PCR is shown as a control.
FIG. 2.
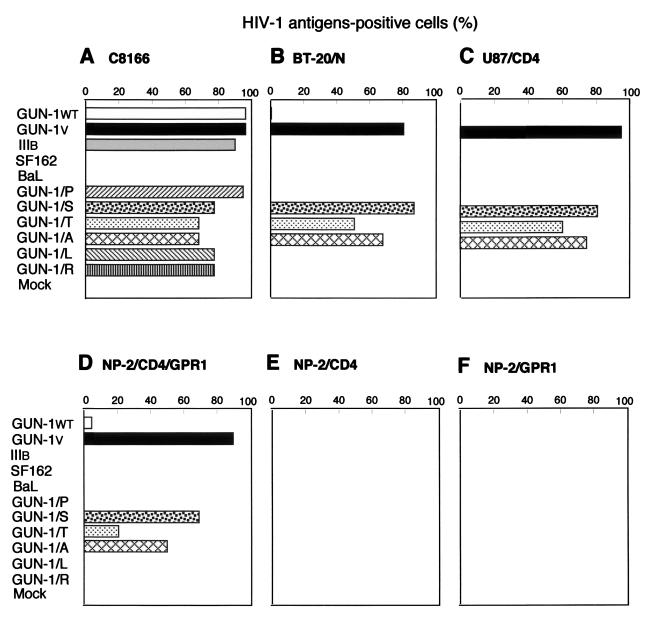
Susceptibilities of various types of cells to HIV-1 strains. C8166 (A), BT-20/N (B), U87/CD4 (C), NP-2/CD4/GPR1 (D), NP-2/CD4 (E), and NP-2/GPR1 (F) cells were infected with HIV-1 strains GUN-1WT, GUN-1V, GUN-1/P, GUN-1/S, GUN-1/T, GUN-1/A, GUN-1/L, GUN-1/R, SF162, BaL, and IIIB. The infections were assessed by detecting HIV-1 antigens by IFA in challenged cells on days 2, 4, and 6 after inoculation. In this figure, results from day 6 are shown.
FIG. 5.
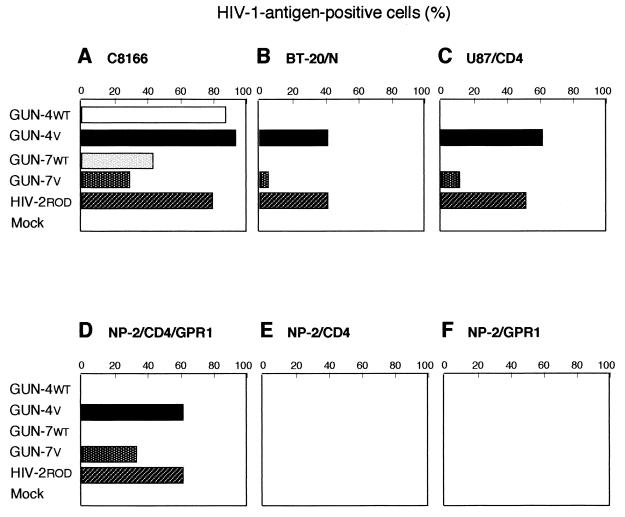
Susceptibilities of various types of cells to HIV-1 and HIV-2 strains. The susceptibilities of C8166 (A), BT-20/N (B), U87/CD4 (C), NP-2/CD4/GPR1 (D), NP-2/CD4 (E), and NP-2/GPR1 (F) cells to the GUN-4WT, the GUN-4V, GUN-7WT, GUN-7V, and HIV-2ROD strains were determined by detection of HIV-1 and HIV-2 antigens in infected cells by using IFA. The cells were harvested for IFA on day 6 after infection.
