Abstract
Equine arteritis virus (EAV) is a positive-stranded RNA virus that synthesizes a 5′- and 3′-coterminal nested set of six subgenomic mRNAs. These mRNAs all contain a common leader sequence which is derived from the 5′ end of the genome. Subgenomic mRNA transcription and genome replication are directed by the viral replicase, which is expressed in the form of two polyproteins and subsequently processed into smaller nonstructural proteins (nsps). During the recent construction of an EAV infectious cDNA clone (pEAV030 [L. C. van Dinten, J. A. den Boon, A. L. M. Wassenaar, W. J. M. Spaan, and E. J. Snijder, Proc. Natl. Acad. Sci. USA 94:991–996, 1997]), a mutant cDNA clone (pEAV030F) which carries a single replicase point mutation was obtained. This substitution (Ser-2429→Pro) is located in the nsp10 subunit and renders the EAV030F virus deficient in subgenomic mRNA synthesis. To obtain more insight into the role of nsp10 in transcription and the nature of the transcriptional defect, we have now analyzed the EAV030F mutant in considerable detail. The Ser-2429→Pro mutation does not affect the proteolytic processing of the replicase but apparently affects the function of nsp10 in transcription. Furthermore, our study showed that EAV030F still produces subgenomic positive and negative strands, albeit at a very low level. Both subgenomic positive-strand synthesis and negative-strand synthesis are equally affected by the Ser-2429→Pro mutation, suggesting that nsp10 plays an important role in an early step of EAV mRNA transcription.
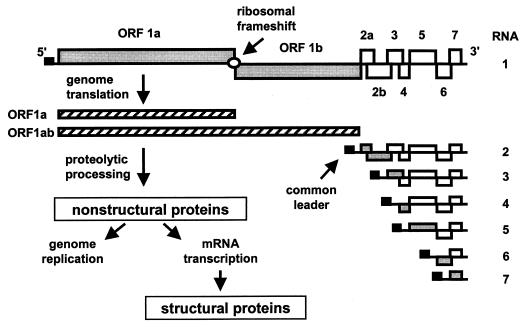
Equine arteritis virus (EAV) is a positive-stranded RNA virus with a genome of 12.7 kb. It is the prototype of the arterivirus family, which has been grouped together with the coronavirus family into the order Nidovirales (9, 18, 28, 43). The taxonomic joining of these two families was based on their presumed evolutionary relationship (15). Although the differences in genome size (13 to 16 kb for arteriviruses and 27 to 32 kb for coronaviruses) and virion architecture seem to argue against an evolutionary link between these virus families, a comparison of their genome organizations and replication strategies makes the evolutionary relationship very obvious (Fig. 1). The nidovirus replicase is encoded by two large open reading frames (ORFs), named ORF1a and ORF1b. The ORF1b reading frame is expressed via ribosomal frameshifting and contains the conserved putative RNA polymerase and helicase domains (reviewed in references 18, 28, and 43). The ORF1a and the ORF1ab polyproteins are extensively processed into a set of nonstructural proteins (nsps) (18, 28, 43). Furthermore, all nidoviruses produce a 3′-coterminal nested set of subgenomic (sg) mRNAs that are used to express the genes downstream of the replicase gene, which mainly encode structural proteins. These sg mRNAs also contain a common 5′ leader sequence, which is derived from the 5′ end of the viral genome (reviewed in references 18, 28, and 43). In view of the common ancestry of their replicases and their similar genome organizations, it has been proposed that coronaviruses and arteriviruses employ essentially similar mechanisms for sg mRNA synthesis (14, 18, 43). Despite extensive studies which mostly involved coronaviruses, the details of this mechanism are poorly understood. UV transcription mapping studies for both coronaviruses and arteriviruses (13, 24, 55) argued against conventional cis splicing as the major mechanism for the joining of the mRNA leader and body. Thus, it is generally accepted that nidovirus sg mRNAs are generated via a discontinuous transcription process, for which several models have been proposed (for recent reviews, see references 7, 18, 28, 43, and 48). These models are based on comparative sequence analysis, experiments using coronavirus- or arterivirus-infected cells, and studies using cloned cDNAs of coronavirus defective interfering RNAs.
FIG. 1.
EAV genome organization and replication strategy. The EAV genome with its genes and the replicase ORF1a and ORF1ab translation products are outlined at the top. The nested set of sg mRNAs is depicted below the genome. The black box represents the common 5′ leader sequence. The ORFs that are translated from each of the sg mRNAs are indicated in grey.
The first model explaining nidovirus transcription was the so-called leader-primed transcription model (5, 27, 45). It was based on the observation that the transcription units in the genome of coronaviruses are preceded by a short conserved sequence element, which is also present at the 3′ end of the leader sequence. It has been demonstrated for coronaviruses that these sequence elements can act as promoters for sg mRNA synthesis (32, 47). Arteriviruses also use a conserved sequence to join the leader and the body of sg mRNAs (12, 14, 16, 20, 33). We will refer to this important regulatory sequence (see also Discussion), and its equivalent in coronaviruses, as the transcription-regulating sequence (TRS). In the case of EAV, the TRS has the sequence 5′ UCAACu 3′ (14, 16), in which the lowercase “u” at position 6 indicates that this nucleotide is conserved in most but not all EAV TRSs (14). The leader-primed transcription model proposes that the TRS at the 3′ end of the leader transcript base pairs with the complement of the body TRS in the negative-stranded template, after which the leader transcript is extended to generate an sg mRNA (4, 5, 27, 32, 45, 47).
Initially, the template for sg mRNA transcription was assumed to be a single, genome-length negative strand (29). The discovery of sg-size negative strands (14, 40, 41) and the fact that these appear to be transcriptionally active (23, 35, 38) was the basis for alternative models (35, 37, 41). According to Sethna et al. (41), sg mRNAs may be replicons. They are generated by leader-primed transcription and then function as templates for the synthesis of sg negative strands, which are subsequently used to amplify the sg mRNAs. However, direct evidence for replication of sg mRNAs has never been obtained: transfection of synthetic mRNAs into coronavirus-infected cells did not result in their amplification (references 10 and 32 and our unpublished data). Still, it could be that transfected sg RNAs are not suitable as templates for replication.