To examine whether GPR1 acts as a coreceptor for the HIV-1 variants, we established several cell lines which stably expressed the GPR1 gene. cDNA containing the coding region of the GPR1 gene was made from total RNA of BT-3 cells by RT-PCR using the specific primers for GPR1. GPR1 DNA was cloned into the EcoRV site of the expression plasmid pcDNA3 (Clontech, Palo Alto, Calif.), which was designated pcDNA3/GPR1. GPR1 DNA was isolated from the pcDNA3/GPR1 plasmid by BamHI and NotI digestion and subcloned into the expression plasmid pMX/puro, which is derived from the pBabe plasmid (20) and has a puromycin resistance gene, using the BamHI and NotI sites. The resultant plasmid was designated pMX-puro/GPR1. The nucleotide sequence of GPR1 in pMX-puro was determined. In the NH2-terminal domain, which shows variable amino acid sequences in each GPCR and contains amino acid sequences highly specific to each GPCR, the amino acid sequence of the cloned GPR1 DNA was almost identical to the reported sequence (23) with one difference: the 11th residue (glutamic acid) in the reported sequence of GPR1 (23) was replaced with lysine in the cloned GPR1. This may be due to the polymorphism of GPR1. There also are several mismatches in the amino acid sequences of GPCRs that have been reported so far. Such mismatches are found among reported sequences of CCR7/EBI1 (3, 36). The homology of amino acid sequences between a given pair of GPCRs reported so far is less than 90% (26). Therefore, the DNA fragments we cloned were considered to be GPR1.
GPR1-transduced cell lines were prepared as follows. The plasmid harboring the genes for the receptor of ecotropic murine leukemia virus (MuLV) and hygromycin resistance was introduced into NP-2/CD4 cells by DNA transfection using LipofectAMINE (GIBCO BRL), and hygromycin-resistant cells were selected as previously described (19). These cells were infected with the ecotropic MuLV pseudotypes containing puromycin resistance and GPR1 genes, which had been produced by BOSC23 cells (ATCC CRL 11554) (21) transfected with pMX-puro/GPR1 plasmid, and the puromycin-resistant cells were selected by cultivation in medium containing 2 μg of puromycin/ml for 1 to 2 weeks. The resultant cells were designated NP-2/CD4/GPR1. Over 80% of these GPR1-transduced cells were positive for CD4 (data not shown). NP-2/CD4/CCR5 and NP-2/CD4/CXCR4 cells (19) had been established in a way similar to that described above. NP-2/GPR1 cells were established as follows. NP-2 cells were infected with the amphotropic MuLV pseudotypes containing puromycin resistance and GPR1 genes, which had been produced by φNX-A cells (16) transfected with pMX-puro/GPR1 plasmid, and the puromycin-resistant cells were selected by cultivation in medium containing 2 μg of puromycin/ml for 1 to 2 weeks.
We examined the susceptibility of C8166, BT-20/N, BT-3, U87/CD4, NP-2/CD4/GPR1, NP-2/CD4/CCR5, and NP-2/CD4/CXCR4 cells to various HIV-1 strains (GUN-1WT, GUN-1V, GUN-1/A, GUN-1/L, GUN-1/P, GUN-1/R, GUN-1/S, GUN-1/T, IIIB, SF162, and BaL). To do so, 5 × 104 cells were inoculated with HIV-1 or HIV-2 strains at 37°C for 2 h in 24-well cell culture plates as described elsewhere (24). For each virus, 104 cpm of reverse transcriptase activity was used as an inoculum. Then, the cells were washed twice with 500 μl of phosphate-buffered saline to remove viruses and were then cultured. The susceptibilities of the cells to HIV-1 and HIV-2 strains were examined by detecting the expression of HIV-1 or HIV-2 antigens by indirect immunofluorescence assay (IFA) as described elsewhere (42). Syncytia which formed were microscopically detected after Giemsa staining of the cells.
C8166 cells were highly susceptible to all HIV-1 strains examined except M-tropic (R5) strains, SF162, and BaL (Fig. 2A): more than 90% of the cells became HIV-1 antigen positive, and large syncytia were detected (data not shown). The high susceptibility of C8166 cells to diverse HIV-1 strains is probably explained by the high-level expression of CXCR4 (Fig. 1B). BT-20/N and U87/CD4 cells were susceptible to the GUN-1V, GUN-1/A, GUN-1/S, and GUN-1/T strains: more than 50% of the cells were HIV-1 antigen positive on day 6 (Fig. 2B and C), and syncytia were detected (data not shown). About 1 to 2% of BT-20/N cells were also infected with the T- and M-tropic (dualtropic) GUN-1WT strain (Fig. 2B). BT-20/N and U87/CD4 cells were resistant to the dualtropic GUN-1/P, GUN-1/L, and GUN-1/R strains, the T-tropic IIIB strain, and the M-tropic SF162 and BaL strains, and syncytia were not induced in these cells.