In the model proposed by Sawicki and Sawicki (35, 37), a nested set of sg negative strands is synthesized first. During negative-strand synthesis, the nascent chain is translocated to the leader sequence of the (positive-stranded) genomic template. Following base pairing between the TRS complement in the nascent negative strand and the leader TRS, the negative strand is extended with the antileader sequence. The resulting sg negative-stranded RNA serves as template for the synthesis of the corresponding sg mRNA.
Recently, a full-length infectious cDNA clone (pEAV030) for the arterivirus EAV was constructed in our laboratory (49). This novel tool in nidovirus research allows us to learn more about the RNA and protein sequences involved in EAV replication and transcription. During the construction of the EAV cDNA clone, an interesting mutant (pEAV030F) which carries a single point mutation in the replicase gene was obtained (49). The mutation (Ser-2429→Pro) is located in the nsp10 part of the ORF1ab protein, between a putative metal-binding motif and the helicase domain (Fig. 2A) (15, 21). It dramatically affects sg mRNA synthesis while leaving genome replication unaffected. The EAV030F phenotype showed for the first time that nidovirus genome replication and sg mRNA transcription are partially separate processes. This intriguing observation raised a number of questions about the nature of the EAV030F defect at the level of sg mRNA synthesis and implications for the transcription models. Therefore, the EAV030F mutant was subjected to a more detailed analysis.
FIG. 2.
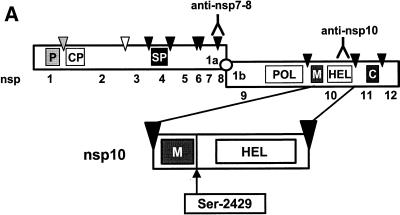
(A) Processing scheme of the EAV replicase ORF1ab protein. The grey, white, and black triangles depict the cleavage sites of the ORF1a-encoded papainlike nsp1 cysteine protease (P), the nsp2 cysteine protease (CP), and the nsp4 serine protease (SP), respectively. The other boxes represent the polymerase domain (POL), the helicase domain (HEL), a putative metal-binding domain (M), and a C-terminal domain conserved in all nidoviruses (C). Also indicated are the nomenclature for the nsps, the location of Ser-2429 in nsp10, and the approximate positions of the epitopes for the anti-nsp7-8 and anti-nsp10 antisera. (B) Immunoprecipitation analysis of cells infected with EAV (lanes I), cells transfected with EAV030F (lanes F) or EAV030H (lanes H), or mock-transfected cells (lanes M). Cells were labeled from 6 to 8, 8 to 10, or 10 to 12 h. The results of the immunoprecipitation analysis using antisera directed against nsp10 (anti-nsp10) and nsp7-8 (anti-nsp7-8; see panel A) are shown.
MATERIALS AND METHODS
Cells and viruses.
In all experiments, baby hamster kidney cells (BHK-21; ATCC CCL10) were used; the cells were grown in BHK-21 medium (Life Technologies) containing 10 mM HEPES, 10% tryptose, and 5% fetal calf serum. For all infection experiments, the EAV Bucyrus strain (19) was used. Both infection and transfection experiments were performed at 39.5°C (46).
Full-length EAV cDNA clones.
The construction of the full-length EAV cDNA clones pEAV030H, pEAV030F, and pEAV030SGA has been described previously (49, 50). The pEAV030H construct is a wild-type EAV cDNA clone containing an engineered HindIII restriction site at genome position 6973. The mutant cDNA clone pEAV030F contains a point mutation (T→C) at nucleotide (nt) 7508 in the ORF1b region, which changes replicase Ser-2429 to Pro. In mutant clone pEAV030SGA the conserved Ser-Asp-Asp core motif (residues 2236 to 2238) of the presumed RNA polymerase domain in nsp9 was changed into Ser-Gly-Ala by mutating nt 6932 to 6937 from 5′-GACGAC-3′ to 5′-GGCGCC-3′.
Construction of CAT gene-containing EAV vectors.
Construction of the chloramphenicol acetyltransferase (CAT) reporter gene-containing constructs pEAV030HCAT7 and pEAV030HTAC7 has been described elsewhere (49). In pEAV030HCAT7, the CAT gene was inserted upstream of EAV ORF7, which encodes the nucleocapsid protein. In negative control construct pEAV030HTAC7, the CAT gene was inserted at the same position but in the antisense orientation. To obtain similar constructs containing the Ser-2429→Pro mutation, the EcoRI-XhoI fragment (nt 11488 to 12845) of pEAV030F was replaced by the EcoRI-XhoI fragment of pEAV030HCAT7 or pEAV030HTAC7, giving rise to construct pEAV030FCAT7 or pEAV030FTAC7, respectively.
In vitro RNA transcription and transfection.
Plasmid DNA of pEAV030H and its derivatives was linearized by using restriction endonuclease XhoI. Full-length in vitro transcripts were generated with T7 RNA polymerase under previously described conditions (14), using nucleoside triphosphate and capping analog [m7G(5′)ppp(5′)G; Life Technologies], each at a concentration of 1 mM. The in vitro-transcribed RNA was introduced into BHK-21 cells by electroporation as described by van Dinten et al. (49).
Protein labeling and immunoprecipitation.
Infected or transfected cells were starved for 15 min in methionine- and cysteine-free medium, and proteins were labeled for 2 h with 200 μCi of [35S]methionine and [35S]cysteine (Expre35S35S label; NEN). Cell lysis and immunoprecipitation of labeled proteins were performed as described by de Vries et al. (17). The EAV replicase-specific rabbit antisera used in this study have been described previously (44, 51). Proteins were separated in sodium dodecyl sulfate (SDS)-containing 12.5% polyacrylamide gels.
Isolation and analysis of viral RNA.