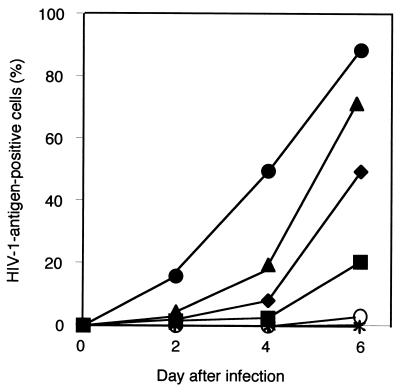
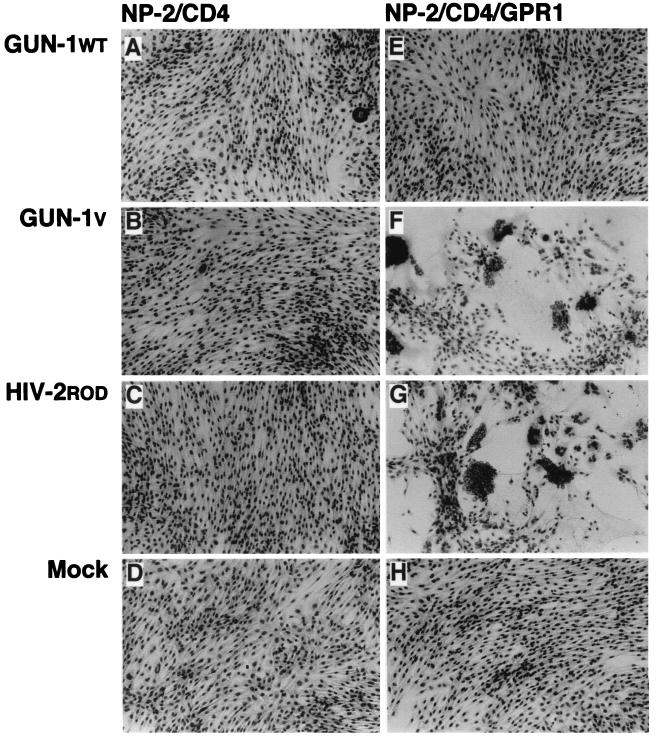
NP-2/CD4 cells were completely resistant to all HIV-1 strains examined under our assay conditions (Fig. 2E). In contrast, NP-2/CD4/GPR1 cells were highly susceptible to HIV-1 strains GUN-1V, GUN-1/A, GUN-1/S, and GUN-1/T that infect the brain-derived cells: more than 50% of the cells became HIV-1 antigen positive on day 6 after infection with the GUN-1V, GUN-1/A, and GUN-1/S strains, and 20% were antigen positive following infection with the GUN-1/T strain (Fig. 2D and 3). Syncytia were induced in NP-2/CD4/GPR1 cells by GUN-1V (Fig. 4F) and HIV-2ROD (Fig. 4G). Syncytia were also induced in NP-2/CD4/GPR1 cells by the GUN-1/A, GUN-1/S, and GUN-1/T strains (data not shown). NP-2/CD4/GPR1 cells were not susceptible to HIV-1 strains GUN-1/L, GUN-1/P, GUN-1/R, IIIB, SF162, and BaL: less than 0.2% of the cells were HIV-1 antigen positive on day 6 (Fig. 2D). NP-2/CD4/GPR1 cells were susceptible to the GUN-1WT strain: 2% of the cells were HIV-1 antigen positive on day 6 (Fig. 2D). GUN-1WT could not induce syncytia in NP-2/CD4/GPR1 cells (Fig. 4E). Infection kinetics of NP-2/CD4/GPR1 cells with various HIV-1 strains are shown in Fig. 3, indicating the rapid spread of the infectious viruses in these cells. A small percentage of NP-2/CD4/GPR1 cells were infected with the GUN-1WT strain (Fig. 2D), suggesting that the low level of infection of BT-20/N cells with the GUN-1WT strain may be explained by the inefficient use of GPR1 as a coreceptor for GUN-1WT (Fig. 2B and D).
FIG. 3.
Infection kinetics of NP-2/CD4/GPR1 cells with various HIV-1 strains as detected by IFA. Symbols: ○, GUN-1WT; ●, GUN-1V; ▴, GUN-1/S; ■, GUN-1/T; ⧫, GUN-1/A; ∗, IIIB, GUN-1/L, GUN-1/P, GUN-1/R, and mock infection.
FIG. 4.
Syncytia induced in NP-2/CD4/GPR1 cells. NP-2/CD4 (A, B, C, and D) and NP-2/CD4/GPR1 (E, F, G, and H) cells were infected with strain GUN-1WT (A and E), GUN-1V (B and F), or HIV-2ROD (C and G). Mock infection of NP-2/CD4 (D) and NP-2/CD4/GPR1 (H) cells is also shown. The cells were fixed with methanol and stained with Giemsa on day 4 after infection. Magnification, ×100.
To further confirm these findings, we examined the susceptibilities of NP-2/CD4/GPR1 cells to other HIV-1 variants and to an HIV-2 strain. GUN-4WT and GUN-7WT are T- and M-tropic (i.e., dualtropic) (unpublished results), and the GUN-4V and GUN-7V strains infected the brain-derived cells (38). HIV-1 strains GUN-4WT, GUN-4V, GUN-7WT, and GUN-7V and an HIV-2 strain, HIV-2ROD, efficiently infected C8166 cells: 60% and 20% of the cells were HIV-1 antigen positive on day 6 after infection with the GUN-4V and HIV-2ROD strains and with the GUN-7V strain, respectively (Fig. 5A). GUN-4WT and GUN-7WT infected C8166 cells more efficiently than these virus strains. Both BT-20/N and U87/CD4 cells were susceptible to the GUN-4V, GUN-7V, and HIV-2ROD strains but not to the GUN-4WT and GUN-7WT strains (Fig. 5B and C). NP-2/CD4/GPR1 cells showed susceptibility to the GUN-4V, GUN-7V, and HIV-2ROD strains but not to the dualtropic GUN-4WT and GUN-7WT strains (Fig. 5D). The GUN-4V, GUN-7V, and HIV-2ROD (Fig. 4F), but not the GUN-4WT and GUN-7WT, strains induced syncytia in NP-2/CD4/GPR1 cells (data not shown). The infectivities of HIV-1 and HIV-2 strains were confirmed by IFA: NP-2/CD4/CXCR4 cells were highly susceptible to X4 viruses, while NP-2/CD4/CCR5 cells were susceptible to X4R5 or R5 strains such as SF162 and BaL (data not shown) as described previously (19). NP-2/CD4 cells were resistant to these virus strains (Fig. 5E). These results indicate that GPR1 can act as a coreceptor which supports the infection of the cell-free HIV-1 variants and the cell fusion process induced by them. Moreover, these results strongly support the idea that GPR1 is a coreceptor of the HIV-1 variants and some HIV-2 strains and that it determines the susceptibility of BT-3, BT-20/N, and U87/CD4 brain-derived cells to these viruses.
Using RT-PCR, we estimated the amount of GPR1 mRNA expressed in NP-2/CD4/GPR1 cells as well as in BT-20/N, BT-3, and U87/CD4 cells (Fig. 1B). The amount of CXCR4 mRNA expressed in C8166 cells was also analyzed by RT-PCR using the specific primers for the CXCR4 sequence as a control (Fig. 1B). C8166 cells were highly susceptible to T-tropic HIV-1 strains. As shown in Fig. 1B, CXCR4 mRNA could be detected in 10,000-fold-diluted cDNA of C8166 cells. GPR1 mRNA was detected in 100-fold-, 1,000-fold-, 1,000-fold-, and 10-fold-diluted cDNAs of BT-20/N, BT-3, U87/CD4, and C8166 cells, respectively. Similarly, GPR1 mRNA could be detected in 10,000-fold-diluted cDNA of NP-2/CD4/GPR1 cells. These results indicate that NP-2/CD4/GPR1 cells expressed amounts of GPR1 mRNA comparable to those expressed by BT-3 and U87/CD4 cells. The amounts of GPR1 mRNA expressed in BT-3 and U87/CD4 cells were estimated to be approximately 10 times greater than that expressed in BT-20/N cells.