Cells were labeled with 100 μCi of [3H]uridine per ml of medium in the presence of 20 μg of dactinomycin per ml. Intracellular RNA was isolated by solubilizing cells for 10 min at room temperature with 5% lithium dodecyl sulfate in LET buffer (100 mM LiCl, 1 mM EDTA, 10 mM Tris-HCl [pH 7.4]) containing 20 μg of proteinase K per ml. After shearing of the DNA using a 25 5/8-gauge needle, the lysates were incubated at 42°C for 15 min and extracted with phenol (pH 4.0) and chloroform, and RNA was ethanol precipitated. The radiolabeled RNAs were separated in denaturing 1% agarose gels containing 2.2 M formaldehyde and MOPS buffer {10 mM MOPS [morpholinepropanesulfonic acid (sodium salt); pH 7], 5 mM sodium acetate, 1 mM EDTA}. Gels were fixed in methanol, impregnated with 2% 2,5-diphenyloxazole (PPO) in methanol, rinsed with water, and dried at 60°C. The 3H-labeled RNAs were visualized via fluorography.
To prepare replicative-form RNA, cells were labeled with [3H]uridine from 4 to 12 h after electroporation or infection, and RNA was isolated as described above. The RNA preparations were treated with RNase T1 (Life Technologies) and separated in nondenaturing 1% agarose gels, which were processed as described above.
CAT assays.
At 12 h after transfection, cells were scraped from the dish in TEN buffer (0.04 M Tris-HCl [pH 7.5], 1 mM EDTA, 0.15 M NaCl) and spun down in a microcentrifuge at 13,000 rpm. Subsequently, cells were resuspended in 0.25 M Tris-HCl (pH 7.5) and freeze-thawed three times in an ethanol-dry ice bath. Cellular debris was pelleted by spinning for 5 min at 13,000 rpm in a microcentrifuge, and the supernatants were stored at −80°C.
CAT assays were performed with 2.5 mM acetyl coenzyme A and 0.6 μCi of 14C-labeled chloramphenicol per ml in 0.16 M Tris-HCl (pH 7.5). CAT assays were incubated at 37°C for 16 h and subsequently extracted with ethylacetate. The ethylacetate-chloramphenicol mixture was lyophilized, and the residue was dissolved in 50 μl of ethylacetate and analyzed via thin-layer silica gel chromatography and subsequent autoradiography (3, 34).
RT-PCR.
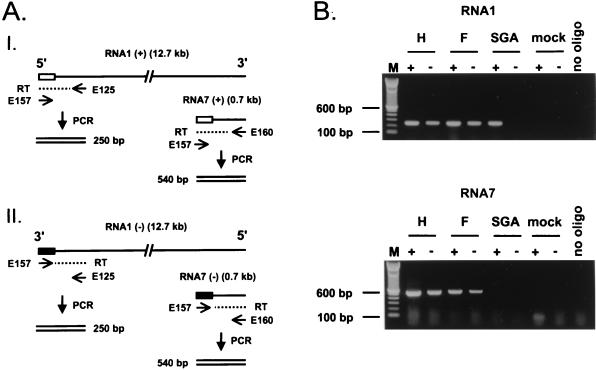
For the negative- and positive-strand-specific reverse transcription (RT)-PCR, we used the approach outlined in Fig. 4A (36). To prime cDNA synthesis on the positive or negative strand of sg RNA7, we used oligonucleotide E160 (5′-CTTACGGCCCTGCTGGAGGCGCAAC-3′; negative sense; genome positions 12623 to 12646) or E157 (5′-CTTGTGGGCCCCTCTCGGTAAATCC-3′; positive sense; genome positions 63 to 89), respectively. To prime cDNA synthesis on the genomic negative strand, we also used oligonucleotide E157, whereas oligonucleotide E125 (5′-CGCCATGCTCACACGCGTCGGGTAAG-3′; negative sense; genome positions 285 to 310) was used for the genomic positive strand. Oligonucleotide E160 or E125 was also used together with oligonucleotide E157 for the subsequent PCR, which consisted of 25 cycles, each comprising 1 min of denaturation at 94°C, 1 min of annealing at 58°C, and 1 min of DNA synthesis at 72°C. The 25 cycles were followed by a 10-min incubation at 72°C.
FIG. 4.
(A) RT-PCR strategy for the detection of positive-stranded (I) and negative-stranded (II) genomic RNA and sg RNA7. Leader and antileader sequence are indicated with white and black boxes, respectively. Oligonucleotides E125 and E160 are directed against the body sequences of RNA1 and sg RNA7, respectively, and are used for priming cDNA synthesis (dashed line) on the positive strands of RNA1 and RNA7. Oligonucleotide E157 is directed against the antileader sequence (black box) and is used for cDNA synthesis on the negative strand. For the PCR, oligonucleotides E157 and E125 are used to amplify regions specific for positive- and negative-stranded RNA1. Oligonucleotides E157 and E160 are used for the PCR specific for positive- and negative-stranded sg RNA7. (B) RT-PCR analysis for the positive and negative strands of RNA1 (top panel) and positive and negative strands of sg RNA7 (bottom panel). The RNA was isolated from cells transfected with EAV030H (lanes H), EAV030F (lanes F), or EAV030SGA (lanes SGA) or from mock-transfected cells. The plus and minus signs indicate specificity of the RT-PCR for positive and negative strands, respectively.
RESULTS
The EAV030F defect in sg mRNA synthesis is not caused by incorrect processing of the replicase.