To examine the CD4 dependency of the infection mediated through GPR1, cells lacking CD4 expression, NP-2/GPR1, were challenged with HIV-1 and HIV-2 strains GUN-1V, GUN-1/A, GUN-1/S, GUN-1/T, GUN-4V, and GUN-7V and HIV-2ROD. NP-2/GPR1 cells were completely resistant to infection with these HIV-1 or HIV-2 strains (Fig. 2F). These results suggest that GPR1 allows HIV-1 and HIV-2 infections to the brain-derived cells in a CD4-dependent manner.
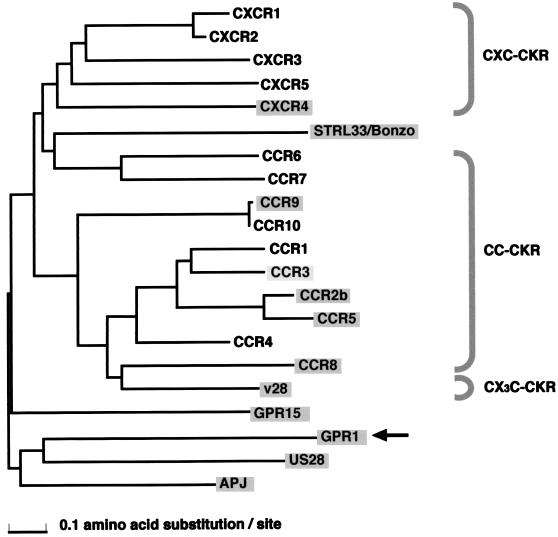
To elucidate the evolutionary relationship among GPR1 and other CKRs, including reported HIV-SIV coreceptors, we constructed a phylogenetic tree for APJ, CCR1, CCR2b, CCR3, CCR4, CCR5, CCR6, CCR7, CCR8, CCR9, CCR10, CXCR1, CXCR2, CXCR3, CXCR4, CXCR5, GPR1, GPR15, v28, and US28 by the N-J method (34) (Fig. 6). A multiple alignment of amino acid sequences was made for the conserved regions of these 16 GPCRs. The conserved regions used to make the multiple alignment were the 4th to 70th, 76th to 96th, 109th to 174th, and 236th to 264th amino acid sites in the case of CXCR4, and similar regions were used for the other GPCRs. The CC-CKRs and CXC-CKRs were clustered separately in this tree. However, GPR1 belonged to neither the cluster of CC-CKRs nor that of CXC-CKRs, suggesting that a ligand for GPR1 is possibly not a typical chemokine. GPR1 is one of the evolutionarily most distant of the known HIV-SIV coreceptors. Because US28 is a viral gene (27) rather than a cellular gene, it can be deleted from this tree. The natural ligand for GPR1 has not been identified. The location of GPR1 observed in this phylogenetic tree suggests that GPR1 is not a typical CKR, and thus various GPCRs in addition to typical CKRs might have a potential to act as coreceptors for HIV-1, HIV-2, or SIV strains.
FIG. 6.
A phylogenetic tree of CKRs and known HIV-SIV coreceptors. The tree was constructed by the N-J method using the multiple alignment of amino acid sequences for the conserved regions. Shadowed GPCRs indicate HIV-SIV coreceptors. The branch of GPR1 is indicated by the arrow.
As described above, GPR1 was specifically expressed in the primary brain-derived fibroblast-like cell strains BT-20/N and BT-3 and the glioma cell line U87/CD4 that are susceptible to the HIV-1 variants and mutants and to HIV-2ROD but not in the cells resistant to them, such as macrophages (Table 1; Fig. 1A). This result suggests that GPR1 may be a coreceptor specific for these HIV-1 and HIV-2 strains. Many syncytia were induced in NP-2/CD4/GPR1 cells positive for HIV-1 or HIV-2 antigens as determined by IFA (Fig. 4F and G and data not shown), indicating that GPR1 acts as a coreceptor and thus supports the fusion process between the envelope protein of HIV-1 and HIV-2 and the cell membrane of target cells.