We have previously described the mutant EAV full-length cDNA clone pEAV030F, which carries a mutation specifying a Ser-2429→Pro substitution in the viral replicase (Fig. 2A) (49). RNA transcribed from this clone was able to replicate efficiently but failed to produce sg mRNAs. Since residue Ser-2429 is located close to the nsp9/10 junction (Glu-2370/Ser-2371) in the ORF1ab polyprotein (50), it was possible that its replacement by Pro influenced the proteolytic processing of this region of the replicase. To test this, BHK-21 cells infected with EAV or transfected with EAV030F or EAV030H RNA were labeled with [35S]methionine and [35S]cysteine for 2 h at 6, 8, or 10 h after electroporation or infection. An immunofluorescence assay showed that an equal percentage (approximately 15%) of the cells had been transfected with EAV030F and EAV030H (data not shown). Immunoprecipitations using 35S-labeled cell lysates and the anti-nsp10 antiserum (51) were performed and analyzed in SDS–12.5% polyacrylamide gels (Fig. 2B). In both EAV030F- and EAV030H-transfected cells, cleaved nsp10 was detected at 8 to 10 h after transfection. nsp2, which is coprecipitated by the anti-nsp10 serum (44, 51), was also observed. There was no difference between the amounts of nsp10 produced in cells transfected with EAV030F or EAV030H. In EAV-infected cells, nsp10 could be detected already at 6 to 8 h postinfection and accumulated to higher levels. This is explained by a higher number of infected cells than present in transfection experiments and the fact that the electroporation procedure delays the onset of virus replication by 2 to 3 h (unpublished observations). We also performed an immunoprecipitation analysis using an antiserum against EAV nsp7-8 (Fig. 2B) (44, 54), which precipitates an extensive set of ORF1a- and ORF1b-encoded processing end products and intermediates. In EAV030F- and EAV030H-transfected cells, the nsp3-12, nsp5-12, nsp3-8, nsp5-8, and nsp5-7 replicase cleavage products and coprecipitated nsp2 were detected at 8 to 10 h after transfection. There was no difference between the amounts of these proteins in EAV030F- or EAV030H-transfected cells. In conclusion, the Ser-2429→Pro mutation in nsp10 did not have any detectable effect on the production of cleaved nsp10 and a number of important other replicase processing products, which makes it unlikely that an nsp10 processing defect is the explanation for the EAV030F phenotype.
EAV030F still produces low levels of sg positive- and negative-strand RNAs.
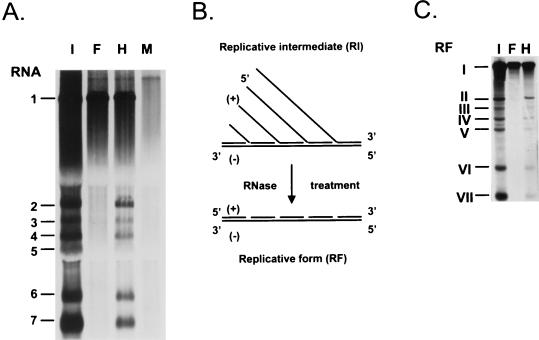
Figure 3A shows the typical accumulation of positive-stranded viral RNAs in BHK-21 cells either infected with EAV or transfected with in vitro-transcribed RNA from the mutant clone pEAV030F or the wild-type clone pEAV030H. Both genomic and sg RNAs were generated in EAV030H-transfected cells. In the cells transfected with pEAV030F RNA, however, only the genomic RNA (RNA1) accumulated. During infection, EAV also produces negative-strand copies of both genomic and sg RNAs (14). It has been suggested that these sg negative strands play an important role as templates for the synthesis of sg mRNAs. Since sg positive-stranded RNAs could not be detected for EAV030F via metabolic labeling, it was important to determine whether this mutant was able to produce any sg negative strands.
FIG. 3.
(A) Analysis of [3H]uridine-labeled intracellular RNA isolated 12 h after infection or transfection from mock-transfected cells (lane M), cells infected with EAV (lane I), or cells transfected with EAV030F RNA (lane F) or EAV030H RNA (lane H). The RNAs were separated in a denaturing 1% agarose gel. (B) Schematic representation of the conversion of RIs into RFs by RNase treatment. (C) Analysis of [3H]uridine-labeled RF RNAs obtained 12 h postinfection or transfection from cells infected with EAV (lane I) and cells transfected with EAV030F RNA (lane F) or EAV030H RNA (lane H). The RFs corresponding to EAV RNA1 to RNA7 are indicated with I to VII, respectively. The RF RNAs were separated in a nondenaturing 1% agarose gel.
In the infected cell, the genomic and sg negative strands of arteriviruses, like those of coronaviruses, are present in double-stranded replicative intermediates (RIs) (14, 35, 39, 41). An RNase treatment can be used to remove the single-stranded portion of the nascent chains in the RIs (Fig. 3B). The double-stranded core structures that remain, the so-called replicative forms (RFs), can be easily separated on nondenaturing agarose gels (14, 35). For wild-type EAV, this procedure yields one genome-length RF and six sg-size RFs, which reflect the presence of genomic as well as sg negative strands (14). In the case of EAV030F, however, only the RF corresponding to the genomic RNA was obtained; sg RFs were not detected (Fig. 3C). Thus, the replicase mutant did not produce detectable amounts of sg negative strands, or there are not enough positive strands to protect the sg negative strands against the RNase treatment. To discriminate between these possibilities, we used an alternative and more sensitive RT-PCR approach to analyze the presence of sg negative strands in EAV030F-transfected cells.
As outlined in Fig. 4A, positive- and negative-strand-specific RT-PCR assays were designed for both the genomic RNA and sg RNA7 (36). Specificity for the genomic and antigenomic template was achieved by using an oligonucleotide complementary to the region just downstream of the leader sequence (E125 [Fig. 4A]), which is lacking in the sg RNAs, as RT and PCR primer for the positive- and negative-strand-specific genomic RT-PCR, respectively. Because the RNAs of EAV form a nested set and all contain the same 5′ and 3′ sequences, absolute specificity for sg RNA7 (or any of the other sg RNAs) cannot be achieved. The oligonucleotides used were complementary to the antileader sequence (E157) and the genomic 3′ end (E160) and primed cDNA synthesis on all negative-stranded RNAs and all mRNAs of EAV, respectively. However, in the PCR, the smallest cDNA molecules, i.e., those derived from positive- or negative-strand RNA7, will have an amplification advantage over the larger ones and will therefore yield the most abundant RT-PCR product. Furthermore, the size of the PCR product, hybridizations with specific probes, and restriction enzyme analysis could be used to confirm the RNA7 specificity of the amplified sequence.
Cells were electroporated with RNA from the wild-type pEAV030H clone or the mutant clones pEAV030F and pEAV030SGA. In the latter clone, the highly conserved Ser-Asp-Asp core motif of the predicted nsp9 RNA polymerase domain was changed to Ser-Gly-Ala, a replacement which has been shown to render EAV030 RNA replication incompetent (50). We used this mutant clone as a negative control for replication and sg RNA synthesis.
RNA was isolated 12 h after transfection and analyzed for the presence of positive and negative strands of RNA1 and RNA7 (Fig. 4B). In cells transfected with pEAV030SGA RNA, only a PCR product derived from positive-strand RNA1 was obtained, which was concluded to result from the presence of input pEAV030SGA transcript in the RNA preparation. The fact that the other RT-PCRs for pEAV030SGA were negative confirmed the specificity of our RT-PCR approach. As expected, RT-PCR products specific for positive- and negative-strand RNA1 were obtained with RNA from cells electroporated with either EAV030H or EAV030F. Furthermore, products derived from positive- and negative-stranded RNA7 were detected for EAV030H. Interestingly, mutant EAV030F also yielded these PCR products, which were not generated in control reactions using RNA from mock-transfected cells or when cDNA was synthesized in the absence of RT primer (lanes labeled “no oligo” in Fig. 4B). The specificity of all PCR products was corroborated by their size, restriction enzyme analysis, and the fact that they hybridized to oligonucleotide probes specific for the amplified regions of RNA1 and RNA7 (data not shown). The amounts of the RNA7-specific RT-PCR products were somewhat lower for EAV030F than for EAV030H, suggesting that the EAV030F mutant produced reduced amounts of both sg positive and negative strands, which could not be detected by metabolic RNA labeling (Fig. 3).
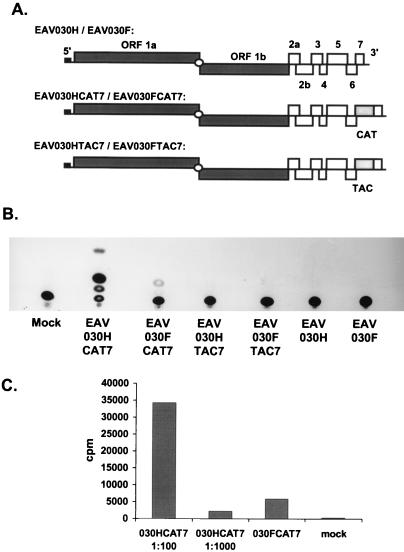
To confirm our RT-PCR data, we used reporter gene constructs pEAV030HCAT7 (49) and pEAV030FCAT7 (Fig. 5A). These are pEAV030H and pEAV030F derivatives in which the CAT gene was inserted just upstream of the ORF7 translation initiation codon. Upon transfection of EAV030HCAT7 RNA, the CAT gene is efficiently expressed from mRNA7, the most abundant sg mRNA (49). The CAT activity in lysates of transfected cells can be monitored very accurately with an enzymatic assay (3, 34), and differences in mRNA7 synthesis will be reflected in the level of CAT expression. The sensitivity of the CAT assay enabled us to compare the levels of mRNA7 transcription by the mutant EAV030FCAT7 and the wild-type EAV030HCAT7 constructs. As negative controls we used pEAV030HTAC7 and pEAV030FTAC7 (Fig. 5A), which contained the CAT gene in the antisense orientation.
FIG. 5.
(A) Schematic representation of the EAV030H and EAV030F clones containing the CAT gene inserted upstream of ORF7 (EAV030HCAT7 and EAV030FCAT7) and the clones containing the CAT gene at the same position in the reverse orientation (EAV030HTAC7 and EAV030FTAC7). (B) CAT assays of lysates obtained from mock-transfected cells and cells transfected with EAV030HCAT7, EAV030FCAT7, EAV030HTAC7, EAV030FTAC7, EAV030H, or EAV030F. (C) Amount of acetylated chloramphenicol (in counts per minute as determined by liquid scintillation counting) for CAT assays using 100- and 1,000-fold dilutions of lysates of EAV030HCAT7-transfected cells or undiluted lysates obtained from cells transfected with EAV030FCAT7 and mock-transfected cells.
Cell lysates were prepared 12 h after electroporation and used for CAT assays, which were analyzed using thin-layer chromatography (Fig. 5B). In the lysates obtained from mock-transfected cells and cells transfected with EAV030H, EAV030F, 030HTAC7, or 030FTAC7 RNA, only background levels of CAT activity were detected. The CAT activity in the wild-type EAV030HCAT7 lysate was very high. For replicase mutant EAV030FCAT7, severely reduced but significant levels of CAT activity were measured in three independent experiments. To quantitate the difference in CAT activity, 10-fold serial dilutions of the EAV030HCAT7 and EAV030FCAT7 lysates were used for CAT assays. The amount of acetylated chloramphenicol was determined by cutting the corresponding spots from the chromatogram and liquid scintillation counting (Fig. 5C). This analysis revealed that the CAT activity in the EAV030FCAT7 lysates was about 500 times lower than that in lysates of EAV030HCAT7-transfected cells. These data clearly confirmed that EAV030FCAT7 still produced sg mRNA7 and extended the RT-PCR data presented in Fig. 4.
EAV030F positive-strand RNA synthesis and negative-strand RNA synthesis are equally impaired.
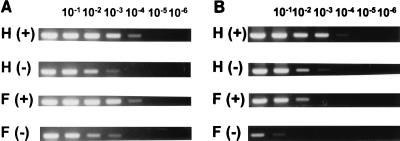
The CAT assays and RT-PCR data described above both indicated that EAV030F still produced small amounts of sg positive and negative strands. To estimate these amounts, and the ratio between sg positive and negative strands, we used the positive- and negative-strand-specific RT-PCR strategy described in Fig. 4A. This time, the RT-PCR was performed on 10-fold serial dilutions (10−1 to 10−6) of the RNA isolated from an equal number of cells transfected with EAV030F or EAV030H (Fig. 6). In this manner, the relative amounts of positive and negative strands could be determined for both genomic RNA and sg RNA7. As can be seen in Fig. 6, EAV030H produced similar amounts of positive-stranded genomic RNA and sg RNA7. The same observation was made for the amounts of negative-stranded genomic RNA and sg RNA7. Assuming that the positive- and negative-strand-specific reactions were equally sensitive, the RT-PCR analysis also revealed that the ratio between positive-stranded and negative-stranded EAV RNAs is approximately 10 to 1 for both genomic and sg RNAs.