It has been shown that a few amino acid changes in the V3 domain affect the use of coreceptors by HIV-1 (40). One-point mutants such as GUN-1/A, GUN-1/S, and GUN-1/T also utilized GPR1 as a coreceptor (Fig. 2D), while dualtropic mutant strains such as GUN-1/P, GUN-1/L, and GUN-1/R did not. As neither T-tropic nor M-tropic HIV-1 strains used GPR1 as a coreceptor, the cell tropism conferred by GPR1 should be independent of the T or M tropism. The results obtained here demonstrate that one-point amino acid changes at the GPGR tip sequences in the V3 domain enable HIV-1 strains to use a phylogenetically distantly related GPCR, GPR1, as a coreceptor. It remains to be examined whether GPR1 functions mainly in BT-3 and BT-20/N cells or U87/CD4 cells as a coreceptor for these HIV-1 variants and mutants and for HIV-2. For this, blocking experiments for GPR1 should be done.
GPR1 was not unanimously expressed among glioma-derived cell lines, such as U87/CD4 and NP-2/CD4. We transduced CD4 into five glioma cell lines: two lines were highly susceptible to T-tropic HIV-1 and two lines could be infected with the HIV-1 variants we describe here. Only NP-2/CD4 cells were resistant to all of the HIV-1 strains we tested (data not shown). Moreover, even from U87/CD4 cells, we could isolate a clonal line that was susceptible to M-tropic virus (unpublished data). These findings indicate that the expression of GPR1 in established cell lines derived from brain tumors is not specific for cell types.
GPR1 was originally cloned from a cDNA library of hippocampus of the human brain and is expressed abundantly there (23). There is a possibility that BT-3 and BT-20/N cells might have been derived from the brain blood vessels. In the brain, the dysfunction of the blood-brain barrier (BBB) can be involved in the development of neurological disorders, which are frequently observed in patients with AIDS (29). However, the precise mechanism by which the BBB is impaired by HIV-1 has not been adequately elucidated. It has been reported that microvascular endothelial cells are susceptible to HIV-1 strains (31). This infection is independent of CD4 and galactosyl ceramide (1). Recently, the involvement of CCR5 in the infection of simian brain endothelial cells with SIV in vitro was reported (12). Some HIV-2, SIV, and primary HIV-1 isolates were reported to infect U87/CD4 cells (9, 44). These isolates are expected to show a cell tropism similar to that of the HIV-1 variants described here and to be present in peripheral blood of HIV-1-infected patients. It remains to be examined whether brain cells like BT-3 or BT-20/N are infected with HIV-1 in vivo or whether these cells are involved in the dysfunction of the BBB.
Acknowledgments
We thank T. J. Schall for kindly supplying us with CKR expression pcDNA3 plasmids. We also thank T. Kumanishi, P. R. Clapham, B. Chesebro, W. S. Pear, and G. P. Nolan for kindly providing us with NP-2 glioma cells, U87 glioma cells, PA317/CD4 cells, BOSC23 cells, and φNX-A cells, respectively.
This work was supported in part by grants-in-aid from the Ministry of Education, Science and Culture and the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Banks W A, Akerstrom V, Kastin A J. Adsorptive endocytosis mediates the passage of HIV-1 across the blood-brain barrier: evidence for a post-internalization coreceptor. J Cell Sci. 1998;111:533–540. doi: 10.1242/jcs.111.4.533. [DOI] [PubMed] [Google Scholar]
- 2.Berthold M, Bartfai T. Modes of peptide binding in G protein-coupled receptors. Neurochem Res. 1997;22:1023–1031. doi: 10.1023/a:1022483027858. [DOI] [PubMed] [Google Scholar]
- 3.Birkenbach M, Josefsen K, Yalamanchili R, Lenoir G, Kieff E. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J Virol. 1993;67:2209–2220. doi: 10.1128/jvi.67.4.2209-2220.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, Gettie A, Ho D D, Mark P A. Primary SIVsm isolates use the CCR5 coreceptor from sooty mangabeys naturally infected in West Africa: a comparison of coreceptor usage of primary SIVsm, HIV-2, and SIVmac. Virology. 1998;246:113–124. doi: 10.1006/viro.1998.9174. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng-Mayer C, Quiroga M, Tung J W, Dina D, Levy J A. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J Virol. 1990;64:4390–4398. doi: 10.1128/jvi.64.9.4390-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choe H, Farzan M, Konkel M, Martin K, Sun Y, Marcon L, Cayabyab M, Berman M, Dorf M E, Gerard N, Gerard C, Sodroski J. The orphan seven-transmembrane receptor Apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J Virol. 1998;72:6113–6118. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Berard N G, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 9.Clapham P R, Blanc D, Weiss R A. Specific cell surface requirements for the infection of CD4-positive cells by human immunodeficiency virus types 1 and 2 and by simian immunodeficiency virus. Virology. 1991;181:703–715. doi: 10.1016/0042-6822(91)90904-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalgleish A G, Beverley P C, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 T4 antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 11.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 12.Edinger A L, Mankowski J L, Doranz B J, Margulies B J, Lee B, Rucker J, Sharron M, Hoffman T L, Berson J F, Zink M C, Hirsch V M, Clements J E, Doms R W. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc Natl Acad Sci USA. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 14.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 16.Grignani F, Kinsella T, Mencarelli A, Valtieri M, Riganelli D, Grignani F, Lanfrancone L, Peschle C, Nolan G P, Pelicci P G. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 1998;58:14–19. [PubMed] [Google Scholar]
- 17.Guyader M, Emerman M, Sonigo P, Clavel F, Montagnier L, Alizon M. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature. 1987;326:662–669. doi: 10.1038/326662a0. [DOI] [PubMed] [Google Scholar]
- 18.Horuk R. Molecular properties of the chemokine receptor family. Trends Pharmacol Sci. 1994;15:159–165. doi: 10.1016/0165-6147(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 19.Jinno A, Shimizu N, Soda Y, Haraguchi Y, Kitamura T, Hoshino H. Identification of the chemokine receptor TER1/CCR8 expressed in brain-derived cells and T cells as a new coreceptor for HIV-1 infection. Biochem Biophys Res Commun. 1998;243:497–502. doi: 10.1006/bbrc.1998.8130. [DOI] [PubMed] [Google Scholar]
- 20.Kitamura T, Onishi M, Kinoshita S, Shibuya A, Miyajima A, Nolan G P. Efficient screening of retroviral cDNA expression libraries. Proc Natl Acad Sci USA. 1995;92:9146–9150. doi: 10.1073/pnas.92.20.9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavau C, Heard J M, Danos O, Dejean A. Retroviral vectors for the transduction of the PML-RARalpha fusion product of acute promyelocytic leukemia. Exp Hematol. 1996;24:544–551. [PubMed] [Google Scholar]
- 22.Liao F, Alkhatib G, Peden K W, Sharma G, Berger E A, Farber J M. STRL33, A novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchese A, Docherty J M, Nguyen T, Heiber M, Cheng R, Heng H H, Tsui L C, Shi X, George S R, O’Dowd B F. Cloning of human genes encoding noble G protein-coupled receptors. Genomics. 1994;23:609–618. doi: 10.1006/geno.1994.1549. [DOI] [PubMed] [Google Scholar]
- 24.Matsushita S, Robert-Guroff M, Rusche J, Koito A, Hattori T, Hoshino H, Javaherian K, Takatsuki K, Putney S. Characterization of human immunodeficiency virus neutralizing monoclonal antibody and mapping of the neutralizing epitope. J Virol. 1988;62:2107–2114. doi: 10.1128/jvi.62.6.2107-2114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKeating J A, Gow J, Goudsmit J, Pearl J H, Mulder C, Weiss R A. Characterization of HIV-1 neutralization escape mutants. AIDS. 1989;3:777–784. doi: 10.1097/00002030-198912000-00001. [DOI] [PubMed] [Google Scholar]
- 26.Murphy P M. The molecular biology of leukocyte chemoattractant receptors. Annu Rev Immunol. 1994;12:593–633. doi: 10.1146/annurev.iy.12.040194.003113. [DOI] [PubMed] [Google Scholar]
- 27.Neote K, DiGregorio D, Mak J Y, Horuk R, Schall T J. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell. 1993;72:415–425. doi: 10.1016/0092-8674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- 28.Oravecz T, Pall M, Norcross M A. Beta-chemokine inhibition of monocytotropic HIV-1 infection. Interference with a postbinding fusion step. J Immunol. 1996;157:1329–1332. [PubMed] [Google Scholar]