FIG. 6.
Semiquantitative RT-PCR analysis to estimate the amounts of positive (+) and negative (−) strands for RNA1 (A) and sg RNA7 (B). In the RT reaction, serial 10-fold dilutions of RNA isolated from cells transfected with EAV030H (rows labeled H) or EAV030F (rows labeled F) were used. The subsequent PCRs (Fig. 4A) were identical for all samples.
Analysis of EAV030F-transfected cells revealed that the amounts of positive- and negative-strand genomic RNA were similar to those produced by EAV030H, as expected in view of our previous results showing that EAV030F and EAV030H genomic RNAs replicate with similar efficiencies (Fig. 3). Analysis of the EAV030F positive- and negative-stranded RNA7 showed that the replicase mutant produced about 100-fold less sg positive- and negative-strand RNA7 than EAV030H. Again an approximately 10-fold-lower level of sg negative-strand RNA compared to positive-strand RNA was observed, suggesting that the ratio between the sg positive and negative strands is the same for EAV030F and EAV030H. These results were confirmed by using RNA samples from an independent, second transfection experiment. From these data it was concluded that sg positive-strand RNA synthesis and negative-strand RNA synthesis are equally impaired by the Ser-2429→Pro mutation in the EAV030F replicase.
DISCUSSION
The pEAV030F defect in sg mRNA synthesis is not caused by incorrect processing of the replicase.
The phenotype of the EAV030F mutant virus, efficient genome replication without significant sg mRNA transcription, could have been caused by aberrant processing of the nsp10 region of the replicase as a result of the Ser-2429→Pro substitution. Proteolytic processing of the replicase can play a crucial role in the regulation of positive-strand RNA virus replication and transcription, as has been demonstrated for alphaviruses and picornaviruses (2, 6, 8, 22, 30, 42). However, our immunoprecipitation studies demonstrated that the nsp9/10 and nsp10/11 sites in the replicase protein are correctly processed (Fig. 2B). The amount of nsp10 produced by EAV030F and its expression in the course of infection, was similar to that generated by the wild-type EAV030H RNA. Immunoprecipitations using an antiserum directed against nsp7-8 revealed that also other major processing steps were unaffected. These data indicate that the amino acid change in nsp10 does not alter the processing of the replicase protein and apparently affects only the function of nsp10 or one of its precursors. The phenotype of the Ser-2429→Pro mutant is unique, as emphasized by a recent nsp10 mutagenesis study in our laboratory. Ser-2429 could be replaced by six other amino acid residues without a noticeable effect on replication or transcription (unpublished data). Mutagenesis of other (conserved) residues in the N-terminal domain of nsp10 resulted in viruses that were replication deficient, showing that this putative metal-binding domain is crucial for viral RNA synthesis. However, none of these mutant viruses displayed the EAV030F phenotype.
EAV030F still produces low levels of sg positive and negative strands.
Despite the fact that mutant EAV030F replicates efficiently, it does not produce sg mRNAs at a level that could be detected via metabolic labeling. During infection, EAV synthesizes a negative-stranded complement of each of its sg mRNAs (14). We wondered if mutant EAV030F was still able to generate normal levels of sg negative strands, even though we were not able to detect any sg positive strands in metabolic labeling experiments. Subgenomic negative strands may play a key role as the templates for sg mRNA synthesis in both arteriviruses and coronaviruses (14, 23, 35, 38, 41). The level at which EAV030F was able to produce sg negative strands would give information about the stage of sg mRNA synthesis that is affected by the mutation in nsp10.
Since we were not able to detect sg negative strands via an RF analysis (Fig. 3), we used the RT-PCR approach (36) as an alternative, more sensitive detection method. Our data show that mutant clone EAV030F produces low levels of sg positive and negative strands. Previously, the detection of very low levels of an undefined EAV030F RT-PCR product in an mRNA7-specific RT-PCR assay was reported (49), but at that time we could not exclude the possibility of an artifact during RT or PCR. The controls used in this study showed that our assay was specific and that the PCR products are indeed derived from positive- and negative-stranded RNA7. Furthermore, the CAT assays performed with construct pEAV030FCAT7 provided independent support for our RT-PCR data (Fig. 5). Low but significant CAT activity was observed in cells transfected with EAV030FCAT7 RNA, confirming that the EAV030F replicase is able to produce low levels of sg mRNA. The CAT activity in cells transfected with EAV030FCAT7 was about 500 times lower than that in cells electroporated with the wild-type EAV030HCAT7, which corresponded to the data obtained with our semiquantitative RT-PCR. The latter analysis showed an approximately 100-fold reduction of sg positive- and negative-strand synthesis in EAV030F-transfected cells. Negative- and positive-strand sg RNA synthesis seemed to be equally impaired in EAV030F, which is reflected in the similar ratios between the amounts of sg positive and negative strands for EAV030F and EAV030H (Fig. 6).
Formally, the very low levels of sg positive and negative strands that were observed in our study could have resulted from EAV030F revertants which arose during the experiment. However, we have shown that EAV030F reverts with a very low frequency (49). Revertants could be isolated only occasionally, at 2 to 4 days after electroporation. Since we (repeatedly) carried out a first cycle analysis at approximately 12 h after electroporation, the presence of revertants which significantly influenced our analysis is considered unlikely.
The role of nsp10 in transcription.
Arteriviruses and coronaviruses are assumed to use similar mechanisms to produce their sg mRNAs (14, 43). Both virus families join the leader to the mRNA bodies at a conserved sequence (the TRS) that is present at the 3′ end of the leader and upstream of the genes in the 3′ part of the genome (18, 28, 43). The arterivirus helicase protein nsp10 (Fig. 2A) contains two of the most conserved domains of the nidovirus replicase (15, 43), which also reside in the same replicase subunit in the case of coronaviruses. Our data indicate that the Ser-2429→Pro mutation in EAV nsp10 dramatically interferes with the synthesis of sg mRNAs but does not block it completely. Furthermore, EAV030F is equally disturbed in both sg positive-strand and negative-strand synthesis. What are the implications of our observations for the role of nsp10 in EAV sg mRNA synthesis?