- 29.Persidsky Y, Stins M, Way D, Witte M H, Weinand M, Kim K S, Bock P, Gendelman H E, Fiala M. A model for monocyte migration through the blood-brain-barrier during HIV-1 encephalitis. J Immunol. 1997;158:3499–3510. [PubMed] [Google Scholar]
- 30.Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 31.Poland S D, Rice G P, Dekaban G A. HIV-1 infection of human brain-derived microvascular endothelial cells in vitro. J Acquired Immune Defic Syndr Hum Retrovirol. 1995;8:437–445. doi: 10.1097/00042560-199504120-00002. [DOI] [PubMed] [Google Scholar]
- 32.Premack B A, Schall T. Chemokine receptors: gateways to inflammation and infection. Nat Med. 1996;2:1174–1178. doi: 10.1038/nm1196-1174. [DOI] [PubMed] [Google Scholar]
- 33.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1998;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou N, Nei N. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 35.Salahuddin S Z, Markham P D, Wang-Staal F, Franchini G, Kalyanaraman V S, Gallo R C. Restricted expression of human T-cell leukemia-lymphoma virus HTLV in transformed human umbilical cord blood lymphocytes. Virology. 1983;129:51–54. doi: 10.1016/0042-6822(83)90395-1. [DOI] [PubMed] [Google Scholar]
- 36.Schweickart V L, Raport C J, Godiska R, Byers M G, Eddy R L, Jr, Shows T B, Gray P W. Cloning of human and mouse EBI1, a lymphoid-specific G-protein-coupled receptor encoded on human chromosome 17q12-q21.2. Genomics. 1994;23:643–650. doi: 10.1006/geno.1994.1553. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu N, Kobayashi M, Liu H-Y, Kido H, Hoshino H. Detection of tryptase TL2 and CD26 antigen in brain-derived cells non-permissive to T-cell line-tropic human immunodeficiency virus type 1. FEBS Lett. 1995;358:48–52. doi: 10.1016/0014-5793(94)01394-g. [DOI] [PubMed] [Google Scholar]
- 38.Shimizu S N, Shimizu N G, Takeuchi Y, Hoshino H. Isolation and characterization of human immunodeficiency virus type 1 variants infectious to brain-derived cells: detection of common point mutations in the V3 region of the env gene of the variants. J Virol. 1994;68:6130–6135. doi: 10.1128/jvi.68.9.6130-6135.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sol N, Ferchal F, Braun J, Pleskoff O, Treboute C, Ansart I, Alizon M. Usage of the coreceptors CCR-5, CCR-3, and CXCR-4 by primary and cell line-adapted human immunodeficiency virus type 2. J Virol. 1997;71:8237–8244. doi: 10.1128/jvi.71.11.8237-8244.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeuchi Y, Akutsu M, Murayama K, Himizu N S, Hoshino H. Host range mutant of human immunodeficiency virus type 1: modification of cell tropism by a single point mutation at the neutralization epitope in the env gene. J Virol. 1991;65:1710–1718. doi: 10.1128/jvi.65.4.1710-1718.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeuchi Y, Inagaki M, Kobayashi N, Hoshino H. Isolation of human immunodeficiency virus from a Japanese hemophilia B patient with AIDS. Jpn J Cancer Res. 1989;78:11–15. [PubMed] [Google Scholar]
- 43.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 44.Tscherning C, Alaeus A, Fredriksson R, Bjorndal A, Deng H, Littman D R, Fenyo E M, Albert J. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology. 1998;241:181–188. doi: 10.1006/viro.1997.8980. [DOI] [PubMed] [Google Scholar]
- 45.Westermark B, Ponten J, Hugosson R. Determinants for the establishment of permanent tissue culture lines from human gliomas. Acta Pathol Microbiol Scand A. 1973;81:791–805. doi: 10.1111/j.1699-0463.1973.tb03573.x. [DOI] [PubMed] [Google Scholar]
- 46.Wong-Staal F, Gallo R C, Chang N T, Ghrayeb J, Papas T S, Lautenberger J A, Pearson M L, Petteway S R, Jr, Ivanoff L, Baumeister K, Whitehorn E A, Rafalski J A, Doran E R, Josephs S J, Starcich B, Livak K J, Patarca R, Haseltine W A, Ratner L. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi K, Byrn R A. Clinical isolates of HIV-1 contain few pre-existing proteinase inhibitor resistance conferring mutations. Biochim Biophys Acta. 1995;1253:136–140. doi: 10.1016/0167-4838(95)00167-1. [DOI] [PubMed] [Google Scholar]