The nsp10 replicase subunit (or its precursors) must be an essential component of the replication and transcription machinery. The protein contains the presumed viral helicase domain (15), which is believed to be indispensable for the proper functioning of the replicase (for a review, see reference 26). However, the Ser-2429→Pro mutation in nsp10 does not appear to disrupt the replicative capabilities of the replicase. The EAV030F genome is replicated very efficiently, suggesting that the EAV030F replicase still recognizes the viral replication signals efficiently and that its processivity is not affected by the mutation. Furthermore, EAV030F still produces low levels of sg positive- and negative-stranded RNAs, and the resulting ratios of sg positive to negative strands are similar to those found for the wild-type replicase. Also, the latter observation is in agreement with the notion that the processivity of the EAV030F replicase is not affected by the mutation in nsp10.
In the context of the transcription model of Sawicki and Sawicki (35, 37), the above findings would suggest that the efficiency with which the small amounts of sg negative strands are used to generate sg positive strands is unchanged. Using the model of Sethna et al. (39), our findings could indicate that the low levels of sg mRNAs which are produced are copied into sg negative strands with normal efficiency. Thus, the EAV030F phenotype suggests that nsp10 plays an important role in an early step in transcription. The protein may directly or indirectly be involved in the process by which the nascent sg strand is translocated during discontinuous transcription, from one TRS on the template to another. However, the TRS is not the only transcription-regulating sequence. RNA sequences surrounding the TRS and genomic sequences immediately downstream of the leader have been reported to be involved (1, 11, 25, 31, 52, 53). nsp10 may be involved in the recognition of these RNA sequences by the transcription complex. Alternatively, nsp10 could interact with other protein factors which recognize these sequences. In the mutant EAV030F nsp10, such interactions may be disturbed, explaining why discontinuous sg RNA synthesis is very inefficient. We are currently investigating with which RNA and protein sequences nsp10 can interact and hope that the properties of the protein with the Ser-2429→Pro mutation will provide additional information about the nature of the EAV030F defect in sg mRNA synthesis.
ACKNOWLEDGMENTS
We thank Richard Molenkamp and Evelyne Bos for stimulating discussions and Jessika Dobbe for technical assistance.
G.v.M. was supported by grant 331-020 from the Council for Chemical Sciences of the Netherlands Organization for Scientific Research.
REFERENCES
- 1.An S, Makino S. Characterizations of coronavirus cis-acting RNA elements and the transcription step affecting its transcription efficiency. Virology. 1998;243:198–207. doi: 10.1006/viro.1998.9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 4.Baker S C, Lai M M C. An in vitro system for the leader-primed transcription of coronavirus mRNAs. EMBO J. 1990;9:4173–4179. doi: 10.1002/j.1460-2075.1990.tb07641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baric R S, Stohlman S A, Lai M M C. Characterization of replicative intermediate RNA of mouse hepatitis virus: presence of leader RNA sequences on nascent chains. J Virol. 1983;48:633–640. doi: 10.1128/jvi.48.3.633-640.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blair W S, Li X, Semler B L. A cellular cofactor facilitates efficient 3CD cleavage of the poliovirus P1 precursor. J Virol. 1993;67:2336–2343. doi: 10.1128/jvi.67.4.2336-2343.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brian D A, Spaan W J M. Recombination and coronavirus defective interfering RNAs. Semin Virol. 1997;8:101–111. doi: 10.1006/smvy.1997.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao X, Wimmer E. Genetic variation of the poliovirus genome with two VPg coding units. EMBO J. 1996;15:23–33. [PMC free article] [PubMed] [Google Scholar]
- 9.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 10.Chang R Y, Hofmann M A, Sethna P B, Brian D A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang R Y, Krishnan R, Brian D A. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J Virol. 1996;70:2720–2729. doi: 10.1128/jvi.70.5.2720-2729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, Kuo L, Rowland R R R, Even C, Faaberg K S, Plagemann P G W. Sequences of 3′ end of genome and of 5′ end of open reading frame 1a of lactate dehydrogenase-elevating virus and common junction motifs between 5′ leader and bodies of seven subgenomic mRNAs. J Gen Virol. 1993;74:643–659. doi: 10.1099/0022-1317-74-4-643. [DOI] [PubMed] [Google Scholar]
- 13.den Boon J A, Spaan W J M, Snijder E J. Equine arteritis virus subgenomic RNA transcription: UV inactivation and translation inhibition studies. Virology. 1995;213:364–372. doi: 10.1006/viro.1995.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.den Boon J A, Kleijnen M F, Spaan W J M, Snijder E J. Equine arteritis virus subgenomic mRNA synthesis: analysis of leader-body junctions and replicative-form RNAs. J Virol. 1996;70:4291–4298. doi: 10.1128/jvi.70.7.4291-4298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.den Boon J A, Snijder E J, Chirnside E D, de Vries A A F, Horzinek M C, Spaan W J M. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vries A A F, Chirnside E D, Bredenbeek P J, Gravestein L A, Horzinek M C, Spaan W J M. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 1990;18:3241–3247. doi: 10.1093/nar/18.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Vries A A F, Chirnside E D, Horzinek M C, Rottier P J M. Structural proteins of equine arteritis virus. J Virol. 1992;66:6294–6303. doi: 10.1128/jvi.66.11.6294-6303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Vries A A F, Horzinek M C, Rottier P J M, de Groot R J. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doll E R, Bryans J T, McCollum W H, Crowe M E W. Isolation of a filterable agent causing arteritis of horses and abortion by mares. Its differentiation from the equine abortion (influenza) virus. Cornell Vet. 1957;47:3–41. [PubMed] [Google Scholar]
- 20.Godeny E K, de Vries A A F, Wang X C, Smith S L, de Groot R J. Identification of the leader-body junctions for the viral subgenomic mRNAs and organization of the simian hemorrhagic fever virus genome: evidence for gene duplication during arterivirus evolution. J Virol. 1998;72:862–867. doi: 10.1128/jvi.72.1.862-867.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris K S, Reddigari S R, Nicklin M J, Hammerle T, Wimmer E. Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J Virol. 1992;66:7481–7489. doi: 10.1128/jvi.66.12.7481-7489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiscox J A, Mawditt K L, Cavanagh D, Britton P. Investigation of the control of coronavirus subgenomic mRNA transcription by using T7-generated negative-sense RNA transcripts. J Virol. 1995;69:6219–6227. doi: 10.1128/jvi.69.10.6219-6227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs L, Spaan W J M, Horzinek M C, van der Zeijst B A M. Synthesis of subgenomic mRNAs of mouse hepatitis virus is initiated independently: evidence from UV transcription mapping. J Virol. 1981;39:401–406. doi: 10.1128/jvi.39.2.401-406.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeong Y S, Repass J F, Kim Y N, Hwang S M, Makino S. Coronavirus transcription mediated by sequences flanking the transcription consensus sequence. Virology. 1996;217:311–322. doi: 10.1006/viro.1996.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadare G, Haenni A L. Virus-encoded RNA helicases. J Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai M M C, Baric R S, Brayton P R, Stohlman S A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc Natl Acad Sci USA. 1984;81:3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai M M C, Patton C D, Stohlman S A. Replication of mouse hepatitis virus: negative-stranded RNA and replicative form RNA are of genome length. J Virol. 1982;44:487–492. doi: 10.1128/jvi.44.2.487-492.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemm J A, Rumenapf T, Strauss E G, Strauss J H, Rice C M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makino S, Joo M. Effect of intergenic consensus sequence flanking sequences on coronavirus transcription. J Virol. 1993;67:3304–3311. doi: 10.1128/jvi.67.6.3304-3311.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makino S, Joo M, Makino J K. A system for study of coronavirus messenger RNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991;65:6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meulenberg J J M, de Meijer E J, Moormann R J M. Subgenomic RNAs of Lelystad virus contain a conserved leader-body junction sequence. J Gen Virol. 1993;74:1697–1701. doi: 10.1099/0022-1317-74-8-1697. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, New York, N.Y.: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sawicki S G, Sawicki D L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawicki S G, Sawicki D L. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv Exp Biol Med. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- 37.Sawicki S G, Sawicki D L. A new model for coronavirus transcription. In: Enjuanes L, Siddell S G, Spaan W J M, editors. Coronaviruses and arteriviruses. New York, N.Y: Plenum Press; 1998. pp. 215–219. [Google Scholar]
- 38.Schaad M C, Baric R S. Genetics of mouse hepatitis virus transcription: evidence that subgenomic negative strands are functional templates. J Virol. 1994;68:8169–8179. doi: 10.1128/jvi.68.12.8169-8179.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sethna P B, Brian D A. Coronavirus genomic and subgenomic minus-strand RNAs copartition in membrane-protected replication complexes. J Virol. 1997;71:7744–7749. doi: 10.1128/jvi.71.10.7744-7749.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sethna P B, Hofmann M A, Brian D A. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J Virol. 1991;65:320–325. doi: 10.1128/jvi.65.1.320-325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sethna P B, Hung S L, Brian D A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc Natl Acad Sci USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirako Y, Strauss J H. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol. 1994;68:1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snijder E J, Meulenberg J J M. The molecular biology of arteriviruses. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 44.Snijder E J, Wassenaar A L M, Spaan W J M. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J Virol. 1994;68:5755–5764. doi: 10.1128/jvi.68.9.5755-5764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spaan W J M, Delius H, Skinner M, Armstrong J, Rottier P J M, Smeekens S, van der Zeijst B A M, Siddell S G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Berlo M F, Horzinek M C, van der Zeijst B A M. Equine arteritis virus-infected cells contain six polyadenylated virus-specific RNAs. Virology. 1982;118:345–352. doi: 10.1016/0042-6822(82)90354-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Most R G, de Groot R J, Spaan W J M. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J Virol. 1994;68:3656–3666. doi: 10.1128/jvi.68.6.3656-3666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Most R G, Spaan W J M. Coronavirus replication, transcription, and RNA recombination. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 11–31. [Google Scholar]
- 49.van Dinten L C, den Boon J A, Wassenaar A L M, Spaan W J M, Snijder E J. An infectious arterivirus cDNA clone: identification of a replicase point mutation which abolishes discontinuous mRNA transcription. Proc Natl Acad Sci USA. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Dinten L C, Rensen S, Spaan W J M, Gorbalenya A E, Snijder E J. Proteolytic processing of the ORF1b-encoded part of the arterivirus replicase is mediated by the nsp4 serine protease and is essential for virus replication. J Virol. 1999;73:2027–2037. doi: 10.1128/jvi.73.3.2027-2037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Dinten L C, Wassenaar A L M, Gorbalenya A E, Spaan W J M, Snijder E J. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J Virol. 1996;70:6625–6633. doi: 10.1128/jvi.70.10.6625-6633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. M. Spaan, and E. J. Snijder. Unpublished data.
- 53.van Marle G, Luytjes W, van der Most R G, van der Straaten T, Spaan W J M. Regulation of coronavirus mRNA transcription. J Virol. 1995;69:7851–7856. doi: 10.1128/jvi.69.12.7851-7856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wassenaar A L M, Spaan W J M, Gorbalenya A E, Snijder E J. Alternative proteolytic processing of the arterivirus ORF1a polyprotein: evidence that nsp2 acts as a cofactor for the nsp4 serine protease. J Virol. 1997;71:9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yokomori K, Banner L R, Lai M M C. Coronavirus mRNA transcription: UV light transcriptional mapping studies suggest an early requirement for a genomic-length template. J Virol. 1992;66:4671–4678. doi: 10.1128/jvi.66.8.4671-4678.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]